Abstract
Candida albicans is frequently detected with heavy infection of Streptococcus mutans in plaque-biofilms from children affected with early-childhood caries, a prevalent and costly oral disease. The presence of C. albicans enhances S. mutans growth within biofilms, yet the chemical interactions associated with bacterial accumulation remain unclear. Thus, this study was conducted to investigate how microbial products from this cross-kingdom association modulate S. mutans build-up in biofilms. Our data revealed that bacterial-fungal derived conditioned medium (BF-CM) significantly increased the growth of S. mutans and altered biofilm 3D-architecture in a dose-dependent manner, resulting in enlarged and densely packed bacterial cell-clusters (microcolonies). Intriguingly, BF-CM induced S. mutans gtfBC expression (responsible for Gtf exoenzymes production), enhancing Gtf activity essential for microcolony development. Using a recently developed nanoculture system, the data demonstrated simultaneous microcolony growth and gtfB activation in situ by BF-CM. Further metabolites/chromatographic analyses of BF-CM revealed elevated amounts of formate and the presence of Candida-derived farnesol, which is commonly known to exhibit antibacterial activity. Unexpectedly, at the levels detected (25–50 μM), farnesol enhanced S. mutans-biofilm cell growth, microcolony development, and Gtf activity akin to BF-CM bioactivity. Altogether, the data provide new insights on how extracellular microbial products from cross-kingdom interactions stimulate the accumulation of a bacterial pathogen within biofilms.
Early childhood caries (ECC) is a highly prevalent and difficult to treat biofilm-dependent oral disease, afflicting mostly underprivileged children worldwide, resulting in an estimated annual expenditures of >$120 billion in the US alone1,2. Children affected with ECC display heavy infection with Streptococcus mutans accompanied by protracted feeding of dietary sugars, such as sucrose3,4,5, leading to rapid accumulation of virulent biofilms that cause rampant destruction of the teeth1,6.
Caries-causing biofilms develop when bacteria interact with dietary sugars and accumulate on tooth surface, forming densely packed cell clusters (or microcolonies) that are firmly adherent and enmeshed in an extracellular matrix of polymeric substances such as exopolysaccharides (EPS)7. EPS, particularly glucans, enhance bacterial adhesion and cohesion, while forming a diffusion-limiting matrix that protects the embedded bacteria and helps to acidify the local microenvironment. These biofilm properties promote the growth of an acidogenic microbiota, and eventually lead to the onset of dental caries8,9,10.
S. mutans is regarded as one of the key etiologic agents of ECC because this pathogen can efficiently catalyze dietary sucrose into extracellular glucans using several exoenzymes (glucosyltransferases or Gtfs) making it a primary EPS producer in the oral cavity, while being both acidogenic and acid-tolerant7. However, S. mutans may not act alone in cariogenic biofilms, as additional organisms may be involved6. Results from clinical studies reveal that C. albicans is frequently detected with high numbers of S. mutans in plaque-biofilms from children with ECC11,12,13,14,15. These findings are intriguing, because this opportunistic fungus usually neither binds well with S. mutans, nor colonizes teeth effectively on its own16,17,18. Rather, C. albicans interacts with commensal (viridans) streptococci and form biofilms on acrylic/mucosal surfaces19,20 to cause oral mucosal infections21,22. However, physical coadhesion of S. mutans and C. albicans is drastically enhanced in the presence of sucrose; these conditions also promote biofilm formation17,23,24,25.
Further in vitro studies have demonstrated that S. mutans-derived Gtfs play a key role by binding avidly to the fungal surface in an enzymatically active form17,26. When sucrose is available, surface-bound exoenzymes produce large amounts of EPS on the fungal surface, which provide enhanced binding sites for S. mutans, thereby promoting their adhesive interactions and crosskingdom biofilm development25. Using a rodent model of the disease, an enhancement of S. mutans levels in plaque-biofilms was observed when co-infected with C. albicans and exposed to a sucrose-rich diet25. Importantly, the virulence was significantly increased, leading to the onset of rampant caries on teeth similar to those found in ECC.
Associations between bacteria and fungi can be antagonistic or cooperative22,27,28. Once together within biofilms, these organisms may cooperate with each other for provision of substrates/metabolites or growth stimulating factors when conditions are conducive for ECC. For example, Candida does not metabolize sucrose efficiently29, and could benefit from cross-feeding of sucrose break-down products (glucose and fructose) by S. mutans30,31. Conversely, the presence of C. albicans dramatically modifies the physical environment of the biofilms by increasing EPS production that is critical for S. mutans accumulation and formation of microcolonies25. Furthermore, C. albicans appears to activate S. mutans genes associated with EPS/fitness25 and competence genes31. However, the manner in which such chemical interactions and secreted molecules stimulate S. mutans growth and accumulation remains unclear.
Thus, this study investigates whether extracellular microbial products derived from S. mutans-C. albicans biofilm interactions modulate the bacterial population build-up within biofilms. Our data revealed that bacterial-fungal conditioned medium (BF-CM) significantly increased the growth of S. mutans biofilm cells and enhanced microcolony formation through triggering of Gtfs activity via up-regulation of gtfBC. Using a recently developed microcapsule-based nanoculture and a S. mutans gtfB::green fluorescence protein promoter, the data demonstrated that BF-CM stimulated bacterial cells confined within these semipermeable chambers to develop into microcolonies while inducing gtfB expression in situ. Metabolite profiling and chromatographic analyses of BF-CM revealed elevated amounts of formate and the presence of farnesol, a quorum-sensing molecule from C. albicans that is commonly understood to exhibit antibacterial activity. Surprisingly, farnesol levels (25–50 μM) detected in BF-CM enhanced S. mutans cell growth, microcolony development, and Gtfs activity in a manner similar to that observed with BF-CM. However, higher concentrations (>100 μM) of farnesol inhibited S. mutans growth. Thus, farnesol is a potential key modulator in this crosskingdom interaction, and S. mutans growth responds non-monotonically to farnesol concentration. Altogether, this study provides new insights on the chemical interactions between an opportunistic fungus (C. albicans) and an oral pathogen (S. mutans), which could help explain how their association lead to virulent biofilms in ECC.
Results
Bacterial-fungal derived conditioned medium (BF-CM) promotes S. mutans growth and microcolony development
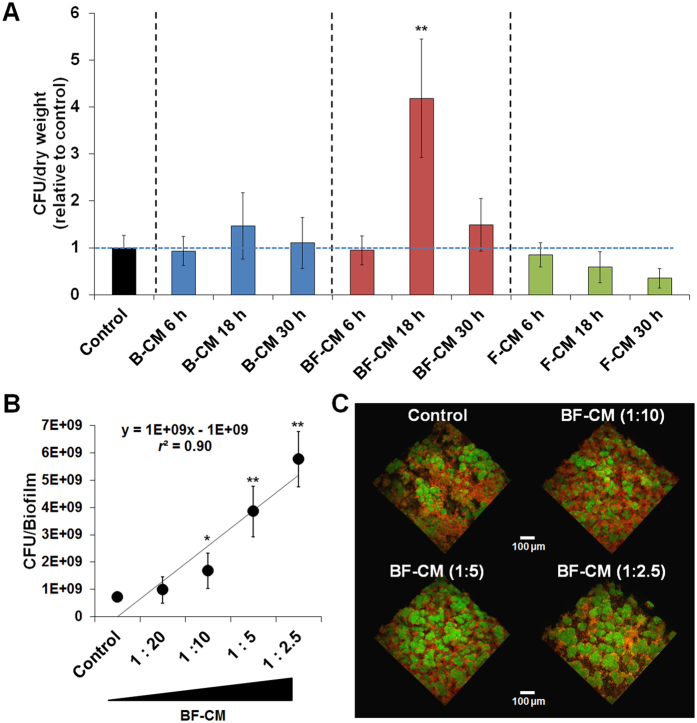
Conditioned medium (CM) was collected and prepared from single-species bacterial (S. mutans; B-CM), single-species fungal (C. albicans; F-CM) or bacterial-fungal (BF-CM) biofilms at three time-points (6, 18 and 30 h) corresponding to active biofilm formation using our sHA model (Figs 1A and S1; also Fig. S2 shows their overall architecture). Initially the influences of various CM preparations on S. mutans biofilm-cells growth were evaluated. The results showed that BF-CM collected at 18 h significantly promoted bacterial accumulation within biofilms compared to control (no supplementation, P < 0.001) (Fig. 1A). In contrast, no significant increase in the bacterial cell population was detected when S. mutans was grown in CM preparations from single-species biofilms or from BF-CM at other time-points. It appears that during the initial phase of active biofilm formation (between 6 h to 18 h) the presence of cross-kingdom metabolites could be enhancing S. mutans cell growth, while at the later time point (30 h) this effect is attenuated possibly due to reduced microbial/metabolic activity. Thus, hereafter further experiments were focused on the bioactivity of BF-CM collected at 18 h on biofilm formation by S. mutans. Using different dilutions of BF-CM, the data showed a clear trend of increasing number of viable S. mutans-biofilm cells (>6-fold increase at highest BF-CM content vs. no supplementation) in a dose-dependent manner (r2 = 0.9; Fig. 1B) suggesting that BF-CM contains bacterial growth-inducing factors.
Figure 1. Influences of conditioned medium on the growth of S. mutans biofilm cells and microcolony development.
(A) Conditioned medium (CM) was collected and prepared from single-species bacterial (B-CM), fungal (F-CM) or bacterial-fungal (BF-CM) biofilms at different time points. S. mutans biofilms were grown on saliva-coated hydroxyapatite (sHA) disc surface in each of the CM preparations (1:5 vol/vol, CM:UFTYE). The viable cells number (colony forming units (CFU)/biofilm) was normalized by dry weight (mg) (n = 8). (B) Dose-response effects of BF-CM on the growth of S. mutans biofilm cells (n = 4). (C) Representative 3D rendering images of biofilms formed with different dilutions of BF-CM and analyzed via multi-photon confocal laser scanning microscopy. S. mutans cells stained with SYTO 9 are depicted in green, while EPS labelled with Alexa Fluor 647 is shown in red. Data represent mean ± SD. The quantitative data were subjected to analysis of variance (ANOVA) in the Tukey’s HSD test for a multiple comparison. Values are significantly different from each other at *P < 0.05 or **P < 0.01.
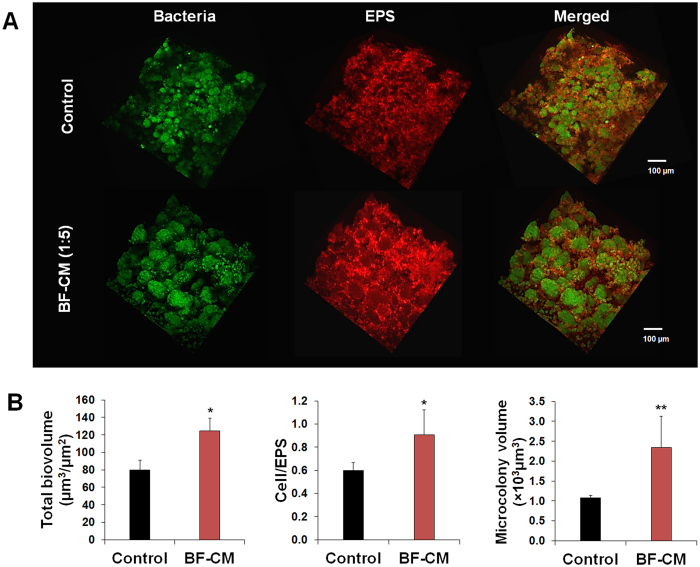
Furthermore, confocal imaging revealed significant alterations in the biofilm three-dimensional (3D) architecture with enhanced bacterial clustering and accumulation when grown in the presence of increased amounts of BF-CM (Fig. 1C). It showed enlarged and densely packed microcolonies (in green) enmeshed in a well-developed EPS matrix (in red) (Fig. 2A). Quantitative computational analyses (COMSTAT) showed significantly higher total biomass and cell/EPS ratio in S. mutans biofilms formed in BF-CM, while the volume of the microcolony was increased by ~2.5 fold (vs. control, P < 0.01) (Fig. 2B). Therefore, BF-CM appears to enhance microcolony development with an elevated carriage of S. mutans cells.
Figure 2. Three-dimensional (3D) architecture and quantitative computational analysis of biofilm formed in BF-CM.
(A) Representative confocal images of S. mutans biofilms grown in BF-CM (1:5vol/vol, CM:UFTYE) and without supplementation (control). The bacterial microcolonies are depicted in green (SYTO 9), while the EPS-matrix is depicted in red (Alexa Fluor 647). (B) Quantitative analysis of total biovolume (biomass), cell/EPS ratio and microcolony volume (size) was performed using COMSTAT. Data represent mean ± SD (n = 8). A pairwise comparison between control and BF-CM was conducted using student’s t-test. Values are significantly different from each other at *P < 0.05 or **P < 0.01.
BF-CM modulates the dynamics of S. mutans-derived glucosyltransferases (Gtfs) activity
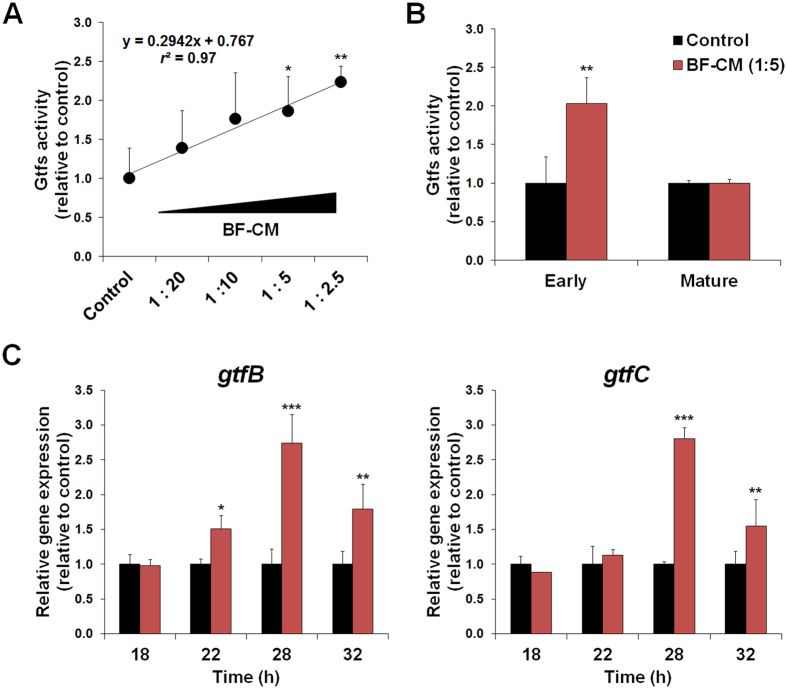
Since microcolony development is linked with the activity of Gtf exoenzymes32,33, the impact of BF-CM on total Gtfs activity and gtf gene expression during S. mutans biofilm formation was further investigated (Fig. 3). The biofilms were formed with varying BF-CM dilutions, and the supernatant collected for Gtfs activity as measured by scintillation counting using radiolabeled sucrose. Linear regression analysis showed that Gtfs activity correlated strongly with increasing amounts of extracellular microbial products in BF-CM (r2 = 0.97) (Fig. 3A). Also, temporal effects were examined to confirm whether the inducing effects of BF-CM on Gtfs activity were dependent on the stage of biofilm development. Interestingly, Gtfs activity was significantly enhanced by BF-CM (~2-fold increase vs. control, P < 0.01; Fig. 3B) during the early-stage of biofilm formation (18–28 h). However, no significant effects were observed in matured biofilms (42 h) (Fig. 3B). Hence, exposure to BF-CM can increase Gtfs activity during biofilm initiation, promoting S. mutans ability to form cell clusters and microcolonies onto sHA surface.
Figure 3. Dynamics of glucosyltransferases (Gtfs) activity and gtfBC expression profiles in biofilms formed in BF-CM.
(A) Dose-response effects of BF-CM (diluted in UFTYE, vol/vol) on Gtfs activity. The data were subjected to linear regression analysis and analysis of variance (ANOVA) in the Tukey’s HSD test for a multiple comparison. Correlation coefficient between increasing amounts of BF-CM and Gtfs activity was r2 = 0.97. (B) Temporal effects of BF-CM on Gtfs activity at different stages of biofilm development; early-stage (18–28 h) and matured biofilms (28–42 h). (C) Influence of BF-CM (collected at 18 h) on gtfB and gtfC gene expression by S. mutans biofilms was analyzed at 18, 22, 28, and 32 h of development. A pairwise comparison between control and BF-CM was conducted using student’s t-test. Data represent relative ratio to control (defined as 1); mean ± SD (n = 4). Values are significantly different from each other at *P < 0.05, **P < 0.01 or ***P < 0.001.
Furthermore, additional experiments were conducted to examine the manner in which BF-CM influences the pattern of gtfB and gtfC expression, which encodes the primary Gtfs associated with microcolony development by S. mutans33. Notably, gtfB expression was up-regulated as early as 22 h within biofilms formed with BF-CM, while both gtfB and gtfC were dramatically induced (~3-fold increase vs. control, P < 0.001; Fig. 3C) at 28 h of development. Altogether, the data reveal that BF-CM contains microbial products that are capable of both stimulating bacterial growth and inducing Gtfs activity, consistent with elevated population of bacterial cells (Fig. 1) and development of enlarged microcolonies within biofilms formed in the presence of BF-CM (Fig. 2B).
Microcolony assembly and gtfB expression in situ using a semipermeable nanoculture system
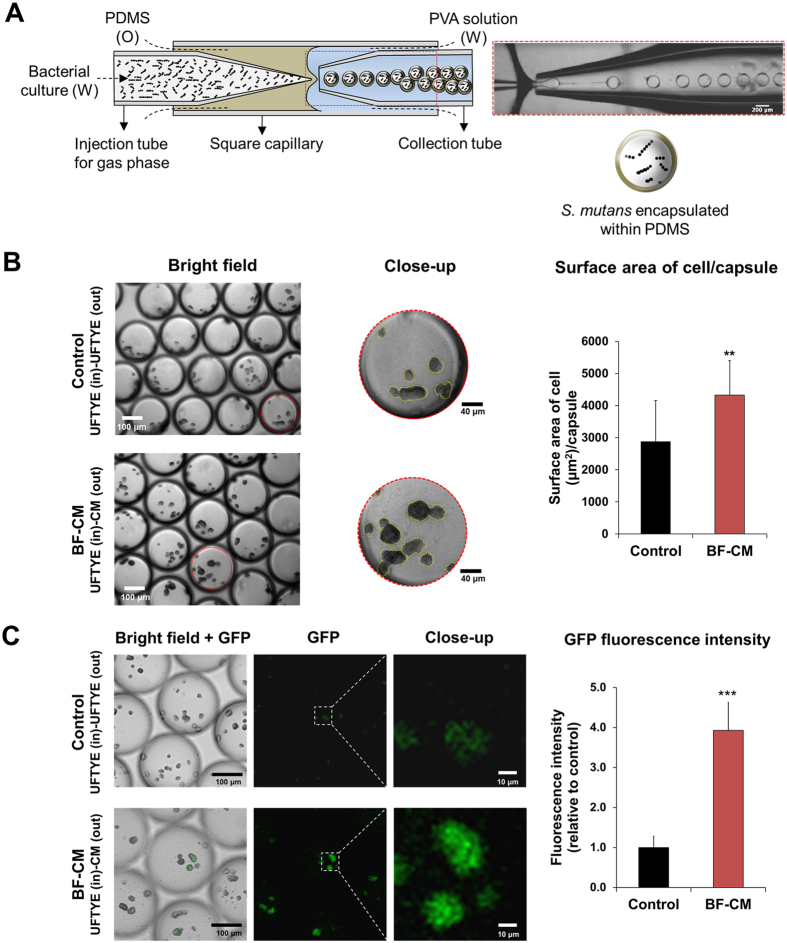
To further demonstrate the biological properties of BF-CM, a newly developed nanoliter-scale culturing system was used, which allows microbial growth within spatially confined yet permeable microcapsules, mimicking biofilm microenvironments. Using a microfluidics fabrication method, defined cell population of S. mutans can be directly inoculated and encapsulated within a physical shell surrounding the bacterial culture (4 to 5 nanoliter) (Fig. 4A). The nanocultures confine microorganisms within a semipermeable polydimethylsiloxane (PDMS) membrane containing growth medium, while allowing transport of small molecules34. Since this platform allows chemical fluxes between internal and external environments, it was determined whether S. mutans cells grown within the nanoculture respond to bioactive molecules in the BF-CM placed outside of the microcapsules.
Figure 4. Microcolony assembly and gtfB expression in situ using a microfluidics-generated nanoculture system.
(A) Defined cell population of S. mutans (~30 cells per nanoculture) was encapsulated in a semipermeable polydimethylsiloxane (PDMS)-based nanoculture (microcapsules). (B) S. mutans cells were inoculated in PDMS nanocultures with culture medium (UFTYE) and seeded in the medium (outside) containing either BF-CM (1:5 vol/vol, CM:UFTYE) or without supplementation (control). The surface area of cell occupying the interior of the capsule was measured by Image J. Data represent mean ± SD (n = 12). (C) Microcolony formation and gtfB expression via S. mutans strain expressing GFP under the control of gtfB promoter; PgtfB::gfp were determined using optical and confocal microscopy. The GFP fluorescent intensity was measured by using Image J. Data represent mean ± SD (n = 20). A pairwise comparison between control and BF-CM was conducted using student’s t-test. Values are significantly different from each other at **P < 0.01, ***P < 0.001.
S. mutans cells were encapsulated in PDMS nanocultures with culture medium, and then seeded in the same medium (outside) containing BF-CM or without supplementation (control). Microcolony formation and in situ gtfB expression (using PgtfB::gfp strain) within the microcapsules were assessed via optical and confocal microscopy. Consistent with the findings using de facto biofilms, the microcolonies appeared visually larger in the nanocultures of S. mutans placed in BF-CM, while the surface area of cell occupying the interior of the capsule was significantly higher than control (P < 0.01; Fig. 4B). Future studies shall include quantitative measurements of the size (e.g. diameter and volume) of the microcolonies within each of the microcapsule. Furthermore, gtfB expression by these bacterial microcolonies was significantly enhanced, showing ~4-fold increase as determined via measurement of fluorescence intensity of green fluorescence protein (GFP) (vs. control, P < 0.001) (Fig. 4C). Together, these observations suggest that BF-CM harbors small molecules, which are capable of diffusing through the PDMS shell and induce the resident bacterial cells to promote microcolony growth with enhanced gtfB expression.
Chemical characterization of conditioned medium
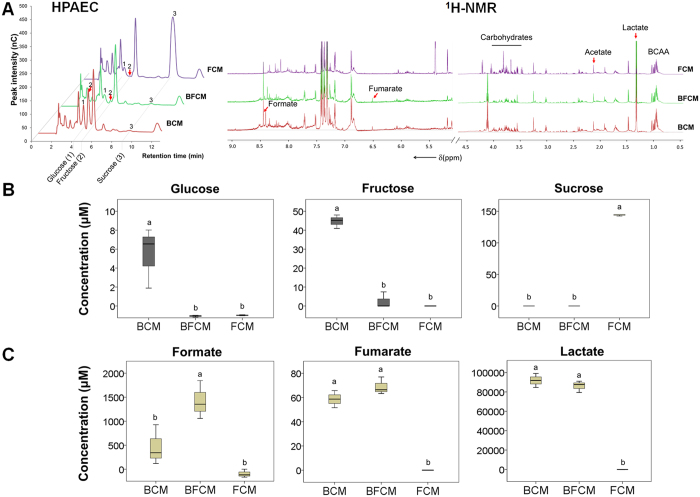
Our data suggest that conditioned medium from bacterial-fungal biofilm interaction contains specific S. mutans-growth inducing and Gtfs-activating molecules that could be critical for enhanced bacterial accumulation observed in previous in vitro and in vivo studies24,25,35. Therefore, the composition of carbohydrates and metabolites in BF-CM as well as in the conditioned medium from single-species biofilms (B-CM and F-CM) was characterized using colorimetric, high-performance anion-exchange chromatography (HPAEC) and 1H nuclear magnetic resonance (1H-NMR) methods (Fig. 5A). Since S. mutans efficiently metabolizes sucrose36,37, this carbohydrate was almost completely utilized in the conditioned medium from single-species bacterial (B-CM) or bacterial-fungal (BF-CM) biofilms (Fig. 5B). In contrast, most of the carbohydrate remained in the medium from single-species fungal biofilm (F-CM), consistent with C. albicans inefficient utilization of sucrose29. Fructose derived from the breakdown of sucrose was detected at high concentration in S. mutans conditioned medium. However, there was a significant difference (14.7-fold, P < 0.01) between the concentration of fructose contained in B-CM (44 μM) and BF-CM (3 μM), indicating active fructose metabolism in S. mutans-C. albicans biofilms.
Figure 5. The composition of carbohydrates and metabolites in the conditioned medium (CM).
CM was collected and prepared from single-species bacterial (B-CM), fungal (F-CM) or bacterial-fungal (BF-CM) biofilms as described in Materials and Methods. (A) HPAEC chromatograms of carbohydrate profile and 1H-NMR analysis for metabolites (organic acids shown). (B) The concentrations of glucose, fructose and sucrose as the main carbohydrates in undiluted CM. (C) The concentrations of formate, fumarate and lactate as the main metabolites in undiluted CM. Data represent mean ± SD (n = 3). The data were subjected to analysis of variance (ANOVA) in the Tukey’s HSD test for a multiple comparison. The letters (a and b) denote significate differences (P < 0.01).
Changes in the metabolite composition and content in the conditioned medium may reflect chemical interactions between S. mutans and C. albicans within biofilms. Hence, the profile of extracellular metabolites (e.g. organic acids, alcohols, sugar alcohols, amino acids) in BF-CM, B-CM and F-CM was determined (Figs 5C and S3). The results of metabolite composition analyses revealed that lactate was the major metabolite detected in BF-CM and B-CM (Figs 5C and S3). Notably, the formate concentration was significantly increased in BF-CM (vs. others, P < 0.01).
After initial chemical characterization, further biological evaluation was conducted to determine whether the bacterial/microcolony-growth inducing factor could be a functional protein. Since heat treatment, digestion with proteinase K, and combination of both did not alter the biological properties of BF-CM but promoted S. mutans cell growth compared to non-treated BF-CM (P > 0.05) (Fig. S4), the extracellular microbial factors appeared to be non-proteinaceous or non-enzymatic molecules that were small and diffusible. In parallel, BF-CM (treated or non-treated, as described above) unexpectedly inhibited C. albicans morphogenesis, blocking the transition from yeast to hyphal forms in a dose-dependent manner (Fig. S5). Since farnesol is a small, diffusible quorum-sensing (QS) molecule secreted by C. albicans, and is well-known to inhibit the yeast-to-hyphae transition38, additional chromatographic analyses were performed to confirm whether farnesol could be detected in BF-CM (Fig. 6). BF-CM or F-CM contained detectable amounts of farnesol, with an estimated concentration of 23.3 and 107.6 μM, respectively based on HPLC quantification (Fig. 6). As expected, farnesol was undetected in B-CM since S. mutans cannot produce and secrete this molecule.
Figure 6. Quantification of farnesol in the conditioned medium (CM).
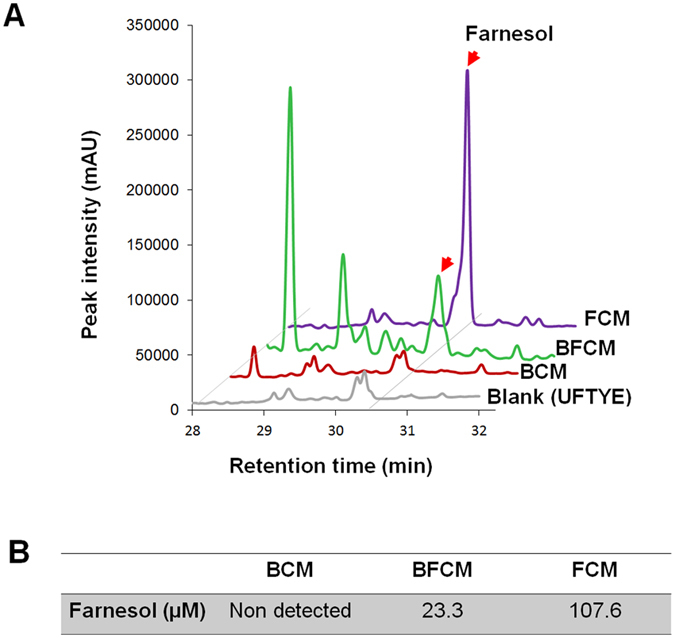
(A) HPLC profile of farnesol was obtained from CM extracted with EtOAc. The peak at 30.5 min is of farnesol (red arrows). Conditioned medium (CM) was collected and prepared from single-species bacterial (B-CM), fungal (F-CM) or bacterial-fungal (BF-CM) biofilms at 18 h. (B) Biological concentration of farnesol detected in the CM.
Low concentrations of farnesol enhance the growth of S. mutans biofilm cells and Gtfs activity
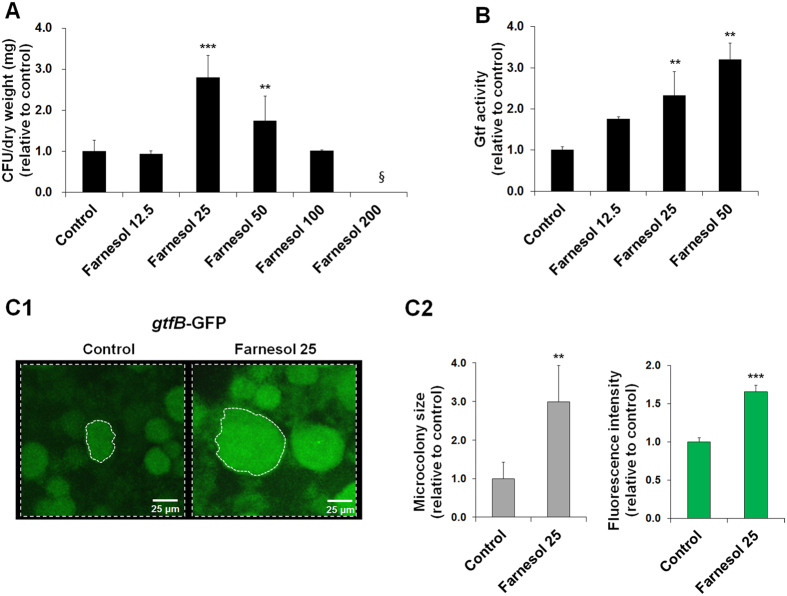
The identification of farnesol in BF-CM is intriguing because this fungal signaling molecule also exhibits antibacterial activity against S. mutans biofilms at high concentrations (>1 mM)39. Thus, the biological effects of farnesol at a range of concentrations (12.5, 25 and 50 μM) akin to that found in BF-CM was investigated. To be consistent with the experiments using BF-CM, the influences of farnesol on both S. mutans biofilm formation and the Gtfs activity were examined. Surprisingly, the results revealed that farnesol at these low concentrations actually induced the growth of S. mutans biofilm cells (Fig. 7A), and concomitantly enhanced Gtfs activity in a dose-dependent manner (vs. control; Fig. 7B). In contrast, bacterial growth was inhibited by farnesol at high concentrations (>100 μM; Fig. 7A).
Figure 7. Influences of farnesol at levels found in BF-CM on Gtfs activity and S. mutans biofilm formation.
(A) Influence of farnesol at different concentrations (0–200 μM) on the S. mutans biofilm cell growth. Data represent mean ± SD (n = 4). The data were subjected to analysis of variance (ANOVA) in the Tukey’s HSD test for a multiple comparison. §Indicates non-detected. (B) Gtfs activity in S. mutans biofilms grown in the presence of farnesol at a range of concentrations (12.5, 25 and 50 μM). Data represent mean ± SD (n = 6). The data were subjected to analysis of variance (ANOVA) in the Tukey’s HSD test for a multiple comparison. (C1) Microcolony development and gtfB expression via PgtfB::gfp S. mutans in biofilms formed on sHA surface in the presence of farnesol at 25 μM. Fluorescence images were obtained using confocal laser scanning microscopy. (C2) The microcolony size and GFP fluorescence intensity were measured by COMSTAT and Image J. Data represent mean ± SD (n = 4). A pairwise comparison between control and farnesol was conducted using student’s t-test. Values are significantly different from each other at **P < 0.01 or ***P < 0.001.
Since the amount of farnesol detected in BF-CM (~25 μM) promoted the growth of S. mutans and the Gtfs activity, further experiments were conducted to confirm whether farnesol can stimulate microcolony development and gtfB expression (via PgtfB::gfp). Strikingly, farnesol induced the formation of enlarged microcolonies with elevated expression of gtfB (Fig. 7C) in a manner similar to that observed with BF-CM. Together, our data suggest that farnesol may be an important molecule mediating this cross-kingdom interaction, with a potential role in regulating S. mutans accumulation and microcolony development within cariogenic biofilms.
Discussion
Our findings reveal that extracellular factors derived from the association between S. mutans and C. albicans stimulates bacterial growth within the biofilms. Gtfs activity via up-regulation of gtf genes, particularly gtfB, is triggered by secreted fungal factors, which in turn enhances microcolony formation by S. mutans. The insoluble glucans produced by GtfB can provide bacterial binding sites for S. mutans accumulation, while serving as a structural scaffold that are essential for microcolony development33,40,41. The microcolonies also facilitate the formation of localized acidic microenvironments that promote acid-dissolution of the adjacent tooth enamel10. Interestingly, elevated GtfB levels in human saliva42 and increased amounts of insoluble glucans in plaque-biofilms43,44 have been associated with caries activity in ECC-affected children. Furthermore, S. mutans defective in gtfB is unable to form microcolonies32, to infect teeth or to cause severe carious lesions in rodent caries model45. Thus, it is plausible that the presence of C. albicans factors in plaque-biofilms might play an important role in gtfB activation in situ, and thereby in the enhanced S. mutans virulence and development of ECC in vivo, which can be further elucidated in future longitudinal clinical studies.
Interestingly, the biochemical characterization using a combination of heat treatment and digestion with proteinase K, and chromatographic analyses indicate that farnesol may be a key chemical modulator. The role of farnesol in mediating C. albicans yeast-to-hyphae transition while exhibiting antibacterial activity has long been appreciated38,39. However, S. mutans growth responds non-monotonically to farnesol concentration. At low farnesol levels (~25 μM) found in the conditioned medium from co-cultured biofilms, it enhances bacterial cell growth and triggers gtfB expression/activity within biofilms, which correlated with microcolony development by S. mutans. In contrast, high concentrations (>100 μM) of farnesol inhibited S. mutans growth, consistent with previous observations of its antibacterial effects39. However, the presence of S. mutans appeared to reduce farnesol production by C. albicans (Fig. 6), which may have contributed to hyphal formation as typically observed in co-species biofilms25 (Fig. S2). In addition, farnesol can be incorporated into S. mutans cell membrane due to its fatty acid-like structure. We have previously shown that farnesol can be detected in the bacterial cell membrane of S. mutans biofilm cells46 as well as planktonic cells following treatment (Fig. S6). These observations suggest a well-controlled mechanism to maintain farnesol at levels that promote a symbiotic fungal-bacterial relationship, and open intriguing questions as to how cell communication prevent S. mutans and C. albicans from killing or inhibiting each other.
Nonetheless, the interactions with S. mutans in co-species biofilms also provide some advantages to C. albicans. The characterization of extracellular sugars and metabolites present in the conditioned media revealed complementary metabolic activity within co-species biofilms. Consistent with the C. albicans inability to efficiently metabolize sucrose29, the fungus formed sparse biofilms on sHA surface when grown alone with minimal carbohydrate utilization. However, the presence of S. mutans and its ability to rapidly breakdown sucrose (releasing glucose and fructose) drastically enhanced C. albicans growth within co-species biofilms, accompanied by increased utilization of glucose and fructose. Interestingly, enhanced fructose metabolism can also promote C. albicans hyphal morphogenesis47,48. In line with the metabolic activity of carbohydrates, lactate (primarily) as well as small amounts of formate and fumarate were produced, which, in turn, would favor S. mutans and C. albicans growth within cariogenic biofilms, as these microbes are both acidogenic and acid-tolerant49,50. This metabolic cooperation together with modulation of farnesol production provides an effective mechanism that resolves potential antagonism, while promoting co-existence and enhancing S. mutans accumulation via Gtf activation.
The findings that S. mutans growth stimulation and Gtf activation through bacterial-fungal interactions are achieved within biofilms via secreted molecules also raise new questions. While farnesol at low levels clearly induces bacterial growth and gtfB expression simultaneously, the underlying molecular mechanisms are unclear. Recently, Peng et al.51 have demonstrated that c-di-AMP intracellular signaling in S. mutans directly regulates gtfB expression. Thus, an intriguing concept may arise where by an extracellular QS molecule from a fungus may trigger intracellular signaling of a bacterial pathogen for enhanced expression of a virulence factor (GtfB) associated with dental caries7. Whether other S. mutans genes, like sigX (which is induced in the presence of C. albicans)31, are affected or not by farnesol in a dose-dependent manner shall be elucidated by assessing the entire S. mutans transcriptome response.
At the same time, the complexity of this bacterial-fungal association exists in many different contexts. Although this study is focused on the influence of the C. albicans on S. mutans accumulation within biofilms, the interactions between these microbes are mutually beneficial. How S. mutans and C. albicans interplay within co-species biofilms to promote fungal and bacterial growth while modulating yeast-to-hyphae transition remains unclear as other factors may be involved. For example, S. mutans-derived mutanobactin A, competence-stimulating peptide and trans-2-decenoic acid may act as cross-kingdom signaling molecules that regulate C. albicans growth and morphogenesis52,53,54. Further analysis on the identity and function of other signaling and small diffusible molecules could reveal a complex and multifaceted role. It is intriguing to think that a highly regulated interplay between cooperative and competitive interactions occurs to allow a symbiotic relationship between these microbes. Thus, it will be interesting to investigate how these different chemical interactors modulate S. mutans and/or C. albicans physiology using our microcapsule-based nanoculture system. This analytical platform enables the selective diffusion of small molecules while assessing changes in the morphology of the microcolonies, which can help elucidate how community behavior can be modified by sensing secreted molecules under physical confinement55,56.
In summary, this study provides new insights on the chemical interactions between an opportunistic fungus and an oral pathogen, which in part help to explain the in vivo and clinical findings showing enhanced S. mutans carriage in the presence of C. albicans in plaque-biofilms associated with ECC. Using a microcapsule-based nanoculture system, S. mutans cells confined within these chambers responded to chemical interactors (without physical contact between the microbes) that diffused through the PDMS shell. Further chemical analysis identified farnesol as a potential key modulator in this cross-kingdom interaction that, at low concentrations, stimulates bacterial growth and gtfB activity, leading to enlarged microcolonies containing densely packed S. mutans cells. These observations also suggest new strategies for manipulating this pathogenic bacterial-fungal interaction. For example, high concentration of farnesol can be used to inhibit both S. mutans growth and C. albicans transition to hyphae, which could lessen the virulence of this cross-kingdom biofilm. In addition, the available evidence prompts the possibility of incorporating antifungal therapy in the treatment of ECC.
Materials and Methods
Microbial strains and growth conditions
Streptococcus mutans UA159 serotype c (an established virulent cariogenic dental pathogen) and Candida albicans SC5314 (a well-characterized fungal strain) were used to generate single- or co-species biofilms. A S. mutans strain expressing green fluorescence protein (GFP) under the control of the gtfB promoter (PgtfB::gfp) (a gift from Dr. Jose Lemos at College of Dentistry, University of Florida) was used for in situ gtfB expression analysis. For inoculum preparation, C. albicans and S. mutans cells were grown to mid-exponential phase (optical density at 600 nm (OD600) of 0.65 and 0.5, respectively) in ultrafiltered (10-kDa molecular-mass cutoff membrane; Millipore, MA, USA) tryptone-yeast extract broth (UFTYE; 2.5% tryptone and 1.5% yeast extract at pH 5.5 and pH 7.0 for C. albicans and S. mutans) with 1% (wt/vol) glucose at 37 °C and 5% CO2 as described previously17,25.
Single and co-species biofilm model
Biofilms were formed using our established saliva-coated hydroxyapatite (sHA) disc model as detailed previously25 (also see Fig. S1). Briefly, sHA discs were vertically suspended in a 24-well plate using a custom-made disc holder32, and inoculated with approximately 2 × 106 (CFU/ml) of S. mutans or 2 × 104 (CFU/ml) of C. albicans in 2.8 ml (per well) UFTYE (pH 7.0) containing 1% (wt/vol) sucrose at 37 °C under 5% CO2. For co-species biofilms, S. mutans (2 × 106 CFU/ml) and C. albicans (2 × 104 CFU/ml) were added to the inoculum (in 2.8 mL (per well) UFTYE (pH 7.0) containing 1% (wt/vol) sucrose); this proportion of the microorganisms is similar to that found in saliva samples from children with ECC11,25. The culture medium was changed twice daily at 8 am and 6 pm until the end of the experimental period. The pH of spent culture medium was measured daily at each medium change using a standard pH electrode (Thermo Scientific, Waltham, MA, USA). Representative single or co-species biofilm 3D architecture is shown in Fig. S2.
Preparation of biofilm-derived conditioned medium
The conditioned medium was collected and prepared from each of the biofilms as detailed in Fig. S1. Briefly, the biofilm and the respective surrounding culture medium were collected at 6, 18 and 30 h, homogenized via sonication and centrifuged at 5,500 × g for 10 min at 4 °C. The supernatant was filtered through 0.2 μm-pore-size membrane filter (ultra-low protein binding, surfactant-free cellulose acetate, Nalgene, Rochester, NY, USA), checked for S. mutans and C. albicans contamination via microscopic observation/plating on blood agar and adjusted to pH 7.0. The cell-free conditioned medium from single-species bacterial (B-CM) or fungal (F-CM) and bacterial-fungal (BF-CM) biofilms were stored at −20 °C.
Treatments using biofilm-derived conditioned medium
The biological activity of each preparation of the conditioned medium on the growth of S. mutans biofilm cells was assessed using the sHA biofilm model25,32,33. Samples of B-CM, F-CM or BF-CM obtained at different time-points (6, 18 and 30 h) were serially diluted with UFTYE (1:2.5, 1:5, 1:10 and 1:20, CM:UFTYE, vol/vol). Thereafter, sucrose was added to each of the culture medium preparations at a final concentration of 1% (wt/vol), and the pH adjusted to 7.0. Each sHA disc was inoculated with 2 × 106 (CFU/ml) of S. mutans in 2.8 ml (per well) of UFTYE with or without B/F/BF-CM supplementation, and the biofilm formed at 37 °C under 5% CO2. Following 42 h of incubation, the biofilms were collected and analyzed by means of multi-photon confocal microscope and microbiological analysis for 3D biofilm architecture and bacterial viability as described previously32,33,57.
Quantitative biofilm analysis
The biofilms formed in each condition were examined using confocal microcopy combined with quantitative computational analysis and microbiological assays. Briefly, bacterial cells were stained with 2.5 μM SYTO 9 green-fluorescent nucleic acid stain (485/498 nm; Molecular Probes Inc., Eugene, OR, USA), while EPS was labelled with 1 μM Alexa Fluor 647-dextran conjugate (647/668 nm; Molecular Probes Inc.)25,33,57. The imaging was performed using multi-photon Leica SP5 microscope with 20 × LPlan (numerical aperture, 1.05) water immersion objective (Leica Microsystems, Buffalo Grove, IL, USA). The excitation wavelength was 780 nm, and the emission wavelength filter for SYTO 9 was a 495/540 OlyMPFC1 filter, while the filter for Alexa Fluor 647 was an HQ655/40M-2P filter. The confocal images were analyzed using COMSTAT version 1 (available as free download at http://www.imageanalysis.dk), written as scripts for MATLAB software (Mathworks, Natick, MA, USA) to calculate the biovolume and size (diameter and height) of microcolonies33. The biovolume represents the overall biomass occupied by bacterial cells or EPS within intact biofilms, which provide direct measurement of their amounts and ratios across the biofilm depth (from disc surface to fluid phase)33. Amira 5.4.1 software (Visage Imaging, San Diego, CA, USA) was used to create 3D renderings to visualize the overall architecture of the biofilms.
Furthermore, a separate set of biofilms was used for standard microbiological analysis. Briefly, the biofilm was removed from sHA discs and homogenized via water bath sonication followed by probe sonication (30 s pulse at an output of 7 W; Branson Sonifier 150, Branson Ultrasonics, Danbury, CT, USA) as described previously25,33; the sonication procedure does not kill bacterial cells, while providing optimum dispersal and maximum recoverable counts. The homogenized suspension was used to determine the total number of viable cells via plating on blood agar using an automated spiral plater (CFU per biofilm)39.
Glucosyltransferases (Gtfs) activity
The Gtfs activity was measured in each of the conditioned medium and biofilm culture supernatant. An aliquot (100 μl) of the medium (adjusted pH 6.5) was mixed with 100 μl of a [14C-glucose]-sucrose substrate (0.2 μCi/ml; 200 mM of sucrose, 40 μM dextran 900, and 0.02% NaN3 in buffer consisting of 50 mM KCl, 1 mM CaCl2, and 0.1 mM MgCl2 at pH 6.5) to a final concentration of 100 mM (200 μl final volume). The reaction mixture was incubated at 37 °C with rocking for 4 h. After incubation, the radio-labeled glucans were quantified by scintillation counting as described elsewhere58. Gtfs activity was measured by the incorporation of [14C-glucose] from labeled sucrose into glucans. One unit of Gtfs was defined as the amount of enzyme needed to incorporate 1 μM of glucose into glucan over the reaction period58.
Assessment of gtf genes expression in biofilms
The dynamics of S. mutans gtf genes expression following the exposure of BF-CM (collected at 18 h) was evaluated at specific-time points (see below) over the course of biofilm formation. RNA was extracted and purified using protocols optimized for biofilms formed in vitro59. Briefly, disc sets were incubated in RNALater (Applied Biosystems/Ambion, Austin, TX, USA) and the biofilm at 18, 22, 28 and 32 h was removed from the sHA discs. The RNAs were purified and DNAse treated on a column using the Qiagen RNeasy Micro kit (Qiagen, Valencia, CA, USA). The RNAs (RNA integrity number of 9.5 or above; as determined using Bioanalyzer, Agilent Technologies Inc., Santa Clara, CA, USA) were then subjected to a second DNaseI treatment with Turbo DNase (Applied Biosystems/Ambion) and purified using the Qiagen RNeasy MinElute Cleanup kit (Qiagen). Then, we performed RT-qPCR to measure the expression profiles of gtfB and gtfC, which are associated with S. mutans microcolony development33. Briefly, cDNAs were synthesized using 0.5 μg of purified RNA and the BioRad iScript cDNA synthesis kit (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The resulting cDNAs were amplified with a Bio-Rad CFX96 using previously published specific primers33. A standard curve was used to transform the critical threshold cycle (Ct) values to relative numbers of cDNA molecules. Comparative expression was calculated by normalizing each gene of interest to the 16S rRNA60.
Microfluidics-derived nanoculture system
To examine microcolony growth and in situ gtfB expression simultaneously, a gtfB-green fluorescent promoter strain was encapsulated in a recently developed polydimethylsiloxane (PDMS)-based nanoculture system34. Briefly, a glass-capillary microfluidic device with hydrodynamic flow-focusing and co-flowing geometry was constructed to deliver three fluid phases and generate monodisperse water/oil/water (W/O/W) PDMS microemulsion droplets (100–200 μm in diameter), which were allowed to polymerize at 37 °C to form the nanocultures. The three fluid phases were delivered to the microfluidic device through polyethylene tubing (Scientific Commodities, Lake Havasu, AZ, USA) attached to syringes (SGE) that were driven by positive displacement syringe pumps (Harvard Apparatus, Holliston, MA, USA). The inner aqueous phase consisted of defined numbers of S. mutans (~30 cells per nanoculture) suspended in UFTYE (a typical low-molecule weight medium) containing 2.5% (wt/vol) sucrose, which was sufficient to reproducibly promote microcolony formation without any influences on the bacterial cell growth. The middle phase consisted of PDMS (Dow Corning Co., Auburn, MI, USA) with 25% (wt/wt) low viscosity silicone oil (50cSt, Thermo Fisher Scientific) and 10% (wt/wt) curing agent. The outside phase comprises 5% (wt/wt) polyvinyl alcohol aqueous solution (PVA, 87–89% hydrolyzed, average MW = 13000–23000, Aldrich). Nanoculture formation was monitored with a high-speed camera (Phantom, Vision Research, Wayne, NJ, USA) attached to an inverted microscope (Eclipse TE200, Nikon, Tokyo, Japan). The nanocultures were collected in a solution of 0.85% NaCl, UFTYE or UFTYE with BF-CM containing 2.5% (wt/vol) sucrose. The S. mutans within the nanocultures were grown at 37 °C in the presence of 5% CO2 for 24 or 48 h. Images were acquired using confocal laser scanning microscope (SP5-FLIM inverted, Leica Microsystems) or fluorescence microscopy (E600 widefield, Nikon). The fluorescence intensity of GFP was measured using Image J 1.43.
Data are publicly available through the Gulf of Mexico Research Initiative information & Data Cooperative (GRIDC) at http://data.gulfresearchinitiative.org (doi:10.7266/N78G8HQC and doi:10.7266/N7D50K1B).
Chemical characterization of conditioned medium
The amount of sucrose, glucose and fructose were quantified using high-performance anion-exchange chromatography (HPAEC) equipped with an electrochemical detector (ED50; Dionex, Sunnyvale, CA, USA). A 25 μl portion of aliquot was injected onto a CarboPac PA-1 pellicular anion-exchange column (Dionex), and isocratically separated by 150 mM NaOH (eluent A) at 1 ml/min for 12 min. From the HPAEC chromatograms, the intensities of individual peaks were compared to standard corresponding to glucose, fructose and sucrose, and the peak areas were calculated to determine the amount of respective products. The biofilm-derived metabolites were identified and quantified through 1H nuclear magnetic resonance (1H-NMR). Briefly, sample (180 μl) was added to 20 μl of D2O containing 2,2-Dimethyl-2-silapentane-1-sulfonic acid (DSS, final concentration of 0.25 mM, Cambridge Isotope Limited). All spectra were acquired on a Bruker Avance III HD NMR spectrometer using a triple resonance inverse (TXI) 3 mm probe (Bruker Biospin, Billerica, MA, USA). For high-throughput processing, a Bruker Samplejet was used for sample handling. The pulseprogram (noesypr1d) took the shape of the first transient of a 2 dimensional NOESY of the form RD-90-t-90-tm-90-ACQ (RD = relaxation delay, t = small time delay between pulses, tm = mixing time and ACQ = acquisition). The water signal was saturated using continuous irradiation during RD and tm. The spectra were acquired using 76 K data points over a 14 ppm spectral width. Thirty two scans were performed with 1-s interscan (relaxation) delay and 0.1-s mixing time. Raw free induction decays from the 1H-NMR acquisition was processed and profiled using Chenomx NMR suite 8.0 (Chenomx, Alberta, Canada). 1H-NMR data was evaluated using targeted profiling strategy61 that allows quantification of metabolite data in the sample. The significance was determined by direct comparison with concentration in the blank solution (original UFTYE medium).
Heat and proteolytic enzyme treatments of the conditioned medium
Each of the conditioned medium (B-CM, F-CM and BF-CM) was subjected to treatments with heat or proteolytic enzyme. Briefly, conditioned medium was adjusted from pH 4.2 to pH 7.5 (as optimum pH of proteinase K). Subsequently pH adjusted conditioned medium was treated with 100 μg/ml proteinase K (Sigma-Aldrich) at 37 °C for 2 h and/or heat at 85 °C for 30 min62. The bioactivity (cell growth and Gtfs activity) of each of the treated-conditioned medium was tested as described above.
Farnesol analysis and bioactivity
To determine whether C. albicans-derived farnesol can be detected in the conditioned medium, we used established chromatographic analyses38. Farnesol in the conditioned medium was extracted with ethyl acetate with pH adjustment (pH 4) or reverse-phase Sep-Pak column (Mega Bond Elut fresh cartridge, C18, Agilent Technologies, Inc., Santa Clara, CA, USA) without pH adjustment and detected on thin-layer chromatography (TLC) or high performance liquid chromatography (HPLC). Farnesol (trans, trans-farnesol) was purchased from Sigma-Aldrich. Farnesol on TLC plates (Slica gel 60F254, Merck, Darmstadt, Germany) was identified through Rf value of standard with detection accomplished with a hand-held UV lamp, iodine vapor and vanillin-sulfuric acid63, while farnesol was detected at 196 nm with retention time at 30.5 min using a HPLC system equipped with a Shimadzu LC-20 solvent delivery unit and SPD-M20A photodiode array detector (Shimadzu, Kyoto, Japan). A mobile phase consisted of acetonitrile and H2O was delivered under gradient condition on a C18 column (Kinetex C18, 5 μm, 4.6 internal diameter × 250 mm, Phenomenex, Torrance, CA, USA) at a flow rate of 1 ml/min with column temperature set at 40 °C, and injection volume of 20 μl (adapted from Hornby et al.38). The bioactivity of farnesol on S. mutans cell growth, Gtfs activity and microcolony development was performed as described in the previous sections.
Statistical analysis
Data represent mean ± standard deviations (SD) from at least 3 distinct experiments. The quantitative data were subjected to analysis of variance (ANOVA) in the Tukey’s HSD test for a multiple comparison. A pairwise comparison was conducted using student’s t-test.
Additional Information
How to cite this article: Kim, D. et al. Candida albicans stimulates Streptococcus mutans microcolony development via cross-kingdom biofilm-derived metabolites. Sci. Rep. 7, 41332; doi: 10.1038/srep41332 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported in part by the National Institute for Dental and Craniofacial Research (NIDCR) grants DE025220 and DE018023 (H.K.). Imaging experiments were performed in the PennVet Imaging Core Facility on instrumentation supported by NIH S10RR027128, the School of Veterinary Medicine, the University of Pennsylvania, and the Commonwealth of Pennsylvania. This research was made possible in part by a grant from the Gulf of Mexico Research Initiative, by NSF Grant No. DMR-1120901 (Penn MRSEC), and PIRE-1545884 (D.L., K.J.S. and T.H.R.N). T.H.R.N was supported by the Postdoctoral Fellowship for Academic Diversity Program (University of Pennsylvania). The authors thank Dr. Bruce R. Hamaker (Whistler Center for Carbohydrate Research, Purdue University) for his support on HPAEC analysis by B.-H.L. The authors are also grateful to Dr. Geelsu Hwang, Dr. Yuan Liu and Dr. Yong Li for helpful discussions during the manuscript preparation.
Footnotes
The authors declare no competing financial interests.
Author Contributions D.K. and H.K. conceived the experiments; D.K., A.S., T.H.R.N., B.-H.L. and V.S.F. performed the experiments and analyzed the results; A.W., R.M.M., K.J.S., D.L. and H.K. contributed reagents/materials/analysis tools; D.K. and H.K. co-wrote the paper. All authors discussed the results and revised the manuscript.
References
- Dye B. A., Thornton-Evans G., Li X. & Iafolla T. J. Dental caries and sealant prevalence in children and adolescents in the United States, 2011–2012. NCHS Data Brief. 191, 1–8 (2015). [PubMed] [Google Scholar]
- Kassebaum N. J. et al. Global burden of untreated caries: a systematic review and metaregression. J. Dent. Res. 94, 650–658 (2015). [DOI] [PubMed] [Google Scholar]
- Berkowitz R. J., Turner J. & Hughes C. Microbial characteristics of the human dental caries associated with prolonged bottle feeding. Arch. Oral Biol. 29, 949–951 (1984). [DOI] [PubMed] [Google Scholar]
- Palmer C. A. et al. Diet and caries-associated bacteria in severe early childhood caries. J. Dent. Res. 89, 1224–1229 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisotto T. M., Steiner-Oliveira C., Silva C. M., Rodrigues L. K. & Nobre-dos-Santos M. Early childhood caries and mutans streptococci: a systematic review. Oral Health Pre. Dent. 8, 59–70 (2010). [PubMed] [Google Scholar]
- Hajishengallis E., Parsaei Y., Klein M. I. & Koo H. Advances in the microbial etiology and pathogenesis of early childhood caries. Mol. Oral Microbiol. 10.1111/omi.12152 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen W. H. & Koo H. Biology of Streptococcus mutans-derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms. Caries Res. 45, 69–86 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh P. D., Moter A. & Devine D. A. Dental plaque biofilms: communities, conflict and control. Periodontol. 2000. 55, 16–35 (2011). [DOI] [PubMed] [Google Scholar]
- Takahashi N. & Nyvad B. The role of bacteria in the caries process: ecological perspectives. J. Dent. Res. 90, 294–303 (2011). [DOI] [PubMed] [Google Scholar]
- Koo H., Falsetta M. L. & Klein M. I. The exopolysaccharide matrix: A virulence determinant of cariogenic biofilm. J. Dent. Res. 92, 1065–1073 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carvalho F. G., Silva D. S., Hebling J., Spolidorio L. C. & Spolidorio D. M. Presence of mutans streptococci and Candida spp. in dental plaque/dentine of carious teeth and early childhood caries. Arch. Oral Biol. 51, 1024–1028 (2006). [DOI] [PubMed] [Google Scholar]
- Raja M., Hannan A. & Ali K. Association of oral candidal carriage with dental caries in children. Caries Res. 44, 272–276 (2010). [DOI] [PubMed] [Google Scholar]
- Yang X. Q. et al. Genotypic distribution of Candida albicans in dental biofilm of Chinese children associated with severe early childhood caries. Arch. Oral Biol. 57, 1048–1053 (2012). [DOI] [PubMed] [Google Scholar]
- Klinke T., Guggenheim B., Klimm W. & Thurnheer T. Dental caries in rats associated with Candida albicans. Caries Res. 45, 100–106 (2011). [DOI] [PubMed] [Google Scholar]
- Qiu R., Li W., Lin Y., Yu D. & Zhao W. Genotypic diversity and cariogenicity of Candida albicans from children with early childhood caries and caries-free children. BMC Oral Health. 15, 144, 10.1186/s12903-015-0134-3 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson H. F., Lala, H. C. & Shepherd M. G. Coaggregation of Streptococcus sanguis and other streptococci with Candida albicans. Infect. Immun. 58, 1429–1436 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoire S. et al. Role of glucosyltransferase B in interactions of Candida albicans with Streptococcus mutans and with an experimental pellicle formed on hydroxyapatite surfaces. Appl. Environ. Microbiol. 77, 6357–6367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H. & Bowen W. H. Candida albicans and Streptococcus mutans: a potential synergistic alliance to cause virulent tooth decay in children. Future Microbiol. 9, 1295–1297 (2014). [DOI] [PubMed] [Google Scholar]
- Jenkinson H. F. & Douglas L. J. Candida interactions with bacterial biofilms in Polymicrobial diseases (eds Brogden K. A. & Guthmiller J. M.) 357–373 (ASM Press, 2002). [PubMed] [Google Scholar]
- Diaz P. I. et al. Synergistic interaction between Candida albicans and commensal oral streptococci in a novel in vitro mucosal model. Infect. Immun. 80, 620–632 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thein Z. M., Seneviratne C. J., Samaranayake Y. H. & Samaranayake L. P. Community lifestyle of Candida in mixed biofilms: a mini review. Mycoses 52, 467–475 (2009). [DOI] [PubMed] [Google Scholar]
- Xu H. et al. Streptococcal co-infection augments Candida pathogenicity by amplifying the mucosal inflammatory response. Cell. Microbiol. 16, 214–231 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branting C., Sund M. L. & Linder L. E. The influence of Streptococcus mutans on adhesion of Candida albicans to acrylic surfaces in vitro. Arch. Oral Biol. 34, 347–353 (1989). [DOI] [PubMed] [Google Scholar]
- Metwalli K. H., Khan S. A., Krom B. P. & Jabra-Rizk M. A. Streptococcus mutans, Candida albicans, and the human mouth: a sticky situation. PLoS Pathog. 9, e1003616, 10.1371/journal.ppat.1003616 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falsetta M. L. et al. Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo. Infect. Immun. 82, 1968–1981 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang G., Marsh G., Gao L., Waugh R. & Koo H. Binding force dynamics of Streptococcus mutans-glucosyltransferase B to Candida albicans. J. Dent. Res. 94, 1310–1317 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales D. K. & Hogan D. A. Candida albicans interactions with bacteria in the context of human health and disease. PLoS Pathog. 6, e1000886, 10.1371/journal.ppat.1000886 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey-Klett P. et al. Bacterial-fungal interactions: Hyphens between agricultural, clinical, environmental, and food microbiologists. Microbiol. Mol. Biol. Rev. 75, 583–609 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson P. R., Huber M. A. & Bennett J. E. Role of maltase in the utilization of sucrose by Candida albicans. Biochem. J 291, 765–771 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNab R. & Lamont R. J. Microbial dinner-party conversations: the role of LuxS in interspecies communication. J. Med. Microbiol. 52, 541–545 (2003). [DOI] [PubMed] [Google Scholar]
- Sztajer H. et al. Cross-feeding and interkingdom communication in dual-species biofilms of Streptococcus mutans and Candida albicans. ISME J 8, 2256–2271 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H., Xiao J., Klein M. I. & Jeon J. G. Exopolysaccharides produced by Streptococcus mutans glucosyltransferases modulate the establishment of microcolonies within multispecies biofilms. J. Bacteriol. 192, 3024–3032 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J. et al. The exopolysaccharide matrix modulates the interaction between 3D architecture and virulence of a mixed-species oral biofilm. PLoS Pathog. 8, e1002623, 10.1371/journal.ppat.1002623 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niepa T. H. R. et al. Microbial nanoculture as an artificial microniche. Sci. Rep. 6, 30578, 10.1038/srep30578 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira-Cenci T. et al. The effect of Streptococcus mutans and Candida glabrata on Candida albicans biofilms formed on different surfaces. Arch. Oral Biol. 53, 755–764 (2008). [DOI] [PubMed] [Google Scholar]
- Ajdić D. et al. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA. 99, 14434–14439 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paes-Leme A. F., Koo H., Bellato C. M., Bedi G. & Cury J. A. The role of sucrose in cariogenic dental biofilm formation-new insight. J. Dent. Res. 85, 878–887 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornby J. M. et al. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl. Environ. Microbiol. 67, 2982–2992 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H. et al. Inhibition of Streptococcus mutans biofilm accumulation and polysaccharide production by apigenin and tt-farnesol. J. Antimicrob. Chemother. 52, 782–789 (2003). [DOI] [PubMed] [Google Scholar]
- Banas J. A. & Vickerman M. M. Glucan-binding proteins of the oral streptococci. Crit. Rev. Oral Biol. Med. 14, 89–99 (2003). [DOI] [PubMed] [Google Scholar]
- Lynch D. J., Fountain T. L., Mazurkiewicz J. E. & Banas J. A. Glucan-binding proteins are essential for shaping Streptococcus mutans biofilm architecture. FEMS Microbiol. Lett. 268, 158–165 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacca-Smith A. M. et al. Salivary glucosyltransferase B as a possible marker for caries activity. Caries Res. 41, 445–450 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattos-Graner R. O., Smith D. J., King W. F. & Mayer M. P. Water-insoluble glucan synthesis by mutans streptococcal strains correlated with caries incidence in 12- to 30-month-old children. J. Dent. Res. 79, 1371–1377 (2000). [DOI] [PubMed] [Google Scholar]
- Parisotto T. M. et al. Can insoluble polysaccharide concentration in dental plaque, sugar exposure and cariogenic microorganisms predict early childhood caries? A follow-up study. Arch. Oral Biol. 60, 1091–1097 (2015). [DOI] [PubMed] [Google Scholar]
- Yamashita Y., Bowen W. H., Burne R. A. & Kuramitsu H. K. Role of the Streptococcus mutans gtf genes in caries induction in the specific-pathogen-free rat model. Infect. Immun. 61, 3811–3817 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J. G. et al. Influences of trans-trans farnesol, a membrane-targeting sesquiterpenoid, on Sterptococcus mutans physiology and survival within mixed-species oral biofilms. Int. J. Oral Sci. 3, 98–106 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke H. A. & Chang C. A. Physiological effects of sucrose substitutes and artificial sweeteners on growth pattern and acid production of glucose-grown Streptococcus mutans strains in vitro. Z. Naturforsch. C. 31, 245–251 (1976). [DOI] [PubMed] [Google Scholar]
- Han T. L., Cannon R. D. & Villas-Bôas S. G. The metabolic basis of Candida albicans morphogenesis and quorum sensing. Fungal. Genet. Biol. 48, 879–889 (2011). [DOI] [PubMed] [Google Scholar]
- Burne R. A. & Marquis R. E. Biofilm acid/base physiology and gene expression in oral bacteria. Methods Enzymol. 337, 403–415 (2001). [DOI] [PubMed] [Google Scholar]
- Klinke T. et al. Acid production by oral strains of Candida albicans and lactobacilli. Caries Res. 43, 83–91 (2009). [DOI] [PubMed] [Google Scholar]
- Peng X., Zhang Y., Bai G., Zhou X. & Wu H. Cyclic di-AMP mediates biofilm formation. Mol. Microbiol. 99, 945–959 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosz L. M., Deng D. M., van der Mei H. C., Crielaard W. & Krom B. P. Streptococcus mutans competence-stimulating peptide inhibits Candida albicans hypha formation. Eukaryot. Cell. 8, 1658–1664 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyner P. M. et al. Mutanobactin A from the human oral pathogen Streptococcus mutans is a cross-kingdom regulator of the yeast-mycelium transition. Org. Biomed. Chem. 8, 5486–5489 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vílchez R. et al. Streptococcus mutans inhibits hyphal formation by the fatty acid signaling molecule trans-2-decenoic acid (SDSF). ChemBioChem. 11, 1552–1562 (2010). [DOI] [PubMed] [Google Scholar]
- Drescher K., Shen Y., Bassler B. L. & Stone H. A. Biofilm streamers cause catastrophic disruption of flow with consequences for environmental and medical systems. Proc. Natl. Acad. Sci. USA. 110, 4345–4350 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvanitis M. & Mylonakis E. Fungal-bacterial interactions and their relevance in health. Cell. Microbiol. 17, 1442–1446 (2015). [DOI] [PubMed] [Google Scholar]
- Klein M. I., Xiao J., Heydorn A. & Koo H. An analytical tool-box for comprehensive biochemical, structural and transcriptome evaluation of oral biofilms mediated by mutans streptococci. J. Vis. Exp. 47, e2512, 10.3791/2512 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H., Rosalen P. L., Cury J. A., Park Y. K. & Bowen W. H. Effects of compounds found in propolis on Streptococcus mutans growth and on glucosyltransferase activity. Antimicrob. Agents Chemother. 46, 1302–1309 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cury J. A. & Koo H. Extraction and purification of total RNA from Streptococcus mutans biofilms. Anal. Biochem. 365, 208–214 (2007). [DOI] [PubMed] [Google Scholar]
- Koo H. et al. Influence of apigenin on gtf gene expression in Streptococcus mutans UA159. Antimicrob. Agents Chemother. 50, 542–546 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weljie A. M., Newton J., Mercier P., Carlson E. & Slupsky C. M. Targeted profiling: quantitative analysis of 1H NMR metabolomics data. Anal. Chem. 78, 4430–4442 (2006). [DOI] [PubMed] [Google Scholar]
- Westwater C., Balish E. & Schofield D. A. Candida albicans-conditioned medium protects yeast cells from oxidative stress: a possible link between quorum sensing and oxidative stress resistance. Eukaryot. Cell. 4, 1654–1661 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shchepin R. et al. Quorum sensing in Candida albicans: Probing Farnesol’s mode of action with 40 natural and synthetic farnesol analogs. Chem. Biol. 10, 743–750 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.