ABSTRACT
Citrobacter rodentium is a murine pathogen used to model intestinal infections caused by the human diarrheal pathogens enterohemorrhagic and enteropathogenic Escherichia coli. During infection, bacteria use two-component systems (TCSs) to detect changing environmental cues within the host, allowing for rapid adaptation by altering the expression of specific genes. In this study, 26 TCSs were identified in C. rodentium, and quantitative PCR (qPCR) analysis showed that they are all expressed during murine infection. These TCSs were individually deleted, and the in vitro and in vivo effects were analyzed to determine the functional consequences. In vitro analyses only revealed minor differences, and surprisingly, type III secretion (T3S) was only affected in the ΔarcA strain. Murine infections identified 7 mutants with either attenuated or increased virulence. In agreement with the in vitro T3S assay, the ΔarcA strain was attenuated and defective in colonization and cell adherence. The ΔrcsB strain was among the most highly attenuated strains. The decrease in virulence of this strain may be associated with changes to the cell surface, as Congo red binding was altered, and qPCR revealed that expression of the wcaA gene, which has been implicated in colanic acid production in other bacteria, was drastically downregulated. The ΔuvrY strain exhibited increased virulence compared to the wild type, which was associated with a significant increase in bacterial burden within the mesenteric lymph nodes. The systematic analysis of virulence-associated TCSs and investigation of their functions during infection may open new avenues for drug development.
KEYWORDS: Citrobacter, attaching/effacing pathogens, bacterial pathogenesis, gastrointestinal infection, host-pathogen interactions, mouse models, two-component regulatory systems
INTRODUCTION
Citrobacter rodentium is a murine-restricted pathogen that is the causative agent of transmissible colonic hyperplasia (1). Together with enteropathogenic and enterohemorrhagic Escherichia coli (EPEC and EHEC), C. rodentium belongs to the attaching and effacing (A/E) family of intestinal pathogens (2). These bacteria share numerous virulence factors, including those responsible for the formation of typical A/E lesions, characterized by intimate attachment to intestinal epithelial cells, localized effacement of microvilli, and the formation of actin-rich pedestals beneath sites of bacterial adherence. EPEC causes diarrhea that kills several hundred thousand children each year in the developing world (3). EHEC is commonly associated with outbreaks of foodborne diarrheal illness in the developed world (4). The similarities between EPEC, EHEC, and C. rodentium infections have resulted in C. rodentium being widely used and recognized as a surrogate model to study intestinal infections caused by the human-restricted pathogens EPEC and EHEC (2, 5, 6).
Following oral inoculation of mice with C. rodentium, bacteria initially colonize a patch within the cecum. A few days later, C. rodentium can be found in the distal colon. This tissue tropism and infection pattern is similar to what is observed during human EHEC infections, where bacteria initially colonize Peyer's patches in the ileum before colonizing the colon (7). Once present in the colon, C. rodentium colonizes to high levels (>109 CFU/g of tissue) between 7 and 14 days postinfection in most mouse strains (8). In some mouse strains, such as C57BL/6, NIH Swiss, and BALB/c, C. rodentium causes self-limited infection and the mice clear the infection by 21 to 28 days postinfection (8). In contrast, other murine strains, such as C3H/HeJ, C3H/HeOuJ, and FVB, experience significantly more hyperplasia and suffer high levels of mortality between 6 and 10 days postinfection (8, 9). Both the nonlethal and lethal murine models are used to provide insight into the infection process and pathogenesis of A/E pathogens.
Two-component systems (TCSs) are used by bacteria to detect changes in their environment and promote an adaptive response to survive (10, 11). The typical TCS consists of a membrane-bound histidine kinase (HK) sensor and a cytoplasmic response regulator (RR) that usually acts as a transcriptional regulator. TCS genes can be encoded as pairs with the RR and HK genes as part of a single transcriptional unit, or the RR and HK genes can be unlinked within the chromosome. In addition, orphan RR genes that lack a cognate HK have been identified. The HK senses specific ligands or environmental cues, which result in the autophosphorylation of a conserved histidine residue of the HK cytoplasmic domain. The phosphoryl group is subsequently transferred to a specific aspartate residue in the RR. This affects its DNA-binding properties and results in changes in the transcription of specific genes. Activation of a TCS has been shown to affect the expression of a few to hundreds of genes, impacting either specific or multiple processes (12–14). Many TCSs affect bacterial metabolism, in addition to functions that may be associated with environmental persistence (15). Other TCSs have been directly implicated in bacterial virulence. For example, TCSs such as Salmonella enterica PhoPQ and Bordetella pertussis BvgAS are known master regulators of virulence (16, 17).
The number of TCSs varies between bacterial species; members of the Enterobacteriaceae family typically possess 20 to 30 TCSs (18). In E. coli, most studies have focused on the analysis of TCS gene and regulon expression under different in vitro conditions (19–22). In contrast, most in vivo studies do not examine TCS expression patterns and instead use a genetic approach to identify TCSs that affect virulence. For example, in uropathogenic E. coli the BarA/UvrY, CpxRA, KguSR, and OmpR/EnvZ TCSs affect in vivo virulence, although their in vivo expression patterns remain unknown (23–27). In EPEC and EHEC, several TCSs, including CpxRA, FusKR, PhoBR, QseBC, and QseFE, have been implicated in the regulation of virulence genes in vitro and, more recently, in vivo for QseBC and QseFE (28–33). In this study, using C. rodentium infection of mice, we assessed the relative expression of each C. rodentium RR gene during in vitro growth and in vivo infection of C3H/HeJ and C57BL/6J mice. In addition, we deleted each TCS in C. rodentium and analyzed the effect both in vitro and in vivo in an effort to determine the functional consequences of these gene deletions. This work is the first systematic study that combines RR gene expression analysis and TCS gene deletions during in vivo infection of a native host using an A/E pathogen and provides valuable data for future characterization of C. rodentium TCSs.
RESULTS
Global analyses of TCS gene expression in vitro and in vivo.
The TCSs encoded by C. rodentium are listed in Table S4 in the supplemental material. To assess expression of the 26 putative RR genes in vitro, quantitative PCR (qPCR) was performed using cDNA prepared from C. rodentium cells grown to mid-log phase in both LB and Dulbecco's modified Eagle's medium (DMEM). Relative expression levels were normalized to the expression of the control gene rpoD, and data were analyzed using the 2−ΔCT method (34). Similar results were obtained when 16S was used as the control gene (data not shown). As shown in Fig. 1A, the expression level of the 26 RR genes was heterogeneous and spread over 4 logs. Notably, whereas all RR genes were significantly expressed in DMEM, the expression of five RR genes (creB, fusR, pmrA, qseF, and yedW) was below the detection limit in the LB-grown samples, indicating that they are not significantly expressed under this growth condition (Fig. 1A). In addition, several genes were differentially expressed under the two in vitro growth conditions: expression of baeR, ompR, and ypdB was increased by at least 2-fold, and expression of dpiA, phoP, rcsB, uvrY, and zraR was decreased by at least 2-fold in DMEM compared to that in LB (Fig. 1A). Overall, these data show that several RR genes are differentially expressed during growth in LB and DMEM and that all 26 RR genes are expressed during growth in DMEM.
FIG 1.
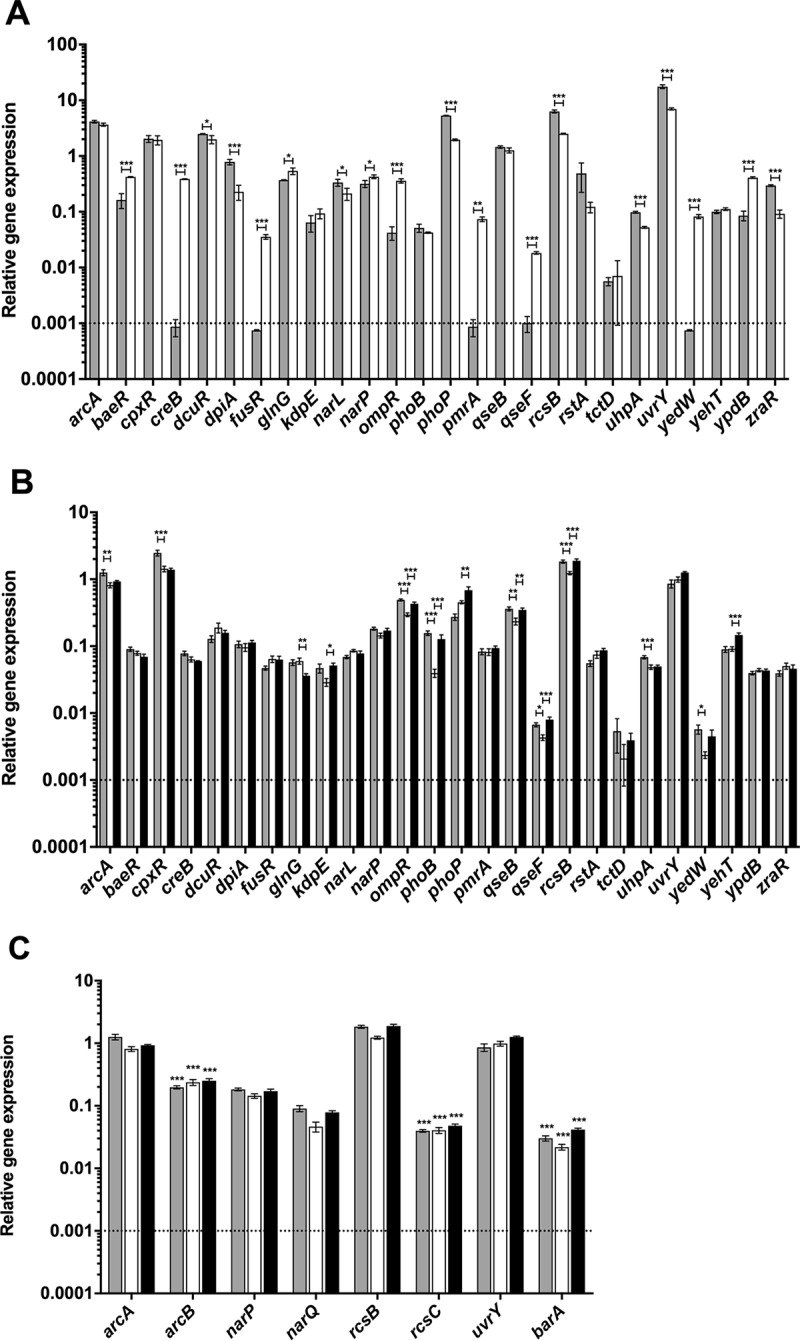
Relative expression of two-component system genes of C. rodentium in vivo and in vitro. Real-time RT-PCR was performed to detect expression levels of each RR gene relative to the primary sigma factor, rpoD. Dotted lines indicate the limit of detection of the assay. (A) Total RNA was prepared from C. rodentium cultures grown in DMEM (gray bars) and LB broth (open bars). (B) Total RNA was prepared from the terminal centimeter of the colon of infected C3H/HeJ mice on day 4 (gray bars) and day 9 (open bars) postinfection and C57BL/6J mice on day 9 (black bars) postinfection. An analysis of variance (ANOVA) followed by Tukey's post hoc analysis was used to determine statistical significance for both in vitro and in vivo growth (***, P < 0.001; **, P < 0.01; *, P < 0.05). (C) Expression levels of the RR genes arcA, narP, rcsB, and uvrY and their unlinked cognate HK sensor genes, arcB, narQ, rcsC, and barA, were measured by real-time RT PCR using total RNA samples described for panel B. An unpaired t test was used to determine statistical significance between each HK sensor gene and its cognate RR from the same in vivo condition (***, P < 0.001).
To assess the expression of the 26 C. rodentium RR genes during intestinal infection, qPCR was performed using cDNA prepared from total RNA isolated from the distal colons of infected mice at day 4 and 9 postinfection. We assessed RR expression in both innately susceptible C3H/HeJ and more resistant C57BL/6J mice, since we hypothesized that the intestinal environment would differ significantly between these two models. Robust and reproducible C. rodentium gene expression signals were obtained from C3H/HeJ mice at days 4 and 9 postinfection, whereas in C57BL/6J mice, reproducible C. rodentium gene expression data were only obtained from the day 9 postinfection samples. This is likely due to the comparatively lower loads of C. rodentium in C57BL/6J mouse colons during early infection (35). To ensure that the expression measured was the result of the specific amplification of transcripts from C. rodentium, control qPCR experiments were performed on cDNA prepared from total RNA isolated from uninfected C3H/HeJ and C57BL/6J mice using all TCS primer pairs, except cpxR and rpoD, which we previously validated using the same criteria (36). As expected, all primer pairs were unable to amplify a product from uninfected murine samples, indicating that the signals detected are C. rodentium specific and not the result of unwanted amplification of microbiota or murine genes (data not shown).
Notably, all 26 RR genes were significantly expressed during infection of both mouse strains and at both time points examined in C3H/HeJ mice (Fig. 1B). Overall, there was a large range in the expression levels of the different RR genes in infected mice, with a 100- to 1,000-fold difference between the most and the least expressed genes. Under all infection conditions, the most highly expressed RR genes were arcA, cpxR, rcsB, and uvrY, which were all expressed at levels similar to rpoD (Fig. 1B). Importantly, several RR genes were differentially expressed between the C3H/HeJ and C57BL/6J murine strains at day 9 postinfection (ompR, phoB, qseF, rcsB, yehT, glnG, phoP, qseB, and kdpE). Additionally, several RR genes were differentially expressed at the two time points of infection in C3H/HeJ mice (cpxR, ompR, phoB, rcsB, uhpA, arcA, qseB, qseF, and yedW). These data indicate that there are differences in RR gene expression between the lethal and nonlethal murine infection models and at different time points of infection.
Although most TCS genes are organized in operons, the arcA-arcB, narP-narQ, rcsB-rcsC, and uvrY-barA TCSs are part of different transcriptional units in the C. rodentium genome. As shown in Fig. 1C, as assessed by qPCR, all unlinked HK genes were significantly expressed in vivo. Most HK sensor genes were expressed at lower levels than their respective RR genes, which is not unprecedented, since lower HK than RR transcript levels were also observed in vivo for the linked cpxRA TCS (36).
Assessment of the roles of C. rodentium TCSs in bacterial virulence.
To systematically assess the roles of the C. rodentium TCSs in bacterial virulence during intestinal infection, we used sacB gene-based allelic exchange to generate bacterial strains bearing individual in-frame deletions for each of the 26 TCSs in C. rodentium. For those TCSs where the HK and RR genes are expressed as an operon, both the HK and RR genes were deleted. In the case of unpaired TCSs, only the RR was deleted. The primers used to generate the deletion strains are provided in Table S3. Prior to testing our strains in a murine infection, we subjected our mutants to a panel of in vitro tests to analyze carbohydrate assimilation, growth rate, oxygen requirements, and stress tolerance. The ΔdpiAB and ΔphoPQ strains were unable to use citrate as a secondary carbon source, the ΔarcA strain was no longer able to catalyze the decarboxylation of ornithine into putrescine, and the ΔarcA and ΔcpxRA strains were slightly more susceptible to alkaline pH (data not shown). Other than the differences listed above, the deletion strains behaved similarly to the wild type and then were used to assess the contribution of each TCS to C. rodentium virulence in C3H/HeJ mice.
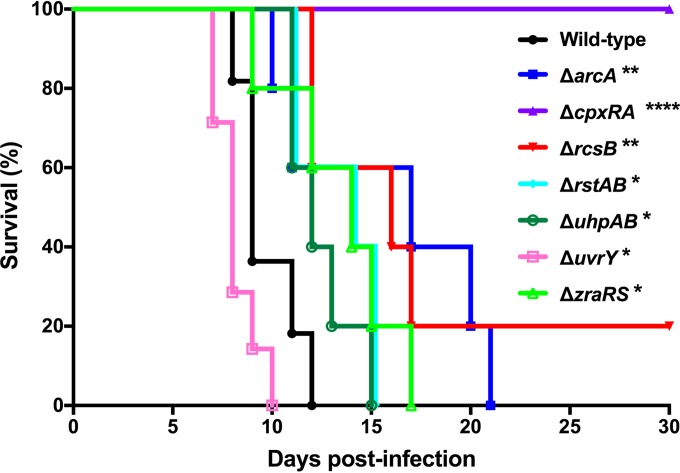
Of the 26 TCS knockout strains tested, 19 were not significantly different from wild-type C. rodentium in terms of mean survival time following infection (Fig. S1). Consistent with our previous data (36), the ΔcpxRA deletion mutant was highly attenuated for virulence, with 100% survival of mice infected with this strain (Fig. 2). Five additional TCS deletion strains (ΔarcA, ΔrcsB, ΔrstAB, ΔuhpAB, and ΔzraRS) were also significantly attenuated for virulence; mice infected with these strains displayed delayed mortality, with some mice surviving infection in the case of the ΔrcsB strain (Fig. 2). Surprisingly, deletion of uvrY resulted in a C. rodentium strain with increased virulence. Mice infected with this strain had a significantly shorter mean survival time than mice infected with wild-type C. rodentium (Fig. 2).
FIG 2.
Survival of female C3H/HeJ mice after infection with C. rodentium. Animals were orally gavaged with 2 × 108 to 3 × 108 CFU of C. rodentium wild type or a TCS deletion strain. Animals were monitored for 30 days postinfection as described in Materials and Methods. The data are pooled from two independent experiments with 5 to 7 mice per group. The log rank (Mantel-Cox) method was used to determine statistical significance (****, P < 0.0001; **, P < 0.01; *, P < 0.05).
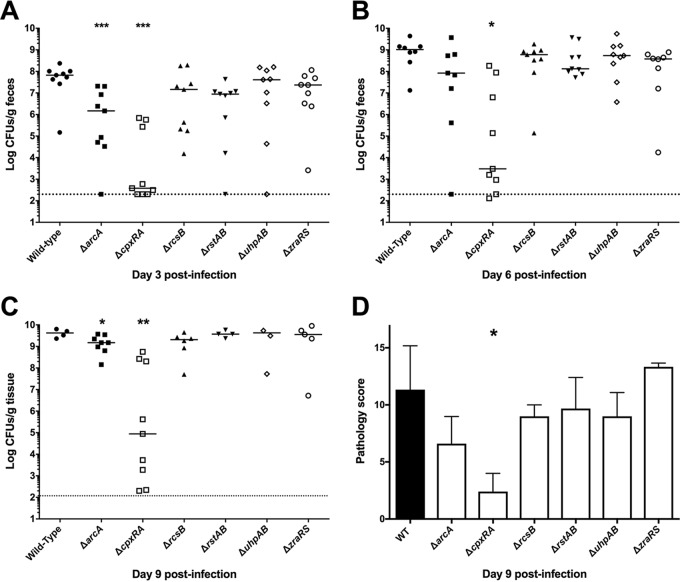
To further investigate the attenuation of the ΔarcA, ΔrcsB, ΔrstAB, ΔuhpAB, and ΔzraRS strains, we monitored the shedding of C. rodentium in the feces, indicative of total bacterial burden, at days 3, 6, and 9 postinfection as well as overall histopathology at day 9 postinfection (Fig. 3). The already-described wild-type and ΔcpxRA strains were included in these analyses as controls (36). As expected, wild-type C. rodentium loads increased swiftly and steadily during the course of infection, reaching a peak of between 109 and 1010 CFU/g of feces by day 9 postinfection. In contrast, and in agreement with our previous data, the ΔcpxRA strain displayed delayed colonization with lower levels of colonization at all time points and greater variability in bacterial burden than the wild-type strain (36). Of the other TCS deletion strains analyzed, only the ΔarcA mutant displayed significantly lower colonization levels than the wild-type strain (Fig. 3A to C). The other attenuated strains colonized mice to levels similar to that of wild-type C. rodentium at all time points, suggesting that the attenuation of these strains is unrelated to decreased or delayed bacterial colonization. Additionally, the only TCS deletion strain with significantly different pathology scores from the wild-type strain on day 9 postinfection was the ΔcpxRA strain, as we previously described (36) (Fig. 3D).
FIG 3.
Bacterial burden and pathology scoring in female C3H/HeJ mice infected with hypovirulent mutants of C. rodentium. Susceptible mice were orally gavaged with 2 × 108 to 3 × 108 CFU of C. rodentium wild type or a TCS deletion strain, and fecal burden of C. rodentium was assessed at days 3 (A), 6 (B), and 9 (C) postinfection by plating on MacConkey agar and counting CFU. At day 9, due to significant illness manifestation, in the absence of fecal matter, colon was homogenized and plated. By day 9 postinfection, several of the mice infected with the wild type and the less attenuated TCS deletion strains had succumbed to infection, leading to fewer samples analyzed at this time point in those groups. Dotted lines indicate the limit of detection of the assay. The data are pooled from two experiments with 4 to 5 mice per group per experiment. A Mann-Whitney test was used to determine statistical significance (***, P < 0.001; **, P < 0.01; *, P < 0.05). (D) The last 0.5 cm of the colon of infected mice was stained by hematoxylin and eosin, and histological scoring was performed in a blinded fashion by a board-certified veterinary pathologist, as described in reference 55. An unpaired t test was used to determine statistical significance between each TCS deletion strain and the wild-type strain (*, P < 0.05).
Virulence-associated phenotype of C. rodentium ΔarcA strain.
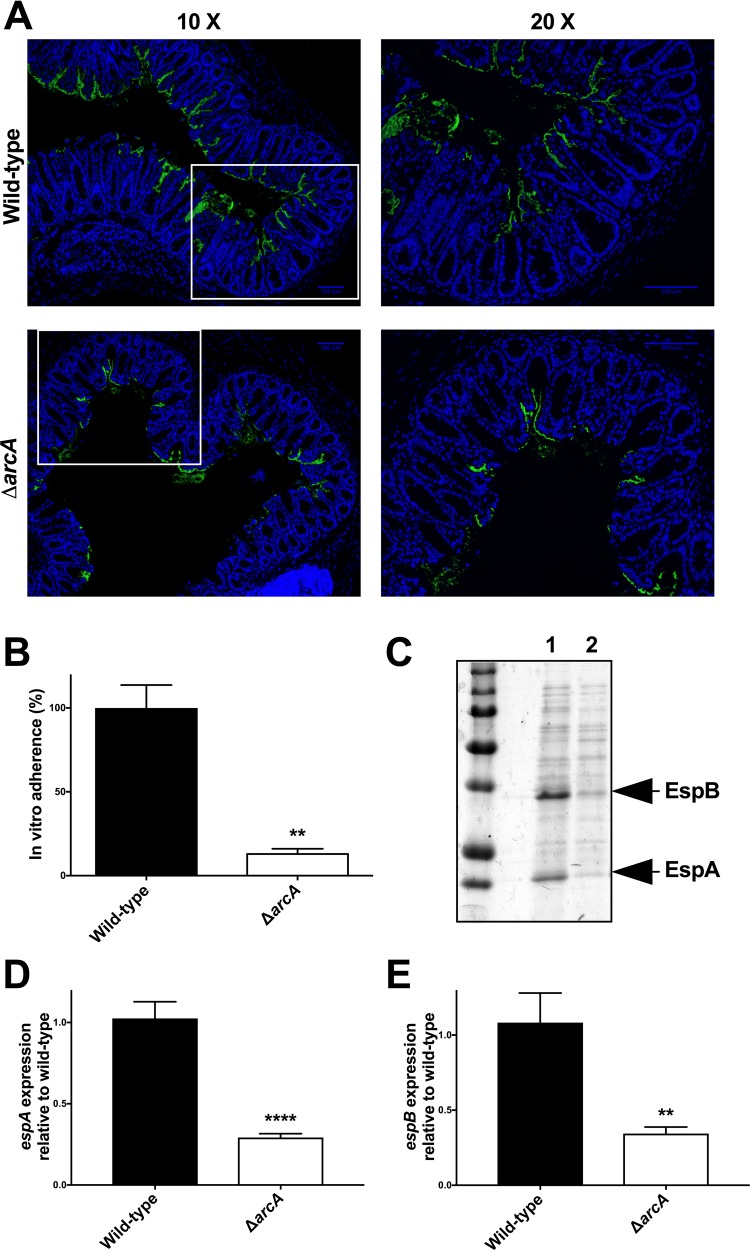
Figure 2 demonstrates that the three TCSs with the greatest influence on C. rodentium virulence are ArcAB, CpxRA, and RcsBC. We previously characterized the ΔcpxRA mutant (36). We next sought to perform a more detailed analysis of the consequences of arcA deletion on C. rodentium virulence. To further investigate the colonization defect of this strain (Fig. 3), we examined the localization of the bacteria in vivo in colonic cross sections taken from mice at day 9 postinfection (Fig. 4A). At this time point, wild-type C. rodentium localized mainly to the colonic mucosal surface, in some cases extending more deeply into the crypts, and with a minor proportion found within the lumen (Fig. 4A, top). In agreement with the bacterial load data (Fig. 3), staining of tissue sections revealed smaller amounts of C. rodentium present in sections of mice infected with the ΔarcA strain compared to mice infected with the wild-type strain (Fig. 4A). However, the C. rodentium strain that was present in the ΔarcA strain-infected colon sections localized to the mucosal surface, similar to what was seen for the wild-type bacteria (Fig. 4A, bottom). We also tested the ΔarcA strain in an in vitro HeLa cell adherence assay and found it to be significantly deficient for in vitro cell adherence (Fig. 4B). C. rodentium adherence in vitro and colonization in vivo are both strongly enhanced by the secretion of effectors through the bacterium's type III secretion system (T3SS). Therefore, we tested the ΔarcA strain for T3SS function using an in vitro secretion assay. As shown in Fig. 4C and S2, T3S was severely affected in the ΔarcA strain but functional in all other TCS deletion strains. Furthermore, qPCR analysis demonstrated significant decreases in the expression of the T3SS translocator genes espA and espB in the arcA mutant (Fig. 4D and E). Thus, we propose that the C. rodentium ΔarcA strain is attenuated for virulence because of a defect in the regulation of the T3SS, leading to a significant impairment in its ability to colonize the host.
FIG 4.
Attenuation of C. rodentium ΔarcA strain. (A) Localization of C. rodentium wild-type and ΔarcA strains in distal colon samples of day 9 infected susceptible mice. Sections were stained with DAPI (blue) and anti-Citrobacter LPS (green). Panels on the right show a zoomed-in view of the boxed areas in the panels on the left. Scale bars, 100 μm. (B) In vitro adherence assay. HeLa cells were infected with C. rodentium wild-type and ΔarcA strains for 8 h. After extensive washing, the samples were fixed and stained with DAPI and anti-Citrobacter LPS. The total number of bacteria per HeLa cell was counted, and adherence was expressed as a percentage of wild-type adherence levels. An unpaired t test was used to determine statistical significance (**, P < 0.01). (C) Total secreted proteins of the C. rodentium wild-type (lane 1) and ΔarcA (lane 2) strain were prepared, separated on a 10% SDS-PAGE gel, and stained with Coomassie blue. The positions of two of the major T3SS-secreted proteins, EspA and EspB, are indicated with arrows to the right of the gel. (D and E) Expression levels of the espA (D) and espB (E) genes by C. rodentium wild-type and ΔarcA strains were measured by real-time RT-PCR. Data were normalized to rpoD expression levels. Statistical significance was assessed by performing an unpaired t test (****, P < 0.0001; **, P < 0.01).
Virulence-associated phenotype of C. rodentium ΔrcsB strain.
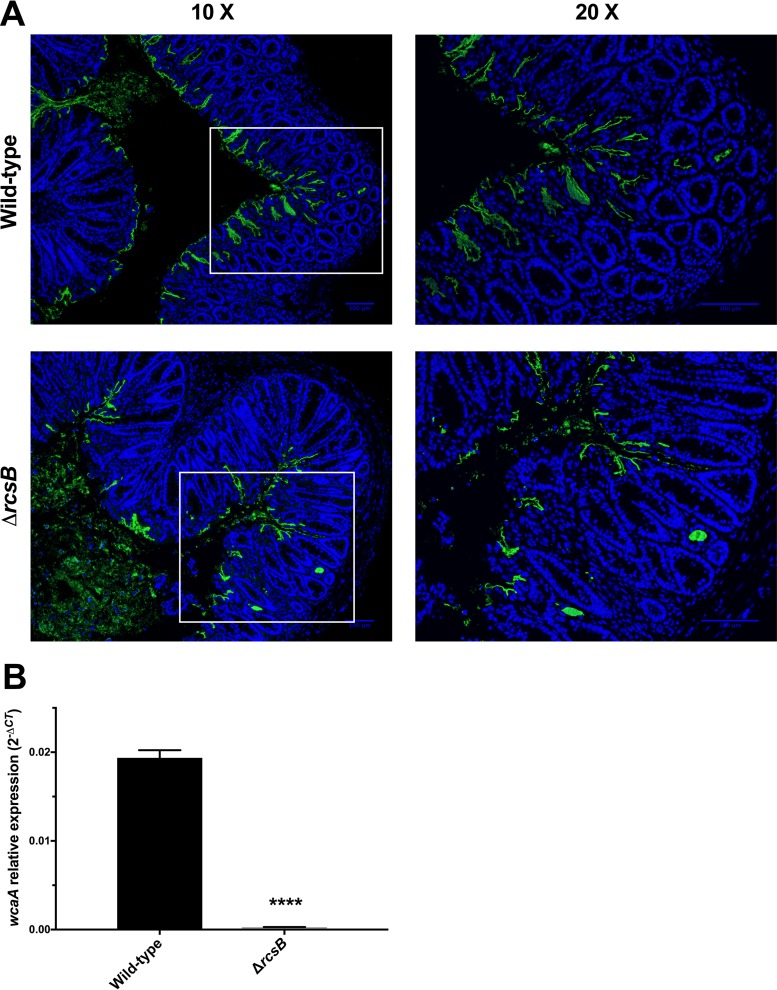
We next further investigated the attenuation of the C. rodentium ΔrcsB strain that had a significant virulence defect, but unlike the ΔarcA strain, it did not display a significant difference in intestinal colonization levels. As described above, we examined the localization of the bacteria, in comparison with wild-type C. rodentium, within infected colon cross sections (Fig. 5A). As described above, wild-type C. rodentium localized to the colonic mucosal surface, with some bacteria extending more deeply into the crypts and a minor proportion of cells within the lumen (Fig. 5A, top). In contrast, a higher proportion of the C. rodentium ΔrcsB strain localized to the intestinal lumen, with a lower proportion than the wild type at the mucosal surface and/or penetrating into the crypts (Fig. 5A, bottom). In contrast to the ΔarcA mutant, the C. rodentium ΔrcsB mutant was not deficient in T3SS function (Fig. S2).
FIG 5.
Attenuation of C. rodentium ΔrcsB strain. (A) Localization of C. rodentium wild-type and ΔrcsB strains in distal colon samples of day 9 infected susceptible mice. Sections were stained with DAPI (blue) and anti-Citrobacter LPS (green). Panels on the right show a zoomed-in view of the boxed areas in the panels on the left. Scale bars, 100 μm. (B) Total RNA was extracted from wild-type- and ΔrcsB strain-infected mouse colons, and real-time RT PCR was performed to detect levels of the wcaA gene relative to the primary sigma factor, rpoD. An unpaired t test was used to determine statistical significance (****, P < 0.0001).
A recent report indicated that the EHEC ΔrcsB strain displays diminished binding of the dye Congo red, suggesting alterations in its surface structures (37). We found that the C. rodentium ΔrcsB strain had increased Congo red binding under all growth conditions tested (Table S5). While the Congo red binding data are different from the results reported for EHEC, both results suggest alterations to surface structures in these strains. In other bacteria, rcsB is known to be required for the regulation of the wca operon, in which wcaA is the first gene of the operon coding for a glycosyl transferase implicated in the production of the capsular component colanic acid (38). To test if the wca operon was affected by deletion of rcsB in C. rodentium, we examined expression of wcaA in the wild-type and ΔrcsB strains using qPCR. We found the ΔrcsB strain to be strikingly deficient in wcaA expression (Fig. 5B). Although this requires further investigation, we hypothesize that the virulence defect in the C. rodentium ΔrcsB strain is related to alterations in surface structures, potentially including colanic acid.
Investigation of increased virulence of the C. rodentium ΔuvrY strain.
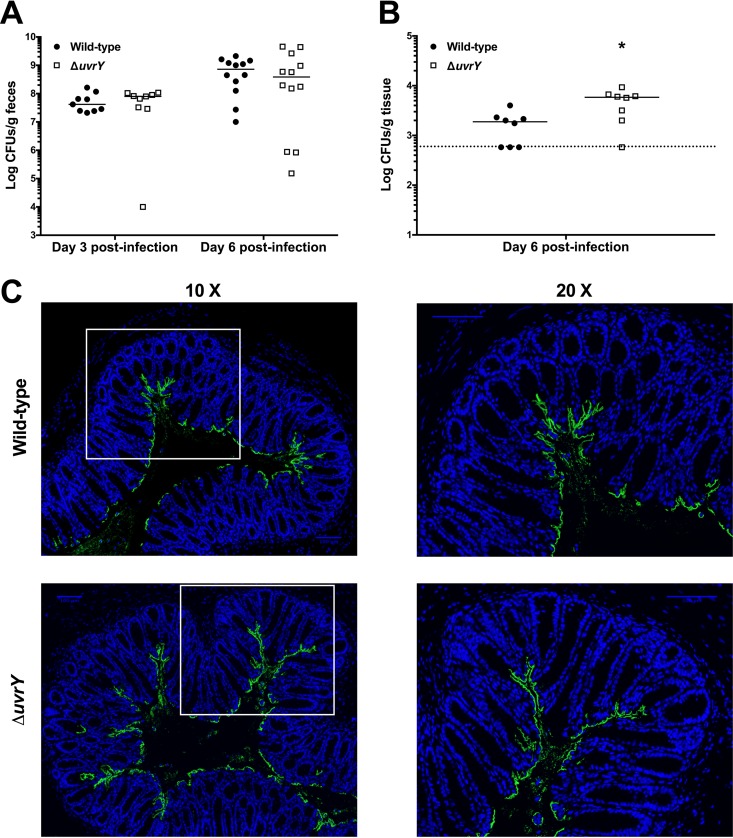
In addition to the investigations described above for the most attenuated TCS deletion strains, we also wanted to perform a more detailed characterization of the increased virulence of the C. rodentium ΔuvrY strain. We analyzed the colonization of the ΔuvrY strain at days 3 and 6 postinfection and found that fecal loads were not significantly different in mice infected with the ΔuvrY strain compared to wild-type C. rodentium (Fig. 6A). Thus, the increased virulence of the ΔuvrY strain is not due to increased bacterial replication and/or survival in the host intestinal tract. However, we did note a significant increase in the amount of C. rodentium present in the mesenteric lymph nodes of mice infected with the ΔuvrY strain (Fig. 6B), indicating an enhanced ability of the ΔuvrY strain to translocate from the intestine to this site or an enhanced ability to resist bacterial killing once translocated. We examined the localization of the bacteria, in comparison with wild-type C. rodentium, within infected colon cross sections (Fig. 6C). Because of the increased mortality at early time points in ΔuvrY strain-infected mice, we assessed this at day 6 postinfection. At this time point, wild-type C. rodentium localized mainly to the colonic mucosal surface, with less bacteria extending into the crypts than was observed at day 9 postinfection (compare Fig. 6C to Fig. 4A and 5A). The localization of the C. rodentium ΔuvrY strain was indistinguishable from that of wild-type bacteria (Fig. 6C). In addition, the ΔuvrY strain was indistinguishable from wild-type C. rodentium in the T3SS assay (Fig. S2), indicating that the enhanced virulence of the C. rodentium ΔuvrY strain is unlikely to be attributed to enhanced intestinal colonization, cell adherence, or T3S.
FIG 6.
Increased virulence of C. rodentium ΔuvrY strain. Susceptible mice were orally gavaged with 2 × 108 to 3 × 108 CFU of the C. rodentium wild-type or ΔuvrY strain. (A) Fecal bacterial burden was assessed at days 3 and 6 postinfection. The data are pooled from two experiments (day 3) and 3 experiments (day 6) with 4 to 5 mice per group per experiment. (B) Mesenteric lymph node bacterial burden was assessed at day 6 postinfection by plating on MacConkey agar and enumerating CFU. The data are pooled from two experiments with 4 mice per group per experiment. A Mann-Whitney test was used to determine statistical significance (*, P < 0.05). (C) Localization of C. rodentium wild type and ΔuvrY strains in distal colon samples of day 6 infected susceptible mice. Sections were stained with DAPI (blue) and anti-Citrobacter LPS (green). Panels on the right show a zoomed-in view of the boxed areas in the panels on the left. Scale bars, 100 μm.
DISCUSSION
TCSs that typically consist of a membrane-bound HK sensor and a cytoplasmic RR are key players in bacterial adaptation to environmental changes (11). During infection, enteric pathogens need to adapt to various environments encountered along the intestinal tract and, thus, rely on TCSs for successful colonization (39). Here, we combined a systematic in vivo study analyzing TCS gene expression with infection experiments that identify the TCSs that are necessary for C. rodentium colonization and/or virulence. This murine pathogen is used as a surrogate model to study the human-restricted pathogens EHEC and EPEC (6). In a previous study, we showed that the C. rodentium ΔcpxRA strain was avirulent, since all mice infected with this strain survived the infection (36). Our current results show that in addition to CpxRA, there are 5 additional TCS knockout strains with attenuated virulence (Fig. 2). In addition, we found that the ΔuvrY strain has enhanced virulence compared to the wild type. At the infectious dose used in this study, deletion of the other TCSs did not affect the survival of infected mice. Recently, Moreira et al. uncovered a role in C. rodentium virulence for the QseBC and QseFE TCSs that respond to the catecholamines epinephrine and norepinephrine in the gut (33). However, the effect on mouse survival was only observed using a much lower infectious dose than the one used in the current study. We anticipate that using lower infectious doses will reveal additional TCSs with subtle effects on virulence. Out of the TCSs that most impact C. rodentium colonization or virulence, it is striking that two of them (i.e., CpxRA and RcsBC) are involved in envelope stress responses, indicating, perhaps unsurprisingly, that the C. rodentium envelope is subjected to extensive stress in the murine intestinal tract. It is also interesting that the TCSs revealed to have the most striking roles in virulence in our studies were also the ones found to be most highly expressed in vivo during intestinal infection (Fig. 1B).
So far, few studies performed a systematic screening of all TCSs of enteric bacteria to identify the ones involved in colonization and/or virulence (40, 41). PhoPQ and EnvZ/OmpR were the only two TCSs found to be critical for Yersinia pestis resistance to host innate immunity in a mouse model of plague (41, 42). Neither of these two TCSs were identified in our study. While the reasons for this dissimilarity are not known, we speculate that it is be due to differences in the host-associated lifestyles of these two pathogens. In contrast, our data identifying CpxRA, ArcAB, and RcsBC as TCSs that are important for C. rodentium colonization and/or virulence are in perfect agreement with the study by Lasaro et al., which found that mutation of the same TCSs produced strong colonization defects for the MP1 E. coli murine commensal strain (40). In the current study, we also identified effects of the ZraRS, RstAB, and UhpAB TCSs on C. rodentium infection, although these TCSs appear to have a more modest role in colonization and/or virulence than CpxRA, ArcAB, and RcsBC. ZraRS has been previously linked to zinc and lead sensing and has recently also been linked to the cell envelope stress response to zinc (43), making it the third TCS that is likely involved in the cell envelope stress response during in vivo infection. RstAB has been shown to be essential for infection of chickens by avian pathogenic E. coli (44). Finally, UhpAB is involved in glucose-6-phosphate transport in E. coli. Future studies are required to investigate if this TCS detects the different levels of glucose-6-phosphate present in the jejunum, duodenum, and colon during infection (45).
Deletion of the C. rodentium arcA RR gene results in both a colonization defect and attenuated virulence (Fig. 4). The ArcAB TCS is known to control the transition from aerobic growth to microaerobic or anaerobic growth in response to the redox status of the ubiquinone electron carriers (46). Therefore, a functional ArcAB TCS appears to be required for bacterial adaptation to the anaerobic environment of the mouse large intestine. ArcA is also a global RR controlling transcription of a plethora of genes, including virulence genes. Remarkably, our data revealed that the ΔarcA strain is the only strain with a defect in T3S (Fig. 4; see also Fig. S2 in the supplemental material). Therefore, it is plausible that the virulence defect of the ΔarcA strain is due to a reduced ability of this strain to translocate effector proteins, including the Tir protein, in intestinal epithelial cells.
Deletion of the rcsB RR gene results in attenuation of virulence exemplified by a delay in mouse death and the survival of some mice infected with this mutant (Fig. 2). As for the CpxRA TCS, the RcsBC TCS is also associated with envelope stress. The RcsC receptor responds to outer membrane and peptidoglycan-related stresses (47). The RcsB-induced genes affect the cell envelope properties, including the presence of the colanic acid capsule and curli (47). In agreement, we found that the wcaA gene, coding for a glycosyl transferase involved in colanic acid synthesis, is poorly expressed in this mutant. In addition, enhanced binding of Congo red to the cell surface of this strain suggests that the cell surface of this mutant is perturbed. Therefore, it is plausible that the ΔrcsB mutant is either more susceptible to the host innate defenses or more easily recognized by the host immune system.
Deletion of the C. rodentium uvrY RR gene results in a more rapid death of infected mice and increased bacterial load in the mesenteric lymph nodes (Fig. 6). The BarA/UvrY TCS controls the transcription of the small RNA genes csrB and csrC that interact with the RNA-binding protein CsrA and, in turn, prevent its interaction with its mRNA targets (48, 49). By antagonizing CsrA, BarA/UvrY regulates carbon metabolism, biofilm formation, motility, and virulence in proteobacteria (50). The fact that inactivation of a TCS results in increased virulence is not unprecedented; inactivation of the CovRS (CsrRS) TCS in the Gram-positive pathogen Streptococcus pyogenes results in the upregulation of various virulence factors, including the hyaluronic acid capsule, streptolysin S, streptokinase, and the cysteine protease SpeB (51). In light of this study, we hypothesize that C. rodentium UvrY also acts as an indirect negative regulator of virulence genes.
TCSs are the main signal transduction systems in bacteria and are absent from mammalian cells. As such, they are promising targets for the development of novel antimicrobial drugs (52, 53). The TCSs we have identified in this study impact intestinal colonization and/or virulence of C. rodentium, a mouse pathogen that serves as a model for human pathogens such as EHEC and other Shiga toxin-producing E. coli. Since the use of classical antibiotics to treat these infections is controversial, due to the risk of increased Shiga toxin production and release, antivirulence approaches such as targeting TCSs may provide novel treatment options. Studies on the TCSs identified here, including genetic complementation of the mutants, further elucidation of the mechanisms underlying the virulence effects, and investigation as to their conservation of function in related human pathogens will be important future directions to pursue.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All strains and plasmids used in this study are listed in Table S1 in the supplemental material. Bacteria were routinely cultured at 37°C with aeration (200 rpm) in Luria-Bertani broth (LB; 1% [wt/vol] tryptone, 0.5% [wt/vol] yeast extract, 1% [wt/vol] NaCl) or in N minimal medium adjusted to pH 7.5 and supplemented with 0.2% glucose and 1 mM MgCl2. For minimal growth conditions, bacteria were cultured in M9 medium supplemented with 0.2% glucose, 2 mM MgSO4, and 0.1 mM CaCl2. When appropriate, LB was supplemented with chloramphenicol (Cm; 30 μg/ml) or dl-diaminopimelic acid (DAP; 50 μg/ml).
RNA extraction and cDNA synthesis.
Total RNA was extracted from in vitro- and in vivo-harvested samples using TRIzol reagents according to the manufacturer's instructions. Contaminating DNA was removed using TURBO DNase I from the TURBO DNA-free kit (Ambion). The absence of contaminating DNA was confirmed by qPCR using rpoD primers (Table S2). The concentration and purity of extracted RNA were analyzed by measuring the absorbances at 260 and 280 nm using a NanoDrop ND-2000 spectrophotometer (Thermo Scientific). First-strand cDNA synthesis was performed on 100 ng total RNA collected from C. rodentium cells grown in vitro or on 1 μg total RNA isolated from the mouse colon samples using Superscript III (Life Technologies) as specified by the manufacturer. As a negative control, a reaction mixture without Superscript III (NRT) was included for each sample.
Primer design for in vivo qPCR.
C. rodentium-specific primers were designed using Primer-BLAST software (54). For each gene, primers and sizes of the expected amplicons are listed in Table S2. Primers were screened for the unintended amplification of Mus musculus, Escherichia coli, and other Citrobacter species. cDNA isolated from uninfected C3H/HeJ and C57BL/6J mice or a pure culture of C. rodentium was used to test the specificity of the primer pairs.
Quantitative reverse transcription-PCR (qRT-PCR).
qPCRs were performed in a Rotor-Gene 3000 thermal cycler (Corbett Research) using the Maxima SYBR green qPCR kit (Thermo Scientific) according to the manufacturers' instructions. All reactions were performed in triplicate. The NRT samples were used as negative controls to ensure that the samples were free of genomic DNA. Relative expression levels of genes were calculated by normalizing the threshold cycle (CT) of the assayed genes to the CT of the endogenous control rpoD gene. The comparative CT method (2−ΔΔCT or 2−ΔCT) was used to calculate relative quantification of gene expression as described in reference 34. The primary sigma factor rpoD (σ70) was chosen as an internal control, because expression of rpoD in E. coli was shown to be independent of the growth phase (55). Results are expressed as the means ± standard deviations (SD) from three experiments performed in triplicate for each gene transcript. A threshold cycle value of 40, which is the limit of detection of our assay, was assigned to samples that did not emit fluorescence during a qPCR cycle and is denoted by a dashed line in our results.
In vivo C. rodentium infections.
All animal experiments were performed under conditions specified by the Canadian Council on Animal Care and were approved by the McGill University Animal Care Committee. C57BL/6J and C3H/HeJ mice were purchased from the Jackson Laboratory (Bar Harbor, ME, USA) and maintained in a specific-pathogen-free facility at McGill University. Four-week-old mice were orally gavaged with C. rodentium strains. For oral inoculations, bacteria were grown for 17.5 h in 3 ml of LB with aeration. Mice were infected by oral gavage of 0.1 ml of overnight culture containing 2 × 108 to 3 × 108 CFU of C. rodentium. The infectious dose was verified by plating of serial dilutions of the inoculum on MacConkey agar (Difco). For RNA analysis, three mice were infected for each time point. C3H/HeJ mice were sacrificed on day 4 and day 9 postinfection, and C57BL/6J mice were sacrificed on day 9 postinfection. The terminal centimeter of distal colon was collected from infected mice, homogenized in 1 ml of TRIzol using a polytron homogenizer (Kinematica), and stored at −80°C. For survival analysis, the mice were monitored daily and were euthanized if they met any of the following clinical endpoints: 20% body weight loss, hunching and shaking, inactivity, and body condition score of <2. To determine colonization levels of mice, mice were euthanized on day 6 or 9 postinfection and bacterial loads were enumerated in the entire colon after weighing and homogenization in 1 ml of sterile phosphate-buffered saline (PBS) using a polytron homogenizer. Homogenates were serially diluted in sterile PBS, and 0.1-ml aliquots of each serial dilution were plated on MacConkey agar. Plates containing between 30 and 300 colonies were counted. When bacterial loads were low, leading to the undiluted sample plate having <30 colonies, the number of colonies on this plate was counted. For histological analysis, the last 0.5 cm of the colon of infected mice was fixed in 10% neutral buffered formalin, processed, cut into 3-μm sections, and stained with hematoxylin and eosin. An expert veterinary pathologist performed histological analysis in the histology facility at the Life Sciences Complex in McGill University.
Construction of deletion strains.
The bacterial strains and plasmids used in this study are listed in Table S3. DH5α and DH5αλpir were routinely used for genetic manipulations. DNA purification, cloning, and transformation were performed according to standard procedures (56). The C. rodentium deletion strains were generated by sacB gene-based allelic exchange (57). Genomic DNA from C. rodentium was used as a template to PCR amplify the upstream (primer pairs 1 and 2; Table S3) and downstream (primer pairs 3 and 4; Table S3) sequences of a TCS operon or RR gene. The upstream and downstream regions of the uvrY gene were then used directly in an overlap PCR. For all other constructs, the resultant PCR products corresponding to the upstream and downstream regions of the TCS or RR genes were treated with XhoI, NheI, or KpnI as appropriate (Table S3), purified, and ligated together. An aliquot of the ligation mixtures was used as a template to PCR amplify the entire product using primer pairs 1 and 4 (Table S3). The resultant PCR products were treated with the appropriate restriction enzymes, found within the sequences of primer pairs 1 and 4 (Table S3), and then ligated into pRE112 that had been treated with the same restriction enzymes, generating the suicide vectors used for homologous recombination. The sequences of these plasmids were verified by sequencing (Genome Québec). The suicide vectors were conjugated into wild-type C. rodentium using E. coli χ7213 as the donor strain. Integration of the plasmid into the chromosome was selected for by plating bacteria on LB agar supplemented with Cm. Cm-resistant transformants of C. rodentium were then plated on peptone agar containing 5% sucrose to isolate colonies that were sucrose resistant. The resultant colonies were also tested for Cm sensitivity. Gene deletions were verified by PCR using primer pairs 1 and 4 (Table S3).
Adherence assays.
In vitro adherence assays of Citrobacter rodentium were performed as previously described (58). Briefly, HeLa cells were seeded at a concentration of 5.0 × 104 cells/coverslip and incubated for 24 h before infection. Cells were incubated for 30 min in 1 ml of DMEM (Life Technologies), supplemented with 2% fetal bovine serum. Cells were infected with an overnight culture of C. rodentium DBS100 or the ΔarcA strain at a multiplicity of infection (MOI) of 1:100 for 8 h at 37°C with 5% CO2. Coverslips were washed 3 times with PBS to remove nonadherent bacteria and fixed in 2.5% paraformaldehyde for 15 min. Monolayers were then washed 3 times for 5 min each with PBS and permeabilized with PBS supplemented with 0.1% Triton X-100. Coverslips were blocked in PBS containing 0.1% Triton X-100 and 2% bovine serum albumin (BSA) overnight at 4°C. Cells were then incubated with anti-Citrobacter LPS rabbit polyclonal antibody for 1 h, followed by three washes with PBS containing 0.1% Triton X-100 and incubation with anti-rabbit Alexa 488 and 4′,6-diamidino-2-phenylindole (DAPI) for 30 min. Coverslips were subsequently mounted in Prolong Gold and imaged on a Zeiss Axiovert 200M microscope with a Zeiss Axiocam monochrome camera. Ten fields of view were imaged per sample, and the number of C. rodentium organisms per cell was calculated. The adherence of the wild-type strain was set at 100%.
Fluorescence microscopy.
C3H/HeJ mice were infected with C. rodentium as described above. Mice were sacrificed on day 6 postinfection (ΔuvrY strain) and day 9 postinfection (ΔrcsB and ΔarcA strains). The third most distal centimeter of colon was fixed in 10% neutral buffered formalin, paraffin embedded, and cut into 4-μm sections. The paraffin-embedded samples were deparaffinized in xylene twice for 5 min, followed by a rehydration in a gradient of ethanol and water: 100% ethanol twice for 5 min, 95% ethanol for 5 min, 70% ethanol for 5 min, and distilled water for 5 min. Antigen recovery was completed by boiling the slides in a solution of citric acid and sodium citrate for 10 min. The slides were washed with PBS containing 0.2% Tween 20 and blocked in PBS containing 0.2% Tween 20, 3% BSA, and 10% fetal bovine serum (FBS) for 1 h at 37°C and then incubated with anti-Citrobacter LPS rabbit polyclonal antibody in PBS containing 0.2% Tween 20 and 3% BSA at 4°C for 3 h. Slides were then incubated in anti-rabbit Alexa 488 (Life Technologies) in PBS containing 0.2% Tween 20 and 3% BSA for 1 h at 37°C, followed by 5 min in DAPI (Sigma) in PBS containing 0.2% Tween 20 and 3% BSA. Slides were then mounted in Prolong Gold (Life Technologies) and on a Zeiss Axiovert 200M microscope with a Zeiss Axiocam monochrome camera.
Secretion assays.
Bacterial secretion assays were performed as described elsewhere (36). Briefly, overnight cultures of bacteria were subcultured 1:50 into 6-well tissue culture plates containing DMEM (Gibco) and incubated for 6 h at 37°C with 5% CO2. After incubation, whole cells were separated from the culture medium by centrifugation. The supernatants were transferred to clean tubes, and the bacterial pellets, now referred to as whole-cell lysates, were resuspended in electrophoresis sample buffer (ESB; 0.0625 M Tris-HCl [pH 6.8], 1% [wt/vol] SDS, 10% glycerol, 2% [vol/vol] 2-mercaptoethanol, 0.001% [wt/vol] bromphenol blue), boiled, and stored at −20°C. Contaminating cells in the supernatant were removed by centrifugation (13,000 × g, 2 min). The proteins in the supernatant, now referred to as secreted proteins, were precipitated with 10% (vol/vol) 6 M trichloroacetic acid. Precipitated secreted proteins were collected by centrifugation (13,000 × g, 30 min at 4°C), washed with ice-cold acetone, air dried, and resuspended in ESB. Protein samples were boiled and aliquots were separated on 10% SDS-PAGE gels and visualized by Coomassie blue staining or transferred onto polyvinylidene difluoride membranes for Western blot analyses using anti-DnaK (Stressgen) or anti-EspB (clone 2A11) antibodies.
Congo red binding.
Congo red binding assays were used to monitor putative changes in the structures present on the surface of C. rodentium colonies. Briefly, bacteria were inoculated onto LB or YESCA (1% [wt/vol] Casamino Acids, 0.1% [wt/vol] yeast extract, 2% [wt/vol] agar) medium supplemented with Congo red (0.002% [wt/vol]) and incubated for 16 h at 37°C aerobically or anaerobically (59). The next day, colonies that were red were determined to have bound Congo red and those that remained white did not.
Supplementary Material
ACKNOWLEDGMENTS
We thank Eugene Kang for the gift of cDNA and Yannick D. N. Tremblay for helpful discussion.
This work was supported by grants from the Fonds de Recherche du Québec–Nature et Technologies (FRQNT 2013-PR-165926) to H.L.M., S.G., and F.D. and from the Canadian Institutes of Health Research (CIHR, MOP-15551) to H.L.M. and S.G. S.G. is supported by a Senior Chercheur-Boursier award from the Fonds de Recherche du Québec–Santé (FRQS). J.-L.T. was supported by a Hugh Burke fellowship from the McGill Faculty of Medicine.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00654-16.
REFERENCES
- 1.Barthold SW, Coleman GL, Jacoby RO, Livestone EM, Jonas AM. 1978. Transmissible murine colonic hyperplasia. Vet Pathol 15:223–236. doi: 10.1177/030098587801500209. [DOI] [PubMed] [Google Scholar]
- 2.Mundy R, MacDonald TT, Dougan G, Frankel G, Wiles S. 2005. Citrobacter rodentium of mice and man. Cell Microbiol 7:1697–1706. doi: 10.1111/j.1462-5822.2005.00625.x. [DOI] [PubMed] [Google Scholar]
- 3.Chen HD, Frankel G. 2005. Enteropathogenic Escherichia coli: unravelling pathogenesis. FEMS Microbiol Rev 29:83–98. doi: 10.1016/j.femsre.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 4.Nataro JP, Kaper JB. 1998. Diarrheagenic Escherichia coli. Clin Microbiol Rev 11:142–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Law RJ, Gur-Arie L, Rosenshine I, Finlay BB. 2013. In vitro and in vivo model systems for studying enteropathogenic Escherichia coli infections. Cold Spring Harb Perspect Med 3:a009977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins JW, Keeney KM, Crepin VF, Rathinam VA, Fitzgerald KA, Finlay BB, Frankel G. 2014. Citrobacter rodentium: infection, inflammation and the microbiota. Nat Rev Microbiol 12:612–623. doi: 10.1038/nrmicro3315. [DOI] [PubMed] [Google Scholar]
- 7.Wiles S, Clare S, Harker J, Huett A, Young D, Dougan G, Frankel G. 2004. Organ specificity, colonization and clearance dynamics in vivo following oral challenges with the murine pathogen Citrobacter rodentium. Cell Microbiol 6:963–972. doi: 10.1111/j.1462-5822.2004.00414.x. [DOI] [PubMed] [Google Scholar]
- 8.Vallance BA, Deng W, Jacobson K, Finlay BB. 2003. Host susceptibility to the attaching and effacing bacterial pathogen Citrobacter rodentium. Infect Immun 71:3443–3453. doi: 10.1128/IAI.71.6.3443-3453.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borenshtein D, Nambiar PR, Groff EB, Fox JG, Schauer DB. 2007. Development of fatal colitis in FVB mice infected with Citrobacter rodentium. Infect Immun 75:3271–3281. doi: 10.1128/IAI.01810-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parkinson JS, Kofoid EC. 1992. Communication modules in bacterial signaling proteins. Annu Rev Genet 26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 11.Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu Rev Biochem 69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 12.Oshima T, Aiba H, Masuda Y, Kanaya S, Sugiura M, Wanner BL, Mori H, Mizuno T. 2002. Transcriptome analysis of all two-component regulatory system mutants of Escherichia coli K-12. Mol Microbiol 46:281–291. doi: 10.1046/j.1365-2958.2002.03170.x. [DOI] [PubMed] [Google Scholar]
- 13.Zhou L, Lei XH, Bochner BR, Wanner BL. 2003. Phenotype microarray analysis of Escherichia coli K-12 mutants with deletions of all two-component systems. J Bacteriol 185:4956–4972. doi: 10.1128/JB.185.16.4956-4972.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raivio TL, Leblanc SK, Price NL. 2013. The Escherichia coli Cpx envelope stress response regulates genes of diverse function that impact antibiotic resistance and membrane integrity. J Bacteriol 195:2755–2767. doi: 10.1128/JB.00105-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beier D, Gross R. 2006. Regulation of bacterial virulence by two-component systems. Curr Opin Microbiol 9:143–152. doi: 10.1016/j.mib.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 16.Dalebroux ZD, Miller SI. 2014. Salmonellae PhoPQ regulation of the outer membrane to resist innate immunity. Curr Opin Microbiol 17:106–113. doi: 10.1016/j.mib.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beier D, Gross R. 2008. The BvgS/BvgA phosphorelay system of pathogenic Bordetellae: structure, function and evolution. Adv Exp Med Biol 631:149–160. doi: 10.1007/978-0-387-78885-2_10. [DOI] [PubMed] [Google Scholar]
- 18.Mizuno T. 1997. Compilation of all genes encoding two-component phosphotransfer signal transducers in the genome of Escherichia coli. DNA Res 4:161–168. doi: 10.1093/dnares/4.2.161. [DOI] [PubMed] [Google Scholar]
- 19.Bergholz TM, Wick LM, Qi W, Riordan JT, Ouellette LM, Whittam TS. 2007. Global transcriptional response of Escherichia coli O157:H7 to growth transitions in glucose minimal medium. BMC Microbiol 7:97. doi: 10.1186/1471-2180-7-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Partridge JD, Sanguinetti G, Dibden DP, Roberts RE, Poole RK, Green J. 2007. Transition of Escherichia coli from aerobic to micro-aerobic conditions involves fast and slow reacting regulatory components. J Biol Chem 282:11230–11237. doi: 10.1074/jbc.M700728200. [DOI] [PubMed] [Google Scholar]
- 21.Partridge JD, Scott C, Tang Y, Poole RK, Green J. 2006. Escherichia coli transcriptome dynamics during the transition from anaerobic to aerobic conditions. J Biol Chem 281:27806–27815. doi: 10.1074/jbc.M603450200. [DOI] [PubMed] [Google Scholar]
- 22.Landstorfer R, Simon S, Schober S, Keim D, Scherer S, Neuhaus K. 2014. Comparison of strand-specific transcriptomes of enterohemorrhagic Escherichia coli O157:H7 EDL933 (EHEC) under eleven different environmental conditions including radish sprouts and cattle feces. BMC Genomics 15:353. doi: 10.1186/1471-2164-15-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palaniyandi S, Mitra A, Herren CD, Lockatell CV, Johnson DE, Zhu X, Mukhopadhyay S. 2012. BarA-UvrY two-component system regulates virulence of uropathogenic E. coli CFT073. PLoS One 7:e31348. doi: 10.1371/journal.pone.0031348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Debnath I, Norton JP, Barber AE, Ott EM, Dhakal BK, Kulesus RR, Mulvey MA. 2013. The Cpx stress response system potentiates the fitness and virulence of uropathogenic Escherichia coli. Infect Immun 81:1450–1459. doi: 10.1128/IAI.01213-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagamatsu K, Hannan TJ, Guest RL, Kostakioti M, Hadjifrangiskou M, Binkley J, Dodson K, Raivio TL, Hultgren SJ. 2015. Dysregulation of Escherichia coli alpha-hemolysin expression alters the course of acute and persistent urinary tract infection. Proc Natl Acad Sci U S A 112:E871–E880. doi: 10.1073/pnas.1500374112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cai W, Wannemuehler Y, Dell'anna G, Nicholson B, Barbieri NL, Kariyawasam S, Feng Y, Logue CM, Nolan LK, Li G. 2013. A novel two-component signaling system facilitates uropathogenic Escherichia coli's ability to exploit abundant host metabolites. PLoS Pathog 9:e1003428. doi: 10.1371/journal.ppat.1003428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwan WR. 2009. Survival of uropathogenic Escherichia coli in the murine urinary tract is dependent on OmpR. Microbiology 155:1832–1839. doi: 10.1099/mic.0.026187-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macritchie DM, Ward JD, Nevesinjac AZ, Raivio TL. 2008. Activation of the Cpx envelope stress response down-regulates expression of several locus of enterocyte effacement-encoded genes in enteropathogenic Escherichia coli. Infect Immun 76:1465–1475. doi: 10.1128/IAI.01265-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pacheco AR, Curtis MM, Ritchie JM, Munera D, Waldor MK, Moreira CG, Sperandio V. 2012. Fucose sensing regulates bacterial intestinal colonization. Nature 492:113–117. doi: 10.1038/nature11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chekabab SM, Jubelin G, Dozois CM, Harel J. 2014. PhoB activates Escherichia coli O157:H7 virulence factors in response to inorganic phosphate limitation. PLoS One 9:e94285. doi: 10.1371/journal.pone.0094285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Njoroge J, Sperandio V. 2012. Enterohemorrhagic Escherichia coli virulence regulation by two bacterial adrenergic kinases, QseC and QseE. Infect Immun 80:688–703. doi: 10.1128/IAI.05921-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reading NC, Torres AG, Kendall MM, Hughes DT, Yamamoto K, Sperandio V. 2007. A novel two-component signaling system that activates transcription of an enterohemorrhagic Escherichia coli effector involved in remodeling of host actin. J Bacteriol 189:2468–2476. doi: 10.1128/JB.01848-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moreira CG, Russell R, Mishra AA, Narayanan S, Ritchie JM, Waldor MK, Curtis MM, Winter SE, Weinshenker D, Sperandio V. 2016. Bacterial adrenergic sensors regulate virulence of enteric pathogens in the gut. mBio 7:e00826-16. doi: 10.1128/mBio.00826-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.Diez E, Zhu L, Teatero SA, Paquet M, Roy MF, Loredo-Osti JC, Malo D, Gruenheid S. 2011. Identification and characterization of Cri1, a locus controlling mortality during Citrobacter rodentium infection in mice. Genes Immun 12:280–290. doi: 10.1038/gene.2010.76. [DOI] [PubMed] [Google Scholar]
- 36.Thomassin J-L, Giannakopoulou N, Zhu L, Gross J, Salmon K, Leclerc J-M, Daigle F, Le Moual H, Gruenheid S. 2015. The CpxRA two-component system is essential for Citrobacter rodentium virulence. Infect Immun 83:1919–1928. doi: 10.1128/IAI.00194-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen CY, Nguyen LH, Cottrell BJ, Irwin PL, Uhlich GA. 2016. Multiple mechanisms responsible for strong Congo-red-binding variants of Escherichia coli O157:H7 strains. Pathog Dis 74:ftv123. doi: 10.1093/femspd/ftv123. [DOI] [PubMed] [Google Scholar]
- 38.Stevenson G, Andrianopoulos K, Hobbs M, Reeves PR. 1996. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J Bacteriol 178:4885–4893. doi: 10.1128/jb.178.16.4885-4893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cameron EA, Sperandio V. 2015. Frenemies: signaling and nutritional integration in pathogen-microbiota-host interactions. Cell Host Microbe 18:275–284. doi: 10.1016/j.chom.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lasaro M, Liu Z, Bishar R, Kelly K, Chattopadhyay S, Paul S, Sokurenko E, Zhu J, Goulian M. 2014. Escherichia coli isolate for studying colonization of the mouse intestine and its application to two-component signaling knockouts. J Bacteriol 196:1723–1732. doi: 10.1128/JB.01296-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reboul A, Lemaitre N, Titecat M, Merchez M, Deloison G, Ricard I, Pradel E, Marceau M, Sebbane F. 2014. Yersinia pestis requires the 2-component regulatory system OmpR-EnvZ to resist innate immunity during the early and late stages of plague. J Infect Dis 210:1367–1375. doi: 10.1093/infdis/jiu274. [DOI] [PubMed] [Google Scholar]
- 42.Oyston PC, Dorrell N, Williams K, Li SR, Green M, Titball RW, Wren BW. 2000. The response regulator PhoP is important for survival under conditions of macrophage-induced stress and virulence in Yersinia pestis. Infect Immun 68:3419–3425. doi: 10.1128/IAI.68.6.3419-3425.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petit-Hartlein I, Rome K, de Rosny E, Molton F, Duboc C, Gueguen E, Rodrigue A, Coves J. 2015. Biophysical and physiological characterization of ZraP from Escherichia coli, the periplasmic accessory protein of the atypical ZraSR two-component system. Biochem J 472:205–216. doi: 10.1042/BJ20150827. [DOI] [PubMed] [Google Scholar]
- 44.Gao Q, Ye Z, Wang X, Mu X, Gao S, Liu X. 2015. RstA is required for the virulence of an avian pathogenic Escherichia coli O2 strain E058. Infect Genet Evol 29:180–188. doi: 10.1016/j.meegid.2014.11.022. [DOI] [PubMed] [Google Scholar]
- 45.Watanabe M, Goto H, Matsushima M, Shimono R, Kihara T. 1983. Distribution of glucose-6-phosphatase activity in mice studied by in vitro whole-body autoradiography. J Histochem Cytochem 31:1426–1429. doi: 10.1177/31.12.6313801. [DOI] [PubMed] [Google Scholar]
- 46.Alvarez AF, Rodriguez C, Georgellis D. 2013. Ubiquinone and menaquinone electron carriers represent the yin and yang in the redox regulation of the ArcB sensor kinase. J Bacteriol 195:3054–3061. doi: 10.1128/JB.00406-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Majdalani N, Gottesman S. 2005. The Rcs phosphorelay: a complex signal transduction system. Annu Rev Microbiol 59:379–405. doi: 10.1146/annurev.micro.59.050405.101230. [DOI] [PubMed] [Google Scholar]
- 48.Liu MY, Gui G, Wei B, Preston JF III, Oakford L, Yuksel U, Giedroc DP, Romeo T. 1997. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J Biol Chem 272:17502–17510. doi: 10.1074/jbc.272.28.17502. [DOI] [PubMed] [Google Scholar]
- 49.Liu MY, Romeo T. 1997. The global regulator CsrA of Escherichia coli is a specific mRNA-binding protein. J Bacteriol 179:4639–4642. doi: 10.1128/jb.179.14.4639-4642.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vakulskas CA, Potts AH, Babitzke P, Ahmer BM, Romeo T. 2015. Regulation of bacterial virulence by Csr (Rsm) systems. Microbiol Mol Biol Rev 79:193–224. doi: 10.1128/MMBR.00052-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heath A, DiRita VJ, Barg NL, Engleberg NC. 1999. A two-component regulatory system, CsrR-CsrS, represses expression of three Streptococcus pyogenes virulence factors, hyaluronic acid capsule, streptolysin S, and pyrogenic exotoxin B. Infect Immun 67:5298–5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bem AE, Velikova N, Pellicer MT, Baarlen P, Marina A, Wells JM. 2015. Bacterial histidine kinases as novel antibacterial drug targets. ACS Chem Biol 10:213–224. doi: 10.1021/cb5007135. [DOI] [PubMed] [Google Scholar]
- 53.Stephenson K, Hoch JA. 2002. Two-component and phosphorelay signal-transduction systems as therapeutic targets. Curr Opin Pharmacol 2:507–512. doi: 10.1016/S1471-4892(02)00194-7. [DOI] [PubMed] [Google Scholar]
- 54.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. 2012. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jishage M, Ishihama A. 1995. Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of sigma 70 and sigma 38. J Bacteriol 177:6832–6835. doi: 10.1128/jb.177.23.6832-6835.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 57.Schauer DB, Falkow S. 1993. The eae gene of Citrobacter freundii biotype 4280 is necessary for colonization in transmissible murine colonic hyperplasia. Infect Immun 61:4654–4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sit B, Crowley SM, Bhullar K, Lai CC, Tang C, Hooda Y, Calmettes C, Khambati H, Ma C, Brumell JH, Schryvers AB, Vallance BA, Moraes TF. 2015. Active transport of phosphorylated carbohydrates promotes intestinal colonization and transmission of a bacterial pathogen. PLoS Pathog 11:e1005107. doi: 10.1371/journal.ppat.1005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hammar M, Bian Z, Normark S. 1996. Nucleator-dependent intercellular assembly of adhesive curli organelles in Escherichia coli. Proc Natl Acad Sci U S A 93:6562–6566. doi: 10.1073/pnas.93.13.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.