Hereditary spastic paraplegias are heterogeneous neurodegenerative disorders characterized by progressive lower limb spasticity. Estrada-Cuzcano et al. report that mutations in the lysosomal transport ATPase gene ATP13A2 – previously associated with Kufor-Rakeb syndrome – cause HSP, and present the first data implicating impaired autophosphorylation of ATP13A2 in the pathogenesis of ATP13A2-associated disease.
Keywords: hereditary spastic paraplegia (HSP), neuronal ceroid lipofuscinosis, Kufor-Rakeb syndrome, parkinsonism, lysosomes
Abstract
Hereditary spastic paraplegias are heterogeneous neurodegenerative disorders characterized by progressive spasticity of the lower limbs due to degeneration of the corticospinal motor neurons. In a Bulgarian family with three siblings affected by complicated hereditary spastic paraplegia, we performed whole exome sequencing and homozygosity mapping and identified a homozygous p.Thr512Ile (c.1535C > T) mutation in ATP13A2. Molecular defects in this gene have been causally associated with Kufor-Rakeb syndrome (#606693), an autosomal recessive form of juvenile-onset parkinsonism, and neuronal ceroid lipofuscinosis (#606693), a neurodegenerative disorder characterized by the intracellular accumulation of autofluorescent lipopigments. Further analysis of 795 index cases with hereditary spastic paraplegia and related disorders revealed two additional families carrying truncating biallelic mutations in ATP13A2. ATP13A2 is a lysosomal P5-type transport ATPase, the activity of which critically depends on catalytic autophosphorylation. Our biochemical and immunocytochemical experiments in COS-1 and HeLa cells and patient-derived fibroblasts demonstrated that the hereditary spastic paraplegia-associated mutations, similarly to the ones causing Kufor-Rakeb syndrome and neuronal ceroid lipofuscinosis, cause loss of ATP13A2 function due to transcript or protein instability and abnormal intracellular localization of the mutant proteins, ultimately impairing the lysosomal and mitochondrial function. Moreover, we provide the first biochemical evidence that disease-causing mutations can affect the catalytic autophosphorylation activity of ATP13A2. Our study adds complicated hereditary spastic paraplegia (SPG78) to the clinical continuum of ATP13A2-associated neurological disorders, which are commonly hallmarked by lysosomal and mitochondrial dysfunction. The disease presentation in our patients with hereditary spastic paraplegia was dominated by an adult-onset lower-limb predominant spastic paraparesis. Cognitive impairment was present in most of the cases and ranged from very mild deficits to advanced dementia with fronto-temporal characteristics. Nerve conduction studies revealed involvement of the peripheral motor and sensory nerves. Only one of five patients with hereditary spastic paraplegia showed clinical indication of extrapyramidal involvement in the form of subtle bradykinesia and slight resting tremor. Neuroimaging cranial investigations revealed pronounced vermian and hemispheric cerebellar atrophy. Notably, reduced striatal dopamine was apparent in the brain of one of the patients, who had no clinical signs or symptoms of extrapyramidal involvement.
Introduction
Hereditary spastic paraplegias (HSPs) are heterogeneous neurodegenerative disorders characterized by progressive spasticity of the lower limbs due to axonopathy of the corticospinal neurons. Clinically, HSPs are classified into pure and complicated forms, the latter being associated with additional neurological or non-neurological symptoms. Mutations in over 60 genes are implicated in HSP, although these only explain ∼30–70% of cases depending on the mode of inheritance (Finsterer et al., 2012; Fink, 2013; Crow et al., 2014; Lo Giudice et al., 2014; Schule et al., 2016). Recent gene discoveries have revealed a genetic overlap between HSP and a spectrum of neuronopathies and axonopathies (Peeters et al., 2013), supporting their clinical similarity and common pathomechanistic background.
ATP13A2 (OMIM 610513) is a lysosomal P5-type transport ATPase, for which the transported substrate remains unidentified (van Veen et al., 2014). ATP13A2 undergoes catalytic autophosphorylation on Asp508 within the conserved P-type ATPase motif, which is regulated by phosphatidic acid and phosphatidylinositol(3,5)bisphosphate (Holemans et al., 2015; Martin et al., 2015). ATP13A2 offers cellular protection to heavy metal ions (Mn2+, Fe3+, Zn2+) (Gitler et al., 2009; Schmidt et al., 2009; Tan et al., 2011; Covy et al., 2012; Park et al., 2014; Rinaldi et al., 2015; Martin et al., 2016), α-synuclein (Gitler et al., 2009; Schultheis et al., 2013; Kong et al., 2014) and mitochondrial toxicity (Grunewald et al., 2012; Gusdon et al., 2012; Holemans et al., 2015; Park et al., 2015; Martin et al., 2016). All of these functions depend on the capability of ATP13A2 to undergo autophosphorylation (Gitler et al., 2009; Schmidt et al., 2009; Tan et al., 2011; Covy et al., 2012; Park et al., 2014; Rinaldi et al., 2015).
Mutations in ATP13A2 have been initially associated with Kufor-Rakeb syndrome (KRS) (Ramirez et al., 2006), an autosomal recessive form of juvenile-onset parkinsonism that is typically accompanied by cognitive impairment, supranuclear gaze palsy and signs or symptoms of pyramidal degeneration. Reports of brain iron accumulation in the putamen and caudate present in some patients have also led to classification of KRS as a form of neurodegeneration with brain iron accumulation (NBIA) (Behrens et al., 2010; Schneider et al., 2010). Subsequent studies have broadened the phenotypic spectrum of biallelic ATP13A2 mutations to include also milder cases without pyramidal or cognitive involvement (Di Fonzo et al., 2007). Additionally, single heterozygous ATP13A2 variants may represent risk factors for early- and late-onset parkinsonism (Djarmati et al., 2009; Rakovic et al., 2009; Fei et al., 2010; Park et al., 2015).
More recently, ATP13A2 mutations were identified in a single kindred with neuronal ceroid lipofuscinosis (NCL) (Farias et al., 2011; Bras et al., 2012; Schultheis et al., 2013), histologically defined by the presence of vacuolated lymphocytes, cytoplasmic positive autofluorescent lipopigments and fingerprint-like profiles in nerve and muscle biopsy (Tome et al., 1985). The clinical presentation in this family showed strong overlap with KRS and was characterized by combined extrapyramidal, pyramidal and cerebellar involvement, accompanied by cognitive deficits, peripheral neuropathy and slow vertical eye movements (Bras et al., 2012).
Here, we report that mutations in ATP13A2 cause complicated HSP type SPG78, further expanding the clinical and genetic spectrum of ATP13A2-associated disorders, which are commonly characterized by lysosomal and mitochondrial dysfunction. We propose that loss of ATP13A2 autophosphorylation activity contributes to the loss of ATP13A2 function and disease aetiology.
Materials and methods
Clinical and imaging evaluation
All patients received a systematic neurological and electrophysiological assessment to evaluate affection of multiple neurological systems by movement disorder specialists.
Routine brain MRI including T1-, T2, diffusion-weighted images (DWI) and fluid attenuated inversion recovery T2 (FLAIR) images were performed in all five cases and reviewed by neuroradiologists and neurologists. Dopamine transporter scintigraphy (DaTSCAN) with 185 MBq 123I-Ioflupan was performed in Patient HIH21480.II.3. Images were acquired with a dual-headed SPECT camera (E.CAM+, Siemens Healthcare) at 180 min after bolus injection of 185 MBq 123I-FP-CIT (GE Healthcare Buchler). Image reconstruction was performed using an iterative algorithm (2D Ordered Subset Expectation Maximization, three iterations, eight subsets). Images were interpreted both visually and quantitatively with the Hermes Brass software package (Version 3.4.4, Hermes Medical Solutions, Stockholm, Sweden).
Written informed consent was obtained from all patients and the study was approved by the respective local institutional review boards.
Whole exome sequencing and homozygosity mapping
Genomic DNA was isolated from peripheral blood according to standard procedures. The exomes of Patients HSP84.II.1, HSP84.II.3, HIH21480.II.3 and HIH22132.II.1 were captured by Agilent’s SureSelect Enrichment solution. Paired‐end sequencing on Hiseq2000/2500 platforms yielded ∼6 Gb of raw data, with 90% of the targets covered at least 20×. The sequencing reads were aligned to hg19 with the Burrows-Wheeler aligner (Li and Durbin, 2009). Variant calling and realignment was done by GATK (McKenna et al., 2010), subsequent annotation and filtering were performed with GenomeComb package for genomic data analysis (Reumers et al., 2012) (for Patients HSP84.II.1 and HSP84.II.3) and Genesis (Gonzalez et al., 2015) (for Patients HIH22132.II.1 and HIH21480.II.3).
Homozygosity mapping was performed using SNPs extracted from whole exome sequencing data using the HOMWES tool in GenomeComb (Kancheva et al., 2015).
Sanger sequencing and genetic validation
Identified variants were validated by bidirectional Sanger sequencing. The PCR products were purified with ExoSAP-IT® (USB), directly sequenced with a BigDye® Terminator v.3.1 kit (Applied Biosystems) and electrophoretically separated on an ABI3730xl DNA Analyzer (Applied Biosystems).
Co-segregation analysis of the variants with the disease was performed for all available family members. The novel mutations were screened in unrelated control individuals.
Genetic nomenclature
Mutations were described according to the Human Genome Variation Society nomenclature; transcript ATP13A2 (NM_001141973.2) was used as a reference.
DNA constructs
The hATP13A2‐pDONR221 was previously described (Holemans et al., 2015). Via Gateway subcloning ATP13A2 was transferred into pcDNA6-GFP-tag (N-terminal) or pcDNA3 vectors. p.Phe177Leu, p.Thr512Ile, p.Gly528Arg, p.Gln1135* mutations were introduced via QuickChange® Mutagenesis (Stratagene) and verified by Sanger DNA sequencing.
Western blotting
Western blotting was performed as previously described (Vandecaetsbeek et al., 2009). Membranes were probed with anti-ATP13A2 rabbit polyclonal antibody (1:5000, A3361, Sigma). Detection was performed via enhanced chemiluminescence (Pierce), which was visualized by a Bio-Rad ChemiDoc™ MP imager.
Cell cultures
Control, Thr512Ile and Phe851Cysfs*6 fibroblasts were maintained in BME media (Sigma), supplemented with 100 U/ml penicillin, 100 µg/ml streptomycin, at 37°C, 5% CO2. HeLa and COS-1 cells were maintained in Dulbecco’s modified Eagle medium containing 10% foetal calf serum, 4 mM GlutaMAXTM-I supplement, 100 U/ml penicillin, 100 µg/ml streptomycin and 0.1 mM minimal essential medium non-essential amino acids (Invitrogen) at 37°C in an atmosphere of air with 5% CO2.
Immunocytochemistry and fluorescence microscopy
Fibroblasts, COS-1 or HeLa cells were seeded on glass-bottomed dishes and their lysosomes and mitochondria were visualized with LysoTracker® or MitoTracker® probes according to the manufacturer’s instructions (Life Technologies). Additionally, control and Thr512Ile fibroblasts were seeded on 12 mm diameter cover slips and 48 h later fixed with ice-cold methanol for 20 min at −20°C. Subsequently, the cells were stained with anti-LAMP1 rabbit polyclonal antibody (1:500, ab24170, Abcam), anti-TOMM20 mouse monoclonal antibody (1:500, ab56783, Abcam), and secondary anti-rabbit IgG Alexa Fluor® 488 and anti-mouse IgG Alexa Fluor® 594 antibodies (1:500, Life Technologies).
HeLa cells were transiently transfected with GFP-N-ATP13A2 WT or Phe177Leu, Thr512Ile, Gln1135* using Lipofectamine® LTX reagent (Life Technologies), cultured on glass-bottomed dishes for 48 h, and stained with LysoTracker® (Life Technologies).
HeLa cells were transiently co-transfected with GFP-N-ATP13A2 WT or mutants Phe177Leu, Thr512Ile and Gln1135* together with pCI-mRuby-KDEL (ER marker) using Lipofectamine® LTX reagent (Life Technologies). Cells were cultured on glass-bottomed dishes for 48 h and fluorescent images were taken using a Zeiss LSM700 confocal microscope.
Lysosomal area and mitochondrial morphology
Lysosomal area ratio was measured in control and patient-derived LysoTracker®-stained fibroblasts (Thr512Ile-HSP) using ImageJ software (Schneider et al., 2012). The transmitted light image was used to manually delineate the cell borders, and for each cell the area covered with lysosomes was segmented based on automatic intensity thresholding (Huang method). Lysosomal area ratio was calculated by dividing the lysosomal area by the total cell area for each cell. More than 50 cells per genotype were analysed.
To compare mitochondrial morphology between control and Thr512Ile-HSP ATP13A2 we quantitatively assessed mitochondrial morphology in fibroblasts stained with MitoTracker®. Mitochondria were automatically detected using an ImageJ script with a customized segmentation procedure, which combines global (Triangle method) and local (Phansalkar method) automatic intensity thresholding, size-selective Watershed algorithm and Boolean operations on the binary masks. The shape factor (circularity) of over 30 000 individual mitochondrial segments originating from more than 200 cells was measured.
Lysosomal and mitochondrial homeostasis
Thr512Ile-HSP, Phe851Cysfs*6 and control fibroblasts were seeded in 12-well plates and allowed to adhere overnight at 37°C. For mitochondrial functionality, cells were stained with MitoTracker® Deep RED FM (1 µM) or tetra-methylrhodamine (TMRM, 1 µM) for 30 min. Pretreatment with carbonyl cyanide m-chlorophenyl hydrazone (CCCP, 1 µM) for 1 h prior to staining was used as a positive control (Supplementary Fig. 1A). For lysosomal functionality cells were stained with either LysoTracker® RED (1 µM) for 1 h or DQ-BSA (1 µM) for 2 h. Lysosomal function was monitored by the addition of Bafilomycin A1 (Baf A1, 10 nM) at least 1 h prior to staining (Supplementary Fig. 1B). Measurements for lysosomal and mitochondrial mass were normalized to cell size whereby forward scatter was used as a cell-size index. All samples were detected using an Attune flow cytometer (Becton Dickinson Immunocytometry systems).
Transmission electron microscopy of human fibroblasts
Thr512Ile-HSP and control fibroblast primary cultures grown on glass cover slips were fixed in 4% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.2) for 4 h at room temperature followed by fixation over night at 4°C. After washing, cells were subsequently dehydrated through a graded ethanol series, including a bulk staining with 1% uranyl acetate at the 50% ethanol step followed by embedding in Spurr’s resin.
Ultrathin sections of a gold interference colour were cut using an ultra-microtome (Leica EM UC6), followed by a post-staining in a Leica EM AC20 for 40 min in uranyl acetate at 20°C and for 10 min in lead stain at 20°C. Sections were collected on Formvar-coated copper slot grids. Grids were viewed with a JEM 1400plus transmission electron microscope (JEOL) operating at 60 kV.
Membrane fractionation
COS-1 cells were transiently transfected with GFP-N-ATP13A2 WT or Phe177Leu, Thr512Ile, Gly528Arg, Gln1135* using GeneJuice® (Novagen) according to the manufacturer’s protocol. Forty-eight hours post-transfection, cells were harvested and fractionated as previously described (Vandecaetsbeek et al., 2009). To prepare total membrane fractions, the lysates were first cleared from the nuclear fraction (1000g, 10 min, 4°C) and then centrifuged to collect the total membranes (200 000g, 35 min, 4°C). Fractions were suspended in 0.25 M sucrose with SIGMAFAST™ protease inhibitor cocktail (Sigma). Protein concentration was determined with the Q‐bit fluorimetric method (Life Technologies).
Proteasome inhibition
COS-1 cells were transiently transfected with GFP-N-ATP13A2 WT or Phe177Leu, Thr512Ile, Gly528Arg, Gln1135* using GeneJuice® (Novagen) according to the manufacturer’s protocol. MG132 (M7449, Sigma-Aldrich) was dissolved in dimethyl sulphoxide (DMSO) solution buffer and used at 20 mM for 6 h. DMSO was used as a control vehicle.
Autophosphorylation assay
Forty micrograms of COS-1 membranes were added to a final volume of 95 μl of EP reaction buffer (17 mM HEPES pH 6.5, 160 mM KCl, 2 mM MgCl2, 1 mM DTT, 5 mM NaN3). The reaction was initiated by addition of γ-32P-ATP (7.5 μCi) in a final ATP concentration of 5.125 µM and stopped after 60 s by adding 400 μl ice-cold stop solution (20% trichloroacetic acid, 10 mM phosphoric acid). After precipitation on ice for 20 min, samples were centrifuged at 20 000g (30 min, 4°C). The pellet was washed twice with 400 μl ice-cold stop solution and dissolved in sample buffer (10% LDS, 10 mM NaH2PO4, 0.01% SDS, 10 mM 2-mercaptoethanol, 0.5 mg/ml bromophenol blue) prior to acidic electrophoresis, as previously described (Sorensen et al., 2012).
Statistics
Analysis of lysosomal and mitochondrial quantification and function in control versus Thr512Ile-HSP or Phe851Cysfs*6 patient-derived fibroblasts was performed using a two-sample t-test. Relative expression levels and autophosphorylation in ATP13A2 WT and mutants were analysed using an ANOVA test with post hoc Bonferroni correction. All tests were two-tailed and the level of significance was set at P < 0.05.
Results
Identification of HSP-associated mutations in ATP13A2
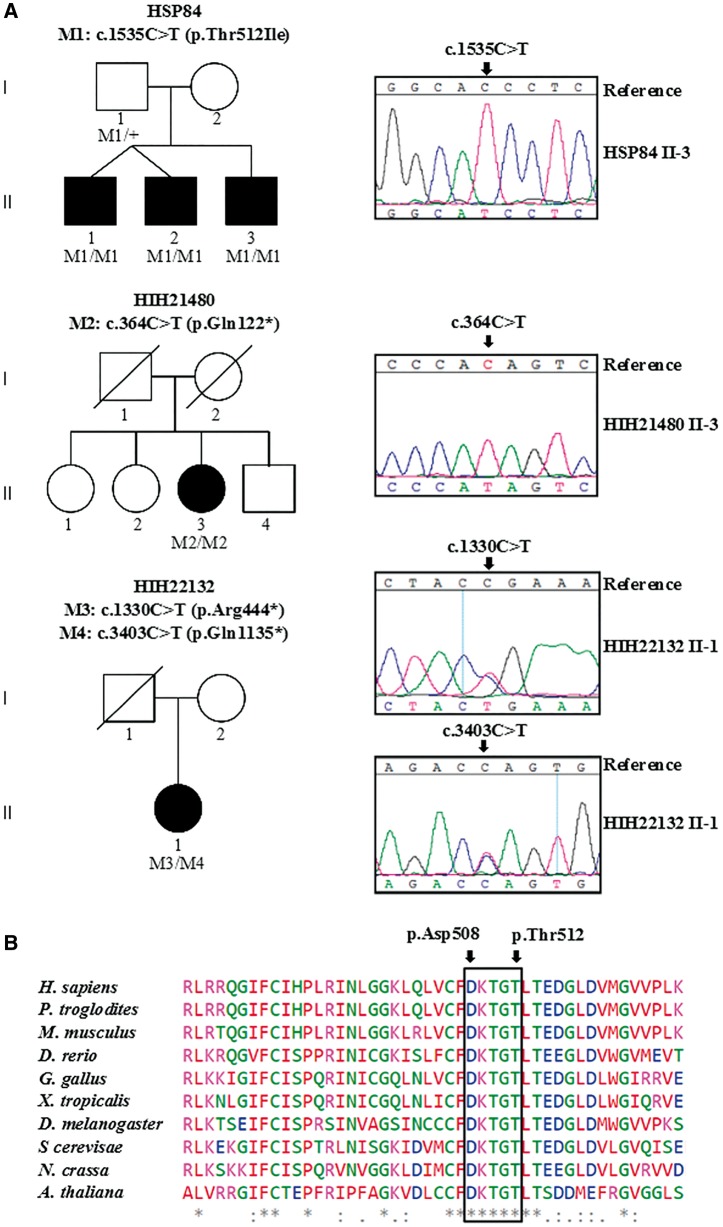
We studied a Bulgarian family with three affected brothers (Family HSP84, Fig. 1A) presenting an adult-onset HSP phenotype, complicated by mild cognitive impairment, cerebellar ataxia, and axonal motor and sensory polyneuropathy (Table 1). Whole exome sequencing was performed on Patients HSP84.II.1 and HSP84.II.3, followed by whole exome sequencing-based homozygosity mapping. A total of 75 shared autozygous regions were identified, encompassing 111 Mb of genomic DNA. The whole exome sequencing data were filtered for homozygous variants located within these regions with an impact on protein structure (non-synonymous and/or affecting mRNA splicing) and ≤5% frequency in dbSNP135 (Sherry et al., 2001), EVS6500 [Exome Variant Server, NHLBI Exome Sequencing Project (ESP), Seattle, WA (URL: http://evs.gs.washington.edu/EVS/) (August 2014)], 1000 Genomes (Genomes Project et al., 2015), and in-house exome/genome databases (Gonzalez et al., 2015). Only one missense variant, c.1535C > T (p.Thr512Ile) in ATP13A2 fulfilled these criteria. This variant co-segregated with the disease (Fig. 1A) and was absent in 94 ethnically matched controls. The p.Thr512Ile substitution (p.Thr512Ile-HSP) targets a highly conserved amino acid (GERP score = 5.6) (Cooper et al., 2005), affecting a residue in the autophosphorylation motif (Fig. 1B), and is predicted to be deleterious by SIFT (Kumar et al., 2009), MutationTaster2 (Schwarz et al., 2014) and PolyPhen-2 (Adzhubei et al., 2010). Surprisingly, the same mutation had previously been described to cause KRS in a sporadic Lebanese patient (p.Thr512Ile-KRS; nomenclature in original publication: NM_022089.2: c.1550C > T, p.Thr517Ile) (Dehay et al., 2012; Grunewald et al., 2012; Usenovic et al., 2012).
Figure 1.
Novel mutations in ATP13A2. (A) Graphic representation of three autosomal recessive families with complicated HSP in which ATP13A2 mutations were identified. Sanger sequencing traces confirming the presence of the mutations are presented and their segregation in available family members is demonstrated. The symbol (+) represents wild-type. Blackened symbols indicate affected individuals and symbols with slashes depict deceased individuals. (B) Amino acid conservation of ATP13A2 polypeptide in different species, the black frame delimits the autophosphorylation motif (508DKTGT), catalytic conserved residues Asp508 and Thr512 in human ATP13A2 are indicated by black arrows. Below the alignment, asterisk indicates fully conserved, colon indicates strongly similar, and full stop indicates weakly similar amino acids.
Table 1.
Clinical characteristics of ATP13A2 HSP patients
| Study ID | HSP84.II.1 | HSP84.II.3 | HSP84.II.4 | HIH21480.II.3 | HIH22132.II.1 |
|---|---|---|---|---|---|
| Mutation (cDNA) | 1535C > T/1535C > T | 1535C > T/1535C > T | 1535C > T/1535C > T | 364C > T/364C > T | 1330C > T/3403C > T |
| Mutation (protein) | Thr512Ile/Thr512Ile | Thr512Ile/Thr512Ile | Thr512Ile/Thr512Ile | Gln122*/Gln122* | Arg444*/Gln1135* |
| Gender | M | M | M | F | F |
| Origin | Bulgaria | Bulgaria | Bulgaria | Serbia, non-consanguineous background | Bosnia, non-consanguineous background |
| Age at onset (y) | 30 | 33 | 30 | 36 | 32 |
| Age at examination (y) | 50 | 40 | 50 | 47 | 39 |
| Disease severity | |||||
| Landmarks of disability (max 4) | 4 | 3 | 4 | 4 | 3 |
| SARA (max 100) | 20 (50 y) | 8 (40 y) | 21 (50 y) | 28 (47 y) | 22 (39 y) |
| Dependency on walking aid after n years | 13 | 8 | 13 | 2 | 5 |
| Loss of ambulation after n years | 18 | n.a. | 18 | 8 | n.a. |
| Presenting symptom | Gait disorder, dysarthria | Gait disorder, dysarthria | Gait disorder, dysarthria | Spastic paraplegia, neurogenic bladder dysfunction, mild dysarthria | Gait disorder |
| Cognitive deficits | Slight verbal memory deficit | None | Slight verbal memory deficit | Severe dementia | Severe frontotemporal dementia |
| Behavioural and psychiatric symptoms | None | None | None | Labile motivation | Aggression, acoustic hallucinations |
| Pyramidal and peripheral motor system | |||||
| UL/LL spasticity | −/+ | −/+ | −/+ | −/+ | −/+ |
| UL/LL weakness | −/+ | −/+ | −/+ | −/+ | (+)/+ |
| Increased tendon reflexes UL/LL | +/+ | +/+ | +/+ | −/+ | +/+ |
| Muscle atrophy | − | − | − | − | − |
| Babinski sign | + | + | + | − | + |
| Extrapyramidal motor system | |||||
| Brady-/hypokinesia | − | − | − | − | (+) |
| Dystonia | − | − | − | − | − |
| Tremor | − | − | − | − | (+) Resting |
| Spinocerebellar system | |||||
| Oculomotor disturbance | + | + | + | + | + |
| Dysarthria | + | + | + | + | + |
| Limb/gait ataxia | +/+ | +/+ | +/+ | +/− | +/− |
| Brainstem | |||||
| Slow saccades | − | − | − | − | − |
| Supranuclear gaze palsy | − | − | − | Vertical | Horizontal and vertical |
| Sensory system | |||||
| Surface sensation deficit | + | + | + | − | − |
| Vibration/joint position deficit | +/− | +/− | +/− | −/− | −/n.d. |
| Bladder | Voiding | Normal | Normal | Urge incontinence | Urge incontinence |
| Rectum | Normal | Normal | Normal | Normal | Normal |
| Other | Divergent strabism | ||||
| Imaging | Cerebellar > cortical atrophy | Mild cortical atrophy Periventricular white matter changes ‘Ear of the lynx’ signa | Cerebellar > cortical atrophy | Cerebellar > cortical / mesencephalic atrophy Thin corpus callosum Hydrocephalus Periventricular white matter changes | Cerebellar > cortical atrophy ‘Ear of the lynx’ signa |
| Nerve conduction studies | Axonal motor and sensory polyneuropathy | Normal | Axonal motor and sensory polyneuropathy | Mixed axonal-demyelinating motor polyneuropathy | Mild axonal sensory neuropathy |
| MEP UL/LL | Normal / prolonged CMCT | ||||
| SEP Med/Tib | Normal / normal | ||||
| VEP | Prolonged | Normal | Prolonged | ||
| AEP | Prolonged | Prolonged | Prolonged | ||
| Harding Classification | Complicated HSP | Complicated HSP | Complicated HSP | Complicated HSP | Complicated HSP |
AEP = auditory evoked potential; CMCT = central motor conduction time; LL = lower limb; Med = median nerve; MEP = motor evoked potential; n.d. = not determined; SARA = scale for the assessment and rating of ataxia; SEP = sensory evoked potential; SPRS = spastic paraplegia rating scale; Tib = tibial nerve; UL = upper limb; VEP = visually evoked potential.
Landmarks of disability (Schule et al., 2006): 1, able to walk > 500 m without walking aid; 2, able to walk > 500 m with walking aid; 3, able to walk < 500 m with walking aid; 4, not able to walk.
The Medical Research Council (MRC) Scale for Muscle Strength was used to evaluate weakness.
T2 hyperinsensity of the anterior forceps of the corpus callosum (Riverol et al., 2009).
To obtain additional genetic evidence on the causal link between ATP13A2 and HSP, we screened a cohort of 795 index cases with phenotypes overlapping the clinical manifestation in Family HSP84. This cohort comprised cases with pure or complicated HSP, ataxia and axonal neuropathy (Supplementary Table 1). To test the specificity of ATP13A2 mutations for this phenotypic spectrum, we additionally screened 1985 patients with autosomal recessive non-overlapping neurological and non-neurological phenotypes including cases with late-onset Alzheimer’s disease, amyotrophic lateral sclerosis, epileptic encephalopathy, cardiomyopathy, deafness and ocular phenotypes (Gonzalez et al., 2015).
Among the 795 cases with overlapping phenotypes we identified two additional families with autosomal-recessive complicated HSP and ATP13A2 mutations, while no biallelic mutations were found in the non-overlapping neurological and non-neurological phenotypes. A homozygous c.364C > T (p.Gln122*) variant was found in a Serbian family (HIH21480). This nonsense mutation introduces a premature stop codon in the fifth exon of ATP13A2. In the Bosnian HIH22132 family, two nonsense mutations [c.1330C > T (p.Arg444*) and c.3403C > T (p.Gln1135*)] were confirmed by Sanger sequencing (Fig. 1A).
Clinical findings
The disease presentation in all five cases was dominated by an adult onset (mean 32 years) combined pyramidal-cerebellar syndrome that led to the clinical diagnosis of complicated HSP, which progressed rapidly (Table 1). Cognitive deficits were present in four of five cases but ranged from very mild deficits to advanced dementia with fronto-temporal characteristics (Patient HIH22132.II.1). Supranuclear gaze palsy was noted in two cases. Nerve conduction studies revealed predominantly axonal sensory-motor peripheral neuropathy in four out of five individuals. Only one of the five cases showed clinical indication of extrapyramidal involvement in the form of very mild bradykinesia and slight resting tremor (Patient HIH22132.II.1) that didn’t interfere with the activities of daily living.
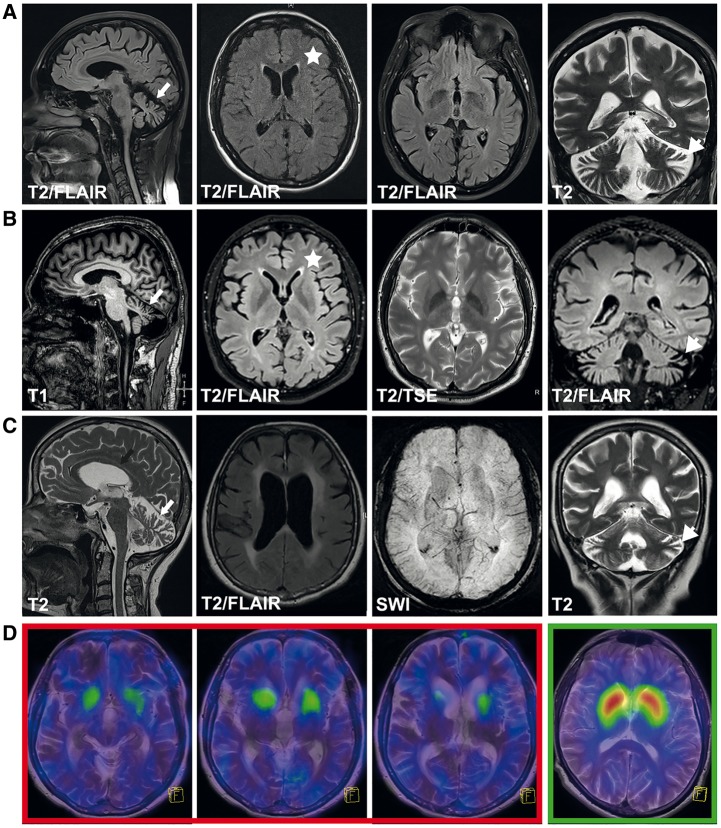
The most striking finding on MRI was a progressive vermian and hemispheric cerebellar atrophy (Fig. 2A–C), accompanied by a less pronounced cortical atrophy and mild periventricular white matter changes in some cases. In two cases (Patients HSP84.II.3 and HIH22132.II.1) an ‘ear of the lynx’ sign (Riverol et al., 2009) and T2 hyperintensities of the anterior fornix of the corpus callosum were noted (Fig. 2A and B). In one case (Patient HIH21480.II.3) thinning of the corpus callosum, periventricular white matter changes and a hydrocephalus internus were additionally present (Fig. 2C). Prompted by the identification of ATP13A2 mutations, we performed a DaTSCAN (123I-FP-CIT) in Patient HIH21480.II.3 to evaluate presynaptic dopamine transporter density in the striatum (Fig. 2D). Although Patient HIH21480.II.3 showed no clinical signs or symptoms of extrapyramidal involvement, the DaTSCAN revealed a drastic decrease of dopamine transporter density, most pronounced in the putamen.
Figure 2.
Cranial imaging in ATP13A2 HSP cases. Routine MRI in Patients HIH22132.II.1 (A) (disease duration 7 years), HSP84.II.3 (B) (disease duration 7 years), and HIH21480.II.3 (C) (disease duration 11 years). Images demonstrate vermian (white arrow) as well as hemispheric cerebellar atrophy (white arrowheads) that was progressive over time in Patient HIH22132.II.1 (not shown). Generalized cortical atrophy was present in all cases. In Patients HIH22132.II.1 (A) and HSP84.II.3 (B) an ‘ear of the lynx’ sign (T2 hyperintensity of the anterior fornix of the corpus callosum, marked by an asterisk) was present, while periventricular white matter changes were more pronounced in Patient HIH21480.II.3 (C). The latter case also demonstrated thinning of the corpus callosum (C, marked by black arrow). (D) Transversal fused 123I-FP-CIT SPECT and T2-weighted MRI images of three representative sections of the striatum in Patient HIH21480.II.3 (red box) and a representative section of the striatum from a healthy adult (green box, far right). Patient HIH21480.II.3 demonstrates severely reduced striatal dopamine.
Functional evaluation of HSP-associated ATP13A2 mutations
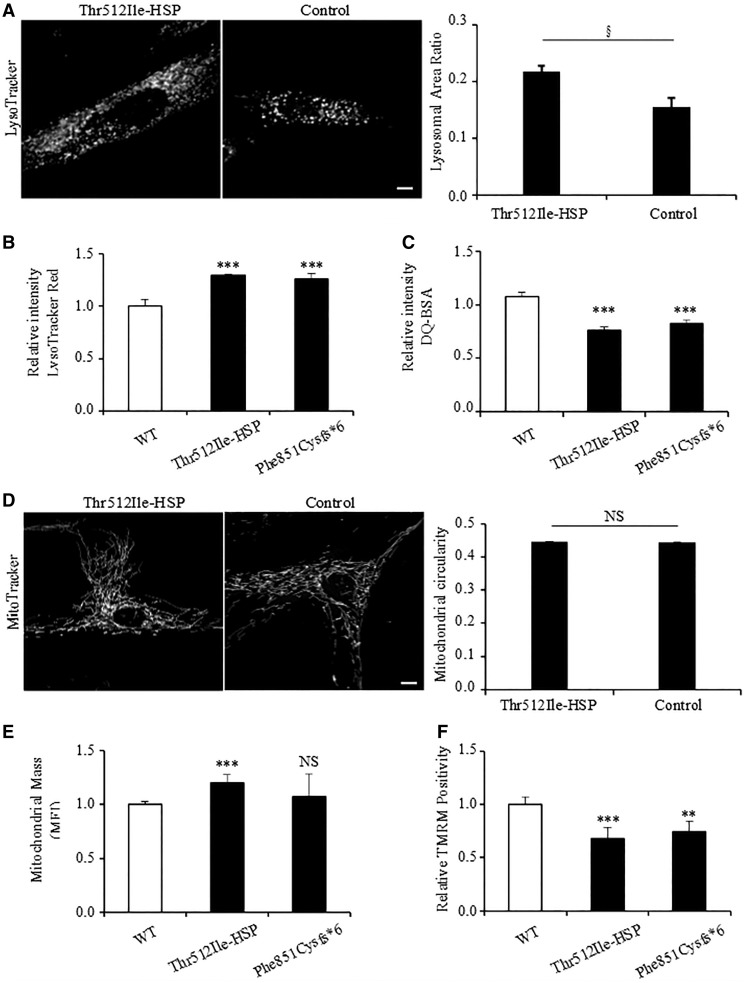
In fibroblasts of a known KRS patient, the p.Thr512Ile-KRS mutation affects both lysosomal function and mitochondrial network integrity (Dehay et al., 2012; Grunewald et al., 2012; Usenovic et al., 2012). Since we now identified the same mutation in patients with complicated HSP (p.Thr512Ile-HSP), we evaluated fibroblasts isolated from Patient HSP84.II.2. Lysosomes in patient and control fibroblasts were visualized by the lysosomal markers LysoTracker® (Fig. 3A) and LAMP1 (Supplementary Fig. 2). Compared to controls, we observed a significant increase in the amount of lysosomes per cell (Fig. 3A), as seen before in fibroblasts from the Thr512Ile-KRS patient (Dehay et al., 2012). Furthermore, via LysoTracker® Red-based flow cytometry we detected an increase in lysosomal mass in Thr512Ile-HSP compared to control fibroblasts (Fig. 3B). The lysosomal functionality was also compromised, as there was a significant reduction in the proteolytic activity, which was assessed by the capability of processing the DQ-BSA substrate into a fluorescent product (Fig. 3C). A similar lysosomal phenotype was also observed in fibroblasts derived from a KRS patient carrying a known truncating p.Phe851Cysfs*6 mutation in ATP13A2 (Fig. 3B and C) (Crosiers et al., 2011).
Figure 3.
Functional characterization of ATP13A2 mutant proteins. (A) Lysosomes of control and patient-derived (Patient HSP84.II.2) fibroblasts were stained with LysoTracker® (left), along with quantification of the lysosomal area ratio (lysosomal area/total cell area) (right). (B) Lysosome function was analysed using Lysotracker® Red and (C) DQ-BSA. DQ-BSA requires enzymatic cleavage in acidic lysosomal compartments to generate a highly fluorescent product. (D) Mitochondria of control and Patient HSP84.II.2-derived fibroblasts were stained with MitoTracker®, and mitochondrial circularity is shown on the right. (E) We used the mitochondrial-specific dye MitoTracker® Deep RED FM, which binds mitochondrial membrane independently of the membrane potential, and thus staining intensity has been considered an index of mitochondrial mass (MFI, mean fluorescence intensity). (F) Tetra-methylrhodamine (TMRM, 1 µM) treatment for quantifying changes in mitochondrial membrane potential. For A and D data are represented by average ± standard error of the mean. For B, C, E and F data are represented by average ± SD. §t-test P = 0.009, **P < 0.05; ***P < 0.001; NS = not significant. Scale bar = 10 µm.
The mitochondrial network in the Thr512Ile-HSP and control fibroblasts was visualized using the mitochondrial markers MitoTracker® and TOMM20 (Fig. 3D and Supplementary Fig. 2). In contrast to the findings of Grunewald et al. (2012), mitochondrial fragmentation or swelling was not markedly present in patient fibroblasts. Also a quantitative comparison of mitochondrial circularity did not reveal differences in mitochondrial morphology between patient-derived and control cells. However, fibroblasts carrying p.Thr512Ile-HSP mutation displayed a significant increase in mitochondrial mass with an overall reduction in mitochondrial membrane potential (Fig. 3E and F), in line with a previous report (Grunewald et al., 2012).
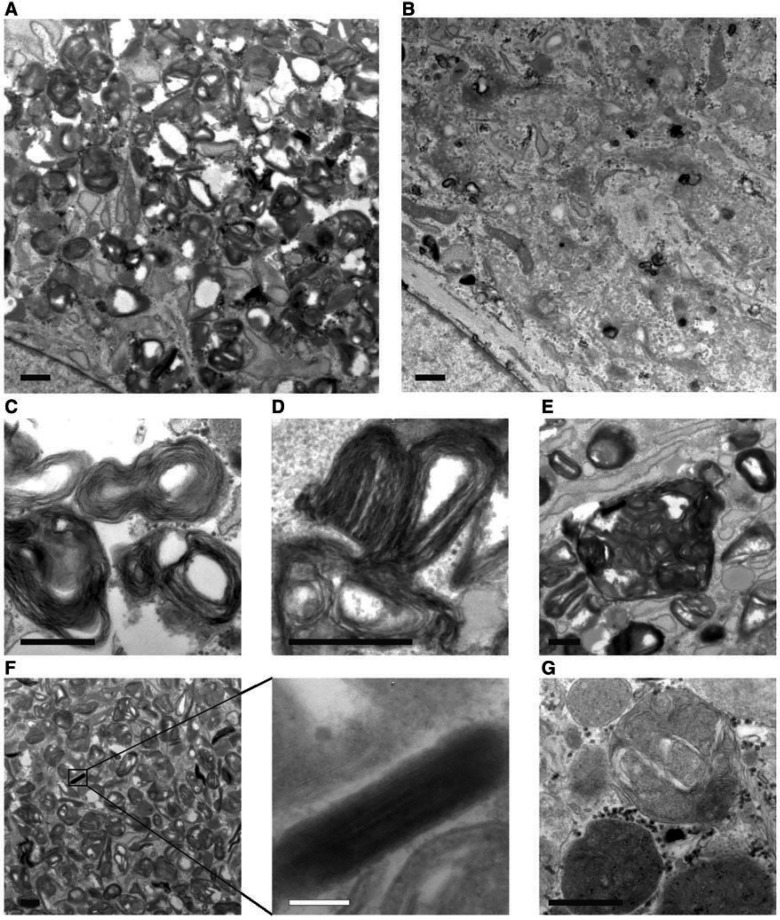
Ultrastructural analysis of Thr512Ile-HSP fibroblasts confirmed an excessive accumulation of lysosomes compared to control cells (Fig. 4A and B). Strikingly, the majority of the lysosomes in Thr512Ile-HSP cells contained abnormal storage material consisting of whirls and stacks of membranes (Fig. 4C–F), a lysosomal content that was almost never observed in control cells (Fig. 4G).
Figure 4.
Ultrastructure of cultured Thr512Ile-HSP fibroblasts. Lysosomes are excessively present in Thr512Ile-HSP fibroblasts (A) compared to controls (B). The majority of lysosomes contain aberrant storage material consisting of whirls and stacks of membranes (C), in some cases forming fingerprint bodies (D) or large clusters of membrane containing vesicles (E). In addition, cytoplasmic inclusions of tightly packed membranes were frequently observed (F). In contrast, the content of lysosomes in control fibroblasts consisted of a normal mix of material (G). Scale bars = 500 nm, except in F = 100 nm.
Known recessive mutations in ATP13A2 causing KRS are considered to be loss-of-function alleles. Some of them lead to aberrant transcripts, which are destroyed by nonsense-mediated mRNA decay (NMD), while others impair protein stability, leading to mislocalization of ATP13A2 to the endoplasmic reticulum and proteasomal degradation (Ramirez et al., 2006; Park et al., 2011, 2014; Dehay et al., 2012; Podhajska et al., 2012; Usenovic et al., 2012). p.Gln122* and p.Arg444* are predicted to be null alleles that are most likely prone to NMD, whereas the p.Gln1135* may potentially escape NMD. This mutation lies in the last exon and likely leads to a truncated protein lacking 40 C-terminal amino acid residues. In contrast, according to in silico predictions the missense p.Thr512Ile mutation may have little impact on protein folding/stability (Phyre2; Kelley et al., 2015).
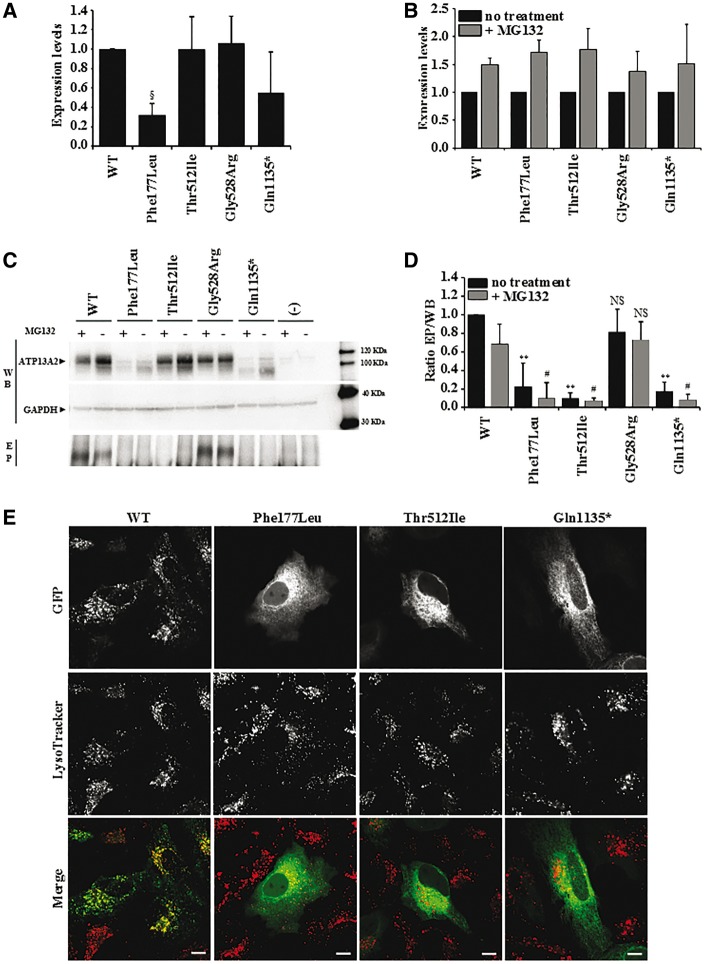
To assess stability of the ATP13A2 mutants in vitro, COS-1 cells were transiently transfected with constructs allowing expression of wild-type, Thr512Ile or Gln1135* followed by analysis of the levels of mutant proteins. We compared the HSP mutants to previously identified KRS mutant with compromised (Phe182Ile) ATP13A2 stability or early-onset parkinsonism mutant with preserved (Gly528Arg) ATP13A2 stability (Podhajska et al., 2012) (Fig. 5A). All wild-type and mutant ATP13A2 forms were successfully expressed, although reduced protein levels were observed for Phe177Leu, in line with an earlier report (Podhajska et al., 2012) (Fig. 5A). Putative degradation of the ATP13A2 mutants through the proteasomal pathway was evaluated using the proteasomal inhibitor MG132 (Fig. 5B). Protein levels of all ATP13A2 variants increased upon MG132 treatment, suggesting partial breakdown by the proteasome. However, the per cent difference in expression levels between non-treated and MG132 conditions were similar for wild-type as for all mutant constructs.
Figure 5.
Expression levels and subcellular localization of wild-type and mutant ATP13A2. (A) Relative expression levels of ATP13A2 WT, Phe177Ile, Thr512Ile, Gly528Arg and Gln1135*, which were normalized to wild-type (WT) in COS-1 cells. (B) Expression levels of ATP13A2 WT, Phe177Ile, Thr512Ile, Gly528Arg and Gln1135* in COS-1 cells following MG132 treatment were normalized to the levels without MG132 treatment. (C) Semi-quantitative western blotting of ATP13A2 WT, Phe177Ile, Thr512Ile, Gly528Arg and Gln1135* overexpressed in COS-1 cells, which were either treated (+) or not treated (−) with proteasome inhibitor MG132. EP (phospho-enzyme) represents the autoradiogram displaying the relative autophosphorylation levels of wild-type and mutant ATP13A2. (D) Graph bar representing the ratio of the autophosphorylation and expression levels (EP/WB). For A, B and D data are represented by average ± SD. ANOVA test with post hoc Bonferroni correction, §P = 0.01, **P < 0.001 with respect to wild-type with no treatment; #P < 0.001 with respect to wild-type treated with MG132. NS = not significant. (E) Subcellular localization of GFP‐N‐ATP13A2 WT, Phe177Ile, Thr512Ile and Gln1135* mutant proteins, transiently expressed in HeLa cells and stained with LysoTracker®. Scale bar = 10 µm.
As the p.Thr512Ile substitution targets a residue within the autophosphorylation motif (Fig. 1B), we questioned its effect on autophosphorylation activity. We compared the autophosphorylation levels of wild-type and disease mutants in total membrane fractions of transiently transfected COS-1 cells (Fig. 5C and D). In contrast to wild-type and Gly528Arg, the Thr512Ile protein was unable to undergo autophosphorylation, although it was expressed to comparable levels (Fig. 5A and D). In addition, we could not detect an autophosphorylation signal for Gln1135* (Fig. 5C) even when we increased the loading to reach comparable protein levels of wild-type and Gln1135* (data not shown). No signal was observed for the Phe177Leu mutant, but this can be attributed to the weak expression levels and/or a loss of autophosphorylation activity (Fig. 5D).
Finally, we evaluated the subcellular localization of the GFP‐N‐ATP13A2 mutants transiently expressed in HeLa cells (Fig. 5E and Supplementary Fig. 4). We first confirmed that wild-type ATP13A2 co-localizes with LysoTracker®-labelled vesicles, whereas the KRS mutant Phe177Ile is mislocalized to the endoplasmic reticulum (Podhajska et al., 2012). The two novel HSP mutants were also mislocalized to the endoplasmic reticulum, as we observed co-localization of mutant proteins with the endoplasmic reticulum marker pCI-mRuby-KDEL (Supplementary Fig. 3). This suggests that, besides protein instability and inactivity, also mistargeting contributes to the loss-of-function of ATP13A2 in complicated HSP and KRS.
Discussion
Clinical spectrum of ATP13A2-related diseases
Loss of ATP13A2 function has been linked to KRS, an early-onset atypical form of Parkinson’s disease with dementia that has been considered among the NBIA syndromes (Micheli et al., 2006; Ramirez et al., 2006; Djarmati et al., 2009; Rakovic et al., 2009), and the lysosomal storage disorder NCL (Bras et al., 2012). Here, we report three novel nonsense and one missense mutations in ATP13A2 in three families diagnosed with complicated HSP for which we demonstrate loss of functionality. Clinically, the HSP phenotype associated with ATP13A2 mutations (SPG78) is reminiscent of the phenotype described for SPG11/15 and SPG48, with spastic paraplegia, cerebellar ataxia, cognitive deficits and axonal neuropathy being common features in both SPG78 and SPG11/15/48. Of note, SPG11/15 and SPG48 also frequently display extrapyramidal disease manifestations (Anheim et al., 2009; Schicks et al., 2011; Hirst et al., 2016). The ‘ear of the lynx’ sign (T2 hyperintensity of the anterior fornix of the corpus callosum), an imaging feature described for SPG11 (Riverol et al., 2009) and SPG48 (Hirst et al., 2016), was also identified in two of five SPG78 cases in our study. However, the age of onset appears to be later in SPG78, neither lenticular nor retinal abnormalities were noted in SPG78, and thinning of the corpus callosum, a hallmark imaging finding in SPG11, 15 and 48 was present in only one of our cases.
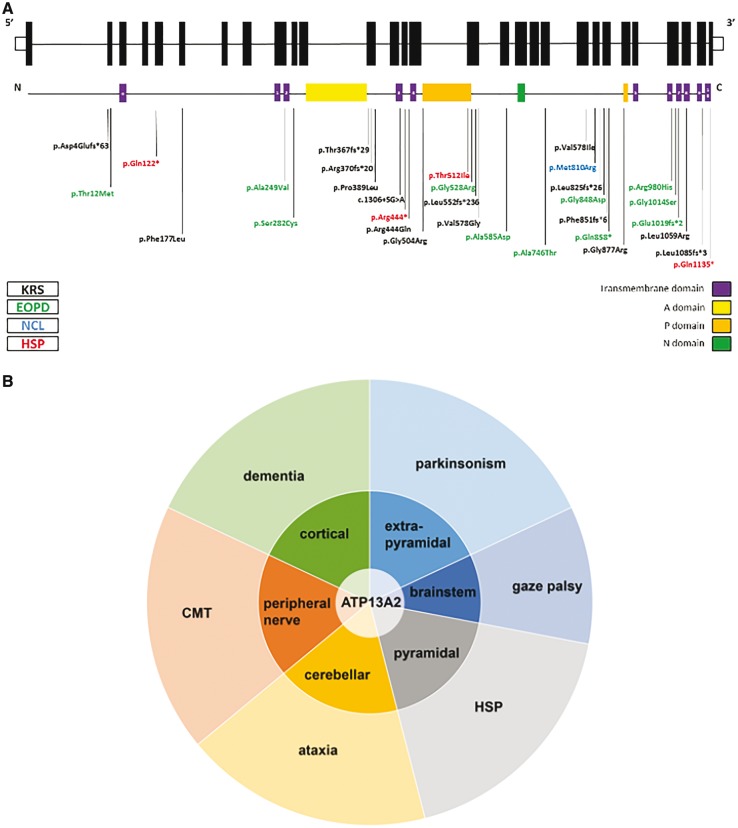
When considered from a clinical diagnostic standpoint, the phenotypes associated with ATP13A2 mutations—KRS, NCL and HSP—appear independent, but this standpoint is now challenged by the discovery of a common genetic aetiology, i.e. loss-of-function mutations in overlapping regions or even identical sites in ATP13A2 (Fig. 6A).
Figure 6.
Clinical spectrum of ATP13A2 mutations. (A) Schematic representation of the genomic and protein lay out of ATP13A2 and location of currently known disease-associated mutations. Topology of ATP13A2 represents 10 transmembrane domains (1–10) and an additional membrane-associated segment in the N-terminus (a). In black, mutations associated with KRS; in green, mutations associated with early-onset parkinsonism (EOPD); in blue, mutation associated with NCL; in red, mutations identified in this study, which are associated with complicated HSP. (B) Schematic representation of the neuronal systems affected by ATP13A2 dysfunction and resulting signs and symptoms apparent on a clinical level. Loss of ATP13A2 function in the cortex, extrapyramidal system, brainstem, pyramidal system, cerebellum, and the peripheral nerves leads to a variety of signs and symptoms including dementia, parkinsonism, vertical gaze palsy, spasticity, ataxia, and peripheral neuropathy. Predominance of extrapyramidal involvement or spasticity has led to the classification of ATP13A2-related disease as parkinsonism or HSP, respectively. We hypothesize that other carriers of ATP13A2 mutations might display predominance of other ATP13A2-dependent systems and therefore manifest as cerebellar ataxia, dementia or Charcot–Marie–Tooth disease.
KRS, HSP and NCL were each defined before their respective genetic basis was uncovered. Despite being multisystemic and thus clinically complex disorders, they were classified according to their most striking feature. The prominent parkinsonism seen in KRS families due to mutations in ATP13A2, led to the classification as an early-onset Parkinson’s syndrome (PARK9) (Micheli et al., 2006; Ramirez et al., 2006; Djarmati et al., 2009; Rakovic et al., 2009). Additionally, the KRS phenotype is considered to be part of the NBIA spectrum of disease due to the presence of iron accumulation in the putamen and caudate of at least a subset of cases (Schneider et al., 2013). The discovery of lipopigment inclusions in another family with ATP13A2 mutations, which were also seen in mice and dogs with ATP13A2 loss of function (Farias et al., 2011; Schultheis et al., 2013), led to the classification as NCL (CLN12). Here, we report that in three families with loss of ATP13A2 function lower-limb predominant spastic paraparesis is by far the most prominent clinical feature and thus, the disease was classified as an HSP and termed SPG78 (preliminary) (Gray et al., 2015). Extrapyramidal involvement, the lead clinical symptom in KRS, was subclinical (Patient HIH21480.II.3), subtle (Patient HIH22132.II.1) or even absent (Family HSP84) in our cases. Why the clinical consequences of ATP13A2 mutations diverge remains unclear. Of interest, the same p.Thr512Ile substitution that causes a complicated HSP phenotype has previously been identified in a Lebanese KRS patient (Dehay et al., 2012; Grunewald et al., 2012; Usenovic et al., 2012), suggesting that the mutation type is not the sole determinant of the clinical phenotype. The Lebanese KRS patient presented with rapidly progressive parkinsonism, supranuclear gaze palsy, perioral myokymia, and cognitive decline at the age of 44 years. Notably, as described in previously reported KRS patients (Williams et al., 2005) and personal observation in the original Chilean family (Ramirez et al., 2006), this patient also had pyramidal involvement with marked spasticity and hyperreflexia, most pronounced in the lower limbs, but no ataxia. It is clear that much is yet to be learned about which additional factors—genetic or environmental—contribute to the individual presentation of disease in these families.
Nevertheless, under the surface of the seemingly disparate clinical diagnostic classification of ATP13A2-related disease into the categories of parkinsonism/NBIA, NCL or HSP, the phenotypic features associated with ATP13A2 deficiency actually converge. Affection of various regions and/or systems of the nervous system (cortical, pyramidal, extrapyramidal, brainstem, cerebellar and peripheral) by loss of ATP13A2 function translates phenotypically into a multidimensional spectrum of associated signs and symptoms including dementia, spasticity, parkinsonism, disturbance of the saccadic eye movement system, ataxia, and peripheral neuropathy (Fig. 6B). The quantitative expression, not the qualitative presence of these features has historically led us to determine diagnosis. While this approach may still be useful in clinical practice for pragmatic reasons, rigid diagnostic classification systems may hamper identification of the underlying genetic cause. Thinking outside the box of diagnostic classifications is needed to recognize genes associated with more fluid, multidimensional phenotypes like ATP13A2, PNPLA6 (Synofzik et al., 2014), VCP (Kazamel et al., 2016), BICD2 (Neveling et al., 2013; Oates et al., 2013; Peeters et al., 2013), SPG11 (Guidubaldi et al., 2011; Daoud et al., 2012; Montecchiani et al., 2016), DYNC1H1 (Vissers et al., 2010; Weedon et al., 2011; Harms et al., 2012), TUBB4 (Erro et al., 2015), SYNE1 (Synofzik et al., 2016) and others. We predict that the increasing use of unbiased and ‘modular’ methods for phenotyping, e.g. by application of the Human Phenotype Ontology (Kohler et al., 2014), in combination with unbiased genetic screening methods such as whole exome and whole genome sequencing will uncover many more Mendelian disease spectra crossing historically defined diagnostic categories in the future.
Several ATP13A2 HSP and KRS mutants are mistargeted, unstable and/or inactive
As in previous studies (Dehay et al., 2012; Grunewald et al., 2012; Podhajska et al., 2012; Usenovic et al., 2012), we confirm that the loss-of-function of ATP13A2 in disease mutants relates to a combination of protein instability and abnormal intracellular localization. Moreover, here we provide the first biochemical evidence demonstrating that HSP- and some KRS-causing ATP13A2 mutations impair autophosphorylation activity, at least in part explaining their loss-of-function. All P-type transport ATPases couple the autophosphorylation reaction to the vectorial transport of (a) ligand(s) over biological membranes, indicating that the impaired autophosphorylation of ATP13A2 might reflect defective substrate transport. Alternatively, like other P-type ATPases (Pierre and Xie, 2006), ATP13A2 may play a scaffolding role in signalling complexes, which might be regulated by autophosphorylation. Defects in one or both functionalities might be disease-causing.
The p.Thr512Ile mutation (HSP and KRS) impairs ATP13A2 autophosphorylation activity, which together with mistargeting of the mutant protein contributes to its loss of function. Thr512 is part of the highly conserved autophosphorylation motif (508D-K-T-G-T), which is present in all P-type transport ATPases. Thr512 is positioned in the catalytic centre in between the autophosphorylation site Asp508 and the Mg2+ binding residue Thr510. Similar to the Thr512Ile mutation in ATP13A2, the Thr512Ala mutation of the equivalent residue in the Ca2+-pump SERCA1a (Thr355), one of the archetypical P-type ATPases, also impairs autophosphorylation and substrate occlusion (McIntosh et al., 1999), notably without affecting protein stability (Maruyama et al., 1989).
The loss-of-autophosphorylation activity of the Gln1135* mutant (HSP) might indicate that the C-terminus of ATP13A2 is functionally important, as is the case for other P-type ATPases (Vandecaetsbeek et al., 2009). However, the low protein abundance and failure to exit the endoplasmic reticulum, rather suggests that this mutant is improperly folded and efficiently degraded. Indeed, the truncation of ATP13A2 at the border of the cytosolic end of transmembrane segment M10 might disturb the membrane topology.
Lysosomal and mitochondrial dysfunctions are common features in HSP and KRS
Why ATP13A2 loss-of-function is implicated in a spectrum of neuronal disorders remains unclear, but the deficiency of this late endo-/lysosomal P-type transporter triggers lysosomal abnormalities (Chen et al., 2011; Park et al., 2011; Dehay et al., 2012; Grunewald et al., 2012; Podhajska et al., 2012; Usenovic and Krainc, 2012; Kong et al., 2014). Fibroblasts derived from the Thr512Ile KRS patient are marked by general lysosomal dysfunction as observed by the accumulation of lysosomes, impaired autophagy, reduced lysosomal proteolysis and increased lysosomal pH (Dehay et al., 2012; Gusdon et al., 2012; Ramonet et al., 2012; Usenovic et al., 2012). Here, we confirm in fibroblasts of unrelated HSP and KRS patients that LAMP1-positive organelles accumulate, correlating with a reduction in their proteolytic activity. Thus, an increased number of LAMP1-positive organelles may be a hallmark of ATP13A2 dysfunction. Ultrastructural analysis revealed deposition of abnormal lysosomal storage material in the form of lipid whorls and stacks of membranes. Interestingly, a similar lysosomal phenotype has previously been reported for three other subtypes of HSP: SPG11 (spatacsin), SPG15 (ZFYVE26/spastizin) and SPG48 (AP5Z1/AP-5). In all three, accumulation of LAMP1-positive organelles was described in primary patient fibroblasts (Vantaggiato et al., 2013; Chang et al., 2014; Renvoise et al., 2014; Hirst et al., 2016). Loss of SPG11/SPG15 results in the depletion of free lysosomes and an accumulation of autolysosomes due to impaired lysosome reformation, an mTOR dependent pathway (Chang et al., 2014). Furthermore, fibroblasts of patients with mutations in the SPG15- or SPG48 gene display multilamellar accumulation of membranous material similar to those we observe in ATP13A12 mutant fibroblasts (Renvoise et al., 2014; Hirst et al., 2016). These findings therefore suggest a molecular link between these four HSP subtypes that is particularly interesting in light of the clinical and imaging overlap (see above).
Neurons depend heavily on mitochondrial clearance pathways to preserve mitochondrial health, which on itself rests on efficient lysosomal function. Like lysosomal dysfunction, impaired mitochondrial function is implicated in NCL (Pezzini et al., 2011; Vidal-Donet et al., 2013), HSP (Casari et al., 1998; Hansen et al., 2002; Patel et al., 2002; Verny et al., 2011; Arif et al., 2013) or KRS (Grunewald et al., 2012; Park et al., 2014), and several studies connect ATP13A2 to mitochondrial clearance (Grunewald et al., 2012; Gusdon et al., 2012; Holemans et al., 2015; Martin et al., 2016). Mitochondrial fragmentation and impaired ATP production were previously described in the Thr512Ile-KRS fibroblasts (Grunewald et al., 2012). Although we did not observe significant changes in the morphology of the mitochondrial network in the fibroblasts of either the HSP or KRS patients, the reduction in the mitochondrial membrane potential suggests impaired mitochondrial functionality that may result in lower ATP levels. Moreover, the increased mitochondrial mass might point to an impaired mitochondrial clearance, which might be connected to the ATP13A2-mediated lysosomal dysfunction. In fact, ATP13A2 has been implicated in autophagy pathways (Dehay et al., 2012; Grunewald et al., 2012; Podhajska et al., 2012; Usenovic et al., 2012) and also the activation of ATP13A2 by the phospholipids phosphatidic acid and phosphatidylinositol(3,5)bisphosphate suggests a role in autophagy (Holemans et al., 2015; Martin et al., 2015). Interestingly, genetic defects in the pathways controlling phospholipid homeostasis, like phosphatidic acid, have been previously implicated in HSP. Mutations in two phospholipases (DDHD1 and DDHD2) cause SPG28 and SPG52 HSP subtypes, respectively. Defects in these enzymes lead to lipid accumulation and disturbed intracellular trafficking (Schuurs-Hoeijmakers et al., 2012; Tesson et al., 2012; Gonzalez et al., 2013).
In conclusion, loss of ATP13A2 function causes a combination of lysosomal and mitochondrial dysfunction that affects multiple neuronal populations. This translates to a spectrum of neurological disorders ranging from complicated HSP to KRS. We propose that patients with complicated HSP should be routinely tested for ATP13A2 mutations, in particular if complicated by progressive cognitive and cerebellar dysfunction.
Funding
This work was supported in part by the Research Fund of the University of Antwerp (project #TOP-BOF-29069 to A.J.), the Fund for Scientific Research–Flanders (grants #G054313N, G078414N, G0D7713N to A.J.), the Bulgarian Ministry of Education and Science (project #DTK-02/67, #DFNI-B02/3 to A.J., I.T.), the Tom Wahlig Foundation, Jena, Germany (to A.J., I.T.), the Michael J. Fox Foundation (to P.V.G), the Interuniversity Attraction Poles of the Belgian Science Policy Office (P7/13 to P.V.G.), and the KU Leuven (OT/13/091 and C16/15/073 to P.V.G.). The German team was supported by the European Union within the 7th European Community Framework Programme through funding for the NEUROMICS network (F5-2012-305121 to L.S.), the E-Rare Networks NEUROLIPID (01GM1408B to R.S.) and PREPARE (01GM1607 to M.S.), and a Marie Curie International Outgoing Fellowship (grant PIOF-GA-2012-326681 to R.S. and L.S.), as well as the Center for Clinical Research (IZKF) Tübingen (grant 1970-0-0 to R.S.) and the Else Kröner Fresenius Stiftung (grant to M.S.). Research reported in this publication was further supported by the National Institute of Neurological Disorders and Stroke of the National Institutes of Health under Award Number R01NS072248 (to S.Z. and R.S.). A.E.C., T.H. and S.v.V. are supported by fellowships from the Flanders Research Foundation (FWO). C.K. is the recipient of a career development award from the Hermann and Lilly Schilling Foundation.
Supplementary material
Supplementary material is available at Brain online.
Table 2.
ATP13A2 mutations in HSP families
| Genomic change | cDNA change | Protein change | Functional consequence | Genesis allele frequency | EVS allele frequency | Phast Cons | GERP | Poly Phen-2 | SIFT | Mutation Taster | dbSNP ID |
|---|---|---|---|---|---|---|---|---|---|---|---|
| chr1: 17 320 323G > A | 1535C > T | Thr512Ile | missense | G = 5438 | G = 12772 | 1.000 | 5.630 | 1.000 | D (0.001) | D | - |
| chr1: 17 331 300G > A | 364C > T | Gln122* | nonsense | G = 7242 | G = 13006 | 0.010 | 3.200 | n.a. | n.a. | D | - |
| chr1: 17 322 757G > A | 1330C > T | Arg444* | nonsense | G = 7234 | G = 13006 | 0.031 | 2.350 | n.a. | n.a. | D | - |
| chr1: 17 312 841G > A | 3403C > T | Gln1135* | nonsense | G = 6318 | G = 12702 | 1.000 | 4.7000 | n.a. | n.a. | D | - |
Mutation nomenclature is based on GRCh37 / hg19 and transcript NM001141973.1.
D = disease causing; n.a. = not applicable.
Supplementary Material
Glossary
Abbreviations
- HSP
hereditary spastic paraplegia
- KRS
Kufor-Rakeb syndrome
- NCL
neuronal ceroid lipofuscinosis
References
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods 2010; 7: 248–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anheim M, Lagier-Tourenne C, Stevanin G, Fleury M, Durr A, Namer IJ, et al. SPG11 spastic paraplegia: a new cause of juvenile parkinsonism. J Neurol 2009; 256: 104–8. [DOI] [PubMed] [Google Scholar]
- Arif B, Kumar KR, Seibler P, Vulinovic F, Fatima A, Winkler S, et al. A novel OPA3 mutation revealed by exome sequencing: an example of reverse phenotyping. JAMA Neurol 2013; 70: 783–7. [DOI] [PubMed] [Google Scholar]
- Behrens MI, Bruggemann N, Chana P, Venegas P, Kagi M, Parrao T, et al. Clinical spectrum of Kufor-Rakeb syndrome in the Chilean kindred with ATP13A2 mutations. Mov Disord 2010; 25: 1929–37. [DOI] [PubMed] [Google Scholar]
- Bras J, Verloes A, Schneider SA, Mole SE, Guerreiro RJ. Mutation of the parkinsonism gene ATP13A2 causes neuronal ceroid-lipofuscinosis. Hum Mol Genet 2012; 21: 2646–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casari G, De Fusco M, Ciarmatori S, Zeviani M, Mora M, Fernandez P, et al. Spastic paraplegia and OXPHOS impairment caused by mutations in paraplegin, a nuclear-encoded mitochondrial metalloprotease. Cell 1998; 93: 973–83. [DOI] [PubMed] [Google Scholar]
- Chang J, Lee S, Blackstone C. Spastic paraplegia proteins spastizin and spatacsin mediate autophagic lysosome reformation. J Clin Invest 2014; 124: 5249–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CM, Lin CH, Juan HF, Hu FJ, Hsiao YC, Chang HY, et al. ATP13A2 variability in Taiwanese Parkinson’s disease. Am J Med Genet B Neuropsychiatr Genet 2011; 156B: 720–9. [DOI] [PubMed] [Google Scholar]
- Cooper GM, Stone EA, Asimenos G, Program NCS, Green ED, Batzoglou S, et al. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res 2005; 15: 901–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covy JP, Waxman EA, Giasson BI. Characterization of cellular protective effects of ATP13A2/PARK9 expression and alterations resulting from pathogenic mutants. J Neurosci Res 2012; 90: 2306–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosiers D, Ceulemans B, Meeus B, Nuytemans K, Pals P, Van Broeckhoven C, et al. Juvenile dystonia-parkinsonism and dementia caused by a novel ATP13A2 frameshift mutation. Parkinsonism Relat Disord 2011; 17: 135–8. [DOI] [PubMed] [Google Scholar]
- Crow YJ, Zaki MS, Abdel-Hamid MS, Abdel-Salam G, Boespflug-Tanguy O, Cordeiro NJ, et al. Mutations in ADAR1, IFIH1, and RNASEH2B presenting as spastic paraplegia. Neuropediatrics 2014; 45: 386–93. [DOI] [PubMed] [Google Scholar]
- Daoud H, Zhou S, Noreau A, Sabbagh M, Belzil V, Dionne-Laporte A, et al. Exome sequencing reveals SPG11 mutations causing juvenile ALS. Neurobiol Aging 2012; 33: 839 e5–9. [DOI] [PubMed] [Google Scholar]
- Dehay B, Ramirez A, Martinez-Vicente M, Perier C, Canron MH, Doudnikoff E, et al. Loss of P-type ATPase ATP13A2/PARK9 function induces general lysosomal deficiency and leads to Parkinson disease neurodegeneration. Proc Natl Acad Sci USA 2012; 109: 9611–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fonzo A, Chien HF, Socal M, Giraudo S, Tassorelli C, Iliceto G, et al. ATP13A2 missense mutations in juvenile parkinsonism and young onset Parkinson disease. Neurology 2007; 68: 1557–62. [DOI] [PubMed] [Google Scholar]
- Djarmati A, Hagenah J, Reetz K, Winkler S, Behrens MI, Pawlack H, et al. ATP13A2 variants in early-onset Parkinson’s disease patients and controls. Mov Disord 2009; 24: 2104–11. [DOI] [PubMed] [Google Scholar]
- Erro R, Hersheson J, Ganos C, Mencacci NE, Stamelou M, Batla A, et al. H-ABC syndrome and DYT4: Variable expressivity or pleiotropy of TUBB4 mutations?. Mov Disord 2015; 30: 828–33. [DOI] [PubMed] [Google Scholar]
- Farias FH, Zeng R, Johnson GS, Wininger FA, Taylor JF, Schnabel RD, et al. A truncating mutation in ATP13A2 is responsible for adult-onset neuronal ceroid lipofuscinosis in Tibetan terriers. Neurobiol Dis 2011; 42: 468–74. [DOI] [PubMed] [Google Scholar]
- Fei QZ, Cao L, Xiao Q, Zhang T, Zheng L, Wang XJ, et al. Lack of association between ATP13A2 Ala746Thr variant and Parkinson’s disease in Han population of mainland China. Neurosci Lett 2010; 475: 61–3. [DOI] [PubMed] [Google Scholar]
- Fink JK. Hereditary spastic paraplegia: clinico-pathologic features and emerging molecular mechanisms. Acta Neuropathol 2013; 126: 307–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finsterer J, Loscher W, Quasthoff S, Wanschitz J, Auer-Grumbach M, Stevanin G. Hereditary spastic paraplegias with autosomal dominant, recessive, X-linked, or maternal trait of inheritance. J Neurol Sci 2012; 318: 1–18. [DOI] [PubMed] [Google Scholar]
- Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature 2015; 526: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitler AD, Chesi A, Geddie ML, Strathearn KE, Hamamichi S, Hill KJ, et al. Alpha-synuclein is part of a diverse and highly conserved interaction network that includes PARK9 and manganese toxicity. Nat Genet 2009; 41: 308–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez M, Falk MJ, Gai X, Postrel R, Schule R, Zuchner S. Innovative genomic collaboration using the GENESIS (GEM.app) platform. Hum Mutat 2015; 36: 950–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez M, Nampoothiri S, Kornblum C, Oteyza AC, Walter J, Konidari I, et al. Mutations in phospholipase DDHD2 cause autosomal recessive hereditary spastic paraplegia (SPG54). Eur J Hum Genet 2013; 21: 1214–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray KA, Yates B, Seal RL, Wright MW, Bruford EA. Genenames.org: the HGNC resources in 2015. Nucleic Acids Res 2015; 43: D1079–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald A, Arns B, Seibler P, Rakovic A, Munchau A, Ramirez A, et al. ATP13A2 mutations impair mitochondrial function in fibroblasts from patients with Kufor-Rakeb syndrome. Neurobiol Aging 2012; 33: 1843.e1–7.. [DOI] [PubMed] [Google Scholar]
- Guidubaldi A, Piano C, Santorelli FM, Silvestri G, Petracca M, Tessa A, et al. Novel mutations in SPG11 cause hereditary spastic paraplegia associated with early-onset levodopa-responsive Parkinsonism. Mov Disord 2011; 26: 553–6. [DOI] [PubMed] [Google Scholar]
- Gusdon AM, Zhu J, Van Houten B, Chu CT. ATP13A2 regulates mitochondrial bioenergetics through macroautophagy. Neurobiol Dis 2012; 45: 962–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JJ, Durr A, Cournu-Rebeix I, Georgopoulos C, Ang D, Nielsen MN, et al. Hereditary spastic paraplegia SPG13 is associated with a mutation in the gene encoding the mitochondrial chaperonin Hsp60. Am J Hum Genet 2002; 70: 1328–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms MB, Ori-McKenney KM, Scoto M, Tuck EP, Bell S, Ma D, et al. Mutations in the tail domain of DYNC1H1 cause dominant spinal muscular atrophy. Neurology 2012; 78: 1714–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J, Madeo M, Smets K, Edgar JR, Schols L, Li J, et al. Complicated spastic paraplegia in patients with AP5Z1 mutations (SPG48). Neurol Genet 2016; 2: e98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holemans T, Sorensen DM, van Veen S, Martin S, Hermans D, Kemmer GC, et al. A lipid switch unlocks Parkinson’s disease-associated ATP13A2. Proc Natl Acad Sci USA 2015; 112: 9040–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kancheva D, Atkinson D, De Rijk P, Zimon M, Chamova T, Mitev V, et al. Novel mutations in genes causing hereditary spastic paraplegia and Charcot-Marie-Tooth neuropathy identified by an optimized protocol for homozygosity mapping based on whole-exome sequencing. Genet Med 2016; 18: 600–7. [DOI] [PubMed] [Google Scholar]
- Kazamel M, Sorenson EJ, McEvoy KM, Jones LK Jr Leep-Hunderfund AN, Mauermann ML, et al. Clinical spectrum of valosin containing protein (VCP)-opathy. Muscle Nerve 2016; 54: 94–9. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 2015; 10: 845–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler S, Doelken SC, Mungall CJ, Bauer S, Firth HV, Bailleul-Forestier I, et al. The Human Phenotype Ontology project: linking molecular biology and disease through phenotype data. Nucleic Acids Res 2014; 42: D966–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong SMY, Chan BKK, Park JS, Hill KJ, Aitken JB, Cottle L, et al. Parkinson’s disease-linked human PARK9/ATP13A2 maintains zinc homeostasis and promotes alpha-Synuclein externalization via exosomes. Hum Mol Genet 2014; 23: 2816–33. [DOI] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 2009; 4: 1073–81. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25: 1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Giudice T, Lombardi F, Santorelli FM, Kawarai T, Orlacchio A. Hereditary spastic paraplegia: clinical-genetic characteristics and evolving molecular mechanisms. Exp Neurol 2014; 261C: 518–39. [DOI] [PubMed] [Google Scholar]
- Martin S, Holemans T, Vangheluwe P. Unlocking ATP13A2/PARK9 activity. Cell Cycle 2015; 14: 3341–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S, van Veen S, Holemans T, Demirsoy S, van den Haute C, Baekelandt V, et al. Protection against mitochondrial and metal toxicity depends on functional lipid binding sites in ATP13A2. Parkinson’s Dis 2016; 2016: 9531917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama K, Clarke DM, Fujii J, Inesi G, Loo TW, MacLennan DH. Functional consequences of alterations to amino acids located in the catalytic center (isoleucine 348 to threonine 357) and nucleotide-binding domain of the Ca2+-ATPase of sarcoplasmic reticulum. J Biol Chem 1989; 264: 13038–42. [PubMed] [Google Scholar]
- McIntosh DB, Woolley DG, MacLennan DH, Vilsen B, Andersen JP. Interaction of nucleotides with Asp(351) and the conserved phosphorylation loop of sarcoplasmic reticulum Ca(2+)-ATPase. J Biol Chem 1999; 274: 25227–36. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010; 20: 1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheli F, Cersosimo MG, Zuniga Ramirez C. Hereditary spastic paraplegia associated with dopa-responsive parkinsonism. Mov Disord 2006; 21: 716–7. [DOI] [PubMed] [Google Scholar]
- Montecchiani C, Pedace L, Lo Giudice T, Casella A, Mearini M, Gaudiello F, et al. ALS5/SPG11/KIAA1840 mutations cause autosomal recessive axonal Charcot-Marie-Tooth disease. Brain 2016; 139(Pt 1): 73–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neveling K, Martinez-Carrera LA, Holker I, Heister A, Verrips A, Hosseini-Barkooie SM, et al. Mutations in BICD2, which encodes a golgin and important motor adaptor, cause congenital autosomal-dominant spinal muscular atrophy. Am J Hum Genet 2013; 92: 946–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oates EC, Rossor AM, Hafezparast M, Gonzalez M, Speziani F, Macarthur DG, et al. Mutations in BICD2 cause dominant congenital spinal muscular atrophy and hereditary spastic paraplegia. Am J Hum Genet 2013; 92: 965–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JS, Blair NF, Sue CM. The role of ATP13A2 in Parkinson’s disease: clinical phenotypes and molecular mechanisms. Mov Disord 2015; 30: 770–9. [DOI] [PubMed] [Google Scholar]
- Park JS, Koentjoro B, Veivers D, Mackay-Sim A, Sue CM. Parkinson’s disease-associated human ATP13A2 (PARK9) deficiency causes zinc dyshomeostasis and mitochondrial dysfunction. Hum Mol Genet 2014; 23: 2802–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JS, Mehta P, Cooper AA, Veivers D, Heimbach A, Stiller B, et al. Pathogenic effects of novel mutations in the P-type ATPase ATP13A2 (PARK9) causing Kufor-Rakeb syndrome, a form of early-onset parkinsonism. Hum Mutat 2011; 32: 956–64. [DOI] [PubMed] [Google Scholar]
- Patel H, Cross H, Proukakis C, Hershberger R, Bork P, Ciccarelli FD, et al. SPG20 is mutated in Troyer syndrome, an hereditary spastic paraplegia. Nat Genet 2002; 31: 347–8. [DOI] [PubMed] [Google Scholar]
- Peeters K, Litvinenko I, Asselbergh B, Almeida-Souza L, Chamova T, Geuens T, et al. Molecular defects in the motor adaptor BICD2 cause proximal spinal muscular atrophy with autosomal-dominant inheritance. Am J Hum Genet 2013; 92: 955–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezzini F, Gismondi F, Tessa A, Tonin P, Carrozzo R, Mole SE, et al. Involvement of the mitochondrial compartment in human NCL fibroblasts. Biochem Biophys Res Commun 2011; 416: 159–64. [DOI] [PubMed] [Google Scholar]
- Pierre SV, Xie Z. The Na,K-ATPase receptor complex: its organization and membership. Cell Biochem Biophys 2006; 46: 303–16. [DOI] [PubMed] [Google Scholar]
- Podhajska A, Musso A, Trancikova A, Stafa K, Moser R, Sonnay S, et al. Common pathogenic effects of missense mutations in the P-type ATPase ATP13A2 (PARK9) associated with early-onset parkinsonism. PLoS One 2012; 7: e39942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakovic A, Stiller B, Djarmati A, Flaquer A, Freudenberg J, Toliat MR, et al. Genetic association study of the P-type ATPase ATP13A2 in late-onset Parkinson’s disease. Mov Disord 2009; 24: 429–33. [DOI] [PubMed] [Google Scholar]
- Ramirez A, Heimbach A, Grundemann J, Stiller B, Hampshire D, Cid LP, et al. Hereditary parkinsonism with dementia is caused by mutations in ATP13A2, encoding a lysosomal type 5 P-type ATPase. Nat Genet 2006; 38: 1184–91. [DOI] [PubMed] [Google Scholar]
- Ramonet D, Podhajska A, Stafa K, Sonnay S, Trancikova A, Tsika E, et al. PARK9-associated ATP13A2 localizes to intracellular acidic vesicles and regulates cation homeostasis and neuronal integrity. Hum Mol Genet 2012; 21: 1725–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renvoise B, Chang J, Singh R, Yonekawa S, FitzGibbon EJ, Mankodi A, et al. Lysosomal abnormalities in hereditary spastic paraplegia types SPG15 and SPG11. Ann Clin Transl Neurol 2014; 1: 379–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumers J, De Rijk P, Zhao H, Liekens A, Smeets D, Cleary J, et al. Optimized filtering reduces the error rate in detecting genomic variants by short-read sequencing. Nat Biotechnol 2012; 30: 61–8. [DOI] [PubMed] [Google Scholar]
- Rinaldi DE, Corradi GR, Cuesta LM, Adamo HP, de Tezanos Pinto F. The Parkinson-associated human P5B-ATPase ATP13A2 protects against the iron-induced cytotoxicity. Biochim Biophys Acta 2015; 1850: 1646–55. [DOI] [PubMed] [Google Scholar]
- Riverol M, Samaranch L, Pascual B, Pastor P, Irigoyen J, Pastor MA, et al. Forceps minor region signal abnormality “ears of the lynx”: an early MRI finding in spastic paraparesis with thin corpus callosum and mutations in the spatacsin gene (SPG11) on chromosome 15. J Neuroimaging 2009; 19: 52–60. [DOI] [PubMed] [Google Scholar]
- Schicks J, Synofzik M, Petursson H, Huttenlocher J, Reimold M, Schols L, et al. Atypical juvenile parkinsonism in a consanguineous SPG15 family. Mov Disord 2011; 26: 564–6. [DOI] [PubMed] [Google Scholar]
- Schmidt K, Wolfe DM, Stiller B, Pearce DA. Cd2+, Mn2+, Ni2+ and Se2+ toxicity to Saccharomyces cerevisiae lacking YPK9p the orthologue of human ATP13A2. Biochem Biophys Res Commun 2009; 383: 198–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 2012; 9: 671–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider SA, Dusek P, Hardy J, Westenberger A, Jankovic J, Bhatia KP. Genetics and pathophysiology of neurodegeneration with brain iron accumulation (NBIA). Curr Neuropharmacol 2013; 11: 59–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider SA, Paisan-Ruiz C, Quinn NP, Lees AJ, Houlden H, Hardy J, et al. ATP13A2 mutations (PARK9) cause neurodegeneration with brain iron accumulation. Mov Disord 2010; 25: 979–84. [DOI] [PubMed] [Google Scholar]
- Schule R, Holland-Letz T, Klimpe S, Kassubek J, Klopstock T, Mall V, et al. The Spastic Paraplegia Rating Scale (SPRS): a reliable and valid measure of disease severity. Neurology 2006; 67: 430–4. [DOI] [PubMed] [Google Scholar]
- Schule R, Wiethoff S, Martus P, Karle KN, Otto S, Klebe S, et al. Hereditary spastic paraplegia: clinicogenetic lessons from 608 patients. Ann Neurol 2016; 79: 646–58. [DOI] [PubMed] [Google Scholar]
- Schultheis PJ, Fleming SM, Clippinger AK, Lewis J, Tsunemi T, Giasson B, et al. Atp13a2-deficient mice exhibit neuronal ceroid lipofuscinosis, limited alpha-synuclein accumulation and age-dependent sensorimotor deficits. Hum Mol Genet 2013; 22: 2067–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuurs-Hoeijmakers JH, Geraghty MT, Kamsteeg EJ, Ben-Salem S, de Bot ST, Nijhof B, et al. Mutations in DDHD2, encoding an intracellular phospholipase A(1), cause a recessive form of complex hereditary spastic paraplegia. Am J Hum Genet 2012; 91: 1073–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods 2014; 11: 361–2. [DOI] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 2001; 29: 308–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen DM, Moller AB, Jakobsen MK, Jensen MK, Vangheluwe P, Buch-Pedersen MJ, et al. Ca2+ induces spontaneous dephosphorylation of a novel P5A-type ATPase. J Biol Chem 2012; 287: 28336–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Synofzik M, Gonzalez MA, Lourenco CM, Coutelier M, Haack TB, Rebelo A, et al. PNPLA6 mutations cause Boucher-Neuhauser and Gordon Holmes syndromes as part of a broad neurodegenerative spectrum. Brain 2014; 137(Pt 1): 69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Synofzik M, Smets K, Mallaret M, Di Bella D, Gallenmuller C, Baets J, et al. SYNE1 ataxia is a common recessive ataxia with major non-cerebellar features: a large multi-centre study. Brain 2016; 139(Pt 5): 1378–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Zhang T, Jiang L, Chi J, Hu D, Pan Q, et al. Regulation of intracellular manganese homeostasis by Kufor-Rakeb syndrome-associated ATP13A2 protein. J Biol Chem 2011; 286: 29654–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesson C, Nawara M, Salih MA, Rossignol R, Zaki MS, Al Balwi M, et al. Alteration of fatty-acid-metabolizing enzymes affects mitochondrial form and function in hereditary spastic paraplegia. Am J Hum Genet 2012; 91: 1051–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tome FM, Brunet P, Fardeau M, Hentati F, Reix J. Familial disorder of the central and peripheral nervous systems with particular cytoplasmic lamellated inclusions in peripheral nerves, muscle satellite cells, and blood capillaries. Acta Neuropathol 1985; 68: 209–17. [DOI] [PubMed] [Google Scholar]
- Usenovic M, Krainc D. Lysosomal dysfunction in neurodegeneration: the role of ATP13A2/PARK9. Autophagy 2012; 8: 987–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usenovic M, Tresse E, Mazzulli JR, Taylor JP, Krainc D. Deficiency of ATP13A2 leads to lysosomal dysfunction, alpha-synuclein accumulation, and neurotoxicity. J Neurosci 2012; 32: 4240–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Veen S, Sorensen DM, Holemans T, Holen HW, Palmgren MG, Vangheluwe P. Cellular function and pathological role of ATP13A2 and related P-type transport ATPases in Parkinson’s disease and other neurological disorders. Front Mol Neurosci 2014; 7: 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandecaetsbeek I, Trekels M, De Maeyer M, Ceulemans H, Lescrinier E, Raeymaekers L, et al. Structural basis for the high Ca2+ affinity of the ubiquitous SERCA2b Ca2+ pump. Proc Natl Acad Sci USA 2009; 106: 18533–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vantaggiato C, Crimella C, Airoldi G, Polishchuk R, Bonato S, Brighina E, et al. Defective autophagy in spastizin mutated patients with hereditary spastic paraparesis type 15. Brain 2013; 136(Pt 10): 3119–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verny C, Guegen N, Desquiret V, Chevrollier A, Prundean A, Dubas F, et al. Hereditary spastic paraplegia-like disorder due to a mitochondrial ATP6 gene point mutation. Mitochondrion 2011; 11: 70–5. [DOI] [PubMed] [Google Scholar]
- Vidal-Donet JM, Carcel-Trullols J, Casanova B, Aguado C, Knecht E. Alterations in ROS activity and lysosomal pH account for distinct patterns of macroautophagy in LINCL and JNCL fibroblasts. PloS One 2013; 8: e55526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers LE, de Ligt J, Gilissen C, Janssen I, Steehouwer M, de Vries P, et al. A de novo paradigm for mental retardation. Nat Genet 2010; 42: 1109–12. [DOI] [PubMed] [Google Scholar]
- Weedon MN, Hastings R, Caswell R, Xie W, Paszkiewicz K, Antoniadi T, et al. Exome sequencing identifies a DYNC1H1 mutation in a large pedigree with dominant axonal Charcot-Marie-Tooth disease. Am J Hum Genet 2011; 89: 308–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DR, Hadeed A, Al-Din AS, Wreikat AL, Lees AJ. Kufor Rakeb disease: autosomal recessive, levodopa-responsive parkinsonism with pyramidal degeneration, supranuclear gaze palsy, and dementia. Mov Disord 2005; 20: 1264–71. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.