ABSTRACT
Staphylococcus aureus is an important opportunistic pathogen and is the etiological agent of many hospital- and community-acquired infections. The golden pigment, staphyloxanthin, of S. aureus colonies distinguishes it from other staphylococci and related Gram-positive cocci. Staphyloxanthin is the product of a series of biosynthetic steps that produce a unique membrane-embedded C30 golden carotenoid and is an important antioxidant. We observed that a strain with an inducible airR overexpression cassette had noticeably increased staphyloxanthin production compared to the wild-type strain under aerobic culturing conditions. Further analysis revealed that depletion or overproduction of the AirR response regulator resulted in a corresponding decrease or increase in staphyloxanthin production and susceptibility to killing by hydrogen peroxide, respectively. Furthermore, the genetic elimination of staphyloxanthin during AirR overproduction abolished the protective phenotype of increased staphyloxanthin production in a whole-blood survival assay. Promoter reporter and gel shift assays determined that the AirR response regulator is a direct positive regulator of the staphyloxanthin-biosynthetic operon, crtOPQMN, but is epistatic to alternative sigma factor B. Taken together, these data indicate that AirSR positively regulates the staphyloxanthin-biosynthetic operon crtOPQMN, promoting survival of S. aureus in the presence of oxidants.
KEYWORDS: S. aureus, staphyloxanthin, transcriptional regulation, AirSR, two-component regulatory systems
INTRODUCTION
Staphylococcus aureus is a leading bacterial agent of hospital- and community-acquired infections ranging from infective endocarditis and osteomyelitis to soft tissue infection (1). S. aureus possesses a vast array of adhesion and virulence factors that allow the organism to infect any part of the body and efficiently evade the immune system. S. aureus colonies are orange to golden (hence the species name, aureus), which was previously used as a distinguishing characteristic and is now recognized as the product of an important virulence factor (2).
The distinctive golden pigment of S. aureus is the biosynthetic product of crtOPQMN and aldH (3, 4). The final product of this six-enzyme-biosynthetic pathway is a membrane-embedded golden C30 triterpenoid carotenoid, α-d-glucopyranosyl-1-O-(4,4′-diaponeurosporen-4-oate)-6-O-(12-methyltetradecanoate), termed staphyloxanthin (STX) (5). STX is an antioxidant (6) and an important virulence factor of S. aureus, promoting intracellular phagocyte survival, and is linked to resistance to reactive oxygen species (ROS) produced by the NADPH oxidase system within the phagocyte phagosome (2).
Currently, it is known that the stress response alternative sigma factor B (SigB; σB) regulates STX production, as a σB deletion mutant is white and lacks STX. This regulation is likely direct, as a consensus σB DNA binding motif was identified in the crt promoter, but direct interaction has not been confirmed (7). Additionally, cold shock protein A (CspA) is a positive regulator, likely in a σB-dependent manner (8). The metabolism of S. aureus is also known to influence pigment production, as mutants with a defective tricarboxylic acid cycle or inability to perform oxidative phosphorylation show increased pigmentation (9, 10). In addition to protein-mediated regulation, the small, stable RNA A (SsrA transfer-messenger RNA [tmRNA]) acts as an antisense RNA, base-pairing in the 5′ untranslated region (UTR) of crtMN to regulate STX production (11).
Two-component signal transduction systems (TCSs) are important regulators of staphylococcal metabolism (12–15) and coordinate expression of virulence factors. Many of the 16 known TCSs, including the quorum-sensing Agr system (16, 17), the regulator of staphylococcal accessory exoproteins SaeRS system (18–20), and autolysin-related locus ArlRS system (21), are well characterized and extensively studied. The oxygen-sensing and redox-signaling AirSR TCS is a global regulator; including regulation of pathways for nitrate respiration and lactose catabolism, as well as virulence factors (12, 14, 15, 22).
During characterization of the several AirSR mutant strains, it was observed that overexpression of the airR response regulator increased colony pigmentation. STX carotenoid is an important staphylococcal antioxidant. Further, it is known that the AirSR TCS phosphorylation relay is activated in the presence of oxygen or mild oxidants (12). Therefore, it was hypothesized that, in the presence of oxidants, the AirSR TCS transcriptionally regulates the STX-biosynthetic operon, crtOPQMN, thereby increasing the antioxidant capacity of the cell. In support of this hypothesis, data are presented demonstrating that the AirSR TCS directly transcriptionally activates the crtOPQMN operon and modulates STX carotenoid production. Moreover, AirSR's regulation of staphyloxanthin influences S. aureus susceptibility to hydrogen peroxide (H2O2) and survival in human whole blood.
RESULTS
Overproduction of AirR enhances the golden color of S. aureus colonies.
The airSR (formerly yhcSR) TCS was initially characterized as an essential operon in the methicillin-resistant S. aureus (MRSA) strain WCUH29 (NCIMB 40771) (23), but it was subsequently determined that the essentiality of the TCS is strain dependent (22). Construction of isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible WCUH29 ΔairS::Pspac-airS and WCUH29 ΔairR::Pspac-airR strains was undertaken before the strain dependence of airSR essentiality was recognized (23, 24). Subsequently, in the absence of the Pspac promoter inducer IPTG, airS expression was increased almost 3-fold over endogenous airS expression, as measured by semiquantitative reverse transcription (RT)-PCR, presumably due to “leaky” Pspac promoter activity (data not shown) (25). The leaky expression prevented IPTG-dependent titration of the growth of each strain.
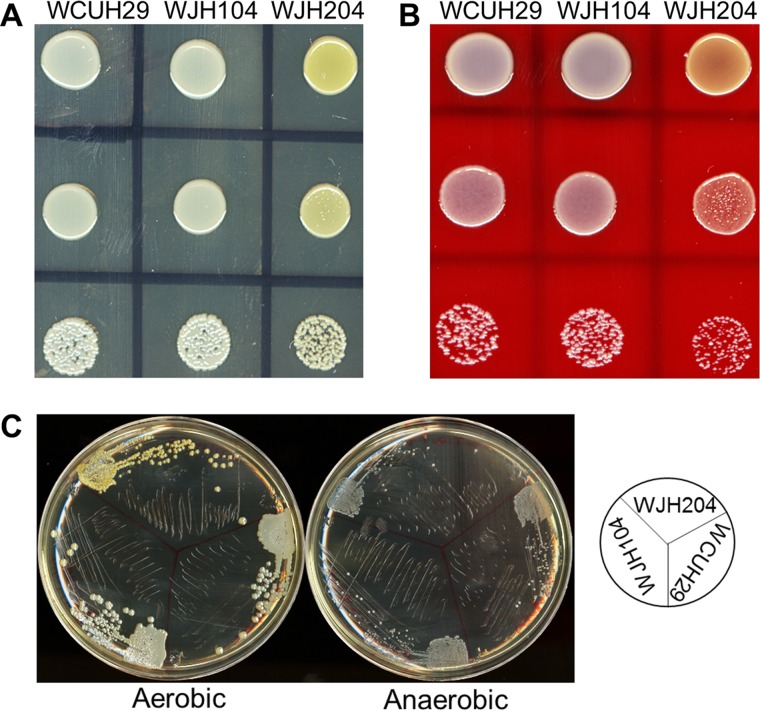
Unexpectedly, it was observed that the golden color of WCUH29 ΔairR::Pspac-airR colonies was remarkably enhanced relative to the wild-type (WT) S. aureus strain WCUH29 and WCUH29 ΔairS::Pspac-airS colonies. This phenotype was observed on both Trypticase soy agar (TSA) plates (Fig. 1A) and sheep's blood agar plates (Fig. 1B). The enhanced golden pigment of the WCUH29 ΔairR::Pspac-airR strain was observed only under aerobic growth conditions; when the bacteria were grown anaerobically, the golden pigment was absent in all the strains (Fig. 1C). The golden pigment of S. aureus is a potent antioxidant (2). Further, oxidants, such as oxygen, are known to activate the AirSR TCS (12), and therefore, it was hypothesized that AirSR regulates the expression of golden pigment in response to the presence of environmental oxidants.
FIG 1.
Overproduction of AirR enhances the golden color of S. aureus. (A and B) Overnight cultures of the strains were serially diluted, and 10 μl of the 10−3, 10−5, and 10−7 dilutions was plated on TSA (A) and 5% sheep's blood agar (B) plates and incubated for 24 h at 37°C. (C) Each strain was streaked on TSA plates and incubated aerobically or anaerobically. The diagram on the right shows the placement of the strains on the plates. WJH104, WCUH29 ΔairS::Pspac-airS; WJH204, WCUH29 ΔairR::Pspac-airR.
The amount of staphyloxanthin produced parallels the amount of AirR produced by S. aureus WCUH29.
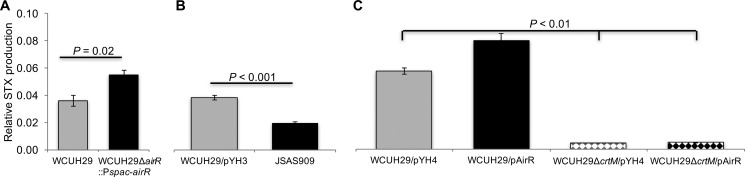
The predominant pigment produced by S. aureus is an alcohol-soluble membrane-embedded golden C30 triterpenoid carotenoid, α-d-glucopyranosyl-1-O-(4,4′-diaponeurosporen-4-oate)-6-O-(12-methyltetradecanoate), termed staphyloxanthin (STX), that is the product of the biosynthetic pathway encoded by crtOPQMN and aldH (3, 4). The initial observational data suggested golden-pigment production was linked to AirSR TCS expression. The amount of golden pigment produced during overproduction of AirR or RNA interference (RNAi) knockdown of the airSR TCS was quantified to support the hypothesis that golden-pigment production was linked to AirR production. The WCUH29 ΔairR::Pspac-airR strain produced 35% more golden pigment than wild-type WCUH29 (Fig. 2A). Furthermore, the JSAS909 strain, expressing an airSR-specific antisense RNA, produced 50% less golden pigment than control WCUH29/pYH3 (Fig. 2B) (23), further suggesting that the AirSR TCS regulates golden-pigment production.
FIG 2.
The AirSR TCS regulates the production of STX. All the strains were cultured for 24 h at 37° with shaking. S. aureus strains were cultured with 200 μM IPTG (A), 500 ng/ml ATc (B), and 250 ng/ml ATc (C). The OD600 was measured after washing. STX was extracted in methanol, and the OD450 was measured. Relative STX values were calculated as follows: OD450/OD600. The data represent the means ± standard errors of the mean (SEM) of the results of at least three independent experiments. JSAS909 is WCUH29 with an airS antisense RNA-expressing plasmid.
To confirm that the golden pigment was indeed staphyloxanthin, a crtM markerless deletion in S. aureus WCUH29 was constructed. As expected, the colonies were devoid of their trademark golden pigment (2). Subsequently, the TetR-regulated empty-control (pYH4) and AirR overproduction (pAirR) plasmids were electroplated into the ΔcrtM strain, resulting in strains WCUH29 ΔcrtM/pYH4 and WCUH29 ΔcrtM/pAirR, respectively. The WCUH29 wild-type empty-control (WCUH29/pYH4) and AirR-overproducing (WCUH29/pAirR) strains (22), along with the new ΔcrtM mutant strains, were cultured with the TetR inducer anhydrotetracycline (ATc) and assayed for golden-pigment production. Consistent with the previous AirR overproduction results, the WCUH29/pAirR strain produced 30% more golden pigment than WCUH29/pYH4 (Fig. 2C). Regardless of a strain's ability to overproduce AirR, golden pigment was absent in the ΔcrtM strains (Fig. 2C). These data indicate that the golden pigment is indeed STX and the AirSR TCS regulates STX production in S. aureus WCUH29.
The AirSR TCS mediates resistance to hydrogen peroxide killing.
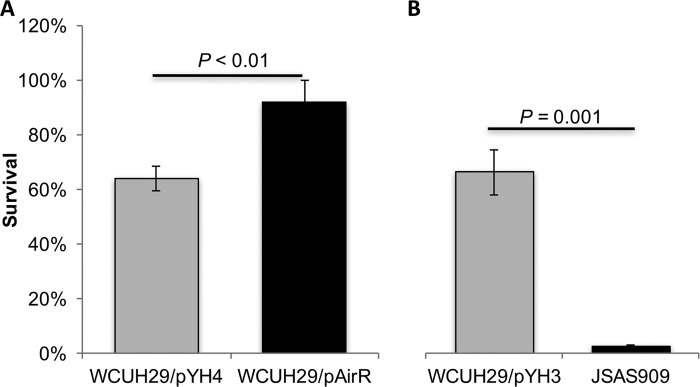
The membrane-bound STX carotenoid protects S. aureus from ROS, such as hydrogen peroxide (2, 6). Therefore, AirSR would be expected to be important for ROS resistance as a potential redox sensor and regulator of STX production. To test this hypothesis and to determine if the AirSR TCS is important for S. aureus resistance to killing by H2O2, the ATc-regulated AirR overproduction strain and airS antisense RNA S. aureus strains were utilized for an H2O2 susceptibility assay. The ATc-induced overproduction of AirR significantly enhanced the survival of WCUH29/pAirR compared to the WCUH29/pYH4 control strain (92% versus 64%) (Fig. 3A). Conversely, depletion of AirSR by RNAi dramatically reduced the ability of JSAS909 to resist H2O2-mediated killing relative to the WCUH29/pYH3 control strain (2% versus 66%) (Fig. 3B). These data indicate that the AirSR TCS is an important mediator of H2O2 resistance and that it contributes to protection from ROS, likely through regulation of STX biosynthesis (2).
FIG 3.
The AirSR TCS is important for H2O2 resistance. All the strains were cultured for 24 h at 37°C with shaking. The S. aureus strains were cultured with 250 ng/ml ATc (A) and 500 ng/ml ATc (B). Approximately 5 × 108 CFU was incubated in sterile PBS with 1.5% hydrogen peroxide at 37°C for 1 h. JSAS909 is WCUH29 with an airS antisense RNA-expressing plasmid. The percent survival was calculated as follows: (number CFUfinal/number CFUinput) × 100. The data represent the means ± SEM of the results of at least three independent experiments.
The AirSR TCS is a positive transcriptional regulator of crtOPQMN but is epistatic to alternative sigma factor B.
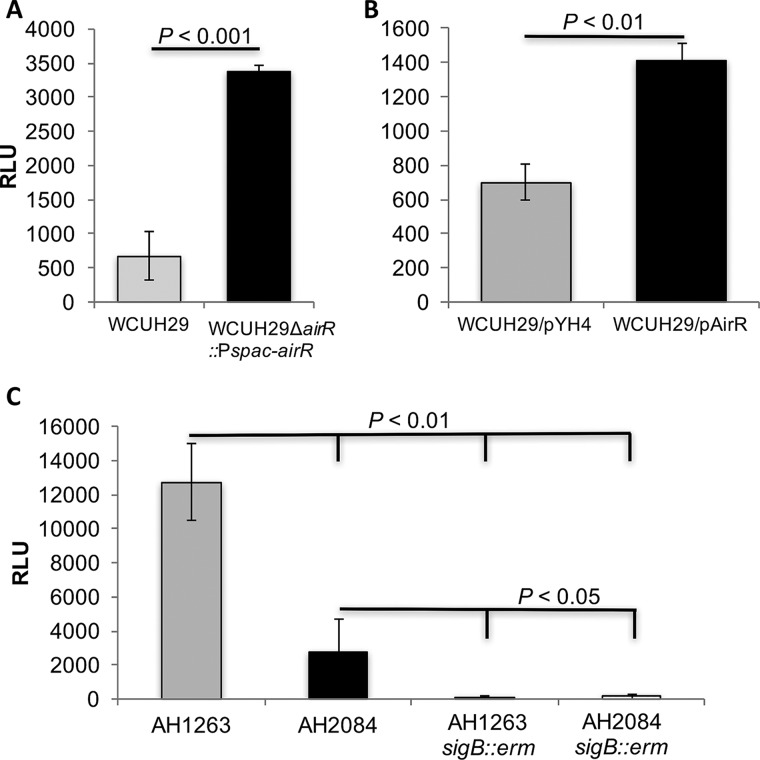
The precursors of staphyloxanthin are the product of the biosynthetic crtOPQMN operon; therefore, a crt promoter-lux (luxABCDE) reporter plasmid was constructed in order to determine the impact of altered AirR levels on crt transcription. In both of the AirR overproduction strains, WCUH29 ΔairR::Pspac-airR and WCUH29/pAirR, the maximum relative luminescence units (RLU) of the crt-lux reporter were 5- and 2-fold higher, respectively, than those of the respective controls (Fig. 4A and B). The AirSR TCS appears to be a positive transcriptional regulator of the crtOPQMN operon.
FIG 4.
The AirSR TCS transcriptionally regulates crtOPQMN and is epistatic to alternative sigma factor B. (A and B) All the strains were cultured overnight without IPTG or ATc. Following overnight growth, the strains were diluted, and 250 μM IPTG (A) and 25 ng/ml ATc (B) were added to the respective cultures. (C) No inducer was added to CA-MRSA USA300 strains AH1263 WT and AH2084 (AH1263 ΔairSR). The data represent the maximum RLU after 2 h of growth. All the strains carry the Pcrt-lux reporter. The RLU were calculated by dividing the luminescence reading by the OD600 reading. The data represent the means ± SEM of the results of at least three independent experiments.
During the course of these investigations, a community-acquired MRSA (CA-MRSA) USA300 ΔairSR mutant was reported (26). The parental wild-type strain and ΔairSR mutant, AH1263 and AH2084, respectively, were kindly provided to our laboratory by Alex Horswill. To further determine if AirSR regulates crt transcription, the crt-lux reporter was introduced into AH1263 and AH2084. The maximum RLU level of the crt-lux reporter was reduced more than 4.5-fold in the AH2084 airSR mutant relative to the AH1263 parental strain (Fig. 4C). These data confirm that the AirSR TCS is a positive transcriptional regulator of the STX-biosynthetic operon, crtOPQMN.
It has been well established that the alternative sigma factor B (SigB) plays an important role in the regulation of crt operon expression (7, 27, 28). To exclude the possibility that downregulation of transcription from the crt promoter is due to an unidentified deficiency of sigB in WCUH29, sigB allelic-replacement mutants were created. The hemolytic activity of the new WCUH29 sigB::erm mutant on sheep's red blood cell agar was dramatically increased compared to the WCUH29 WT control (data not shown). These data are consistent with previous reports and further demonstrate the integrity of the SigB regulatory circuit in WCUH29 (29, 30). To further decipher the transcriptional regulation of crt, sigB allelic mutations were constructed in AH1263 and the airSR mutant, AH2084. The AH1263 sigB::erm strain had dramatically increased hemolytic activity relative to the parental AH1263 (data not shown). The hemolytic activity of AH2084 was slightly reduced compared to that of the AH1263 strain. Consistent with functional SigB regulation, the AH2084 sigB::erm had noticeably increased hemolytic activity relative to the parental AH2084, but it still appeared to be slightly reduced relative to that of the AH1263 sigB::erm mutant (data not shown). These data indicate that the SigB regulatory circuit is intact and functional in AH1263 and derivative strains. Furthermore, airSR and sigB may regulate hemolytic activity independently.
To determine the relationship between AirSR and SigB on transcription from the crt promoter, the crt promoter-lux reporter system was electroporated into AH1263 sigB::erm and AH2084 sigB::erm. The AH1263 sigB::erm strain had a maximum RLU 100-fold less than that of the AH1263 WT, while the maximum RLU was reduced more than 15-fold in the AH2084 sigB::erm relative to the parental AH2084 (Fig. 4C). No difference in maximum RLU was observed between an AH1263 sigB single mutant and an AH2084 airSR sigB double mutant (Fig. 4C). These results indicate that airSR is epistatic to sigB, as AirSR regulation of crt is dependent on the presence of SigB.
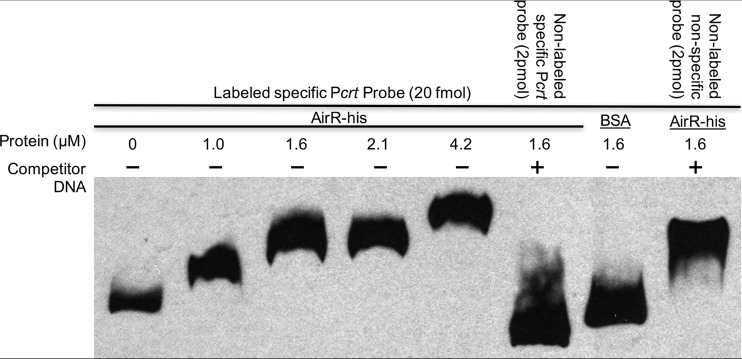
AirR directly binds the promoter region of crtOPQMN.
To determine if AirR directly regulates the crtOPQMN operon, an electrophoretic mobility shift assay (EMSA) was performed with purified AirR-His and a biotinylated Pcrt probe. The electrophoretic mobility of the Pcrt probe during native PAGE was increasingly retarded as the concentration of AirR-His increased (Fig. 5, lanes 1 to 5), indicating that AirR binds the crt promoter probe. To determine the specificity of this binding, 100-fold excess of nonlabeled specific Pcrt probe was added to the reaction mixture, resulting in almost complete elimination of labeled specific Pcrt retardation, indicating that the nonlabeled specific probe competed with the labeled probe for the limited amount of AirR-His (Fig. 5, compare lanes 4 and 6). To further confirm the specificity of the binding, a nonspecific bovine serum albumin (BSA) protein control was added to the reaction mixture (Fig. 5, lane 7), and no apparent probe shift was observed, indicating that the mere presence of protein did not inhibit the mobility of the Pcrt probe. The inclusion of 100-fold excess of a nonlabeled internal fragment of the sspA gene (22) in the reaction mixture did not interfere with the AirR-His–Pcrt probe interaction. These data indicate that the AirR response regulator binds the crtOPQMN promoter and that recognition of the promoter is specific.
FIG 5.
AirR specifically binds the crt promoter region. The electrophoretic mobility of the labeled promoter fragment alone is shown in the first lane. Increasing amounts of AirR (1.0, 1.6, 2.1, and 4.2 μM) were incubated with labeled crt promoter probe. −, absence of competitor DNA; +, addition of 100-fold excess competitor DNA. BSA (1.6 μM) and a nonspecific unlabeled probe (an internal fragment of sspA) were used as nonspecific binding controls. For each reaction, 20 fmol of biotin-labeled crt promoter probe was included. All the reaction mixtures totaled 20 μl.
Loss of staphyloxanthin eliminates AirR-mediated enhanced survival in human blood.
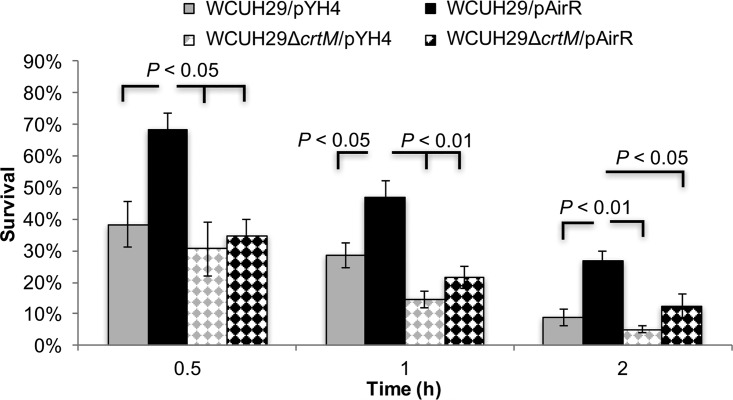
Previously, the AirSR TCS was recognized as an important factor for survival in human blood (22). The apparent global regulatory role of the TCS makes the assessment of AirSR-specific regulation of any virulence factor's contribution to overall S. aureus survival and pathogenesis cumbersome (12, 14, 15, 22). To facilitate the evaluation of a specific virulence factor's contribution to AirSR-mediated survival in blood, the virulence factor of interest is removed from the system by targeted gene deletion and the AirR response regulator is overproduced; in this case, crtM was deleted to prevent STX biosynthesis (reference 22 and data not shown). Using this principle, the significance of AirR-mediated STX production for S. aureus survival in human blood was evaluated. As seen previously, the ATc-induced WCUH29/pAirR strain had significantly enhanced survival in human blood throughout the experiment (Fig. 6). Overproduction of AirR in the STX-less WCUH29 ΔcrtM/pAirR strain was significantly decreased relative the WCUH29/pAirR strain throughout the experiment. Interestingly, there was not a significant statistical difference between the WCUH29/pYH4 control strain and the STX-less vector control strain, WCUH29 ΔcrtM/pYH4, at any time in the experiment, contrary to a previous report (2). Excitingly, there was not a statistical difference in survival between WCUH29 ΔcrtM/pYH4 and WCUH29 ΔcrtM/pAirR at any time in the experiment. Taken together, these data indicate that the enhanced production of STX facilitated by overproduction of AirR is a significant factor contributing to AirR-mediated enhanced survival of S. aureus in human blood (22).
FIG 6.
Absence of STX eliminates the enhanced survival of S. aureus resulting from AirR overproduction. Shown is the percent survival of S. aureus strains in freshly collected heparinized human blood with ATc. Bacteria were cultured overnight with ATc (250 ng/ml), and the following day were diluted, inoculated into blood with ATc (250 ng/ml), and incubated at 37°C in a rotisserie incubator. The percent survival was calculated as follows: (number CFUfinal/number CFUinput) × 100. The data represent the means ± SEM of the results of four independent experiments.
DISCUSSION
The AirS sensor kinase uses a [2Fe-2S]-containing GAF domain as a redox sensor, and oxidation of AirS stimulates phosphorylation of the AirR transcriptional-response regulator (12). The STX carotenoid promotes virulence as an antioxidant against exogenous ROS and the neutrophil-mediated oxidative burst that occurs during phagocytosis (2, 31). In this study, we established for the first time that the AirSR TCS is important for the production of STX and resistance to H2O2 under normal atmospheric conditions. Further, we demonstrated that AirSR positively regulates the STX-biosynthetic operon, crtOPQMN, in a direct transcriptional-regulation manner, linking a redox sensor to the production of a protective carotenoid antioxidant. Our data further indicate that the AirSR regulatory system is epistatic to the alternative sigma factor B for transcriptional regulation of the crt operon, as transcription from the crt promoter was essentially the same in a sigB mutant regardless of the presence or absence of airSR.
Regulation of STX production is multifactorial, influenced by alternative virulence factor regulators, antisense RNA, and metabolic processes, as well as oxygen (7–11). Our quest to construct an IPTG-regulated airS and airR S. aureus strain ultimately proved unsuccessful due to high basal Pspac activity, but we were able to observe that a consequence of increased AirR production was increased colony pigmentation. We confirmed that the pigment was indeed STX and that the levels of STX were directly correlated with the amount of AirSR TCS produced by S. aureus. Furthermore, it was determined that the AirSR TCS was important for resisting killing by H2O2. The AirR response regulator is a DNA binding transcription factor, and thus, it was conceivable the AirSR TCS regulated STX production.
Our observations suggested that the AirSR TCS regulated STX production, likely through regulation of the biosynthetic operon, crtOPQMN. In support of AirSR-mediated regulation of STX, reevaluation of our unpublished microarray data found two STX-biosynthetic genes, crtM (dehydrosqualene synthase) and crtN (dehydrosqualene desaturase), to be downregulated 2- to 3-fold at the mid-exponential phase of growth during airS antisense RNA induction when cultured aerobically (Y. Ji, unpublished data). Moreover, transposon disruption of airR in S. aureus Newman reportedly did not alter crt expression at the mid-exponential growth phase when cultured anaerobically (12), consistent with AirSR activation during aerobic growth and its acting as a likely positive regulator of crtOPQMN. Using a Pcrt-lux reporter, we confirmed that the AirSR TCS is a positive regulator of the crtOPQMN biosynthetic operon, as overproduction of AirR in S. aureus WCUH29 increased Pcrt-lux reporter activity and deletion of airSR in S. aureus USA300 decreased reporter activity.
Using a gel shift assay, it was determined that regulation of the biosynthetic operon is direct, as AirR bound the upstream promoter region of crtOPQMN specifically. The AirR DNA recognition motif is posited to be AAATNNAAAATNNNNTT based on the analysis of four AirR-bound promoters (32). Using the upstream promoter regions of 11 genes known to be directly bound by AirR and the motif discovery program MEME (12, 14, 22, 32, 33), we identified the motif (G/T)AA(C/A)ATNNA(C/A)AAAT in all 11 promoters. This updated potential recognition motif maintains the core AAATNNAAAAT sequence previously identified and is consistent with the sequence structure of other NarL-like response regulator binding sequences (34, 35). AirR belongs to the LuxR/NarL family of response regulators.
We previously determined that overproduction of AirR enhanced the survival of S. aureus in human blood and that AirR regulated the sspAB protease genes (22). Deletion of the sspAB gene only minimally decreased survival of the AirR overproduction strain in blood, however, indicating other virulence factors were contributing to the enhanced survival of the AirR overproduction strain, as well. In this study, genetic elimination of STX production by deleting crtM had a significant impact on survival for AirR overproduction, suggesting regulatory mechanisms or mutations that increase the production of AirSR or STX benefit S. aureus survival in blood.
In the absence of oxidants, the AirS [2Fe-2S]+ oxygen sensor is maintained in a reduced form; thus, kinase activity of AirS is inactive and AirR is not phosphorylated (12), and subsequently, AirR does not promote transcription. Conversely, oxygen oxidizes the AirS [2Fe-2S]2+ cluster. The oxidation of AirS results in phosphorylation and effector function activation of AirR, namely, transcriptional activation. As an AirSR-regulated operon, crtOPQMN transcription is increased, ultimately resulting in more STX production and protection from environmental oxidants.
The AirS kinase function is inactivated by strong oxidation, such as in the presence of millimolar concentrations of H2O2 in vitro (12), but modeling of the neutrophil phagosome suggests that the H2O2 concentration is only in the low micromolar range (36). It is therefore conceivable that mild oxidation of the AirS [2Fe-2S] cluster may occur in the phagosomes of neutrophils within an anaerobic wound site and promote AirSR activation and production of STX (37). Oxidation of AirS likely upregulates crt transcription and promotes STX biosynthesis as a way to counteract oxidative killing by the neutrophil. If oxygen were present in the phagosome, it, too, would promote AirSR-mediated upregulation of STX and antioxidant protection. In support of this suggestion, STX has been shown to be important for bacterial survival when S. aureus is cultured with neutrophils (2), and AirSR is important for survival in blood, where neutrophils constitute 50 to 60% of all leukocytes (22, 38). Furthermore, genetic mutations or pharmacological inhibition of the NADPH oxidase pathway in neutrophils eliminated the requirement for STX by S. aureus in both assays (2). Likewise, the data presented here show that the genetic disruption of STX biosynthesis completely eliminated the enhanced survival of S. aureus due to AirR-mediated STX overproduction.
The AirSR TCS is not essential in every S. aureus strain, as previously believed (26), but the importance of this undercharacterized system for regulation of virulence factors and resistance to phagocytosis is becoming more apparent. Investigations of the role of AirSR in responding to and resisting phagocytosis and the subsequent phagocytic oxidative burst are ongoing. It is now apparent that the AirSR TCS is an important modulator of S. aureus antioxidants, virulence factors, and, potentially, survival within phagosomes.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth media.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli DC10B (a gift from T. J. Foster) served as the host for all in vitro recombinant DNA (39). E. coli transformants were selected on brain heart infusion (BHI) (Difco) agar containing erythromycin (100 μg/ml) or Luria-Bertani agar containing ampicillin (100 μg/ml). S. aureus was cultured in Trypticase soy broth (TSB) (Difco) or on TSA at 37°C with appropriate antibiotics. All the bacterial cell cultures were incubated with shaking at 220 rpm. S. aureus transformants were selected on TSA containing chloramphenicol (10 μg/ml) or erythromycin (5 μg/ml). Sheep's blood agar plates were obtained from BD. For anaerobic growth, TSA plates were placed in an oxygen-free nitrogen-hydrogen gas mixture COY chamber (Coy Laboratory Products) and incubated at room temperature.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Source |
|---|---|---|
| Strains | ||
| DC10B | Dam− E. coli that allows direct electroporation of purified plasmid DNA into wild-type S. aureus | 39 |
| WCUH29 | Human clinical MRSA isolate; sigB+ rsbU+ | NCIMB40771 |
| SASJ104 | RN4220 geh::pFF71-airS; airS::tet Tetr Cmr | 23 |
| SASJ204 | RN4220 geh::pFF71-airR; airR::tet Tetr Cmr | 23 |
| WCUH29 ΔairS::Pspac-airS | WCUH29 geh::pFF71-airS; ΔairS Cmr | This study |
| WCUH29 ΔairR::Pspac-airR | WCUH29 geh::pFF71-airR; ΔairR Cmr | This study |
| WCUH29 ΔairR::Pspac-airR/Pcrt-lux | Carries Pcrt-lux reporter; Cmr Kanr | This study |
| WCUH29/pYH3 | WCUH29 antisense control strain with empty pYH3; Ermr | 23 |
| JSAS909 | WCUH29 with pSAS909; Ermr | 23 |
| WCUH29/pYH4 | WCUH29 protein overproduction control with empty pYH4; Ermr | 22 |
| WCUH29/pAirR | WCUH29 with pYH4-airR; Ermr | 22 |
| WCUH29/pYH4/Pcrt-lux | WCUH29/pYH4 with with Pcrt-lux reporter; Ermr Cmr | This study |
| WCUH29/pAirR/Pcrt-lux | WCUH29/pAirR with Pcrt-lux reporter; Cmr Cmr | This study |
| WCUH29 ΔcrtM | WCUH29 with in-frame deletion of crtM | This study |
| WCUH29 ΔcrtM/pYH4 | WCUH29 ΔcrtM with pYH4; Ermr | This study |
| WCUH29 ΔcrtM/pAirR | WCUH29 ΔcrtM with pAirR; Ermr | This study |
| AH1263 | USA300CA-MRSA; Erms (LAC*) | 26 |
| AH2084/JMB1241 | AH1263 ΔairSR | 26 |
| AH1263 sigB::erm | AH1263 with deletion of sigB; Ermr | This study |
| AH2084 sigB::erm | AH2084 with deletion of sigB; Ermr | |
| AH1263/Pcrt-lux | AH1263 with Pcrt-lux reporter; Cmr | This study |
| AH2084/Pcrt-lux | AH2084 with Pcrt-lux reporter; Cmr | This study |
| AH1263 sigB::erm/Pcrt-lux | AH1263 sigB::erm with Pcrt-lux reporter; Cmr Ermr | This study |
| AH2084 sigB::erm/Pcrt-lux | AH2084 sigB::erm with Pcrt-lux reporter; Cmr Ermr | This study |
| Plasmids | ||
| pYH3 | Shuttle vector with a TetR-regulated inducible promoter; Ermr | 23 |
| pSAS909 | pYH3 with airS antisense downstream of TetR promoter; Ampr Ermr | 23 |
| pYH4 | pYH3 with Ampr removed; Ermr | 46 |
| pAirR | airR cloned downstream of pYH4 TetR promoter; Ermr | |
| pCY1006 | agr-promoter-luxABCDE reporter system; Cmr | 44 |
| Pcrt-lux | pCY1006 with crt promoter cloned upstream of luxABCDE in place of agr promoter; Cmr | This study |
| pKOR1 | Temperature-sensitive inducible allelic exchange plasmid for S. aureus; Cmr | 42 |
| pKOR1-crtM | pKOR1 with in-frame crtM upstream/downstream deletion region; Cmr | This study |
Construction of S. aureus WCUH29 ΔairS::Pspac-airS, WCUH29 ΔairR::Pspac-airR, WCUH29 ΔsigB, AH1263 ΔsigB, and AH2084 ΔsigB strains.
S. aureus WCUH29 Pspac-airS and -airR were constructed by ϕ11 phage transduction and selection of the pFF71-airS or -airR chromosomal fragment from SASJ104 and SASJ204, respectively (23, 40). Briefly, SASJ104 and SASJ204 were cultured and used to make separate ϕ11 lysates. The lysates were then used to infect wild-type S. aureus WCUH29, and transductants were selected on TSA with chloramphenicol. The pFF71 plasmid site-specifically integrates into the S. aureus chromosome at the ϕL54α attB site located in the lipase (geh) gene (41). Transductants that were Cmr and lipase negative on Spirit Blue agar plates with lipase reagent (BD) were screened by diagnostic PCR using the PspacFor/AirROE-Rev primer set (listed in Table 2) to specifically identify colonies that had properly recombined the pFF71-airS or -airR chromosomal fragment into the chromosome. Subsequently, airS and airR were deleted from their native loci in the respective strains using pKOR1-airS and pKOR1-airR plasmids (42).
TABLE 2.
Oligonucleotide sequences
| Primer | Sequence |
|---|---|
| Pspacfor | 5′-GCTTGAATTCATTCAGAACGCTCGGTTGCC-3′ |
| AirROE-rev | 5′-TTGGCGCGCCCTATTTTATAGGAATTGTGAATTG-3′ |
| crtM-pKOR1-L-For | 5′-GGGG ACAAGTTTGTACAAAAAAGCAGGCT CAGCAAGTTCCAGTAGATGTCATTG-3′ |
| crtM-pKOR1-L-Rev | 5′-CATACTAGTCCTCCTATATTGAAATGCC-3′ |
| crtM-pKOR1-R-For | 5′-Phos-CATAGAATATAGGTGGTTGAATAATGAAGATTG-3′ |
| crtM-pKOR1-R-Rev | 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTTG ATCTGTCACATCAATATCTATACCG-3′ |
| luxA Rev | 5′-GCTCCAGTAACCATACGG TATC-3′ |
| Pcrt For-lux | 5′-TAGCGAATTCTAATGGTTATGCATCAGGAAGTAAC-3′ |
| Pcrt Rev-lux | 5′-TTCCCGGGCTAAATTGAATCACTCTCAATCATACTGAC-3′ |
| Pcrt For-EMSA | 5′-biotin-CTAATGGTTATGCATCAGGAAGTAAC-3′ |
| Pcrt Rev-EMSA | 5′-biotin-CTAAATTGAATCACTCTCAATCATACTGAC-3′ |
| sigBFor seq | 5′-ATGGCGAAAGAGTCGAAATCAG-3′ |
| sigBRev seq | 5′-AATTGCCGTTCTCTGAAGTCG-3′ |
To determine whether sigB is involved in the regulation of the crt operon by airSR, we created sigB allelic-replacement mutants in wild-type WCUH29 and AH1263 strains, as well as AH2084, an airSR deletion knockout (KO) mutant, using phage transduction, as described previously (23, 40). The sigB mutants were selected with erythromycin and further confirmed by diagnostic PCR and DNA sequencing. The donor sigB allelic-replacement mutant was kindly provided by Knut Ohlsen (43).
Extraction of staphyloxanthin from S. aureus strains.
To determine if AirSR contributed to the production of staphyloxanthin, S. aureus strains were cultured in 5 ml of TSB in a 50-ml conical vial with or without the respective inducer, as indicated: 200 μM IPTG or 500 ng/ml ATc. After 24 h, each culture was pelleted, washed twice in PBS, and resuspended in 5 ml of PBS. The optical density at 600 nm (OD600) of the culture was read in 200-μl duplicates using a 96-well flat-bottom microtiter plate (Sarstedt) and a BioTek Synergy II spectrophotometer. One milliliter of culture was pelleted and resuspended in 1 ml of 100% methanol, with pipetting and vortexing until all large clumps of cells were dispersed. The methanol extractions were placed in a rotisserie incubator and rotated end over end at room temperature for 2 h. The cellular debris was pelleted, and the OD450 (2) for each methanol extraction was measured in 200-μl duplicates. The relative STX levels were calculated as the OD450 divided by the respective OD600 for each sample. The experiment was repeated at least three times.
Hydrogen peroxide survival assay.
To determine the contribution of airSR to the survival of S. aureus when challenged with oxidative stress, overnight cultures were washed with PBS and ∼2 × 108 CFU was incubated at 37°C in a 1.5% H2O2–phosphate-buffered saline (PBS) solution for 60 min (2). Serial dilutions were plated on TSA for enumeration of surviving CFU. Percent survival was calculated as the number of surviving CFU divided by the number of input CFU multiplied by 100 [(number CFUfinal/number CFUinput) × 100].
Construction of a promoter-luxABCDE reporter fusion system.
To investigate if the airSR TCS transcriptionally regulates the promoter of the crtOPQMN operon, we constructed a crt promoter-lux (luxABCDE) reporter using the pCY1006 vector (44), as previously described (45). The upstream crt promoter region was PCR amplified with primers Pcrt For-lux/Pcrt Rev-lux (listed in Table 2), purified, and digested with EcoRI and SmaI (NEB). The pCY1006 plasmid was digested with the same enzymes, and the crt promoter PCR fragment was ligated into the gel-purified vector backbone with T4 DNA ligase (Promega). The reconstructed Pcrt-lux promoter reporter (Pcrt-lux) was confirmed by diagnostic PCR using the promoter-specific forward primer and a plasmid-specific luxA rev reverse primer (listed Table 2). The plasmid was purified from E. coli DC10B (39) and electroporated into the S. aureus strains, as indicated in Table 1.
The Pcrt-lux reporter plasmid was electroporated into several strains to measure the effect of the presence or absence of the AirR response regulator (Table 1). For analysis using strains overproducing AirR, strains WAirR and WJH204, and the respective controls, cultures were grown overnight in TSB at 37°C with shaking overnight. The overnight cultures were diluted 1:300 in TSB with the appropriate inducer, as indicated in the legend for Fig. 4, in a sterile 96-well flat-bottom white microtiter plate (CoStar), sealed with a Breathe-Easy sealing membrane (Electron Microscopy Sciences), and incubated at 37°C with shaking every 15 min in a BioTek Synergy II spectrophotometer. The bioluminescence intensities and optical densities of duplicate cultures were measured after 2 h of growth. The RLU were calculated by dividing the average bioluminescence reading by the average OD600 reading for each strain. The experiment was repeated three times with separate colonies of each strain.
For analysis of Pcrt-lux reporter activity in AH1263 (USA300 CA-MRSA), AH2084 (AH1263 ΔairSR), AH1263 sigB::erm, and AH2084 sigB::erm, cultures were grown overnight in TSB at 37°C with shaking overnight. The overnight cultures were diluted 1:100 in TSB and incubated at 37°C with shaking. The bioluminescence intensity and optical density at 600 nm of each culture were measured in duplicate in a 96-well flat-bottom white microtiter plate (CoStar) using a BioTek Synergy II spectrophotometer. The RLU were calculated by dividing the average bioluminescence reading by the average OD600 reading for each strain. The experiment was repeated three times with separate colonies of each strain.
Electrophoretic mobility shift assay.
DNA for EMSA was amplified from the region upstream of the crtOPQMN operon. The Pcrt-EMSA For/Rev promoter primers listed in Table 2 were purchased with biotin conjugated to their 5′ termini. The dual 5′-biotin-labeled promoter fragment was obtained by high-fidelity PCR. A total of 300 μl of PCR was produced and loaded into a 2% agarose gel. The reaction mixture was electrophoresed and stained with ethidium bromide. The promoter fragment was cut from the gel and purified using the NucleoSpin gel cleanup kit (Macherey-Nagel). The probe was further purified by 5% native Tris-borate EDTA (TBE) PAGE, and the gel was stained with ethidium bromide. The DNA band corresponding to the calculated Pcrt fragment size was cut from the gel and repurified according to the NucleoSpin gel cleanup kit protocol. Lastly, a sample of the purified probe was electrophoresed on a 1.2% agarose gel to verify the size and purity of the biotin-labeled Pcrt probe.
The EMSA was performed using the LightShift Chemiluminescent EMSA kit (Thermo Scientific) essentially as described in the manufacturer's protocol. All the samples contained 1× LightShift binding buffer, 50 ng/μl poly(dI · dC), 2.5% glycerol, and 25 mM acetyl phosphate (Sigma). The labeled probe, AirR-His (22), and, where indicated, nonlabeled specific probe, BSA, and nonspecific nonlabeled probe were all added to the concentrations outlined in the figures, and ultrapure water was added so that all the reaction volumes totaled 20 μl. The reaction mixtures were incubated at room temperature for 20 min, followed by addition of 5 μl of 5× loading buffer to each reaction mixture. Twenty microliters of each reaction mixture was loaded into a prerun 8% TBE native polyacrylamide gel and electrophoresed at 75 V for 3 h at 4°C. The samples were transferred to a nylon membrane, cross-linked, and processed as outlined in the manufacturer's protocol. BioMax light film (Kodak) was used to detect the chemiluminescent reaction.
Blood survival assay.
A human blood survival assay was used to determine the contribution of staphyloxanthin to AirSR-mediated survival in humans. Bacteria were cultured in TSB with appropriate antibiotics overnight, and the inducer ATc was added where indicated. The overnight cultures were washed twice in sterile PBS and resuspended to an OD of 0.14 using a Behring photometer in PBS. Fresh venous human whole blood was collected, using heparin-containing Vacutainer tubes (BD), from outwardly healthy adult donors and then immediately used in the assay. The University of Minnesota Institutional Review Board approved the blood collection. Approximately 5 × 106 CFU in 50 μl of PBS was added to 450 μl of blood per microcentrifuge tube with appropriate antibiotics and ATc where indicated. The microcentrifuge tubes were capped, placed in a rotisserie incubator, and incubated at 37°C with end-over-end mixing. A 20-μl sample was removed from each sample at the indicated time points, serially diluted, and plated on TSA to determine the surviving CFU count for each sample. The percentage of surviving bacteria was calculated as follows: CFUtime point/CFUinitial input × 100.
Gene deletion protocol.
Deletion of crtM was carried out following the pKOR1 allelic-exchange protocol as described previously (42) and primer sets crtM-pKOR1-L-For/Rev and CrtM-pKOR1-R-For/Rev (listed in Table 2). The R-For primer was synthesized with a 5′ phosphate group. Each PCR fragment was purified, and the two fragments were ligated together with T4 DNA ligase (Promega). The ligation product was mixed with BP Clonase according to the manufacturer's instructions, and plasmid pKOR1, incubated at 25°C overnight, was then transformed into E. coli DC10B. The pKOR1-crtM KO plasmid was subsequently transformed into S. aureus WCUH29. White colonies were restreaked to fresh TSA plates, and deletion of crtM was confirmed by diagnostic PCR and absence of colony pigment.
Data analysis.
Independent samples were statistically analyzed using a Student t test, with an alpha level of ≤0.05 considered significant. For data with more than two independent samples, a one-way analysis of variance (ANOVA) with a post hoc Tukey honestly significant difference (HSD) test was used to determine if there was statistical significance between samples, with an alpha level of ≤0.05 considered significant.
ACKNOWLEDGMENTS
We thank Alexander R. Horswill for kindly providing strains AH1263 and AH2084.
This work was partially supported by a grant from the National Institutes of Health (AI057451) and by a grant from the USDA/Minnesota agriculture station. H.G. was supported by China Scholarship Council grant no. 201308220113.
We declare no conflicts of interest.
REFERENCES
- 1.Tong SYC, Davis JS, Eichenberger E, Holland TL, Fowler VG. 2015. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev 28:603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu GY, Essex A, Buchanan JT, Datta V, Hoffman HM, Bastian JF, Fierer J, Nizet V. 2005. Staphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activity. J Exp Med 202:209–215. doi: 10.1084/jem.20050846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim SH, Lee PC. 2012. Functional expression and extension of staphylococcal staphyloxanthin biosynthetic pathway in Escherichia coli. J Biol Chem 287:21575–21583. doi: 10.1074/jbc.M112.343020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pelz A, Wieland K-P, Putzbach K, Hentschel P, Albert K, Götz F. 2005. Structure and biosynthesis of staphyloxanthin from Staphylococcus aureus. J Biol Chem 280:32493–32498. doi: 10.1074/jbc.M505070200. [DOI] [PubMed] [Google Scholar]
- 5.Marshall JH, Wilmoth GJ. 1981. Pigments of Staphylococcus aureus, a series of triterpenoid carotenoids. J Bacteriol 147:900–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clauditz A, Resch A, Wieland K-P, Peschel A, Gotz F. 2006. Staphyloxanthin plays a role in the fitness of Staphylococcus aureus and its ability to cope with oxidative stress. Infect Immun 74:4950–4953. doi: 10.1128/IAI.00204-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kullik I, Giachino P, Fuchs T. 1998. Deletion of the alternative sigma factor σB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J Bacteriol 180:4814–4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katzif S, Lee E-H, Law AB, Tzeng Y-L, Shafer WM. 2005. CspA regulates pigment production in Staphylococcus aureus through a SigB-dependent mechanism. J Bacteriol 187:8181–8184. doi: 10.1128/JB.187.23.8181-8184.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lan L, Cheng A, Dunman PM, Missiakas D, He C. 2010. Golden pigment production and virulence gene expression are affected by metabolisms in Staphylococcus aureus. J Bacteriol 192:3068–3077. doi: 10.1128/JB.00928-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding Y, Liu X, Chen F, Di H, Xu B, Zhou L, Deng X, Wu M, Yang C-G, Lan L. 2014. Metabolic sensor governing bacterial virulence in Staphylococcus aureus. Proc Natl Acad Sci U S A 111:E4981–E4990. doi: 10.1073/pnas.1411077111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, Wu N, Dong J, Gao Y, Zhang X, Shao N, Yang G. 2010. SsrA (tmRNA) acts as an antisense RNA to regulate Staphylococcus aureus pigment synthesis by base pairing with crtMN mRNA. FEBS Lett 584:4325–4329. doi: 10.1016/j.febslet.2010.09.024. [DOI] [PubMed] [Google Scholar]
- 12.Sun F, Ji Q, Jones MB, Deng X, Liang H, Frank B, Telser J, Peterson SN, Bae T, He C. 2012. AirSR, a [2Fe-2S] cluster-containing two-component system, mediates global oxygen sensing and redox signaling in Staphylococcus aureus. J Am Chem Soc 134:305–314. doi: 10.1021/ja2071835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hall JW, Ji Y. 2013. Sensing and adapting to anaerobic conditions by Staphylococcus aureus. Adv Appl Microbiol 84:1–25. doi: 10.1016/B978-0-12-407673-0.00001-1. [DOI] [PubMed] [Google Scholar]
- 14.Yan M, Hall JW, Yang J, Ji Y. 2012. The essential yhcSR two-component signal transduction system directly regulates the lac and opuCABCD operons of Staphylococcus aureus. PLoS One 7:e50608. doi: 10.1371/journal.pone.0050608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan M, Yu C, Yang J, Ji Y. 2011. The essential two-component system YhcSR is involved in regulation of the nitrate respiratory pathway of Staphylococcus aureus. J Bacteriol 193:1799–1805. doi: 10.1128/JB.01511-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Novick RA, Microbiology AS for . 2006. Staphylococcal pathogenesis and pathogenicity factors: genetics and regulation of Gram-positive pathogens. Wiley-Blackwell, Charlottesville, VA. [Google Scholar]
- 17.Novick RP. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol 48:1429–1449. doi: 10.1046/j.1365-2958.2003.03526.x. [DOI] [PubMed] [Google Scholar]
- 18.Liang X, Yu C, Sun J, Liu H, Landwehr C, Holmes D, Ji Y. 2006. Inactivation of a two-component signal transduction system, SaeRS, eliminates adherence and attenuates virulence of Staphylococcus aureus. Infect Immun 74:4655–4665. doi: 10.1128/IAI.00322-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mrak LN, Zielinska AK, Beenken KE, Mrak IN, Atwood DN, Griffin LM, Lee CY, Smeltzer MS. 2012. saeRS and sarA act synergistically to repress protease production and promote biofilm formation in Staphylococcus aureus. PLoS One 7:e38453. doi: 10.1371/journal.pone.0038453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olson ME, Nygaard TK, Ackermann L, Watkins RL, Zurek OW, Pallister KB, Griffith S, Kiedrowski MR, Flack CE, Kavanaugh JS, Kreiswirth BN, Horswill AR, Voyich JM. 2013. Staphylococcus aureus nuclease is an SaeRS-dependent virulence factor. Infect Immun 81:1316–1324. doi: 10.1128/IAI.01242-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fournier B, Klier A, Rapoport G. 2001. The two-component system ArlS-ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Mol Microbiol 41:247–261. doi: 10.1046/j.1365-2958.2001.02515.x. [DOI] [PubMed] [Google Scholar]
- 22.Hall JW, Yang J, Guo H, Ji Y. 2015. The AirSR two-component system contributes to Staphylococcus aureus survival in human blood and transcriptionally regulates sspABC operon. Front Microbiol 6:682. doi: 10.3389/fmicb.2015.00682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun J, Zheng L, Landwehr C, Yang J, Ji Y. 2005. Identification of a novel essential two-component signal transduction system, YhcSR, in Staphylococcus aureus. J Bacteriol 187:7876. doi: 10.1128/JB.187.22.7876-7880.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan F, Lunsford RD, Sylvester D, Fan J, Celesnik H, Iordanescu S, Rosenberg M, McDevitt D. 2001. Regulated ectopic expression and allelic-replacement mutagenesis as a method for gene essentiality testing in Staphylococcus aureus. Plasmid 46:71–75. doi: 10.1006/plas.2001.1526. [DOI] [PubMed] [Google Scholar]
- 25.Zhang L, Fan F, Palmer LM, Lonetto MA, Petit C, Voelker LL, St John A, Bankosky B, Rosenberg M, McDevitt D. 2000. Regulated gene expression in Staphylococcus aureus for identifying conditional lethal phenotypes and antibiotic mode of action. Gene 255:297–305. doi: 10.1016/S0378-1119(00)00325-5. [DOI] [PubMed] [Google Scholar]
- 26.White MJ, Boyd JM, Horswill AR, Nauseef WM. 2014. Phosphatidylinositol-specific phospholipase c contributes to survival of Staphylococcus aureus USA300 in human blood and neutrophils. Infect Immun 82:1559–1571. doi: 10.1128/IAI.01168-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herbert S, Ziebandt A-K, Ohlsen K, Schäfer T, Hecker M, Albrecht D, Novick R, Götz F. 2010. Repair of global regulators in Staphylococcus aureus 8325 and comparative analysis with other clinical isolates. Infect Immun 78:2877–2889. doi: 10.1128/IAI.00088-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palma M, Cheung AL. 2001. σB activity in Staphylococcus aureus is controlled by RsbU and an additional factor(s) during bacterial growth. Infect Immun 69:7858–7865. doi: 10.1128/IAI.69.12.7858-7865.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicholas RO, Li T, McDevitt D, Marra A, Sucoloski S, Demarsh PL, Gentry DR. 1999. Isolation and characterization of a sigB deletion mutant of Staphylococcus aureus. Infect Immun 67:3667–3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bischoff M, Dunman P, Kormanec J, Macapagal D, Murphy E, Mounts W, Berger-Bächi B, Projan S. 2004. Microarray-based analysis of the Staphylococcus aureus σB regulon. J Bacteriol 186:4085–4099. doi: 10.1128/JB.186.13.4085-4099.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rigby KM, DeLeo FR. 2012. Neutrophils in innate host defense against Staphylococcus aureus infections. Semin Immunopathol 34:237–259. doi: 10.1007/s00281-011-0295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun H, Yang Y, Xue T, Sun B. 2013. Modulation of cell wall synthesis and susceptibility to vancomycin by the two-component system AirSR in Staphylococcus aureus NCTC8325. BMC Microbiol 13:286. doi: 10.1186/1471-2180-13-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME Suite: tools for motif discovery and searching. Nucleic Acids Res 37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Been M, Bart MJ, Abee T, Siezen RJ, Francke C. 2008. The identification of response regulator-specific binding sites reveals new roles of two-component systems in Bacillus cereus and closely related low-GC Gram-positives. Environ Microbiol 10:2796–2809. doi: 10.1111/j.1462-2920.2008.01700.x. [DOI] [PubMed] [Google Scholar]
- 35.Maris AE, Sawaya MR, Kaczor-Grzeskowiak M, Jarvis MR, Bearson SMD, Kopka ML, Schröder I, Gunsalus RP, Dickerson RE. 2002. Dimerization allows DNA target site recognition by the NarL response regulator. Nat Struct Biol 9:771–778. doi: 10.1038/nsb845. [DOI] [PubMed] [Google Scholar]
- 36.Winterbourn CC, Hampton MB, Livesey JH, Kettle AJ. 2006. Modeling the reactions of superoxide and myeloperoxidase in the neutrophil phagosome: implications for microbial killing. J Biol Chem 281:39860–39869. doi: 10.1074/jbc.M605898200. [DOI] [PubMed] [Google Scholar]
- 37.Sawyer RG, Spengler MD, Adams RB, Pruett TL. 1991. The peritoneal environment during infection. The effect of monomicrobial and polymicrobial bacteria on pO2 and pH. Ann Surg 213:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Kessel KPM, Bestebroer J, van Strijp JAG. 2014. Neutrophil-mediated phagocytosis of Staphylococcus aureus. Front Immunol 5:467. doi: 10.3389/fimmu.2014.00467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monk IR, Shah IM, Xu M, Tan M-W, Foster TJ. 2012. Transforming the untransformable: application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. mBio 3:e00277–11. doi: 10.1128/mBio.00277-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Novick RP. 1991. Genetic systems in staphylococci. Methods Enzymol 204:587–636. doi: 10.1016/0076-6879(91)04029-N. [DOI] [PubMed] [Google Scholar]
- 41.Lee CY, Buranen SL, Zhi-Hai Y. 1991. Construction of single-copy integration vectors for Staphylococcus aureus. Gene 103:101–105. doi: 10.1016/0378-1119(91)90399-V. [DOI] [PubMed] [Google Scholar]
- 42.Bae T, Schneewind O. 2006. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55:58–63. doi: 10.1016/j.plasmid.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 43.Lorenz U, Hüttinger C, Schäfer T, Ziebuhr W, Thiede A, Hacker J, Engelmann S, Hecker M, Ohlsen K. 2008. The alternative sigma factor sigma B of Staphylococcus aureus modulates virulence in experimental central venous catheter-related infections. Microbes Infect 10:217–223. doi: 10.1016/j.micinf.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 44.Liang X, Zheng L, Landwehr C, Lunsford D, Holmes D, Ji Y. 2005. Global regulation of gene expression by ArlRS, a two-component signal transduction regulatory system of Staphylococcus aureus. J Bacteriol 187:5486–5492. doi: 10.1128/JB.187.15.5486-5492.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng L, Yu C, Bayles K, Lasa I, Ji Y. 2007. Conditional mutation of an essential putative glycoprotease eliminates autolysis in Staphylococcus aureus. J Bacteriol 189:2734–2742. doi: 10.1128/JB.01806-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang J, O'Toole PW, Shen W, Amrine-Madsen H, Jiang X, Lobo N, Palmer LM, Voelker L, Fan F, Gwynn MN, McDevitt D. 2004. Novel chromosomally encoded multidrug efflux transporter MdeA in Staphylococcus aureus. Antimicrob Agents Chemother 48:909–917. doi: 10.1128/AAC.48.3.909-917.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]