Abstract
Background and Objectives:
Strains of Shigella spp. can cause shigellosis, or bacillary dysentery. that is a public health problem worldwide. The aim of this study was to describe the population structure and genetic relatedness of multidrug resistant S. sonnei and S. flexneri isolated during a one year period from children with diarrhea in Tehran, Iran.
Materials and Methods:
A total of 70 Shigella spp. were detected during the study period. Twenty MDR isolates of Shigella spp. were randomly selected and used in this study. Bacterial identification was performed by conventional biochemical and serological and confirmed by molecular method. After antimicrobial susceptibility testing, we used Multilocus sequence typing (MLST) for subtyping isolates.
Results:
We found 14 Shigella sonnei and 6 Shigella flexneri isolates. Results of MLST showed five sequence types (ST) (145, 152, 241, 245, 1502) and BURST analysis revealed the largest number of single locus variant (SLV) and highest frequency (FREQ) for ST152.
ST 152 with nine members was predicted as the founder by BURST. Frequency for ST 1502 and ST 245 was four isolates and the least frequency was seen for ST 241 and 145 with one and two members, respectively. ST 145 and ST 245 were described as singletons in BURST. All isolates with ST145 and ST245 were identified as Shigella flexneri.
Conclusion:
Annual Multi locus sequence typing of MDR Shigella would help us in better understanding of dominant species and comparing our results with the same studies in other countries especially our neighbor countries in source tracking purposes.
Keywords: Shigella, Multilocus sequence typing, Multidrug resistant
INTRODUCTION
Shigella is a genus in the family Entrobacteriaceae that can cause shigellosis with diarrhea, fever and abdominal cramps. Shigellosis may vary from self-limited disease in healthy adults to severe in young children, elderly and immune compromised patients. Severe cases can be associated with seizures in children less than 2 years old, Reiter’s syndrome and Haemolytic Uremic Syndrome (1).
Epidemiological studies reveal 1,000,000 deaths per year caused by shigellosis in world (2). A total of 500000 illnesses have been generated by Shigella in the United States per year whereas shigellosis incidence in six country of Asia was about 100-folds higher than in industrialized nations (2–3).
Different rate of morbidity, mortality and prevalence of Shigella spp. have been observed in various geographical regions.
The most cases of shigellosis usually recover without antibiotic treatment but it can be lethal for people with immunosuppressed systems. Therefore, application of a treatment plan is imperative (2). Treatment challenges may especially occur in severe cases with multidrug resistant (MDR) isolates which are characterized as resistance to more than three antibiotic classes (3–4). Antimicrobial resistance patterns and prevalence of serotypes vary in different geographical regions (5). Bacteria can acquire and disseminate genes for resistance to antibiotics through mobile genetic elements in their evolutionary pathways. The genetic structure of natural population of bacteria changes according to pattern and frequency of recombination and horizontal exchange of these element (4, 6, 7).
Shigella are human adapted E. coli that have obtained the new abilities (8). Virulence and invasion plasmid antigen H (ipaH) genes are located on the chromosome and the large plasmid PINV (9).
There are many typing methods based on phenotypic or genotypic properties of bacteria that are selected based on the epidemiological approaches. MLST method is based on sequencing of at least seven housekeeping genes, assigning the alleles at the seven loci and allocate the sequence type of each isolate. The provided databases are comparable with other data in the world and allow molecular typing of bacteria via internet (10). MLST is a gold standard method for epidemiological studies and was proved to be discriminatory, accurate, portable and reproducible over the years (11).
The aim of this study was to describe the population structure and genetic relatedness of multidrug resistant S. sonnei and S. flexneri isolated during a one year period from children with diarrhea in Tehran, Iran.
MATERIALS AND METHODS
Bacterial strains.
A total of 70 Shigella spp. isolates were detected during study period from 5291 investigated stool samples (1.32 %) and these included S. sonnei (n=61, 87.14%) and S. flexneri. (n=8), (11.43%) based on the serological and molecular methods used for identification. As low as 1.43% of isolates were characterized as S. boydii and no S. dysentriae was recovered from the patients. The majority of isolates were isolated during November and December 2012 and August, September and October 2013 among which 20 Shigella spp. were randomly selected and used in this study. Selection was performed according to frequency of isolation of Shigella spp. in each month and accordingly, 14 (70%) S. sonnei and 6 (30%) S. flexneri were selected (Table 1). Bacterial isolates were identified using both conventional biochemical tests and serology using agglutination with specific A–D antisera (Baharafshan Institute of Research & Development, Tehran, Iran) (12). The identity of isolates was also confirmed by PCR amplification of ipaH, wbgZ and rfcgenes, specific for Shigella spp., S. sonnei and S. flexneri, respectively (13–14).
Table 1.
Bacterial isolates under study
| Date of Isolation | Number of diarrheal stool samples | No. of Shigella spp. isolated | Number of Shigella spp. Selected for MLST analysis |
|---|---|---|---|
| November/2012 | 959 | 30 | 7 |
| December/2012 | 693 | 18 | 4 |
| January/2013 | 292 | 0 | 0 |
| February/2013 | 312 | 2 | 1 |
| March/2013 | 333 | 0 | 0 |
| April/2013 | 356 | 0 | 0 |
| May/2013 | 415 | 0 | 0 |
| June/2013 | 403 | 1 | 1 |
| July/2013 | 353 | 2 | 1 |
| August/2013 | 385 | 4 | 2 |
| September/2013 | 414 | 5 | 2 |
| October/2013 | 357 | 8 | 2 |
| Total | 5291 | 70 | 20 |
The primer sequences used in this study are shown in Table 2 and all of primers were synthesized-by TAGC Company (Tag Copenhagen A/S Kong GeorgsVej 12 DK-2000 Frederiksberg Denmark). PCR was performed in a reaction with total volume of 25 μL, containing 2.5 μL 10x Taq polymerase buffer, 0.3 μL dNTPs (10 mmol.L−1), 1 U Taq DNA polymerase (Fermentas, Lithuania), 0.6 μL MgCl2 (50 mmol.L−1) and 0.3 mol.L−1 of each primer. PCR was done as follows: initial denaturation step at 94°C for 5 min, followed by 30 cycles consisting of denaturation (at 94°C for 1 min), annealing (1 min, separately adjusted for each set of primer pairs according Table 2), extension (at 72°C for 1 min), followed by a final extension step at 72°C for 5 min. Finally PCR products were assessed for specific band on agarose gel (90 volt, 45 min).
Table 2.
Oligonucleotide primers used in this study.
| Primers | Sequence (5’→3’) Annealing | Annealing °C | Amplicon size (bp) |
|---|---|---|---|
| ipaH-F | GTTCCTTGACCGCCTTTCCGATACCGTC | 60 | 619 |
| ipaH-R | GCCGGTCAGCCACCCTCTGAGAGTAC | ||
| wbgZ-F | TCT GAATATGCCCTCTACGCT | 60 | 430 |
| wbgZ-R | GACAGAGCCCGAAGAACCG | ||
| rfc-F | TTTATGGCTTCTTTGTCGG | 60 | 537 |
| rfc-F | CTGCGTGATCCGACCATG | ||
| adkF | ATTCTGCTTGGCGCTCCGGG | 57 | 583 |
| adkR | CCGTCAACTTTCGCGTATTT | ||
| fumCF | TCACAGGTCGCCAGCGCTTC | 57 | 805 |
| fumCR | GTACGCAGCGAAAAAGATTC | ||
| gyrBF | TCGGCGACACGGATGACGGC | 59 | 879 |
| gyrBR | ATCAGGCCTTCACGCGCATC | ||
| icdF | ATGGAAAGTAAAGTAGTTGTTCCGGCA | 58 | 877 |
| icdR | GGACGCAGCAGGATCTGTT | ||
| mdhF | AGCGCGTTCTGTTCAAATGC | 56 | 798 |
| mdhR | CAGGTTCAGAACTCTCTCTGT | ||
| purAF | TCGGTAACGGTGTTGTGCTG | 57 | 816 |
| purAR | CATACGGTAAGCCACGCAGA | ||
| recAF | ACCTTTGTAGCTGTACCACG | 56 | 634 |
| recAR | AGCGTGAAGGTAAAACCTGTG |
Antimicrobial susceptibility testing.
Antimicrobial susceptibility testing was performed for minocycline (30μg), tetracycline (30μg), doxycycline (30μg), ampicillin (10μg), trimethoprim-sulfamethoxazole (25μg), nalidixic acid (30μg), norfloxacin (10μg), ciprofloxacin (5μg), levofloxacin (5μg) and streptomycin (10μg) by disk diffusion method according to CLSI guidelines. E. coli 25922 was used as the control strain.
Multi-locus sequence typing (MLST).
The genomic DNA from Shigella isolates was extracted using Exgene Cell SV kit (Genen all Biotechonolgy Co. Ltd Korea) that were grown in LB broth medium at 37°C overnight.
Seven housekeeping genes: adk (adenylate kinase), fumC (fumaratehydratase), gyrB (DNA gyrase), icd (isocitrate/isopropylmalate dehydrogenase), mdh (malate dehydrogenase), purA (adenylosuccinate dehydrogenase), recA (ATP/GTP binding motif) were amplified using the primers and PCR conditions described in protocols introduced for MLST of E. coli (15).
Primers sequences are shown in Table 2. PCR products were purified and both DNA strands were sequenced using ABI 3730X capillary sequencer (Macrogen, Seoul, Korea).
Data analysis.
Sequenced data were read by Laser-gene 66 software and trimmed according to database of MLST Home and National Center for Biotechnology Information (NCBI) for each housekeeping genes. Sequences were compared with MLST databases of E. coli and were identified allelic profiles and Sequence Types. Data of MLST were analyzed by BURSTalgorithm and clonal complex were determined based on SLV(16).
We used the Minimum spanning tree (MST), a graphical tool according to Prim’s algorithm, to draw a tree using MLST allelic profile data. Phylogenetic analysis was done by MLST Philip program based on neighbor-joining software using sequences of the seven housekeeping genes for each STs.
RESULTS
Bacterial strains.
Twenty isolates of Shigella spp. were selected and used in this study consisting of 14 (70%) S. sonnei and 6 (30%) S. flexneri.
All Shigella spp. harboured ipaH gene specific for Shigella genus with an amplification band of 619bp, while simultaneously all S. sonnei and S. flexneri isolates produced 430bp and 537bp bands related to wbgZ and rfc, respectively, followed by sequencing which confirmed the identity of both species (Table 2). The results of serogrouping were in agreement with molecular identification method.
Antimicrobial susceptibility testing.
The results of antimicrobial susceptibility revealed high prevalence of resistance to streptomycin, trimethoprim-sulfamethoxazole and tetracycline. All isolates were susceptible to quinolones including norfloxacin, levofloxacin and ciprofloxacin. The majority of isolates were multi drug resistant (MDR) with seven patterns of resistance. The result of antimicrobial susceptibility of isolates is shown in Table 3.
Table 3.
Antimicrobial susceptibility testing
| Antibiotics | Sensitive % | Intermediate % | Resistant % |
|---|---|---|---|
| Ampicillin | 76 | 0 | 24 |
| Ciprofloxacin | 98 | 2 | 0 |
| Doxycycline | 4 | 38 | 58 |
| Levofloxacin | 98 | 2 | 0 |
| Minocycline | 38 | 62 | 0 |
| Nalidixic Acid | 80 | 2 | 38 |
| Norfloxacin | 100 | 0 | 0 |
| Streptomycin | 0 | 0 | 100 |
| Tetracycline | 4 | 5 | 96 |
| Trimethoprim | 2 | 0 | 98 |
MLST, BURST, minimum spanning tree and phylogenetic tree.
Electrophoresis of PCR products revealed the bands that specified by 879bp (adk), 805bp (fum), 879bp (gyr), 877bp (icd), 798bp (mdh), 816bp (pur) and 634bp (recA) (Table 2).
A comparison between the E. coli sequences of seven housekeeping genes available at MLST and data obtained in this study, the allelic profiles, STs and ST complexes were determined. A total of five STs (145, 152, 1502, 241, 245) were found for twenty isolates while no new ST was detected in this study (Table 4).
Table 4.
Sequence types, ST complexes and allelic profiles in relation to Shigellasero groups and antimicrobial resistance patterns.
| ST | Code number | ST complex | Species | Resistance profile |
|---|---|---|---|---|
| 152 | 5 | 152 | S. sonnei | Te,sxT,S |
| 152 | 6 | 152 | S. sonnei | Te,sxT,S |
| 152 | 2 | 152 | S. sonnei | Te,sxT,S |
| 152 | 17 | 152 | S. sonnei | Te,D,sxT,S |
| 152 | 12 | 152 | S. sonnei | Te,D,sxT,S |
| 152 | 16 | 152 | S. sonnei | Te,D,sxT,S |
| 152 | 18 | 152 | S. sonnei | Te,D,sxT,S |
| 152 | 15 | 152 | S. sonnei | Te,D,sxT,S,,NA |
| 152 | 9 | 152 | S. sonnei | Te,D,sxT,S,,NA |
| 241 | 8 | 152 | S. sonnei | Te,D,sxT,S,,NA |
| 1502 | 1 | None | S. sonnei | Te,sxT,S |
| 1502 | 11 | None | S. sonnei | Te,D,sxT,S |
| 1502 | 20 | None | S. sonnei | Te,D,sxT,S |
| 1502 | 13 | None | S. sonnei | Te,D,sxT,S,,Am |
| 245 | 3 | 245 | S. flexneri | Te,sxT,S,Am |
| 245 | 10 | 245 | S. flexneri | Te,sxT,S,Am |
| 245 | 4 | 245 | S. flexneri | Te,D,S,Am |
| 245 | 7 | 245 | S. flexneri | Te,sxT,S,,Am |
| 145 | 14 | 243 | S. flexneri | sxT,S,Am,NA |
| 145 | 19 | 243 | S. flexneri | sxT,S |
The largest number of single locus variant (SLV), n=2, and the highest frequency (FREQ) for ST152, n=9, were revealed by BRUST analysis (Table 5). ST 152 and ST 241 together fell in ST complex 152, all of which were identified as S. sonnei and constituted about 50% of total isolates under study (Table 4).
Table 5.
Results of BURST analysis.
| ST | No. of isolates | FREQ | SLv | DLv | TLv | Average Distance |
|---|---|---|---|---|---|---|
| Group | 14 | |||||
| 152 * | 9 | 2 | 0 | 0 | 1.0 | |
| 1502 | 4 | 1 | 1 | 0 | 1.5 | |
| 241 | 1 | 1 | 1 | 0 | 1.5 | |
| Singleton | 6 | |||||
| 145 | 2 | |||||
| 245 | 4 |
Predicted Founder; ST, sequence type; FREQ, Frequency; SLV, Single -Locus Variant; DLV, Double -Locus Variant; TLV, Triple- Locus Variant.
ST 152 with the greatest number of SLV was as predicted founder. Frequency for ST 145 and ST 245 was low. Allelic profile of ST145 and ST245 were more different compared to other isolates so described as singletons in BURST (Table 5). All isolates in ST145 and ST245 were identified as S. flexneri (Table 4). ST 152 was predicted as the founder (ST that has the greatest number of single-locus variants) by BURST with 9 isolates falling in this ST. Frequency for both ST 1502 and ST 245 was four, while one and two isolates fell in ST 241 and 145 respectively. Allelic profile of ST145 and ST245 were more substantially different compared to other isolates so described as singletons in BURST (Table 5). All isolates with these 2 allelic profiles (n=6) were identified as S. flexneri (Table 4).
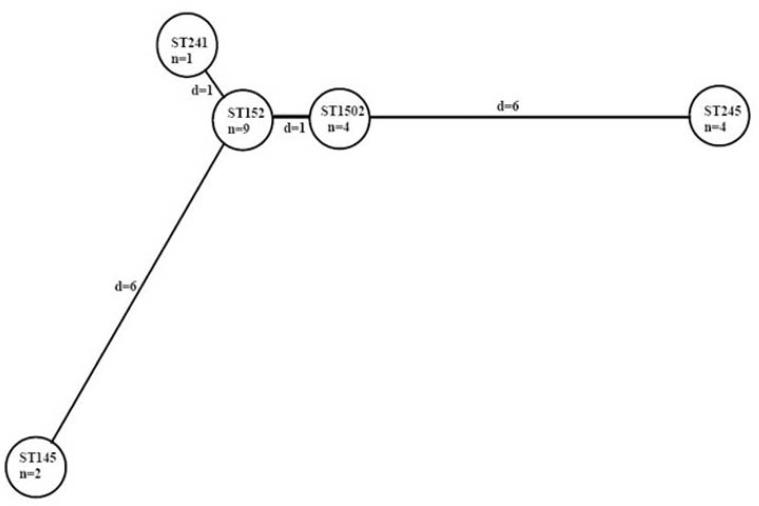
Output of MST indicated five circles (STs) which are linked together. In topological arrangement of MST, each circle represents one ST, also length of lines show distance between sequence types. Analysis of STs according to MST also revealed the highest priority for ST152 with the largest SLV in this study (Fig. 1).
Fig. 1.
Minimum spanning treeprim: Minimum spanning tree with cost, d: distance, n: number, ST: Sequence type. Each ST is represented by a circle.
Phylogenetic analysis also revealed the ST152 as the root of tree that was predicted as ancestor for Shigella isolates of this study.
Similar results of BURST, Minimum spanning tree and phylogenetic tree also emphasize on ST152 as the predicted founder of our isolates (Fig. 1 and Table 5).
DISCUSSION
According to previous studies in Iran, S. flexneri was the most common serogroup during 2001–2006, while the predominant serogroup was S. sonnei during 2008–2012 (17). The present study revealed a higher frequency of S. sonnei, compared to other Shigella species in Iran during 2012–2013, which imitates the infection profile of more developed countries. One reason for the rise in the incidence of S. sonnei can be the improving hygiene level in Iran and the industrialization of the capital city. Although, the increased environmental adaptation of S. sonnei should not be ignored (18–19).
The majority of Shigella isolates (95%) in this study fulfilled the MDR criteria, however the antimicrobial resistance pattern was different among the isolates.
The results of antimicrobial susceptibility assay revealed a high resistance, among the Shigella isolates, against streptomycin, trimethoprim-sulfamethoxazole and tetracycline. Despite the CLSI recommendation for the use of trimethoprim-sulfamethoxazole (TMP) for fecal isolates of Shigella, its use is nowadays limited due to the high resistance of this organism (20). Previous reports in Iran have reported a 92.2% to 94%, resistance to TMP during 2000–2011 (1, 18, 21). A high level resistance to TMP has also been reported from Nepal (81.54%), and the USA (66%) (22–23). The wide distribution of resistance to TMP can be attributed to over prescription or misuse of the antibiotics in clinics (24). All TMP resistant isolates in the current study were also resistant to streptomycin (Table 3). Simultaneous resistance to streptomycin and TMP can probably be related to a 6.3kb plasmid that was investigated by Barman and colleagues (2010) or other mobile resistance genetic elements (25).
As with tetracycline, 10% of the isolates were susceptible to doxycycline. According to the previous studies in Iran and other countries, resistance to tetracycline might be an intrinsic characteristic among Shigella spp, being observed in the majority of clinically isolated strains (18, 26).
Two isolates which were recognized as ST145 (S. flexneri) were susceptible to tetracycline which rules out the intrinsic characteristic of resistance to this antibiotic among S. flexneri strains.
The results showed 25% and 5% resistance to ampicillin among S. flexneri and S. sonnei, respectively. This phenomenon which was also reported by Seidlein and colleagues (2006) reveals a higher ampicillin resistance among S. flexneri compared with S. sonnei isolates (3).
Resistance to nalidixic acid was shown to be 12.5% among S. flexneri isolates and 16.5% among the S. sonnei isolates. Approximately similar results were obtained for both species. Due to the rise in antimicrobial resistance among Shigella isolates it can be concluded that antimicrobial susceptibility testing must be done to warrant the effectiveness of prescribed antimicrobial agents. Fortunately, all isolates in this study were susceptible to fluoroquinolones including; ciprofloxacin, levofloxacin and norfloxacin. Other studies in Iran have revealed similar results for fluoroquinolones (18, 19, 21), except for Gharibi and colleagues (2012) which reported a 4.25% resistance to ciprofloxacin among Shigella isolates in Bushehr province of Iran (27). There also exist some discrete reports on resistance to these drugs in other countries (28). Excessive use of ciprofloxacin as the first line treatment for shigellosis may be the cause of increasing resistance to this drug. Totally, the low rate of resistance to fluoroquinolones makes these drugs a better for the treatment.
Result of MLST Philip was more suitable for this study and ST152 was considered as a root in this tree and the branch lengths were similar to MST.
Our results revealed five ST (145, 152, 241, 245, and 1502) among 20 Shigella isolates with ST 152 comprising the most frequent type which was indicated as predicted founder in this study. ST 152 for Shigella isolates was also reported from other countries of all continents (Table 6). The ST152 as the dominant ST with the MDR phenotype is of great significance which can render the treatment of shigellosis more challenging.
Table 6.
Geographical distribution of Shigella spp. sequence types
| species | Allelic profile | Country/ Continent | Year of isolated | References | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 152 | S. sonnei | 11 | 63 | 7 | 1 | 14 | 7 | 7 | Iran | 2012 | This study |
| 152 | S. sonnei | 11 | 63 | 7 | 1 | 14 | 7 | 7 | China | 2009 | Cao, Y. & Wei, D. (2012) |
| 152 | S. flexneri | 11 | 63 | 7 | 1 | 14 | 7 | 7 | China | 2010 | Cao, Y. & Wei, D. (2012) |
| 152 | S. sonnei | 11 | 63 | 7 | 1 | 14 | 7 | 7 | Germany | 1997 | Cao, Y. & Wei, D. (2012) |
| 152 | S. sonnei | 11 | 63 | 7 | 1 | 14 | 7 | 7 | Asia, Africa America, Europe | 1943–2008 | Inouye et al (2012) |
| 152 | S. sonnei | 11 | 63 | 7 | 1 | 14 | 7 | 7 | Asia+Pacific, Africa America, Europe | Wirth et al., 2006 | |
| 241 | S. sonnei | 11 | 63 | 6 | 1 | 14 | 7 | 7 | China | 1997 | Cao, Y. & Wei, D. (2012) |
| 1502 | S. sonnei | 6 | 63 | 7 | 1 | 14 | 7 | 7 | Asia, Africa America, Europe | 1943–2008 | Inouye et al (2012) |
| 245 | S. flexneri | 6 | 61 | 6 | 11 | 13 | 3 | 50 | Iran | 2012 | This study |
| 245 | S. flexneri | 6 | 61 | 6 | 11 | 13 | 3 | 50 | Asia+Pacific, Africa America, Europe | Wirth et al., 2006 | |
| 245 | S. flexneri | 6 | 61 | 6 | 11 | 13 | 3 | 50 | China | 1983 | Cao, Y. & Wei, D. (2012) |
| 245 | S. flexneri | 6 | 61 | 6 | 11 | 13 | 3 | 50 | Germany | 1997 | Cao, Y. & Wei, D. (2012) |
| 245 | S. flexneri | 6 | 61 | 6 | 11 | 13 | 3 | 50 | Canada | 1982 | Cao, Y. & Wei, D. (2012) |
| 245 | S. flexneri | 6 | 61 | 6 | 11 | 13 | 3 | 50 | Asia, Africa America, Europe | 1943–2008 | Inouye et al (2012) |
| 245 | S.boydii | 6 | 61 | 6 | 11 | 13 | 3 | 50 | China | 2009 | Cao, Y. & Wei, D. (2012) |
| 145 | S. flexneri | 1 | 10 | 1 | 1 | 1 | 1 | 1 | Asia+Pacific, Africa America, Europe | Wirth et al. (2006) | |
| 149 | S. boydii | 6 | 60 | 60 | 3 | 6 | 6 | 3 | Asia+Pacific, Africa America, Europe | Wirth et al. (2006) | |
| 151 | S. sonnei | 11 | 62 | 7 | 1 | 14 | 7 | 7 | Germany | 1997 | Cao, Y. & Wei, D. (2012) |
| 240 | S. flexneri | 6 | 61 | 4 | 11 | 13 | 3 | 50 | Asia+Pacific, Africa America, Europe | Wirth et al., 2006 | |
| 243 | S. boydii | 1 | 4 | 1 | 1 | 1 | 1 | 1 | Asia, Africa America, Europe | 1943–2008 | Inouye et al (2012) |
| 243 | S. boydii, S. flexneri S. dysenteriae | 1 | 4 | 1 | 1 | 1 | 1 | 1 | Asia+Pacific, Africa America, Europe | Wirth et al., 2006 | |
| 248 | S. flexneri | 6 | 74 | 6 | 66 | 13 | 3 | 50 | Asia+Pacific, Africa America, Europe | Wirth et al., 2006 | |
| 250 | S. boydii, S. dysenteriae | 6 | 59 | 60 | 3 | 47 | 3 | 3 | Asia+Pacific, Africa America, Europe | Wirth et al., 2006 | |
| 255 | S. flexneri | 6 | 61 | 4 | 11 | 48 | 3 | 50 | Asia+Pacific, Africa America, Europe | Wirth et al., 2006 | |
| 259 | S. flexneri | 6 | 78 | 6 | 11 | 13 | 3 | 50 | Asia+Pacific, Africa America, Europe | Wirth et al., 2006 | |
| 262 | S. flexneri | 1 | 6 | 1 | 1 | 1 | 1 | 1 | Asia+Pacific, Africa America, Europe | Wirth et al., 2006 | |
| 264 | S. flexneri | 6 | 61 | 68 | 11 | 13 | 3 | 55 | Asia+Pacific, Africa America, Europe | Wirth et al., 2006 | |
| 412 | S.boydii | 95 | 111 | 91 | 99 | 68 | 70 | 76 | Bangladesh | Cao, Y. & Wei, D. (2012) | |
| 626 | S. flexneri | 116 | 61 | 6 | 11 | 1 | 3 | 50 | Taiwan | 1991 | Choi et al (2007) |
| 627 | S. flexneri | 6 | 61 | 6 | 11 | 13 | 97 | 50 | Korea | 2002 | Choi et al (2007) |
| 628 | S. flexneri | 6 | 61 | 6 | 11 | 13 | 3 | 55 | Philippine | 1981–2000 | Choi et al (2007) |
| 629 | S. flexneri | 6 | 145 | 6 | 11 | 13 | 3 | 55 | Korea | Choi et al (2007) | |
| 630 | S. boydii | 6 | 61 | 6 | 11 | 6 | 95 | 7 | Asia+Pacific, ,Africa America, Europe | Wirth et al., 2006 | |
| 631 | S. flexneri | 6 | 74 | 6 | 123 | 13 | 3 | 50 | France | Choi et al (2007) | |
| 632 | S. flexneri | 1 | 10 | 1 | 1 | 1 | 96 | 1 | Taiwan | Choi et al (2007) | |
| 633 | S. flexneri | 6 | 61 | 6 | 11 | 13 | 98 | 50 | Japan | 2006 | Choi et al (2007) |
| 634 | S. flexneri | 6 | 61 | 6 | 123 | 13 | 3 | 50 | China | 2002 | Nie et al., 2006 |
| 651 | S. flexneri | 6 | 149 | 6 | 11 | 13 | 3 | 50 | China | 1943–2008 | Choi et al (2007) |
| 1025 | S. boydii | 6 | 61 | 6 | 174 | 13 | 3 | 50 | Asia, Africa America, Europe | 1943–2008 | Inouye et al (2012) |
| 1504 | S. sonnei | 11 | 63 | 7 | 1 | 14 | 17 | 27 | Asia, Africa, America, Europe | 1943–2008 | Inouye et al (2012) |
| 1505 | S. sonnei | 11 | 63 | 7 | 1 | 14 | 17 | 37 | Asia, Africa America, Europe | 2010 | Inouye et al (2012) |
| 2208 | S. sonnei | 45 | 240 | 192 | 99 | 91 | 159 | 142 | China | Cao, Y. & Wei, D. (2012) | |
ST152 was closely related to ST1502 and ST241 which were two single locus variant for ST152. All the strains in ST152, ST1502 and ST 241 were recognized as S. sonnei. This results is in agreement with the studies by Cao & Wei and Inouye et al. (2012) for ST 241 and ST 1502 (Table 6). In these studies, Shigella spp. were typed to ST152 and ST241 with ST152 comprising the predicted founder which is in consistent with our study, however both S. flexneri and S. sonnei species were included in ST152 in the above mentioned studies (29), which was in contrast with the results of our study.
Two isolates fell in ST145 (ST complex 243) and four isolates in ST245 (ST complex 245), both of which were identified as singletons. In the present study, all strains in ST145 and ST245 were identified as S. flexneri, aphenomenon which was also reported by Wirth et al. (2006). However, ST 245 encompassed S. boydii and S. flexneriin the study of Cao & Wei (2012). This suggests that probably S. flexneri is derived from a distinct parental clone (8).
It is notable that only isolates with ST145 properties were susceptible to tetracycline which suggests the hypothesis that ST specification of the isolates may affect the acquisition and/or expression of antimicrobial resistance properties.
All the STs in the current study have been previously reported in other studies for Shigella and so we did not find any new sequence type which may be related to i) population size or ii) geographic distribution of specific Shigella sequence types or to iii) the Shigella genome stability (2, 15, 29). Although other sequence types of Shigella spp. have been identified throughout the world (Table 6) but there are still insufficient data on the incidence of Shigella STs in different geographical locations which emphasizes the need to annual monitoring and MLST sequence typing of outbreak and sporadic Shigella strains for better understanding of distribution and dominant sequence types in the world.
CONCLUSION
This study was the first report of MLST genotyping of MDR resistant Shigella spp. in Iran which determined five sequence types with ST 152 as the dominant sequence type being composed of S. sonnei isolates. Resistance to ampicillin was most frequently observed in ST245 and susceptibility to tetracycline was only found in ST 145 which could indicate a common mechanism of resistance acquisition. These results emphasized the need to monitor and evaluate the resistance profile change among sequence types for better prevention, control and treatment of shigellosis.
ACKNOWLEDGMENT
This study was supported by Iran University of Medical Sciences with grant number 92-02-134-21592.
REFERENCES
- 1. Todar K. Todar-s online textbook of bacteriology. University of Wisconsin-Madison Department of Bacteriology; 2005. [Google Scholar]
- 2. Heiman KE, Karlsson M, Grass J, Howie B, Kirkcaldy RD, Mahon B, et al. Notes from the field: Shigella with decreased susceptibility to azithromycin among men who have sex with men-United States, 2002–2013. MMWR Morb Mortal Wkly Rep 2014;63: 132– 133 . [PMC free article] [PubMed] [Google Scholar]
- 3. Seidlein L von, Kim DR, Ali M, Lee H, Wang X, Thiem VD, et al. A multicentre study of Shigella diarrhoea in six Asian countries: disease burden, clinical manifestations, and microbiology. PLoS Med 2006;3: e353– e353 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Priest F, Ramos-Cormenzana A, Tindall BJ. Bacterial diversity and systematics. Springer Science & Business Media; 2012. [Google Scholar]
- 5. Pazhani GP, Ramamurthy T, Mitra U, Bhattacharya SK, Niyogi SK. Species diversity and antimicrobial resistance of Shigella spp. isolated between 2001 and 2004 from hospitalized children with diarrhoea in Kolkata (Calcutta), India. Epidemiol Infect 2005;133: 1089– 1095 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rowe-Magnus DA, Guerout A-M, Mazel D. Bacterial resistance evolution by recruitment of super-integron gene cassettes. Mol Microbiol 2002;43: 1657– 1669 . [DOI] [PubMed] [Google Scholar]
- 7. Sire J-M, Macondo EA, Perrier-Gros-Claude J-D, Siby T, Bahsoun I, Seck A, et al. Antimicrobial resistance in Shigella species isolated in Dakar, Senegal (2004–2006). Jpn J Infect Dis 2008;61: 307– 309 . [PubMed] [Google Scholar]
- 8. Choi SY, Jeon Y-S, Lee JH, Choi B, Moon SH, von Seidlein L, et al. Multilocus sequence typing analysis of Shigella flexneri isolates collected in Asian countries. J Med Microbiol 2007;56: 1460– 6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Holt KE, Baker S, Weill F-X, Holmes EC, Kitchen A, Yu J, et al. Shigella sonnei genome sequencing and phylogenetic analysis indicate recent global dissemination from Europe. Nat Genet 2012;44: 1056– 1059 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Spratt BG. Multilocus sequence typing: molecular typing of bacterial pathogens in an era of rapid DNA sequencing and the internet. Curr Opin Microbiol 1999;2: 312– 316 . [DOI] [PubMed] [Google Scholar]
- 11. Inouye M, Conway TC, Zobel J, Holt KE. Short read sequence typing (SRST): multi-locus sequence types from short reads. BMC Genomics 2012; 13: 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Services M. UK Standards for Microbiology Investigations. Bacteriology 2014; B 55: 1– 21. [Google Scholar]
- 13. Toma C, Lu Y, Higa N, Nakasone N, Chinen I, Baschkier A, et al. Multiplex PCR assay for identification of human diarrheagenic Escherichia coli. J Clin Microbiol 2003; 41: 2669– 2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ojha SC, Yean Yean C, Ismail A, Banga Singh K-K. A pentaplex PCR assay for the detection and differentiation of Shigella species. Biomed Res Int 2013; 2013: 412370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 2006;60: 1136– 1151 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol 2004;186: 1518– 1530 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eftekhari N, Bakhshi B, Pourshafie MR, Zarbakhsh B, Rahbar M, Hajia M, et al. Genetic diversity of Shigella spp. and their integron content. Foodborne Pathog Dis 2013;10: 237– 242 . [DOI] [PubMed] [Google Scholar]
- 18. Mardaneh J, Poor SA, Afrugh P. Prevalence of Shigella species and antimicrobial resistance patterns of isolated strains from infected pediatrics in Tehran. Int J Entric Pathog 2013;1: 28– 31 . [Google Scholar]
- 19. Ranjbar R, Dallal MMS, Talebi M, Pourshafie MR. Increased isolation and characterization of Shigella sonnei obtained from hospitalized children in Tehran, Iran. J Health Popul Nutr 2008; 26: 426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Eliopoulos GM, Huovinen P. Resistance to trimethoprim-sulfamethoxazole. Clin Infect Dis 2001;32: 1608– 1614 . [DOI] [PubMed] [Google Scholar]
- 21. Pourakbari B, Mamishi S, Mashoori N, Mahboobi N, Ashtiani MH, Afsharpaiman S, et al. Frequency and antimicrobial susceptibility of Shigella species isolated in Children Medical Center Hospital, Tehran, Iran, 2001–2006. Braz J Infect Dis 2010;14: 153– 157 . [DOI] [PubMed] [Google Scholar]
- 22. Khan S, Singh P, Ansari M, Asthana A. Isolation of Shigella species and their resistance patterns to a panel of fifteen antibiotics in mid and far western region of Nepal. Asian Pacific J Trop Dis 2014;4: 30– 34 . [Google Scholar]
- 23. Toro C, Arroyo A, Sarria A, Iglesias N, Enríquez A, Baquero M, et al. Shigellosis in subjects with traveler’s diarrhea versus domestically acquired diarrhea: implications for antimicrobial therapy and human immunodeficiency virus surveillance. Am J Trop Med Hyg 2015;93: 491– 496 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Iqbal MS, Rahman M, Islam R, Banik A, Amin MB, Akter F, et al. Plasmid-mediated sulfamethoxazole resistance encoded by the sul2 gene in the multidrug-resistant Shigella flexneri 2a isolated from patients with acute diarrhea in Dhaka, Bangladesh. PLoS One 2014; 9: e85338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Barman S, Chatterjee S, Chowdhury G, Ramamurthy T, Niyogi SK, Kumar R, et al. Plasmid-mediated streptomycin and sulfamethoxazole resistance in Shigella flexneri 3a. Int J Antimicrob Agents 2010;36: 348– 351 . [DOI] [PubMed] [Google Scholar]
- 26. Bowen A, Hurd J, Hoover C, Khachadourian Y, Traphagen E, Harvey E, et al. Importation and domestic transmission of Shigella sonnei resistant to ciprofloxacin-United States, May 2014–February 2015. MMWR Morb Mortal Wkly Rep 2015;64: 318– 320 . [PMC free article] [PubMed] [Google Scholar]
- 27. Gharibi O, Zangene S, Mohammadi N, Mirzaei K, Karimi A, Gharibi A, et al. Increasing antimicrobial resistance among Shigella isolates in the Bushehr, Iran. Pak J Biol Sci 2012; 15: 156– 159. [DOI] [PubMed] [Google Scholar]
- 28. Jeon Y La, Nam Y, Lim G, Cho SY, Kim Y-T, Jang J-H, et al. Quinolone-resistant Shigella flexneri isolated in a patient who travelled to India. Ann Lab Med 2012;32: 366– 369 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cao Y, Wei D, Kamara IL, Chen W. Multi-Locus Sequence Typing (MLST) and Repetitive Extragenic Palindromic Polymerase Chain Reaction (REP-PCR), characterization of shigella spp. over two decades in Tianjin China. Int J Mol Epidemiol Genet 2012; 3( 4): 321– 332 . [PMC free article] [PubMed] [Google Scholar]