Significance
Because genome-wide CRISPR/Cas9 libraries are mostly constructed in lentiviral vectors, direct in vivo screening has not been possible as a result of low efficiency in delivery. Here, we examined the piggyBac (PB) transposon as an alternative vehicle to deliver a guide RNA (gRNA) library for in vivo screening. Through hydrodynamic tail vein injections, we delivered a PB-CRISPR library into mouse liver. Rapid tumor formation could be observed in less than 2 mo. By sequencing analysis of PB-mediated gRNA insertions, we identified corresponding genes mediating tumorigenesis. Our results demonstrate that PB is a simple and nonviral choice for efficient in vivo delivery of CRISPR libraries for phenotype-driven screens.
Keywords: CRISPR/Cas9, piggyBac transposon, liver cancer, tumorigenesis, screening
Abstract
CRISPR/Cas9 is becoming an increasingly important tool to functionally annotate genomes. However, because genome-wide CRISPR libraries are mostly constructed in lentiviral vectors, in vivo applications are severely limited as a result of difficulties in delivery. Here, we examined the piggyBac (PB) transposon as an alternative vehicle to deliver a guide RNA (gRNA) library for in vivo screening. Although tumor induction has previously been achieved in mice by targeting cancer genes with the CRISPR/Cas9 system, in vivo genome-scale screening has not been reported. With our PB-CRISPR libraries, we conducted an in vivo genome-wide screen in mice and identified genes mediating liver tumorigenesis, including known and unknown tumor suppressor genes (TSGs). Our results demonstrate that PB can be a simple and nonviral choice for efficient in vivo delivery of CRISPR libraries.
For the last decade, transposon mutagenesis and RNA interference-mediated screens have been the main methods for in vivo screening and validation of cancer genes in mice (1–6). However, because of their low efficiency, these two methods have not been widely used. Recently, CRISPR/Cas9 has been developed as an efficient mutagenesis tool (7–9) and was quickly adapted as a technique for in vivo tumor induction and validation of cancer genes (10–15). By transplanting CRISPR library-transduced cancer cells into immunocompromised mice, several genes involved in growth and metastasis of human lung cancer were identified (16). However, direct in vivo genome-wide CRISPR screening has not been successfully achieved because of the limitations of current lentiviral delivery methods (10, 16). Furthermore, all previous screening strategies suffer from several drawbacks. These screens typically start with an immuno-comprised genetic background or a genetic background carrying multiple pre-engineered mutations, and thus the results may not be applicable to wild-type mice (1, 5). They usually need >1 y to obtain tumors (1, 3, 15). Therefore, there is a strong need for an alternative delivery system that can overcome these shortcomings and be used for direct in vivo CRISPR screening.
For library screening, genomic integration of gRNA constructs is required for later identification of the corresponding gRNAs that mediated the intended phenotypes. In addition to lentivirus/retrovirus, the only good choices to mediate efficient genomic integrations are transposons. Both piggyBac (PB) and Sleeping Beauty (SB) transposons have been used for the delivery of individual gRNAs (15, 17), suggesting they might be adapted to deliver and express a genome-wide single guide RNA (sgRNA) library for high-throughput screening.
Results
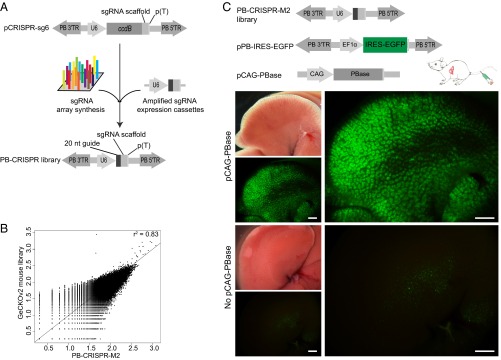
To use PB to deliver and express a genome-wide sgRNA library for high-throughput screening, we constructed three PB vectors (pCRISPR-sg4, pCRISPR-sg5, and pCRISPR-sg6), which all express a sgRNA under control of the human U6 promoter. pCRISPR-sg4 and pCRISPR-sg5 carry puromycin and neo resistance genes, respectively (Fig. S1A), enabling convenient use in cultured cells. PB vectors tend to have multiple copy integrations for inserts <10 kb, and single copy integration for inserts >10 kb (18, 19). To make PB more efficient for in vivo uses, pCRISPR-sg6 was designed to contain minimal sgRNA expression elements without any selectable marker and associated promoter, and thus be more likely to result in multiple copy insertions. The inclusion of the toxic gene ccdB in these vectors ensures that essentially no background colonies can grow during library construction (Fig. 1A).
Fig. S1.
PB-CRISPR vectors and validation by targeting Tet1 and Tet2 in mouse iPS cells. (A) PB-based CRISPR vectors. pCRISPR-sg4, sgRNA-expressing vector with neo gene; pCRISPR-sg5, sgRNA-expressing vector with puromycin gene. (B) pCRISPR-S10, PB plasmid expressing Doxycycline-inducible Cas9; pCRISPR-sg6-Tet1/Tet2, pCRISPR-sg6-based plasmids expressing Tet1 or Tet2 sgRNA. (C) PCR-RFLP analysis of Tet1/Tet2 loci targeted by pCRISPR-sg6-Tet1/Tet2. Expected mutations would eliminate the SacI or EcoRV site in Tet1 and Tet2, respectively. The target regions (∼500 bp) of Tet1 or Tet2 were amplified by PCR. PCR products were digested with corresponding enzymes. Results showed the successful targeting in Tet1-clone 1, Tet1-clone 2, and Tet2-clone 2. (D) Sequencing results of Tet1/Tet2 sgRNA targeted loci. Sequencing results for Tet1-clone 1 revealed a 4-bp deletion in one allele and a 1-bp deletion in another, resulting in elimination of the SacI site. Sequencing results for Tet1-clone 2 revealed mutations in both alleles, with a 3-bp deletion in one and a 1-bp insertion in another, resulting in elimination of the SacI site. Sequencing results for Tet2-clone 2, with an 8-bp deletion in one allele and a 14-bp deletion in another, resulting in elimination of the EcoRV site.
Fig. 1.
PB-CRISPR library construction and in vivo delivery. (A) Work flow of PB-CRISPR library construction. 20 nt guide, guide sequence for chimeric sgRNA; ccdB, a toxin gene for bacteria; PB 3′TR/5′TR, 3′ and 5′ terminal repeat sequence of PB; p(T), poly T terminator sequence; sgRNA scaffold, scaffold sequence for chimeric sgRNA; U6, human U6 promoter. (B) PB-CRISPR-M2 library correlated well (r2 = 0.83) with the GeCKOv2 mouse library in terms of total gRNA distribution, and 95% of sgRNAs in GeCKOv2 can be found in PB-CRISPR-M2. (C) In vivo delivery of PB-CRISPR-M2 library by tail vein injection. pPB-IRES-EGFP, PB plasmid expressing IRES-EGFP. pCAG-PBase expresses CAG promoter-driven PBase. Mice were injected with PB-CRISPR-M2 library, pPB-IRES-EGFP, and pCAG-PBase. Control group was injected without pCAG-PBase. Liver samples were evaluated for GFP expression and used for NGS at 14 d postinjection. (Scale bars: 2 mm.)
After validating PB vector-mediated CRISPR mutagenesis by successfully targeting mouse Tet1 and Tet2 in cultured cells (Fig. S1 B–D), we amplified the sgRNA expression cassettes in the GeCKOv2 genome-scale mouse CRISPR/Cas9 knockout library (20), including 130,209 synthesized sgRNA oligonucleotides targeting all mouse protein coding genes and miRNAs, and cloned into pCRISPR-sg6 to obtain the PB-CRISPR-M2 library (Fig. 1A). The integrity of this PB-CRISPR library was confirmed by deep sequencing, with 95% sgRNAs from the GeCKOv2 mouse library having representation in the PB-CRISPR-M2 library (Fig. 1B). We also constructed a PB sgRNA library by cloning 130,209 synthesized sgRNA oligonucleotides into pCRISPR-sg6, resulting in the PB-CRISPR-M1 library. Because of the simplicity of cloning, genome-wide PB-CRISPR libraries can be constructed rapidly, going from synthesis of oligonucleotides to ready-for-use libraries in a week.
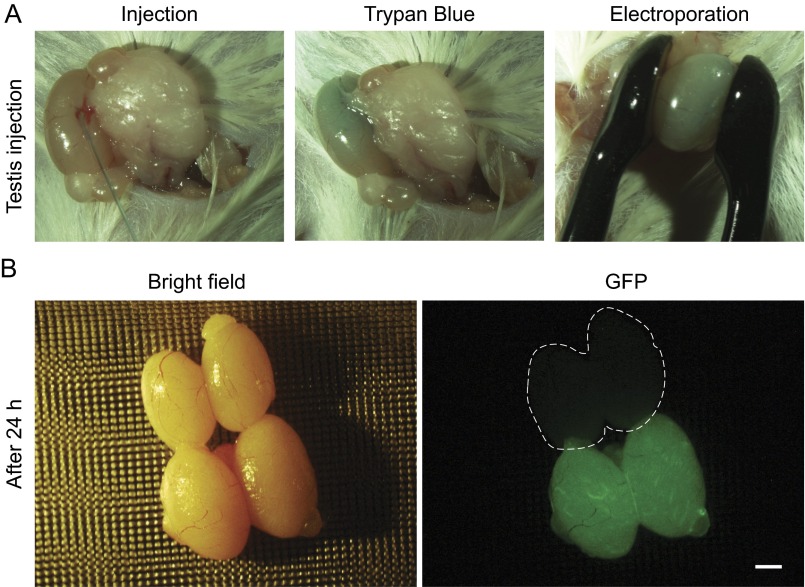
To evaluate the efficiency of delivery into mouse liver, we performed high-pressure tail vein injection of the PB-CRISPR-M2 library, and an EGFP expression plasmid pPB-IRES-EGFP, with or without PB transposase (PBase) overexpression plasmid pCAG-PBase, and analyzed liver samples at day 14 postinjection (Fig. 1C). Strong and uniform GFP fluorescence across the entire liver could be detected when PBase was included (coinjected); in contrast, the control group without PBase (n = 3) had few GFP-positive cells (Fig. 1C). Using deep sequencing to measure sgRNA representation in day 14 liver samples, on average, 89.64 ± 2.79% (n = 3) of library sgRNAs were detected in each liver sample. In addition, we confirmed that PB could be used for efficient transduction of other tissues, such as testis (Fig. S2). These results indicated that PB-mediated in vivo CRISPR delivery is very efficient.
Fig. S2.
Transfection of mouse testis with PB vectors. (A) In vivo transfection of testis with PB vectors by electroporation. Control testis was injected with Trypan blue only. Experiment testis was injected with pPB-IRES-EGFP and pCAG-PBase. (B) Twenty-four hours after electroporation, testes were examined for GFP expression. Dashed line indicates testis without transfection by PB vectors. (Scale bar: 1 mm.)
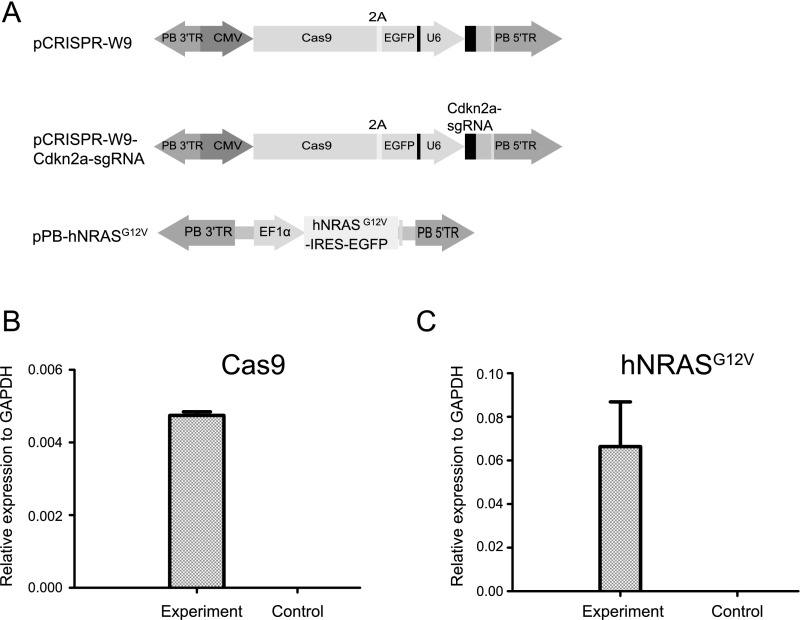
Because liver tumor screens typically require more than a year to obtain tumors (1, 3), we aimed to find a faster scheme to demonstrate the feasibility of PB-CRISPR library screening in wild-type mice. A recent CRISPR validation study showed that gene Cdkn2a sgRNA and Ras oncogene overexpression, with sgRNAs targeting nine other tumor suppressor genes (TSGs) delivered by SB transposon, generated tumors, but only at 20–30 wk after injection (15). We performed tail vein injections to test whether Cdkn2a-sgRNA combined with oncogene NRASG12V overexpression delivered by the PB vectors, pCRISPR-W9-Cdkn2a-sgRNA and pPB-hNRASG12V (Fig. S3), could be used as a sensitized genetic background. We examined the 21 mice injected at day 61, and no tumors were detected (Table 1), whereas Cas9 and NRASG12V expression could be detected by quantitative real-time RT-PCR (qRT-PCR) in liver samples (Fig. S3) from these mice. This result indicated that the sensitized background of Cdkn2a sgRNA/NRASG12V could be ideal for rapid screening within 2 mo, as an additional trigger from the PB-CRISPR library could accelerate tumor formation.
Fig. S3.
Quantitative RT-PCR for transgene expression in mouse liver injected with PB vectors. (A) Schematic maps of PB vectors used in the screening experiment. Mice (n = 3) were injected with pPB-hNRASG12V, pCRISPR-W9-Cdkn2a-sgRNA, and pCAG-PBase. Control mice (n = 3) were injected with saline only. (B) Cas9 expression in mouse liver samples. (C) hNRASG12V expression in mouse liver samples.
Table 1.
PB-CRISPR library screening for tumorigenesis in mouse livers
| Mouse group | pCRISPR-W9-Cdkn2a-sgRNA | pPB-hNRASG12V | PB-CRISPR-M2 library | pCAG-PBase | Tumorigenesis efficiency |
| Control | 12 µg | 12 µg | — | 8 µg | 0/21 (♂, 0%) |
| Screen | 8 µg | 8 µg | 8 µg | 8 µg | 9/27 (♂, 33.3%) |
In addition to the 27 male mice in the screening group, we also performed screening with 20 female mice that were not included in the table. No tumor induction was observed in the 20 female mice at day 61. It is known that male mice are many-fold more likely to develop liver tumors than female mice.
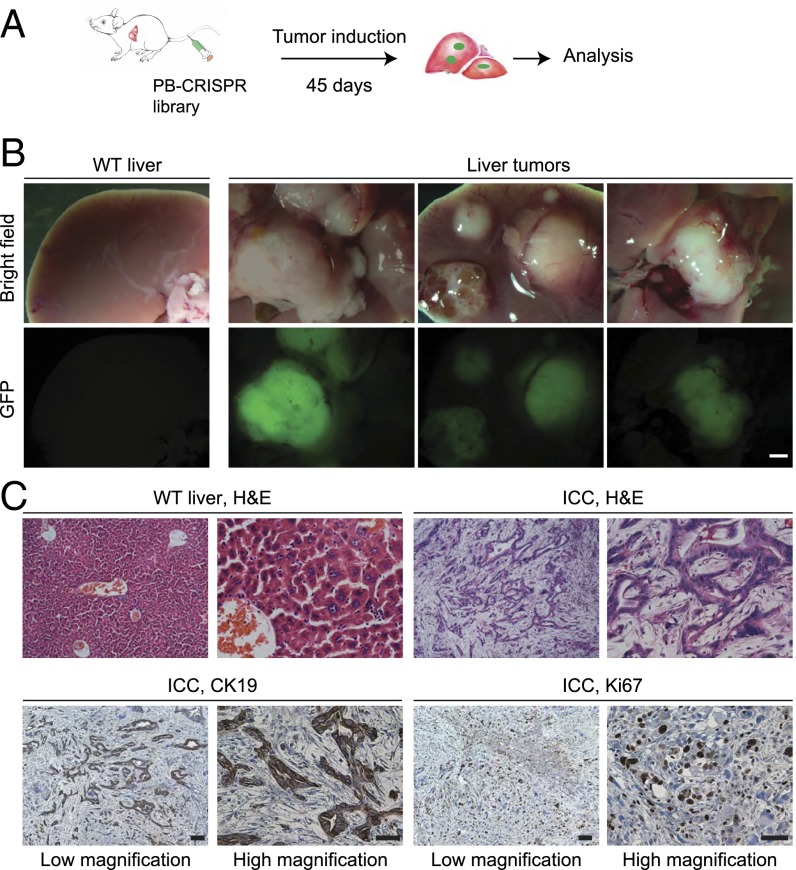
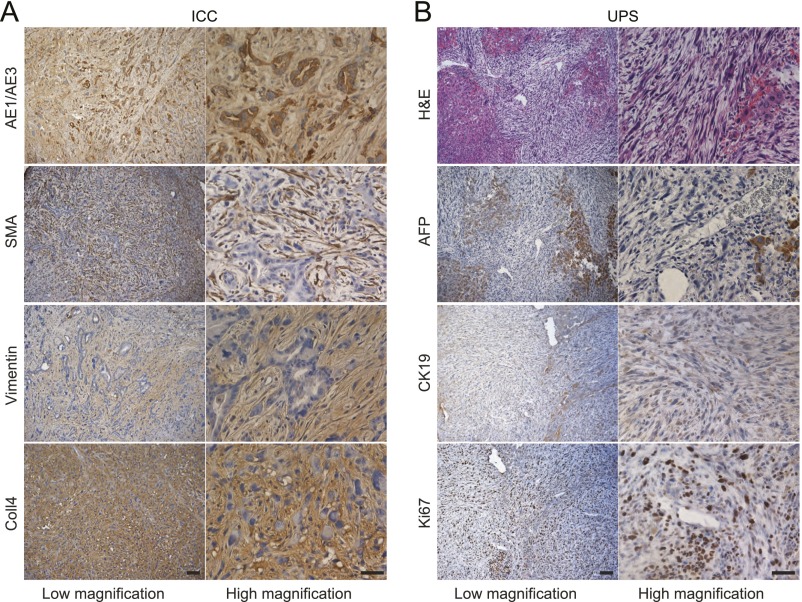
We next conducted a genome-wide screen for liver tumorigenesis through injection with pCRISPR-W9-Cdkn2a-sgRNA, pPB-hNRASG12V, and the PB-CRISPR-M2 library, along with pCAG-PBase (Fig. 2A and Table 1), into 27 mice. All 27 mice injected were examined at 45 d postinjection, when the first mouse in this group died with a tumor. Liver tumors developed in nine of 27 mice, with each mouse containing between one and nine tumors, but no tumors were detected outside the liver. Tumors were readily detected because of their large size (∼5–20 mm) and strong GFP fluorescence (Fig. 2B). Histological analysis by hematoxylin and eosin (H&E) staining and immunohistochemistry showed that most tumors analyzed were intrahepatic cholangiocarcinoma (ICC) (Fig. 2C and Fig. S4), consistent with previous observations that most tumors induced in mouse liver tumor models are ICCs (2, 14, 15). In addition, two tumors appeared to be undifferentiated pleomorphic sarcoma (Fig. S4), which has not been reported in mouse liver cancer models, but suggests that transfection of nonhepatocytes such as stromal cells might have also contributed to liver tumors. The results of rapid tumor formation demonstrated that PB-mediated CRISPR library delivery is practical for in vivo screening in mice.
Fig. 2.
Successful induction of liver tumors in mice using PB-CRISPR library screening. (A) Procedure to conduct a PB-CRISPR screen for genes promoting tumorigenesis in liver. Liver delivery of PB-CRISPR system was carried out with hydrodynamic tail vein injection. (B) Representative liver tumors obtained from the screen. (Scale bar: 2 mm.) (C) Histology and immunohistochemistry analysis of a moderately differentiated ICC. H&E slides show that tumor cells have a tubular growth pattern, in contrast to the normal liver tissue. Tumor cells express CK19 and Ki67. (Scale bars: 100 μm for low magnification, 50 μm for high magnification.)
Fig. S4.
Histology and IHC analysis of representative tumors. (A) A moderately differentiated ICC. Tumor cells express cytokeratin markers AE1/AE3. The surrounding stroma can be identified by SMA, Vimentin, and Collagen-4 (Coll4) staining. (B) A representative undifferentiated pleomorphic sarcoma (UPS). Tumor cells were negative for AFP and CK19 but have high proliferative capacity, as shown by Ki67 staining. (Scale bars: 100 μm for low magnification, 50 μm for high magnification.)
To identify the targets of sgRNAs that had inserted into the tumor genome, we selected 18 tumors for further analysis. We used PCR to amplify sgRNAs from each tumor for next-generation sequencing (NGS). A total of 271 library sgRNAs were identified, with each tumor containing 15.06 ± 7.64 sgRNAs (Dataset S1). The differences in counts for sgRNAs within a tumor suggest some tumors may have a multiclonal origin. Also, the differences in sgRNA content for tumors isolated from one mouse (i.e., tumor 5-1 to tumor 5-8) showed they were clonally unrelated. Among the 271 sgRNAs, the prominent TSG Trp53 was targeted twice, and Cdkn2b, a TSG not previously implicated in mouse liver cancers (21), was targeted in four tumors by three distinct sgRNAs (Table S1). In total, 26 of the 271 sgRNAs were targeting 21 mouse TSG orthologs. Analysis by Fisher’s exact test found these sgRNAs for TSGs were significantly enriched (P < 0.01; Fig. S5 and Dataset S1) (22).
Table S1.
Genes that have been targeted twice or more
| Gene | sgRNA | sgRNA | sgRNA | sgRNA |
| Cdkn2b | B_09614_Cdkn2b | A_09612_Cdkn2b | A_09613_Cdkn2b | B_09614_Cdkn2b |
| Tumor 5–1 | Tumor 5–7 | Tumor 5–9 | Tumor 6–2 | |
| Fxyd4 | B_19120_Fxyd4 | A_19126_Fxyd4 | ||
| Tumor 3 | Tumor 5–7 | |||
| mir-99b | A_63598_mmu-mir-99b | A_63598_mmu-mir-99b | ||
| Tumor 1 | Tumor 5–3 | |||
| Rdh12 | B_44971_Rdh12 | B_44971_Rdh12 | ||
| Tumor 5–3 | Tumor 5–9 | |||
| Tecr | A_53331_Tecr | A_53332_Tecr | ||
| Tumor 5–1 | Tumor 5–6 | |||
| Trp53 | A_56035_Trp53 | A_56035_Trp53 | ||
| Tumor 1 | Tumor 2 |
Fig. S5.
Summary of sgRNA content of 18 tumors. PCR was performed on each tumor for NGS. On average, 15 library sgRNAs were present in each tumor. Among the total of 271 sgRNAs isolated in 18 tumors, 26 sgRNA targeting known TSGs were indicated for the corresponding tumor (two-sided Fisher’s exact test, P < 0.01). Cdkn2b and Trp53 were targeted four and two times, respectively.
Because each tumor in our screen contained multiple copy sgRNA insertions, we tested whether large deletions and translocations that resulted from targeting by two sgRNAs could have made some contribution to tumorigenesis, as suggested by previous reports (13, 23). To survey this possibility, we chose seven tumors (tumors 1, 2, 3, 4-2, 5-4, 5-6, and 5-7) and performed PCR reactions with all possible combinations of primers (Dataset S2). However, no translocations and large deletions in seven tumors were detected. Previous reports suggested that insertional mutagenesis by multiple transposon insertions could contribute to tumorigenesis (1–4). However, considering that the control group was injected with the same amount of PB vectors (Table 1), but did not develop any tumor, tumors obtained from the screen should be largely attributed to library-mediated CRISPR mutagenesis. Taken together, these analyses suggest that identified TSGs could be the main reason for the increased tumorigenesis in the screen.
We next tested sgRNA of the prominent Trp53 to verify whether it would contribute to accelerated tumor formation in our PB delivery system. In the Trp53 group with Cdkn2a-sgRNA, all mice were examined at day 21 postinjection, when the first mouse in this group died of tumors (Fig. 3A and Table S2). Strikingly, 10 of 11 mice injected developed liver tumors, with tumor numbers ranging from a few to >100. To validate Trp53-sgRNA more definitively, we performed injections of Trp53-sgRNA without Cdkn2a-sgRNA. All mice were examined at day 28 postinjection, and eight of 11 mice developed liver tumors (Fig. 3A and Table S2).
Fig. 3.
Validation of sgRNAs for Trp53 and Cdkn2b. (A) Validation of Trp53 and Cdkn2b sgRNAs for liver tumorigenesis in mice. Typical tumors are shown for each group. Histology and immunohistochemistry analyses indicated they were intrahepatic cholangiocarcinomas. In the Trp53 group with Cdkn2a-sgRNA, when mice were examined at day 21 postinjection, 10 of 11 mice had tumors in the liver (P < 0.01, χ2 test). In the Trp53 group without Cdkn2a-sgRNA, eight of 11 mice had liver tumors at 28 d (P < 0.01, χ2 test). In the Cdkn2b group, four of 11 mice developed liver tumors (P < 0.01, χ2 test) at 45 d postinjection. (Scale bars: 2 mm for tumors, 100 μm for H&E, 50 μm for CK19.) (B) Representative Sanger sequencing results of target regions of Trp53 (frameshift indels), Cdkn2b (frameshift indel and nonsense mutation T) in the tumors.
Table S2.
Validated TSGs in liver tumorigenesis
| sgRNA_target gene | pCRISPR-W9 | pCRISPR-W9-Cdkn2a-sgRNA | pPB-hNRASG12V | pCAG-PBase | Tumorigenesis efficiency (%) |
| A_56035_Trp53 | − | + | + | + | 10/11 (♂,90.9%) |
| A_56035_Trp53 | + | − | + | + | 8/11 (♂, 72.7%) |
| B_09614_Cdkn2b | − | + | + | + | 11/11 (♂, 100%) |
| B_09614_Cdkn2b | + | − | + | + | 4/11 (♂, 36.4%) |
| Control group | − | + | + | + | 0/20 (♂,0%) |
We further conducted validation experiments for sgRNA of Cdkn2b, whose tumor suppressor role has not been previously implicated in mouse liver cancers. In the Cdkn2b-sgRNA group with Cdkn2a-sgRNA, at 21 d postinjection, 11 of 11 mice developed liver tumors (Table S2), with tumor numbers in each mouse ranging from hundreds to more, a big increase compared with screening experiments. In the Cdkn2b-sgRNA group, at 45 d postinjection, four of 11 mice developed liver tumors (Fig. 3A and Table S2), with tumor numbers ranging from 1 to 3, indicating that Cdkn2b alone could be a potent TSG in liver tumorigenesis. In addition, mutations in the target regions of Trp53 and Cdkn2b tumors were confirmed (Fig. 3B). Together, these results demonstrate the rapidity and efficiency of PB-CRISPR for in vivo screening and proved that sgRNAs for known and novel TSGs in the screen could be readily recovered.
Discussion
Our PB-CRISPR method has provided an efficient approach to conduct direct in vivo CRISPR library screening, as well as rapid in vivo validation of cancer genes. Compared with previous indirect in vivo screening by transplanting cultured cells (16), our method is much simpler and more likely to reveal relevant TSGs by recapitulating the complexity of the in vivo environment. In this proof-of-principle study, we focused on a fast screening scheme, which by design is more likely to recover mutational events for early tumor occurrence; with longer incubation time or other genetic backgrounds, however, tumors with different mutational profiles should develop in the screening. With the increase of sample numbers, it may be possible to obtain a more complete list of TSGs involved in liver cancer development.
This speed of tumor screening and validation in our study is unprecedented; for example, in the validation experiments for Cdkn2b sgRNA, numerous tumors developed within liver in less than 3 wk. In contrast, similar previous in vivo tumor modeling using CRISPR and SB transposon or pX330 plasmid required a much longer time for tumor formation (14, 15). One possible explanation is that PB mediates very efficient stable transposition in most hydrodynamically injected liver cells (Fig. 1). In the future, combined with other innovative delivery methods, such as nanoparticles and electroporation (12, 24), the extreme simplicity of PB-CRISPR libraries should greatly enhance the already powerful CRISPR weaponry.
Materials and Methods
Plasmids.
All plasmids and libraries constructed in the current study will be deposited to Addgene for distribution. pCRISPR-sg4 and pCRISPR-sg5 and pCRISPR-sg6 were constructed by PCR assembly of the U6-sgRNA expression cassette from pX330 (7), SV40-neo from pIRES2-EGFP (Clontech), puro from pMSCVpuro (BD Biosciences), and ccdB from pStart-K (25) on a PB backbone from pZGs (26). pPB-hNRASG12V was constructed by PCR assembly of NRASG12V amplified from cDNA, and IRES-EGFP from pIRES2-EGFP on a PB backbone from pZGs (26). To construct the pCRISPR-W9 backbone, PB terminal repeats were amplified from pZGs (26) and inserted into pX330 (7), and GFP was added to Cas9 gene with a 2A sequence.
sgRNA targeting individual genes was PCR amplified from oligonucleotide template with primers xcl732/xcl733 (Dataset S2). The purified PCR products were cloned into the BbsI site of pCRISPR-sg6, using the Gibson Assembly method (New England Biolabs), resulting in pCRISPR-sg6-Trp53, and pCRISPR-sg6-Cdkn2b plasmids. All plasmids were confirmed by sequencing. Qiagen EndoFree Plasmid Maxi Kit was used to prepare plasmid DNA for injection.
Library Construction.
To construct the PB-CRISPR-M1 library, we synthesized oligos according to the genome-wide gRNA list (27), amplified sgRNA with primer pair xcl732/xcl733, and cloned them into pCRISPR-sg6 at the BbsI site with the Gibson Assembly method (New England Biolabs). To construct the PB-CRISPR-M2 library, we PCR amplified the U6-sgRNA cassettes from the GeCKOv2 mouse library (20) and cloned them into pCRISPR-sg6. For both the PB-CRISPR-M1 library and PB-CRISPR-M2 library, 10 individual electroporations of 100 µL DH10B competent cells with 20 µL ligation products were carried out. Bacterial cells were plated on a hundred 15-cm dishes to obtain about 107 recombinants; about 80-fold coverage of genome-wide gRNAs was obtained for the PB-CRISPR M1 library, and about 10-fold coverage of genome-wide gRNAs was obtained for the PB-CRISPR M2 library. Bacteria were harvested for maxipreparation of PB-CRISPR libraries with the Endo-free Plasmid Maxi kit (Qiagen).
Test of PB-CRISPR Vectors in Mouse iPS Cells.
Mouse iPS cell line (iPS-ZX11-18-2) used was described previously (28). iPS cells were cultured in embryonic stem cell medium composed of DMEM (Gibco), 15% (vol/vol) FBS (Gibco), 1× penicillin and streptomycin (Gibco), and 1,000 U/mL LIF (Millipore). One million cells were electroporated with 1.5 µg pCRISPR-S10 that expresses Cas9 nuclease, 1.5 µg pCRISPR-sg6-Tet1/Tet2, and 1 µg pCAG-PBase. After electroporation, 1,000 cells were plated in a 10-cm dish. After 10 d, individual clones were picked for further culture and analysis. For PCR-RFLP assay, ∼500-bp DNA fragments around gRNA target sites were amplified using primers, as previously published (29), from genomic DNA of iPS cells (Dataset S2), subjected to restriction endonuclease digestion, and resolved on a 2% (wt/vol) agarose gel.
Mice and Tail Vein Injection.
All mouse experiments in this study were approved by the institutional animal care and use committees at China Agricultural University. Four-week-old CD-1 mice from Charles River were selected for hydrodynamic tail vein injection of the PB-CRISPR library. It was shown that rapid injection of a large volume of DNA solution (∼10% of body weight) via mouse tail vein can achieve efficient gene transfer and expression in vivo, preferentially in the liver (30). We followed a previously described injection protocol (10). The number of animals for screening and validation is derived from experience and confirmed with power analysis, using data from prior, similar-type studies (10, 16). Mice were randomly allocated into different experimental groups. All mice injected were included for analysis. The investigators who assessed mice for tumorigenesis were blinded without knowing whether the animal was from the control or the experiment group.
To examine the in vivo library size after PB mediated delivery, three mice were injected with PB-CRISPR-M2 library, pPB-IRES-EGFP, and pCAG-PBase, at 8 µg each, and three control mice (no pCAG-PBase) were injected with PB-CRISPR-M2 library and pPB-IRES-EGFP, at 8 µg each. DNA was mixed in saline at a volume of 10% body weight. Each injection was finished within 10 s. Liver tissues (∼300 mg) were collected for genomic DNA extraction at day 14 postinjection. sgRNAs were PCR amplified with primers listed in Dataset S2. The purified PCR products were used for NGS.
For in vivo screening, each mouse was injected with pCRISPR-W9-Cdkn2a-sgRNA, pPB-hNRASG12V, PB-CRISPR-M2 library, and pCAG-PBase at 8 µg each in saline at a volume of 10% body weight. Control groups were injected with plasmids, according to Table 1.
For validation experiments, each mouse was injected with corresponding PB-sgRNA, pCRISPR-W9-Cdkn2a-sgRNA (or pCRISPR-W9), pPB-hNRASG12V, and pCAG-PBase at 8 µg each in saline at a volume of 10% body weight. On the day the first mouse in a group died, all mice in the same group were examined. If no mice died in a validation group, all mice were examined at day 45 postinjection. For the control group, mice were examined at day 61 postinjection.
Quantitative RT-PCR Analysis of Expression of Cas9 and hNRASG12V.
Total RNA was isolated from mouse liver, using RNeasy Fibrous Tissue Mini Kit (Qiagen), following the manufacturer's protocol. RNA (2 µg) was reverse transcribed into cDNA, using M-MLV reverse transcriptase (Promega). Quantitative RT-PCR was performed on LightCycler 480 (Roche), using LightCycler 480 SYBR Green I Master (Roche) following the program: preincubation (95 °C, 10 s), amplification (95 °C, 10 s; 60 °C, 10 s; 72 °C, 10 s) 30 cycles, melting curve (95 °C, 5 s; 65 °C, 1 min), cooling (40 °C, 10 s). The primers used to detect the expression of Cas9 and hNRASG12V are displayed in Dataset S2. Gene expression was normalized to the GAPDH.
NGS and Bioinformatics Analysis.
Deep sequencing was used to profile the PB-CRISPR-M2 and GeCKOv2 libraries. After sequencing, we compared normalized read counts of gRNA between the two libraries and calculated Spearman correlation efficiency to measure their similarity (r2 = 0.83; P < 0.001).
To identify sgRNA contents in tumors, ∼100-bp DNA fragments spanning the 20-nt gRNA region of the PB library were PCR amplified from tumor genomic DNA or the library control. Sequencing libraries were constructed with these PCR products, following standard protocols for the Illumina HiSeq2500. Individual libraries from different samples were barcoded and pooled. Sequences of ∼100 bp were demultiplexed from raw data and trimmed into 28-nt gRNA sequences containing sgRNA sequences, which were mapped against index libraries made from the GeCKOv2 library. Fully mapped reads were used to generate gRNA reads list.
To detect mutations in sgRNA target sites, we amplified ∼300 bp DNA, including the gRNA sequence in the center, and performed NGS by Hiseq2500, following standard protocol. BWA aligner was used to map deep sequence data to the mouse genome (mm9) (31). The bam files generated from BWA aligner were sorted and indexed by samtools (32). Mutation variants were called by VarScan.v2.3.9 (33).
Histology and Immunohistochemistry.
Tumors were fixed in 4% (wt/vol) formalin in PBS at 4 °C overnight, paraffin embedded, sectioned at 5 µm, and stained with H&E for pathology. The following antibodies were used for immunostaining: anti-actin, α-smooth muscle antibody, mouse monoclonal clone 1A4 (Sigma, A5228); monoclonal anti-vimentin clone LN-6 (Sigma, V2258); anti-collagen type IV antibody (EMD Millipore Corporation, AB8201); anti-alpha 1 fetoprotein antibody (Abcam, ab46799); purified mouse anti-Ki-67 (BD, 550609); and anti-cytokeratin AE1/AE3 antibody (Abcam, ab115963). The pathologists reading the slides were blinded.
Statistical Analysis.
We generated a list of 1,149 TSG orthologs in mouse genome, using human TSG as comparative information (https://bioinfo.uth.edu/TSGene/) (22). In the PB-CRISPR libraries, there were 6,650 sgRNAs targeting all these mouse TSG orthologs. Out of 271 sgRNAs identified in 18 tumors, 26 sgRNAs targeting 21 mouse TSG orthologs were found to be significantly enriched (P < 0.01) by two-sided Fisher’s exact test.
Supplementary Material
Acknowledgments
We thank Dr. Mei Ling Chen for help with pathology, Yuzhe Wang and Jia Li for help with bioinformatic analysis, and Dr. Anne Boulet and Dr. Kevin Jones for critical reading of the manuscript. This work was supported by the National High Technology Research and Development Program (2013AA102502), Transgenic Research Grant 2014ZX0801014B, and The Project for Extramural Scientists of State Key Laboratory of Agrobiotechnology (Grant 2015SKLAB6-15).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1615735114/-/DCSupplemental.
References
- 1.Bard-Chapeau EA, et al. Transposon mutagenesis identifies genes driving hepatocellular carcinoma in a chronic hepatitis B mouse model. Nat Genet. 2014;46(1):24–32. doi: 10.1038/ng.2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carlson CM, Frandsen JL, Kirchhof N, McIvor RS, Largaespada DA. Somatic integration of an oncogene-harboring Sleeping Beauty transposon models liver tumor development in the mouse. Proc Natl Acad Sci USA. 2005;102(47):17059–17064. doi: 10.1073/pnas.0502974102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keng VW, et al. A conditional transposon-based insertional mutagenesis screen for genes associated with mouse hepatocellular carcinoma. Nat Biotechnol. 2009;27(3):264–274. doi: 10.1038/nbt.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436(7048):221–226. doi: 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
- 5.Zender L, et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell. 2008;135(5):852–864. doi: 10.1016/j.cell.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schramek D, et al. Direct in vivo RNAi screen unveils myosin IIa as a tumor suppressor of squamous cell carcinomas. Science. 2014;343(6168):309–313. doi: 10.1126/science.1248627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sánchez-Rivera FJ, et al. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature. 2014;516(7531):428–431. doi: 10.1038/nature13906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiou SH, et al. Pancreatic cancer modeling using retrograde viral vector delivery and in vivo CRISPR/Cas9-mediated somatic genome editing. Genes Dev. 2015;29(14):1576–1585. doi: 10.1101/gad.264861.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zuckermann M, et al. Somatic CRISPR/Cas9-mediated tumour suppressor disruption enables versatile brain tumour modelling. Nat Commun. 2015;6:7391. doi: 10.1038/ncomms8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maddalo D, et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. 2014;516(7531):423–427. doi: 10.1038/nature13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xue W, et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature. 2014;514(7522):380–384. doi: 10.1038/nature13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weber J, et al. CRISPR/Cas9 somatic multiplex-mutagenesis for high-throughput functional cancer genomics in mice. Proc Natl Acad Sci USA. 2015;112(45):13982–13987. doi: 10.1073/pnas.1512392112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen S, et al. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell. 2015;160(6):1246–1260. doi: 10.1016/j.cell.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koike-Yusa H, Li Y, Tan EP, Velasco-Herrera MdelC, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol. 2014;32(3):267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- 18.Li MA, et al. Mobilization of giant piggyBac transposons in the mouse genome. Nucleic Acids Res. 2011;39(22):e148. doi: 10.1093/nar/gkr764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woltjen K, et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458(7239):766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11(8):783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krimpenfort P, et al. p15Ink4b is a critical tumour suppressor in the absence of p16Ink4a. Nature. 2007;448(7156):943–946. doi: 10.1038/nature06084. [DOI] [PubMed] [Google Scholar]
- 22.Zhao M, Sun J, Zhao Z. TSGene: a web resource for tumor suppressor genes. Nucleic Acids Res. 2013;41(Database issue):D970–D976. doi: 10.1093/nar/gks937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blasco RB, et al. Simple and rapid in vivo generation of chromosomal rearrangements using CRISPR/Cas9 technology. Cell Reports. 2014;9(4):1219–1227. doi: 10.1016/j.celrep.2014.10.051. [DOI] [PubMed] [Google Scholar]
- 24.Platt RJ, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159(2):440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu S, Ying G, Wu Q, Capecchi MR. A protocol for constructing gene targeting vectors: generating knockout mice for the cadherin family and beyond. Nat Protoc. 2008;3(6):1056–1076. doi: 10.1038/nprot.2008.70. [DOI] [PubMed] [Google Scholar]
- 26.Wu S, Ying G, Wu Q, Capecchi MR. Toward simpler and faster genome-wide mutagenesis in mice. Nat Genet. 2007;39(7):922–930. doi: 10.1038/ng2060. [DOI] [PubMed] [Google Scholar]
- 27.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343(6166):84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu S, Wu Y, Zhang X, Capecchi MR. Efficient germ-line transmission obtained with transgene-free induced pluripotent stem cells. Proc Natl Acad Sci USA. 2014;111(29):10678–10683. doi: 10.1073/pnas.1409933111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153(4):910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu F, Song Y, Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999;6(7):1258–1266. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- 31.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koboldt DC, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22(3):568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.