Abstract
Gliomas are primary brain tumors, originating from the glial cells in the brain. In contrast to the more traditional view of glioma as a localized disease, it is becoming clear that global brain functioning is impacted, even with respect to functional communication between brain regions remote from the tumor itself. However, a thorough investigation of glioma-related functional connectomic profiles is lacking. Therefore, we constructed functional brain networks using functional MR scans of 71 glioma patients and 19 matched healthy controls using the automated anatomical labelling (AAL) atlas and interregional Pearson correlation coefficients. The frequency distributions across connectivity values were calculated to depict overall connectomic profiles and quantitative features of these distributions (full-width half maximum (FWHM), peak position, peak height) were calculated. Next, we investigated the spatial distribution of the connectomic profile. We defined hub locations based on the literature and determined connectivity (1) between hubs, (2) between hubs and non-hubs, and (3) between non-hubs. Results show that patients had broader and flatter connectivity distributions compared to controls. Spatially, glioma patients particularly showed increased connectivity between non-hubs and hubs. Furthermore, connectivity distributions and hub-non-hub connectivity differed within the patient group according to tumor grade, while relating to Karnofsky performance status and progression-free survival. In conclusion, newly diagnosed glioma patients have globally altered functional connectomic profiles, which mainly affect hub connectivity and relate to clinical phenotypes. These findings underscore the promise of using connectomics as a future biomarker in this patient population.
Keywords: Functional connectivity, Neuro-oncology, Connectome, Glioma, Network theory, Hubs
Highlights
-
•
Glioma patients have altered connectomic profiles.
-
•
Connectivity between hub and non-hub regions is increased in glioma patients.
-
•
Connectivity between hub regions is decreased in glioma patients.
-
•
Alterations in the connectomic profile relate to clinical phenotypes.
1. Introduction
Gliomas are the most frequently occurring type of primary brain tumors, originating from the glial cells supporting neurons. They can be subdivided according to molecular features, in addition to the commonly used histopathological techniques according to the World Health Organization (WHO) grading system, which classifies them into grades I to IV (Kleihues et al., 2002, Louis et al., 2016). The median overall survival (OS) ranges from seven years in grade II glioma to only fourteen months in grade IV glioma, also termed glioblastoma multiforme (GBM) (Olson et al., 2000, Pignatti et al., 2002, Stupp et al., 2005). Furthermore, OS and progression-free survival (PFS) also depend on age and performance status (Buckner, 2003, Lote et al., 1997).
Glioma has traditionally been viewed as a focal disease. However, it has become increasingly clear that gliomas lead to widespread alterations in functional connectivity, i.e. synchronized activity between brain regions (Aertsen et al., 1989). In glioma patients, a number of studies have used magnetoencephalography (MEG) to investigate whole-brain connectivity patterns. Patients show widespread alterations in functional connectivity; compared to healthy controls, they display higher local connectivity in the lower frequency range and the opposite in the higher frequency range. Global integration is also altered in glioma patients, depending on frequency range and tumor grade (Bartolomei et al., 2006a, Bartolomei et al., 2006b, Bosma et al., 2009, Bosma et al., 2008a, Douw et al., 2010, van Dellen et al., 2012). Moreover, these general network disturbances relate to the symptoms that most patients experience, such as cognitive deficits and epileptic seizures (Derks et al., 2014). Yet, our understanding of the spatial properties of these network disturbances is limited.
Furthermore, the association between connectomic profiles and clinical phenotypes is largely unknown. Connectivity disturbance relate to cognitive functioning (Derks et al., 2014), but the direction of association and specificity of connectomic features is still ambiguous. There is also some evidence that low-grade (i.e. grade II) and high-grade (i.e. grade III and IV) glioma patients show distinct connectivity disturbances (Harris et al., 2014, van Dellen et al., 2012, Zhang et al., 2016), but these investigations concern small cohorts and/or limited investigation of connectomic profiles.
An important next step in research on the interaction between glioma and the functional connectome is the rigorous investigation of so-called hub nodes. Functional brain pathology may occur and spread through the brain according to network-related mechanisms, particularly making use of ‘hubs’ (Aerts et al., 2016, Crossley et al., 2014). Hubs are generally characterized by high information throughput, reflected in the brain by high connectivity or centrality of the region. They are therefore crucial for brain functioning and are intensely used in the healthy brain (Buckner et al., 2009, Crossley et al., 2013, Power et al., 2013). Hypothetically, when (localized) neurological disease occurs, such as glioma, hub regions will eventually be implicated, because all (shortest) routes of communication lead through these areas. In Alzheimer's disease, hub-mediated network failure and its phases has been investigated nicely with computational modeling (de Haan et al., 2012) and in experiments using resting-state functional MRI (fMRI) (Jones et al., 2015) showing a prominent role for hubs in network degeneration. We have previously computationally modeled neurophysiological effects of glioma (van Dellen et al., 2013b), suggesting that hubs indeed have a special role in disease processes, but experimental evidence is lacking.
There is some relevant literature guiding our in-depth analysis of connectomic (hub) profiles in glioma. Several studies using (resting-state) fMRI have specifically focused on subnetworks of the brain, particularly the default mode network (DMN). The DMN, which encompasses the posterior cingulate cortex (PCC), precuneus, medial prefrontal and lateral parietal regions is most active and connected during rest, but also plays a major role in distribution of information processing during active tasks (Anticevic et al., 2012, Buckner and Vincent, 2007, Raichle et al., 2001). In glioma, the DMN seems to show either reduced or increased connectivity strength depending on the specific study, which may or may not correlate with clinical status and/or tumor grade (Esposito et al., 2012, Ghumman et al., 2016, Harris et al., 2014, Tuovinen et al., 2016, Xu et al., 2013, Zhang et al., 2016). However, these studies did not assess the fundamental alterations in patients' full connectomic profiles at diagnosis. Moreover, hub functioning within the context of whole brain connectivity has not been explored yet.
We therefore investigated whole-brain functional connectomic profiles as well as the spatial distribution by investigating hub versus non-hub related connectivity in glioma patients. We expected to find overall shifts in connectomic profiles. Spatially, we expected hub-mediated connectivity alterations in patients. Furthermore, we hypothesized that these disturbances relate to distinct clinical phenotypes.
2. Materials and methods
2.1. Participants
All newly diagnosed glioma patients undergoing preoperative language fMRI at the VUmc CCA Brain Tumor Center Amsterdam between 2006 and July 2015 were eligible for participation. Inclusion criteria were (1) histologically confirmed glioma WHO grade II-IV (grade I was excluded, since these represent mostly non-growing, indolent types of glioma), (2) language fMRI, and (3) structural 3D MRI necessary for co-registration. Exclusion criteria were (1) previous craniotomy, (2) previous chemo- or radiotherapy, and (3) neurological or psychiatric comorbidity, such as cerebrovascular accidents. Karnofsky Performance Status (KPS) is a widely used clinical measure of daily functional impairment in brain tumor patients, ranging from 100 (no symptoms) to 0 (death) (Karnofsky et al., 1948). KPS is measured in steps of 10 units, e.g. the second best score is 90 and not 99. In this study, KPS was summarized categorically as either 90–100 or 70–80 (none of the patients had a KPS lower than 70), with the latter indicating a poorer clinical phenotype. Overall survival (OS) was defined as the number of months between the date of diagnosis and date of death, while progression-free survival (PFS) related to the number of months between date of diagnosis and date of clinical and/or radiological progression as determined by the multidisciplinary tumor board of the VUmc CCA Brain Tumor Center Amsterdam. Patients who had not died at analysis in August 2016 were censored as of the last contact date with their treating neuro-oncologist. Furthermore, age and gender matched healthy controls were used for comparison (Hulst et al., 2012).
The VUmc Medical Ethics Committee approved the study and all subjects provided written informed consent.
2.2. MRI acquisition
Imaging was performed on a 1.5T MR scanner (Siemens Sonata), including an anatomical 3D T1-weighted MPRAGE scan (sequence parameters: TR = 2700 ms, TE = 5.2 ms, TI = 950 ms, 1 mm isotropic resolution, 176 slices). Furthermore, a 3D fluid-attenuated inversion recovery (FLAIR) scan was performed in glioma patients to facilitate tumor masking (TR = 6500 ms, TE = 384 ms, TI = 2200 ms, 1.3 mm isotropic resolution, 160 slices).
In patients, fMRI was performed using a standard echo-planar imaging (EPI) sequence (TR = 2850 ms, TE = 60 ms, 144 volumes, 3.3 mm isotropic resolution, 2 runs of 7 min). The fMRI scan consisted of a language task in which 9 volumes word generation were alternated with 9 volumes rest (imagery of a landscape). No task participation data was available, however, results of this fMRI scan held valuable clinical information and patient were therefore very willing to cooperate.
In controls, EPI was performed during an episodic memory encoding task (Hulst et al., 2012, Van Der Werf et al., 2009), while scanning parameters were comparable to the fMRI collected in patients (TR = 2220 ms, TE = 60 ms, 204 volumes, 3.3 mm isotropic resolution, ~ 7.7 min acquisition). Since this task was different from patients, we also analysed resting-state fMRI data of the same healthy controls, with identical scanning parameters to the patient data (TR = 2850 ms, TE = 60 ms, 190 volumes, 3.3 mm isotropic resolution, ~ 9 min acquisition) (Hulst et al., 2015). It would have been preferable to use the same task in patients and controls, or use resting-state fMRI in both, but these data were not available due to the retrospective nature of the fMRI analyses in the glioma patients. However, previous studies have shown that functional connectivity patterns highly correlate across task states (correlation coefficient between 0.7 and 0.8), supporting the use of these data (Krienen et al., 2014).
2.3. MRI analysis
The imaging processing steps were performed using FSL 5 (FMRIB's software library, http://www.fmrib.ox.ac.uk/fsl). First, non-brain tissue was removed from the 3D T1-weighted images using the Brain Extraction Tool (Smith, 2002), and grey and white matter segmentation was performed using FAST (Zhang et al., 2001). To construct each individual's functional brain network, the Automated Anatomical Labeling (AAL) atlas was used to define 78 cortical regions in each subject (Tzourio-Mazoyer et al., 2002). This atlas was warped from standard space to native space, and masked with each subject's native grey matter mask. In addition to the AAL atlas, 264 regions were defined and the data was analysed in parallel (Power et al., 2011). Furthermore, tumor masks were created manually for each patient, by drawing the tumor on 3D anatomical images slice by slice [LD]. Both contrast enhanced T1-weighted (mainly for high-grade glioma) and FLAIR (mainly for low-grade glioma) images were used to determine which voxels contained tumor. Consequently, AAL regions were excluded on a subject level if fully covered by the tumor mask, since tumor tissue may theoretically alter the BOLD response locally, thereby confounding our network analysis. We therefore excluded these regions from our analysis and used the number of regions overlapping the tumor as a covariate in our analyses. Furthermore, to control for the extent to which the tumor influenced the hub regions, we calculated the number of hub regions that contained tumor and controlled for this variable in our analyses of hub connectivity. In order to exclude partial voluming effects, we calculated this overlap in two ways: (1) by defining overlap as any overlap between the tumor and the AAL hub areas, and (2) by defining overlap if the tumor spread across > 50% of all voxels within the AAL hub area.
2.4. fMRI analysis
Preprocessing of the fMRI data was performed using standard FSL procedures (Smith et al., 2004) included in Melodic (Beckmann et al., 2005), which included discarding the first 5 volumes, motion correction, spatial smoothing (6 mm full width at half maximum Gaussian kernel), and high-pass filtering (100 s cut-off). Following these steps, the functional images were co-registered to the anatomical scans using linear and non-linear co-registration methods (Jenkinson et al., 2002).
Subsequently, only regions with at least 30% of the voxels remaining after registration were included in the analyses using a custom-made fMRI mask based on healthy controls. This custom-made mask was created in order to remove any residual non-brain tissue and to reduce the effect of EPI-distortions, by excluding voxels with signal intensities in the lowest quartile of the robust range. Based on these criteria, ten regions with low signal-to-noise ratios were excluded, mainly in the bilateral orbitofrontal areas. The final atlas therefore segmented the fMRI sequence into 68 grey matter regions for which mean time-series were obtained.
To ascertain that our connectivity results would not be due to motion during fMRI (Van Dijk et al., 2010), we applied strict exclusion criteria for motion: (1) average relative motion ≥ 4 mm, and (2) more than five frame-to-frame movements ≥ 5 mm. Furthermore, average relative motion was calculated per subject to test group differences therein. In order to exclude a confounding effect of frame-to-frame movement on our connectivity analyses we also analysed our patient data after scrubbing time points with > 0.5 mm frame-to-frame displacements. We replicated our main significant results with this method. All connectivity and hub analyses were performed using in-house scripts and the Brain Connectivity Toolbox (Rubinov and Sporns, 2010) in Matlab R2012a (Mathworks, Natick, MA, US). A 68 × 68 connectivity matrix per subject was first created by calculating Pearson correlation coefficients between time series from all 68 regions. The absolute values were then used as a weighted indication of connectivity between all region pairs.
2.5. Connectomic profiles
In order to visualize differences in connectomic profiles, we first created two vectors. Each contained every connectivity value from all matrices in all subjects per group, yielding a patient vector and a control vector of connectivity. Differences between these group-level distributions were tested using Kolmogorov-Smirnov tests for two samples.
Next, we created distribution functions per subject, using the kernel smoothing density function in Matlab. This algorithm does not assume any distribution, but creates equally spaced bins (n = 100) and smoothed frequencies of occurrence according to subjects' individual connectivity values. The advantage of this approach is that no assumption is made regarding to distribution of the values, while being maximally sensitive to between-subject differences in the shape of the underlying distribution, but not the absolute connectivity values.
To further characterize these individual distributions we calculated several measures. First, we calculated full-width half maximum (FWHM), which is the width of the curve measured between those counts that are half the maximum. In other words, FWHM measures the width of the distribution relative to its peak. Higher FWHM indicates a flatter distribution, meaning less variance in the connectivity values on whole brain level. Furthermore, the peak height (i.e. maximum of connectivity occurrences) and peak position (i.e. connectivity value occurring most frequently) were calculated.
2.6. Spatial distribution of the connectomic profile
To investigate the spatial distribution of the connectomic profile we focused on the connectivity between hub regions and non-hub regions. To avoid possible distortion of hubs and/or their locations due to the presence of the tumor in our patient population, we chose to adhere to previously published definitions of hub areas in the brain. The DMN has been amply shown to mainly contain hub areas, using for instance measures of (degree and betweenness) centrality (Buckner et al., 2009, van den Heuvel and Hulshoff Pol, 2010). Other recent studies have also indicated the frontoparietal network (FPN) regions to be hubs, based on their extent of connectivity with other subnetworks in the brain (Cole et al., 2013, Power et al., 2013). Therefore, hubs were defined as regions belonging to the DMN or FPN with selection of these regions from the AAL atlas based on previous work (Tewarie et al., 2013, van Dellen et al., 2013a).
Then, in order to specifically test our hypothesis concerning the spatial distribution of the connectomic profile, we calculated connectivity between hubs, between hubs and non-hubs, and between non-hubs (Fig. 1). All measures were individually normalized for mean intra-individual functional connectivity, since previous work has shown globally altered connectivity in these patients (Bartolomei et al., 2006b, Bosma et al., 2008a, Bosma et al., 2008b), and we were specifically interested in the altered topology of hub connectivity. However, this normalization approach may induce overestimation of differences in hub-hub connectivity, since a larger number of regions is a non-hub. We therefore also tested non-normalized values of hub connectivity.
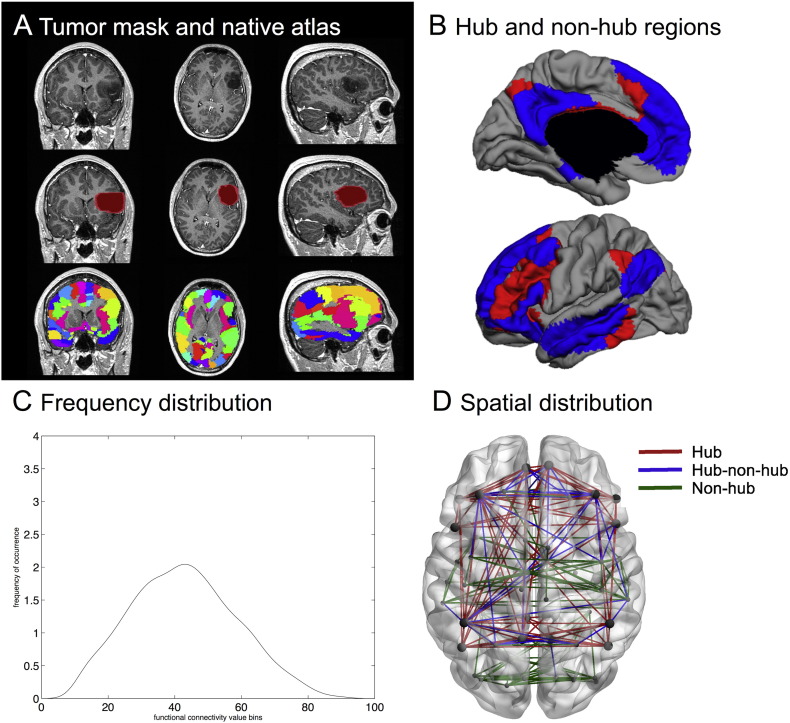
Fig. 1.
Visualization of the analysis pipeline. (A) depicts an exemplar patient MR image in the top row (coronal, axial and sagittal views), with the second row containing the lesion mask and the third row indicating the automated anatomical labelling (AAL) atlas regions in native space. In (B), the hub regions, i.e. those areas belonging to the default mode network (DMN, blue) and the frontoparietal network (FPN, red), are displayed on a surface plot. Grey areas are non-hub regions. (C) depicts our first main outcome parameter, namely frequency distribution. This is a smoothed curve depicting connection strength distribution for a single subject, based on the binning of each element in the adjacency matrix. In (D), the second set of outcome parameters is indicated. The connections between hubs (red), between hubs and non-hubs (blue), and between non-hubs (green) were used to obtain three averages in each subject. Hubs are displayed as larger circles while non-hub regions are represented by the smaller circles. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.7. Second cohort
To attempt to scope the generalizability of our results, a second cohort of glioma patients and healthy controls obtained from a different medical center were used. Data were kindly provided by Dr. M. Raemaekers, Dept. Neurology and Neurosurgery, Brain Center Rudolf Magnus, University Medical Center Utrecht. All images were obtained with a whole body 3.0 Tesla (3T) Philips Achieva MRI scanner (Philips Medical Systems, Best, The Netherlands). The participant's head was held in place with padding. Heartbeat was recorded using a pulse-oximeter placed on the left index finger. Respiration was measured with a pneumatic belt positioned at the level of the abdomen (Birn et al., 2006).
First, a T1-weighted structural image of the whole brain in sagittal orientation was acquired for anatomical reference (3D FFE pulse sequence; acquisition parameters: TR 8.4 ms, TE 3.8 ms; FOV 288 × 288 × 175 mm; voxel size 1 mm isotropic; SENSE p-reduction/s-reduction 2/1.3; flip-angle 8°; 175 slices; scan duration 265.8 s). For functional scans, 3D-PRESTO (Neggers et al., 2008) was used covering the whole brain with the following parameters: TR 22.5 ms; effective TE 32.4 ms; FOV 256 × 224 × 160 mm, voxel size 4 mm isotropic; matrix 64 × 56 × 40; SENSE p-reduction/s-reduction, 1.8/2; flip-angle 10°; scan duration 0.6075 s (for the whole volume). 400 functional images were acquired in sagittal orientation with a foot-head frequency encoding direction. Finally, a PRESTO scan with the same field of view and scan parameters, but with a flip-angle of 27° (called FA27), was acquired in 0.72 s and used in the image coregistration routine (see Section 2.7).
The functional images of the second cohort were first realigned and resliced to the Fa27 of the first scanning session using SPM5 (http://www.fil.ion.ucl.ac.uk/spm/). Then, custom Matlab scripts were used (Aztec, http://www.ni-utrecht.nl/downloads/aztec) for correction of cardio-respiratory artefacts. The correction method used has been described in detail previously (van Buuren et al., 2009). After these corrections, the functional images were high-pass filtered (Gaussian-weighted least squares straight-line fitting, with sigma = 50 s) in FSL, version 5.92 (http://www.fmrib.ox.ac.uk/fsl/) (Smith et al., 2004). Finally, the functional images were skull stripped (Smith, 2002) and normalized by a single scaling factor (grand mean scaling) in FSL. No spatial smoothing was performed on these functional images.
2.8. Statistical analysis
For the statistical analysis, customized scripts in Matlab R2012a (Mathworks, Natick, MA, US) and SPSS 22.0 (IBM Corp, Armonk, NY, US) were employed. Basic differences between patients and healthy controls were investigated using Student's t-tests for independent samples in variables showing a normal distribution (age, average motion), Mann-Whitney U tests for variables not following a normal distribution (number of frame-to-frame movements ≥ 5 mm), and an exact chi-square test for the categorical variable gender. The same tests were used to investigate differences regarding these characteristics between patients based on WHO grade of their glioma, in addition to additional Mann-Whitney U tests for patient-specific non-normally distributed variables (tumor volume, overlap of tumor with hub regions) and chi-square tests for categorical variables (presence of epilepsy, KPS, histopathological type of glioma, and dexamethasone use). To further explore characteristics of our patient cohort, Cox proportional hazards regression analyses were used to investigate OS and PFS in relation to tumor grade, while controlling for KPS, age, and tumor volume.
Connectivity profiles of glioma patients versus healthy controls were first compared using two-sample Kolmogorov-Smirnov tests, which tests the null hypothesis that the summed connectivity profile per group follow the same distribution. We then proceeded to test group differences in the individual quantitative connectivity profile measures (i.e. FWHM, peak position, peak height, hub connectivity, hub-non-hub connectivity, non-hub connectivity) using ANOVAs with age and gender as covariates. Bonferroni correction for multiple comparisons was used on these six tests. Tests of normality showed that three of these measures (FWHM, peak height, and hub-hub connectivity) were not normally distributed (Kolmogorov-Smirnof test for normality p < 0.05). However, ANOVAs are relatively robust to violations of normality in larger datasets (Lumley et al., 2002), while non-parametric tests do not allow for the important corrections for covariates we performed. However, we replicated all main findings using non-parametric tests, to ascertain the validity of our results. To confirm that abovementioned group differences between patients and controls would not be due to the assumption of normality, Mann-Whitney U tests were employed in case of significant ANOVA results.
In order to investigate differences between patients in connectivity profiles depending on tumor grade, general linear modeling took place, using each connectivity measure as the dependent variable, tumor grade as the categorical predictor, and age, tumor volume, KPS, tumor lateralization, and presence of epilepsy as covariates. For the analyses of FWHM and peak characteristics, we controlled for the number of missing regions due to tumor location, while we corrected for tumor overlap with hubs in the hub connectivity analyses. In case of significance, post-hoc testing took place (with Bonferroni correction for multiple comparisons). Again, non-parametric U tests were used to confirm our main findings.
Cox regression analyses were again employed to investigate the relevance of connectivity profiles for OS and PFS. Unfortunately, no non-parametric tests are available for this type of analysis.
A p-value lower than 0.05 (two-tailed) after correction for multiple tests was considered statistically significant. For all main analyses, relevant estimators of effect size are reported.
3. Results
3.1. Subject characteristics
In total, 120 glioma patients were screened for inclusion, but 8 patients were excluded because of neurological comorbidities. Another 41 patients had to be excluded because of poor quality fMRI (n = 30) or extensive motion during functional scans (n = 11). Patient characteristics of the final sample (n = 71) can be found in Table 1.
Table 1.
Subject characteristics.
| Variable | Controls (n = 19) | All patients (n = 71) | WHO grade II patients (n = 41) | WHO grade III patients (n = 17) | WHO grade IV patients (n = 13) |
|---|---|---|---|---|---|
| Age in years (SD) | 43.7 (8.7) | 43.8 (11.2) | 42.5 (9.5) | 42.6 (12.6) | 49.5 (13.4) |
| Number of males (females) | 8 (11) | 46 (25) | 23 (18) | 12 (5) | 11 (2) |
| Number of patients with KPS 90–100 (KPS 70–80)⁎ | NA | 30 (41) | 21 (20) | 8 (9) | 1 (12) |
| Mean motion (SD) | 0.081 (0.039) | 0.089 (0.039) | 0.082 (0.037) | 0.092 (0.029) | 0.110 (0.052) |
| Median number of movements > 5 mm (range) | 0 (0–3) | 0 (0–5) | 0 (0–5) | 0 (0–5) | 1 (0–5) |
| Number of patients with 0/1/>2 missing regions | NA | 44/18/9 | 30/8/3 | 10/4/3 | 4/6/3 |
| Median hub overlap strict/proportional (range)⁎ | NA | 3/2 (0–9/0–6) | 2/2 (0–8/0–6) | 4/3 (0–9/0–6) | 2/2 (0–5/0–5) |
| Mean tumor volume in cm3 (SD) | NA | 685 (545) | 542 (488) | 860 (536) | 909 (574) |
| Number of patients with A/O/OA/GBM | NA | 26/21/11/13 | 19/13/9/0 | 7/8/2/0 | 0/0/0/13 |
| Number of patients with LOH/no LOH/unknown | NA | 14/4/53 | 4/1/36 | 10/3/4 | 0/0/13 |
| Number of patients with left (right) tumor lateralization | NA | 40 (31) | 23 (18) | 9 (8) | 8 (5) |
| Median OS in months (number of censored patients) | NA | 30 (50) | 48 (33) | 38 (14) | 16 (3) |
| Median PFS in months (number of censored patients) | NA | 19.5 (25) | 23 (14) | 51 (10) | 7.5 (1) |
| Number of patients with epilepsy (without) | NA | 53 (18) | 31 (10) | 12 (5) | 10 (3) |
| Number of patients on DEX/not on DEX/unknown | NA | 6/63/2 | 0/39/2 | 1/16/0 | 5/8/0 |
Indicates p < 0.05 difference between tumor grades. WHO = World Health Organization; SD = standard deviation; KPS = Karnofsky performance status; NA = not applicable; A = astrocytoma; O = oligodendroglioma; OA = oligoastrocytoma; GBM = glioblastoma multiforme; LOH = loss of heterozygosity; OS = overall survival; PFS progression free survival; DEX = dexamethasone.
In addition, 19 healthy control subjects were included (see Table 1), after exclusion of 3 subjects with extensive motion during functional scans. Healthy controls did not differ from patients in terms of age (t(88) = 0.584, p = 0.561), gender (χ2 = 3.213, p = 0.112), average motion (t(88) = − 0.801, p = 0.425), or the number of frame-to-frame movements ≥ 5 mm (Mann-Whitney U = 716.5, p = 0.517).
When comparing patients with different WHO tumor grades, there were no differences regarding age (F(2,70) = 2.142, p = 0.124), average motion (F(2,70) = 2.314, p = 0.107), number of movements ≥ 5 mm (Kruskal-Wallis = 3.230, p = 0.199), gender (χ2 = 3.848, p = 0.132), number of missing regions due to tumor overlap (χ2 = 8.061, p = 0.061), or tumor lateralization (χ2 = 0.224, p = 0.947). There was a significant difference in KPS based on grade (χ2 = 7.877, p = 0.015), with all but one GBM patient having KPS 70–80, whereas approximately half of grade II and III patients had KPS 90–100.
Tumor volume was significantly different between grades (F(2,70) = 3.797, p = 0.027), although post-hoc tests comparing two patient groups with different grades, did not specifically show any significance.
Tumor overlap with the hub regions was present in 91% of patients when using the strictest definition, and in 79% of patients when using the proportional measure of hub overlap. For both measures, the overlap of the glioma with hub regions was significantly different between tumor grades (strict definition: Kruskal-Wallis = 8.287, p = 0.016; proportional definition: Kruskal-Wallis = 7.799, p = 0.020), with grade III tumors showing greater overlap of the tumor with the hub regions than the other two groups. In all following analyses, the proportional measure of hub overlap was used as a covariate.
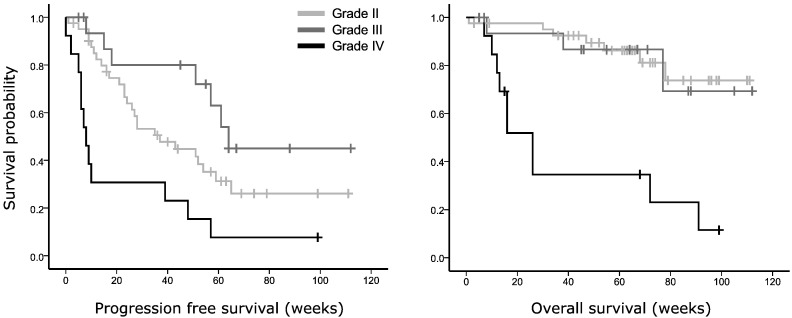
In terms of survival, our cohort showed a significantly shorter OS for GBMs than the two other tumor grades (χ2 = 23.916, p = 0.005, see Fig. 2A), while KPS, age, and tumor volume were non-significant covariates (p = 0.101, p = 0.936 and p = 0.858, resp.). Unexpectedly, the Kaplan-Meier curve showed that grade III tumors had longer PFS than grade II patients in our dataset (χ2 = 14.705, p = 0.006, Fig. 2B), independent of KPS (p = 0.333), age (p = 0.262), and tumor volume (p = 0.996). Upon further investigation, the prognostic beneficial loss of heterozygosity (LOH) at 1p/19q was present in 10 out of 17 grade III glioma, versus 4 of the 41 grade II glioma. Unfortunately, LOH status was not known in most patients, while isocitrate dehydrogenase (IDH) mutation status was unknown in all patients, precluding more in-depth exploration of these important factors in relation to connectomic profiles.
Fig. 2.
Kaplan-Meier survival plots of glioma patients depending on WHO tumor grade. In (A), overall survival is plotted as a function of WHO tumor grade (II, III or IV (GBM), p = 0.004, corrected for Karnofsky performance status (KPS) and tumor volume). (B) Shows progression-free survival per tumor grade (p = 0.007, corrected for KPS and tumor volume).
The second cohort included 23 glioma patients (16 grade II, 5 grade III, and 1 grade IV glioma) and 17 age and gender matched healthy controls.
3.2. Glioma patients show altered connectivity profiles
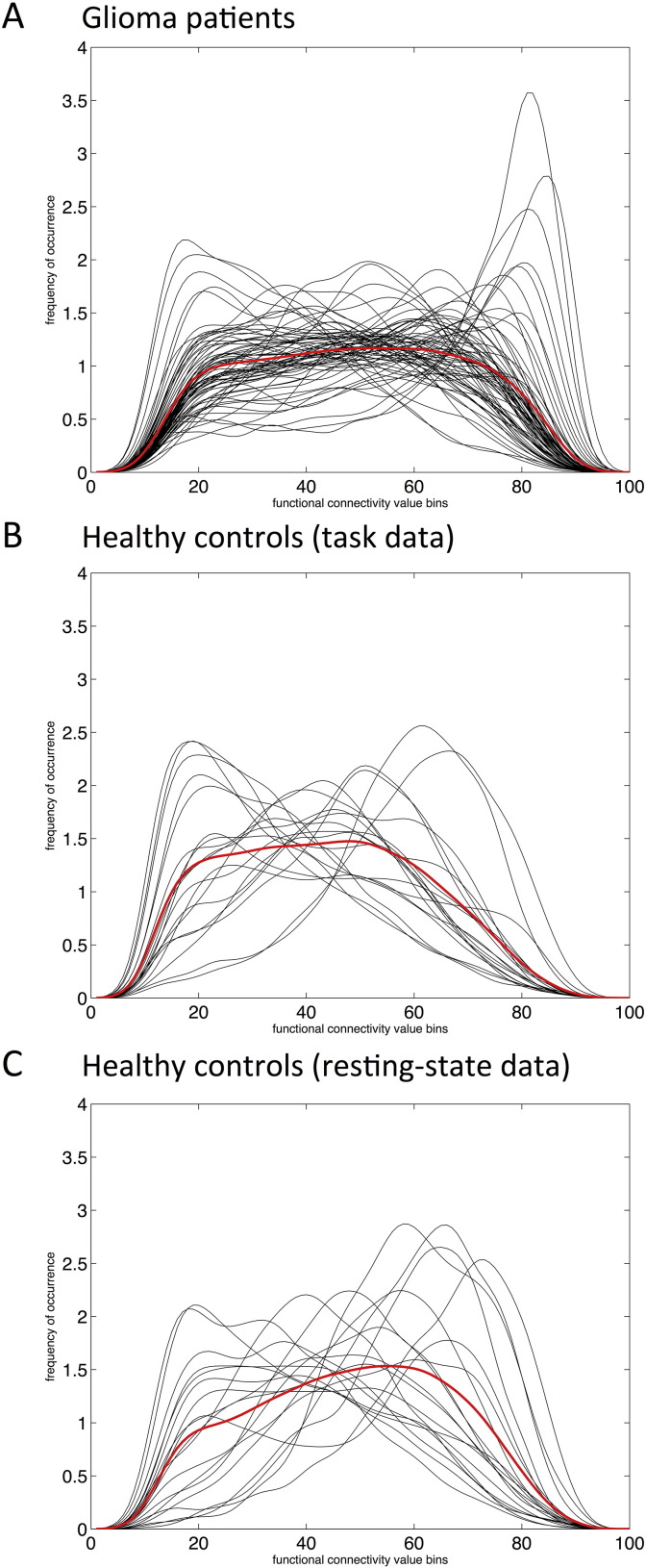
We compared the frequency distribution (FD) of all connectivity values in patients versus controls controls. Patients had significantly different FD compared to controls, both when using the AAL atlas (D = 0.59, p < 0.001) and the 264 region Power atlas (D = 0.08, p < 0.001). Fig. 3 depicts the individually created kernel smoothed density functions of patient and healthy controls.
Fig. 3.
Smoothed connectivity profiles for glioma patients and healthy controls. In all panels, smoothed frequencies of occurrence according to subjects' individual connectivity values (y-axes) were determined for 100 equally spaced bins (x-axes), and depicted in black. Averaged distributions over the entire group are shown in red. (A) Depicts the frequency distribution (FD) of functional connectivity for glioma patients during task-state fMRI (n = 71). In (B), the FD of the healthy controls using task-state fMRI is shown. In (C), the FD of the resting-state data of the same healthy controls is depicted. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To ascertain that differences were robust and not due to the different fMRI tasks, we also compared patients' average FD to that of the same HC group, but in this case using the resting-state data of the controls. Again, there was a significant difference in FD (D = 0.44, p < 0.001). Furthermore, the second, independent cohort showed a comparable difference in FD (D = 0.1165, p < 0.001).
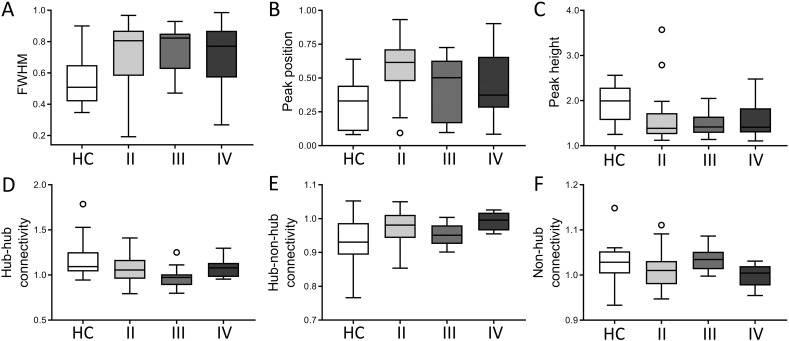
In order to further investigate this shift in connectivity, group differences in full-width half maximum (FWHM), peak position, and peak height of each subjects' kernel smoothed density function were tested using ANOVAs with age and gender as covariates. Patients had significantly higher FWHM and peak position, but lower peak height, than controls (see Table 2 and Fig. 4A), which was confirmed using non-parametric tests (FWHM: Mann-Whitney U = 1057, p < 0.001; peak position U = 1052, p < 0.001, peak height U = 238, p < 0.001). When comparing with the resting-state HC data, results were identical. We therefore proceed only using the task-state data in both populations. After parcellation with the Power atlas, peak height remained significantly different between groups (p = 0.025). In the second cohort, these measures were not significantly different (FWHM p = 0.999, peak position p = 0.350, peak height p = 0.706, all corrected for age and sex).
Table 2.
Group differences in connectivity profiles between patients and controls.
| Measure | Mean HC (SD) | Mean PT (SD) | F-statistic (df) | ƞ2 | p |
|---|---|---|---|---|---|
| FWHM | 0.539 (0.149) | 0.725 (0.191) | 15.752 (1,89) | 0.152 | < 0.001** |
| Peak position | 0.303 (0.180) | 0.527 (0.227) | 15.306 (1,89) | 0.151 | 0.001** |
| Peak height | 1.952 (0.370) | 1.528 (0.406) | 17.607 (1,89) | 0.169 | < 0.001** |
| Hub conn | 1.180 (0.208) | 1.040 (0.126) | 11.270 (1,89) | 0.114 | 0.001** |
| Hub-non-hub conn | 0.931 (0.071) | 0.974 (0.040) | 12.351 (1,89) | 0.129 | 0.001** |
| Non-hub conn | 1.028 (0.439) | 1.013 (0.034) | 2.833 (1,89) | 0.033 | 0.096 |
ANOVAs were corrected for sex, age, and motion, which were not significant in any model. As a measure of effect size, eta squared is reported. **Denotes p < 0.01 (after correction for multiple comparisons). Values in bold represent identical significance when using the 264 region atlas instead of the AAL atlas. HC = healthy controls (n = 19); SD = standard deviation; PT = glioma patients (n = 71), FWHM = full-width half maximum, conn = connectivity.
Fig. 4.
Connectivity profiles per subgroup. This figure depicts boxplots for all connectivity profiles of healthy controls (HC), as well as glioma patients specified according to WHO tumor grade (II, III, and IV). In the top row, the whole-brain measures based on the frequency distribution ((A) full-width half maximum (FWHM), (B) peak position and (C) peak height) are indicated. In the bottom row, average normalized connectivity (D) between hubs, (E) between hubs and non-hubs, and (F) between non-hubs is shown.
3.3. Glioma patients show altered spatial distributions of the connectomic profile
Overall, patients had higher connectivity over all links in their connectome (p < 0.001, corrected for age, gender, and motion). Alterations in hub and non-hub connectivity were subsequently investigated. While normalized connectivity between non-hub regions was not statistically different between groups (p = 0.096, see Table 2), glioma patients had higher hub-non-hub connectivity (p = 0.001) and lower hub connectivity (p = 0.001) than healthy controls (see Fig. 4B). However, non-normalized values of hub-hub connectivity were not significantly different (p = 0.659), suggesting that the spatial connectivity shift was particularly present in connections of non-hubs. Indeed, hub-non-hub connectivity remained significantly increased in patients (p = 0.001), even after correcting for their globally increased level of connectivity. The same patterns of difference were seen when applying the Power atlas to these data (normalized hub-hub connectivity p < 0.001, non-normalized hub-hub connectivity p = 0.637, normalized hub-non-hub connectivity p = 0.056). The result with respect to non-normalized hub-hub connectivity reached near significance in the smaller second cohort (p = 0.073), but hub-non-hub connectivity was not significantly different (p = 0.156).
3.4. Connectomic profiles distinctly relate to tumor grade
We sought to further investigate hub connectivity profiles within the main cohort of glioma patients for all measures differing between patients and controls (see Table 3). Grade II glioma patients showed significantly higher peak position than both grade III and IV patients (see also Fig. 4A). Results were also significant when using non-parametric Kruskal-Wallis testing (test statistic = 6.5, p = 0.039). When looking at the difference in peak position of grade II glioma patients with high (90–100) or low (70–80) KPS, patients with lower KPS tended to have higher, i.e. more disturbed, peak position than patients with higher KPS (t(39) = 1.931, p = 0.061).
Table 3.
Group differences in connectivity profiles between tumor grades.
| Measure | F-statistic (df) |
Apriori statistics |
Post-hoc statistics |
|||
|---|---|---|---|---|---|---|
| ƞ2 | p | Grade | Cohen's d | p | ||
| FWHM | 0.322 (2,60) | 0.010 | 0.726 | NA | NA | NA |
| Peak position | 5.147 (2,60) | 0.137 | 0.009** | II vs III | 0.707 | 0.026* |
| II vs IV | 0.592 | 0.048* | ||||
| Peak height | 0.393 (2,60) | 0.012 | 0.676 | NA | NA | NA |
| Hub conn | 2.990 (2,62) | 0.076 | 0.058 | NA | NA | NA |
| Hub-non-hub conn | 3.513 (2,62) | 0.097 | 0.036* | III vs IV | 1.404 | 0.037* |
ANOVAs were corrected for several covariates (see Statistical analysis section), which were not significant in any model. As measures of effect size, eta squared and Cohen's d are reported. Post-hoc statistics are Bonferroni corrected for multiple comparisons. *Denotes p < 0.05, **denotes p < 0.01. FWHM = full-width half maximum, conn = connectivity; NA = not applicable.
Upon visual inspection of Fig. 4B, we see that grade III glioma patients overall show the most distinct pattern compared to the grade II and grade IV glioma patients, while the latter two groups have largely comparable hub connectivity patterns. However, none of these differences were objectified statistically.
3.5. Connectomic profiles relate to progression-free survival in GBM patients
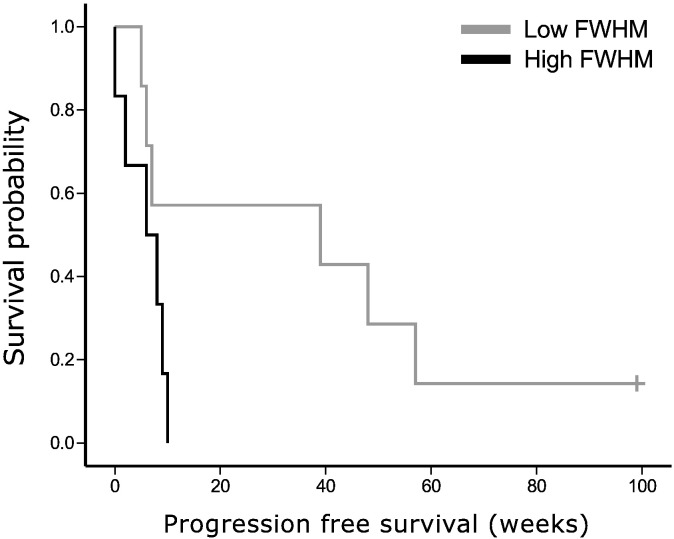
For each tumor grade separately, we performed Cox regression analysis on both PFS and OS. In grade II and III glioma, connectivity profile measures were not significantly related to survival. However, higher FWHM was a significant predictor of shorter PFS in GBM patients (beta = 1536, 95% confidence interval [1.294 1824193], p = 0.042), while using age (p = 0.134), KPS (p = 0.086), and tumor volume (p = 0.175) as covariates (see Fig. 5 for the Kaplan-Meier curve based on the median split FWHM).
Fig. 5.
Progression-free survival relates to connectivity profile in grade IV glioma. Full-width half maximum (FWHM), based on the frequency distribution, was dichotomized using a median split in order to draw this Kaplan-Meier plot of progression-free survival (PFS) in grade IV glioma patients (p = 0.042, while correcting for Karnofsky performance status and tumor volume).
4. Discussion
The brain network of newly diagnosed glioma patients is characterized by shifts in functional connectomic profiles compared to healthy controls, as is shown across cohorts and methods of analysis. The connectomic profile of glioma patients is characterized by a broader and flatter frequency distribution showing less variance in connectivity. On the spatial level, our results confirm that most significant changes occur in connectivity between hub and non-hub regions: patients have increased connectivity between non-hubs and hubs across normalized and non-normalized connectivity values and atlases. However, connectivity between non-hubs is not significantly different.
Hubs are implicated in a variety of neurological diseases including glioma (Aerts et al., 2016, Crossley et al., 2014). A theoretical explanation for the particular redistribution of connectivity in glioma patients relates to cascadic network failure (Stam, 2014). In line with all (interdependent) complex networks (Buldyrev et al., 2010), a final common pathological pathway across neurological diseases may be the global spread of brain network failure. This cascade is hypothetically due to the altered role and eventual loss of hub regions (Crossley et al., 2014, Stam, 2014): when (localized) neurological disease occurs, such as glioma, the inflicted region may develop diminished communication with other brain regions due to disconnection or altered activity patterns. This network dysfunction then spreads to other regions in the vicinity, as they take over functioning from the lesioned region. Next, more and more information is relayed to these regions as well as to hubs higher up in the network hierarchy. In this initial phase of hub overload, connectivity likely increases between non-hub and hub regions, as the latter aggregate more and more information. This increase in hub enlistment may initially support functioning, after which hub-hub connectivity should decrease as hubs start failing due to their increased usage. In Alzheimer's disease, cascadic brain network failure and its phases has been investigated nicely with computational modeling (de Haan et al., 2012) and in experiments using resting-state fMRI (Jones et al., 2015). Furthermore, the abovementioned recent review on the impact of lesions on the brain network also concludes that the damaging effects of for instance glioma may propagate throughout the brain network via hubs, thereby inducing clinical deterioration (Aerts et al., 2016). Our current results corroborate this idea, showing particularly increased connectivity between non-hub and hubs. Longitudinal or computational studies are necessary to further scope network failure over time in glioma patients.
Although glioma patients with different tumor grades all showed the same type of connectivity disturbances compared to healthy controls, our study additionally highlights distinct connectomic profiles relating to clinical phenotype. Peak position was particularly disturbed in low-grade glioma patients, in whom the connectivity value of the peak occurrence was significantly higher than in high-grade glioma patients. In other words, grade II glioma patients most often had connectivity indices higher than the other tumor grades, which was spatially unspecific, as it was not reflected in the hub-related connectivity measures. This increase tended to relate to lower Karnofsky performance status, suggesting a pathological mechanism. Since low-grade glioma grow slowly and in an infiltrative manner, it is possible that these tumors cause even greater connectivity pathology than the high-grade glioma, as has been shown with MEG before (van Dellen et al., 2012).
Interestingly, the grade III gliomas formed a special subgroup in our cohort. First, progression-free survival of our grade III patients was better than expected and even exceeded that of grade II patients. Upon further investigation, loss of heterozygosity at 1p/19q, which is a favorable prognostic factor (Cairncross et al., 1998), was often present in the grade III tumors, suggesting that these patients were representative only of a subgroup of all anaplastic gliomas with relatively favorable prognosis. In terms of connectivity profiles, grade III patients tended to show less disturbances than both grade II and IV glioma patients. This was particularly true with respect to hub-non-hub connectivity, which was within the normal range. Interestingly grade III glioma patients did have the strongest decrease in hub-hub connectivity of all patient subgroups. However, in grade III glioma patients the tumor was more frequently localized in the hub areas, which will certainly have influenced connectivity between hub nodes specifically. The small sample of grade III patients (n = 17) limits our options to further investigate the mechanism underlying connectivity profile shifts in these patients. Future studies will have to elucidate whether the combination of preserved hub-non-hub connectivity but decreased hub connectivity is representative of grade III glioma as a whole, or relates to the favorable aspects of this particular cohort (i.e. 1p/19q LOH, long progression-free survival).
All patients in our cohort were newly diagnosed with glioma. However, one must keep in mind that it is near impossible to determine the exact disease duration in these patients, which could influence connectivity profiles as well as clinical status. A glioma may be present for years, before the patient experiences symptoms and a diagnosis is reached. Our findings, particularly with respect to the differences between tumor grades, were corrected for covariates possibly reflecting differences in disease duration and/or clinical phenotype, such as tumor volume and presence of epilepsy. Therefore, only longitudinal studies taking growth speed into account may delve into the mechanisms of change according to clinical phenotypes. A foreseeable problem in this respect is the tumor treatment (e.g. resection, chemotherapy, radiotherapy) that most if not all glioma patients undergo, possibly confounding the natural course of connectivity shifts.
Some limitations of the current study should be kept in mind. Firstly, fMRI tasks differed between patients and controls in our main analyses. Moreover, areas of the DMN and FPN networks have been found to be hubs consistently across brain states, with differences in connectivity between states being small (James et al., 2015, Krienen et al., 2014). In addition, pre-operative language fMRI has been used in previous work investigating connectivity in brain tumor patients (Esposito et al., 2012). Furthermore, our findings regarding the differences between patient groups are completely independent of the healthy control cohort. However, we attempted to scope the generalizability of our results by including a smaller second cohort of glioma patients. This database was much smaller and did not contain clinical information, but patients (n = 23) and controls (n = 17) did undergo identical resting state fMRI. We observed altered FD in these groups as well, corroborating our main analyses. Furthermore, there were trend-level differences between healthy controls and glioma patients regarding hub connectivity. In addition to the smaller size, this sample was characterized by a different distribution of tumor grades compared to the original cohort, which in addition to limited statistical power may explain some of the non-significant findings. Second, the boundaries of gliomas are challenging to define, due to infiltration of tumor into the healthy tissue at a microscopic scale and the lack of specificity of imaging. Therefore, the study design could neither guarantee exclusion of all regions of the AAL atlas covered by tumor on a subject level, nor complete accuracy of the measurement of overlap between tumor and hubs. Third, due to tumor masking and subsequent exclusion of regions on an individual level, the number of regions taken into account for calculation of connectivity differed between subjects, which may have impacted our results, even though tumor volume, individual tumor overlap with the hubs, and number of regions excluded did not reach any statistical significance. Fourth, glioma patients selected to undergo preoperative language mapping may not be representative of glioma patients in general, as patients with a priori disrupted language or highly aggressive tumors are unlikely to undergo such a work-up. Thus, this sample may be biased towards relatively high functioning patients and/or less acute disease, which is corroborated by patient characteristics, particularly for the grade III glioma.
To conclude, our current results reveal distinct connectomic profiles as compared to healthy subjects. Glioma patients seem to lose structure in their connectivity profile, which is spatially reflected by increased connectivity between periphery and hubs. Furthermore, even in this relatively small sample, we are able to report distinct connectomic profiles for different clinical phenotypes, relating to performance status and survival. This study firstly provides a conceptual framework for connectomic dysfunction in glioma patients, and secondly underlines the promise of using connectomics as a biomarker in this patient population.
Conflict of interest
The authors declare that there are no conflicts of interest.
References
- Aerts H., Fias W., Caeyenberghs K., Marinazzo D. Brain networks under attack: robustness properties and the impact of lesions. Brain. 2016 doi: 10.1093/brain/aww194. [DOI] [PubMed] [Google Scholar]
- Aertsen A.M., Gerstein G.L., Habib M.K., Palm G. Dynamics of neuronal firing correlation: modulation of “effective connectivity”. J. Neurophysiol. 1989;61:900–917. doi: 10.1152/jn.1989.61.5.900. [DOI] [PubMed] [Google Scholar]
- Anticevic A., Cole M.W., Murray J.D., Corlett P.R., Wang X.-J., Krystal J.H. The role of default network deactivation in cognition and disease. Trends Cogn. Sci. 2012;16:584–592. doi: 10.1016/j.tics.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomei F., Bosma I., Klein M., Baayen J.C., Reijneveld J.C., Postma T.J., Heimans J.J., van Dijk B.W., de Munck J.C., de Jongh A., Cover K.S., Stam C.J. Disturbed functional connectivity in brain tumour patients: evaluation by graph analysis of synchronization matrices. Clin. Neurophysiol. 2006;117:2039–2049. doi: 10.1016/j.clinph.2006.05.018. [DOI] [PubMed] [Google Scholar]
- Bartolomei F., Bosma I., Klein M., Baayen J.C., Reijneveld J.C., Postma T.J., Heimans J.J., van Dijk B.W., de Munck J.C., de Jongh A., Cover K.S., Stam C.J. How do brain tumors alter functional connectivity? A magnetoencephalography study. Ann. Neurol. 2006;59:128–138. doi: 10.1002/ana.20710. [DOI] [PubMed] [Google Scholar]
- Beckmann C.F., DeLuca M., Devlin J.T., Smith S.M. Investigations into resting-state connectivity using independent component analysis. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2005;360:1001–1013. doi: 10.1098/rstb.2005.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birn R.M., Diamond J.B., Smith M.A., Bandettini P.A. Separating respiratory-variation-related fluctuations from neuronal-activity-related fluctuations in fMRI. NeuroImage. 2006;31:1536–1548. doi: 10.1016/j.neuroimage.2006.02.048. [DOI] [PubMed] [Google Scholar]
- Bosma I., Douw L., Bartolomei F., Heimans J.J., van Dijk B.W., Postma T.J., Stam C.J., Reijneveld J.C., Klein M. Synchronized brain activity and neurocognitive function in patients with low-grade glioma: a magnetoencephalography study. Neuro-Oncology. 2008;10:734–744. doi: 10.1215/15228517-2008-034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosma I., Douw L., Bartolomei F., Heimans J.J., van Dijk B.W., Postma T.J., Stam C.J., Reijneveld J.C., Klein M. The influence of low-grade glioma on resting state oscillatory brain activity: a magnetoencephalography study. J. Neuro-Oncol. 2008;88:734–744. doi: 10.1007/s11060-008-9535-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosma I., Reijneveld J.C., Klein M., Douw L., van Dijk B.W., Heimans J.J., Stam C.J., Bartolomei F. Disturbed functional brain networks and neurocognitive function in low-grade glioma patients: a graph theoretical analysis of resting-state MEG. Nonlinear Biomed. Phys. 2009;3:9. doi: 10.1186/1753-4631-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner J.C. Factors influencing survival in high-grade gliomas. Semin. Oncol. 2003;30:10–14. doi: 10.1053/j.seminoncol.2003.11.031. [DOI] [PubMed] [Google Scholar]
- Buckner R.L., Vincent J.L. Unrest at rest: default activity and spontaneous network correlations. NeuroImage. 2007;37 doi: 10.1016/j.neuroimage.2007.01.010. 10.1016/j.neuroimage.2007.01.010 1091-6-9. [DOI] [PubMed] [Google Scholar]
- Buckner R.L., Sepulcre J., Talukdar T., Krienen F.M., Liu H., Hedden T., Andrews-Hanna J.R., Sperling R.A., Johnson K.A. Cortical hubs revealed by intrinsic functional connectivity: mapping, assessment of stability, and relation to Alzheimer's disease. J. Neurosci. 2009;29:1860–1873. doi: 10.1523/JNEUROSCI.5062-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buldyrev S.V., Parshani R., Paul G., Stanley H.E., Havlin S. Catastrophic cascade of failures in interdependent networks. Nature. 2010;464:1025–1028. doi: 10.1038/nature08932. [DOI] [PubMed] [Google Scholar]
- Cairncross J.G., Ueki K., Zlatescu M.C., Lisle D.K., Finkelstein D.M., Hammond R.R., Silver J.S., Stark P.C., Macdonald D.R., Ino Y., Ramsay D.A., Louis D.N. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. JNCI J. Natl. Cancer Inst. 1998;90:1473–1479. doi: 10.1093/jnci/90.19.1473. [DOI] [PubMed] [Google Scholar]
- Cole M.W., Reynolds J.R., Power J.D., Repovs G., Anticevic A., Braver T.S. Multi-task connectivity reveals flexible hubs for adaptive task control. Nat. Neurosci. 2013;16:1348–1355. doi: 10.1038/nn.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley N.A., Mechelli A., Vértes P.E., Winton-Brown T.T., Patel A.X., Ginestet C.E., McGuire P., Bullmore E.T. Cognitive relevance of the community structure of the human brain functional coactivation network. Proc. Natl. Acad. Sci. U. S. A. 2013;110:11583–11588. doi: 10.1073/pnas.1220826110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley N.A., Mechelli A., Scott J., Carletti F., Fox P.T., McGuire P., Bullmore E.T. The hubs of the human connectome are generally implicated in the anatomy of brain disorders. Brain. 2014;137:2382–2395. doi: 10.1093/brain/awu132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan W., Mott K., van Straaten E.C.W., Scheltens P., Stam C.J. Activity dependent degeneration explains hub vulnerability in Alzheimer's disease. PLoS Comput. Biol. 2012;8 doi: 10.1371/journal.pcbi.1002582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derks J., Reijneveld J.C., Douw L. Neural network alterations underlie cognitive deficits in brain tumor patients. Curr. Opin. Oncol. 2014;26:627–633. doi: 10.1097/CCO.0000000000000126. [DOI] [PubMed] [Google Scholar]
- Douw L., van Dellen E., de Groot M., Heimans J.J., Klein M., Stam C.J., Reijneveld J.C. Epilepsy is related to theta band brain connectivity and network topology in brain tumor patients. BMC Neurosci. 2010;11:103. doi: 10.1186/1471-2202-11-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito R., Mattei P.A., Briganti C., Romani G.L., Tartaro A., Caulo M. Modifications of default-mode network connectivity in patients with cerebral glioma. PLoS One. 2012;7 doi: 10.1371/journal.pone.0040231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghumman S., Fortin D., Noel-Lamy M., Cunnane S.C., Whittingstall K. Exploratory study of the effect of brain tumors on the default mode network. J. Neuro-Oncol. 2016;128:437–444. doi: 10.1007/s11060-016-2129-6. [DOI] [PubMed] [Google Scholar]
- Harris R.J., Bookheimer S.Y., Cloughesy T.F., Kim H.J., Pope W.B., Lai A., Nghiemphu P.L., Liau L.M., Ellingson B.M. Altered functional connectivity of the default mode network in diffuse gliomas measured with pseudo-resting state fMRI. J. Neuro-Oncol. 2014;116:373–379. doi: 10.1007/s11060-013-1304-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulst H.E., Schoonheim M.M., Roosendaal S.D., Popescu V., Schweren L.J.S., van der Werf Y.D., Visser L.H., Polman C.H., Barkhof F., Geurts J.J.G. Functional adaptive changes within the hippocampal memory system of patients with multiple sclerosis. Hum. Brain Mapp. 2012;33:2268–2280. doi: 10.1002/hbm.21359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulst H.E., Schoonheim M.M., Van Geest Q., Uitdehaag B.M.J., Barkhof F., Geurts J.J.G. Memory impairment in multiple sclerosis: relevance of hippocampal activation and hippocampal connectivity. Mult. Scler. 2015;21:1705–1712. doi: 10.1177/1352458514567727. [DOI] [PubMed] [Google Scholar]
- James G., Hazaroglu O., Bush K.A. A human brain atlas derived via n-cut parcellation of resting-state and task-based fMRI data. Magn. Reson. Imaging. 2015 doi: 10.1016/j.mri.2015.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M., Bannister P., Brady M., Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. NeuroImage. 2002;17:825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- Jones D.T., Knopman D.S., Gunter J.L., Graff-Radford J., Vemuri P., Boeve B.F., Petersen R.C., Weiner M.W., Jack C.R. Cascading network failure across the Alzheimer's disease spectrum. Brain. 2015;139:547–562. doi: 10.1093/brain/awv338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnofsky D.A., Abelmann W.H., Craver L.F. The use of nitrogen mustards in the palliative treatment of carcinoma. Cancer. 1948;1:634–656. [Google Scholar]
- Kleihues P., Louis D.N., Scheithauer B.W., Rorke L.B., Reifenberger G., Burger P.C., Cavenee W.K. The WHO classification of tumors of the nervous system. J. Neuropathol. Exp. Neurol. 2002;61:215–219. doi: 10.1093/jnen/61.3.215. [DOI] [PubMed] [Google Scholar]
- Krienen F.M., Yeo B.T.T., Buckner R.L. Reconfigurable task-dependent functional coupling modes cluster around a core functional architecture. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2014;369 doi: 10.1098/rstb.2013.0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lote K., Egeland T., Hager B., Stenwig B., Skullerud K., Berg-Johnsen J., Storm-Mathisen I., Hirschberg H. Survival, prognostic factors, and therapeutic efficacy in low-grade glioma: a retrospective study in 379 patients. J. Clin. Oncol. 1997;15:3129–3140. doi: 10.1200/JCO.1997.15.9.3129. [DOI] [PubMed] [Google Scholar]
- Louis D.N., Perry A., Reifenberger G., von Deimling A., Figarella-Branger D., Cavenee W.K., Ohgaki H., Wiestler O.D., Kleihues P., Ellison D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- Lumley T., Diehr P., Emerson S., Chen L. The importance of the normality assumption in large public health data sets. Annu. Rev. Public Health. 2002;23:151–169. doi: 10.1146/annurev.publhealth.23.100901.140546. [DOI] [PubMed] [Google Scholar]
- Neggers S.F.W., Hermans E.J., Ramsey N.F. Enhanced sensitivity with fast three-dimensional blood-oxygen-level-dependent functional MRI: comparison of SENSE-PRESTO and 2D-EPI at 3T. NMR Biomed. 2008;21:663–676. doi: 10.1002/nbm.1235. [DOI] [PubMed] [Google Scholar]
- Olson J.D., Riedel E., DeAngelis L.M. Long-term outcome of low-grade oligodendroglioma and mixed glioma. Neurology. 2000;54:1442–1448. doi: 10.1212/wnl.54.7.1442. [DOI] [PubMed] [Google Scholar]
- Pignatti F., van den Bent M., Curran D., Debruyne C., Sylvester R., Therasse P., Afra D., Cornu P., Bolla M., Vecht C., Karim A.B. Prognostic factors for survival in adult patients with cerebral low-grade glioma. J. Clin. Oncol. 2002;20:2076–2084. doi: 10.1200/JCO.2002.08.121. [DOI] [PubMed] [Google Scholar]
- Power J.D., Cohen A.L., Nelson S.M., Wig G.S., Barnes K.A., Church J.A., Vogel A.C., Laumann T.O., Miezin F.M., Schlaggar B.L., Petersen S.E. Functional network organization of the human brain. Neuron. 2011;72:665–678. doi: 10.1016/j.neuron.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power J.D., Schlaggar B.L., Lessov-Schlaggar C.N., Petersen S.E. Evidence for hubs in human functional brain networks. Neuron. 2013;79:798–813. doi: 10.1016/j.neuron.2013.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichle M.E., MacLeod A.M., Snyder A.Z., Powers W.J., Gusnard D.A., Shulman G.L. A default mode of brain function. Proc. Natl. Acad. Sci. U. S. A. 2001;98:676–682. doi: 10.1073/pnas.98.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinov M., Sporns O. Complex network measures of brain connectivity: uses and interpretations. NeuroImage. 2010;52:1059–1069. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Smith S.M. Fast robust automated brain extraction. Hum. Brain Mapp. 2002;17:143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.M., Jenkinson M., Woolrich M.W., Beckmann C.F., Behrens T.E., Johansen-Berg H., Bannister P.R., De Luca M., Drobnjak I., Flitney D.E., Niazy R.K., Saunders J., Vickers J., Zhang Y., De Stefano N., Brady J.M., Matthews P.M. Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage. 2004;23:S208–S219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- Stam C.J. Modern network science of neurological disorders. Nat. Rev. Neurosci. 2014;15:683–695. doi: 10.1038/nrn3801. [DOI] [PubMed] [Google Scholar]
- Stupp R., Mason W.P., van den Bent M.J., Weller M., Fisher B., Taphoorn M.J., Belanger K., Brandes A.A., Marosi C., Bogdahn U., Curschmann J., Janzer R.C., Ludwin S.K., Gorlia T., Allgeier A., Lacombe D., Cairncross J.G., Eisenhauer E., Mirimanoff R.O. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- Tewarie P., Schoonheim M.M., Stam C.J., van der Meer M.L., van Dijk B.W., Barkhof F., Polman C.H., Hillebrand A. Cognitive and clinical dysfunction, altered MEG resting-state networks and thalamic atrophy in multiple sclerosis. PLoS One. 2013;8 doi: 10.1371/journal.pone.0069318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuovinen N., de Pasquale F., Caulo M., Caravasso C.F., Giudice E., Miceli R., Ingrosso G., Laprie A., Santoni R., Sabatini U. Transient effects of tumor location on the functional architecture at rest in glioblastoma patients: three longitudinal case studies. Radiat. Oncol. 2016;11:107. doi: 10.1186/s13014-016-0683-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N., Landeau B., Papathanassiou D., Crivello F., Etard O., Delcroix N., Mazoyer B., Joliot M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. NeuroImage. 2002;15:273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- van Dellen E., Douw L., Hillebrand A., Ris-Hilgersom I.H., Schoonheim M.M., Baayen J.C., De Witt Hamer P.C., Velis D.N., Klein M., Heimans J.J., Stam C.J., Reijneveld J.C. MEG network differences between low- and high-grade glioma related to epilepsy and cognition. PLoS One. 2012;7 doi: 10.1371/journal.pone.0050122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Buuren M., Gladwin T.E., Zandbelt B.B., van den Heuvel M., Ramsey N.F., Kahn R.S., Vink M. Cardiorespiratory effects on default-mode network activity as measured with fMRI. Hum. Brain Mapp. 2009;30:3031–3042. doi: 10.1002/hbm.20729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dellen E., de Witt Hamer P.C., Douw L., Klein M., Heimans J.J., Stam C.J., Reijneveld J.C., Hillebrand A. Connectivity in MEG resting-state networks increases after resective surgery for low-grade glioma and correlates with improved cognitive performance. NeuroImage Clin. 2013;2:1–7. doi: 10.1016/j.nicl.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dellen E., Hillebrand A., Douw L., Heimans J.J., Reijneveld J.C., Stam C.J. Local polymorphic delta activity in cortical lesions causes global decreases in functional connectivity. NeuroImage. 2013;83:524–532. doi: 10.1016/j.neuroimage.2013.06.009. [DOI] [PubMed] [Google Scholar]
- van den Heuvel M.P., Hulshoff Pol H.E. Exploring the brain network: a review on resting-state fMRI functional connectivity. Eur. Neuropsychopharmacol. 2010;20:519–534. doi: 10.1016/j.euroneuro.2010.03.008. [DOI] [PubMed] [Google Scholar]
- Van Der Werf Y.D., Altena E., Schoonheim M.M., Sanz-Arigita E.J., Vis J.C., De Rijke W., Van Someren E.J.W. Sleep benefits subsequent hippocampal functioning. Nat. Neurosci. 2009;12:122–123. doi: 10.1038/nn.2253. [DOI] [PubMed] [Google Scholar]
- Van Dijk K.R.A., Hedden T., Venkataraman A., Evans K.C., Lazar S.W., Buckner R.L. Intrinsic functional connectivity as a tool for human connectomics: theory, properties, and optimization. J. Neurophysiol. 2010;103:297–321. doi: 10.1152/jn.00783.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Ding S., Hu X., Yang K., Xiao C., Zou Y., Chen Y., Tao L., Liu H., Qian Z. Reduced efficiency of functional brain network underlying intellectual decline in patients with low-grade glioma. Neurosci. Lett. 2013;543:27–31. doi: 10.1016/j.neulet.2013.02.062. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Brady M., Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans. Med. Imaging. 2001;20:45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- Zhang H., Shi Y., Yao C., Tang W., Yao D., Zhang C., Wang M., Wu J., Song Z. Alteration of the intra- and cross- hemisphere posterior default mode network in frontal lobe glioma patients. Sci. Rep. 2016;6:26972. doi: 10.1038/srep26972. [DOI] [PMC free article] [PubMed] [Google Scholar]