Significance
Effective prevention strategies are urgently needed to control the spread of HIV-1. A critical barrier to developing such strategies is the lack of understanding of the host antiviral defenses that control HIV-1 replication in the mucosa at the site of entry. Here, we characterized viruses from matched donor and recipient pairs to determine whether transmitted HIV-1 strains exhibit traits that increase their transmission fitness. Characterizing 300 limiting dilution-derived isolates, we identified several properties that enhance virus replication in the face of a vigorous innate immune response, of which resistance to type 1 IFNs is the most important. These results provide new insights into the HIV-1 transmission process and define possible new targets for AIDS prevention and therapy.
Keywords: mucosal HIV-1 transmission, type I interferons, innate immunity, HIV-1 transmission pairs, HIV-1 transmission fitness
Abstract
Sexual transmission of HIV-1 is an inefficient process, with only one or few variants of the donor quasispecies establishing the new infection. A critical, and as yet unresolved, question is whether the mucosal bottleneck selects for viruses with increased transmission fitness. Here, we characterized 300 limiting dilution-derived virus isolates from the plasma, and in some instances genital secretions, of eight HIV-1 donor and recipient pairs. Although there were no differences in the amount of virion-associated envelope glycoprotein, recipient isolates were on average threefold more infectious (P = 0.0001), replicated to 1.4-fold higher titers (P = 0.004), were released from infected cells 4.2-fold more efficiently (P < 0.00001), and were significantly more resistant to type I IFNs than the corresponding donor isolates. Remarkably, transmitted viruses exhibited 7.8-fold higher IFNα2 (P < 0.00001) and 39-fold higher IFNβ (P < 0.00001) half-maximal inhibitory concentrations (IC50) than did donor isolates, and their odds of replicating in CD4+ T cells at the highest IFNα2 and IFNβ doses were 35-fold (P < 0.00001) and 250-fold (P < 0.00001) greater, respectively. Interestingly, pretreatment of CD4+ T cells with IFNβ, but not IFNα2, selected donor plasma isolates that exhibited a transmitted virus-like phenotype, and such viruses were also detected in the donor genital tract. These data indicate that transmitted viruses are phenotypically distinct, and that increased IFN resistance represents their most distinguishing property. Thus, the mucosal bottleneck selects for viruses that are able to replicate and spread efficiently in the face of a potent innate immune response.
Understanding the host and viral factors that influence HIV-1 transmission may aid the development of an effective AIDS vaccine. In 2015, ∼2 million individuals were newly infected with HIV-1, the great majority of whom acquired the virus by sexual routes (1). Although a number of factors, such as high donor viral loads, genital inflammation, altered mucosal microbiota, and recipient gender, are known to increase the infection risk (2–4), virus transmission across intact mucosal surfaces is inherently inefficient, with only a small fraction (less than 1%) of unprotected sexual exposures leading to productive infection (5–8). This inefficiency is exemplified by a stringent population bottleneck, in which only one or a limited number of variants from the diverse quasispecies of the transmitting donor establish the new infection (9). Transmitted viruses are not usually the most abundant strains in the genital secretions of infected donors (10), and analyses of viral sequences from 137 matched donor and recipient pairs indicated that viruses with a more ancestral genotype are preferentially transmitted (11). These data suggested that mucosal transmission selects for variants with enhanced transmission fitness (11). However, the viral properties that contribute to this transmission fitness have not been defined.
For obvious reasons, viruses cannot be collected from, or studied in, humans at the time of transmission. However, by sequencing plasma virion RNA (vRNA) in the first few weeks following transmission, it is possible to enumerate and infer the genome(s) of the virus(es) that established the infection (9, 12–14). In the absence of adaptive immune pressures, HIV-1 diversifies in a random fashion, with viral sequences exhibiting a Poisson distribution of mutations and a star-like phylogeny that coalesces to an inferred consensus sequence. This consensus sequence represents the genome of the virus that initiated the infection, termed the transmitted founder (TF) virus (9). Single genome amplification (SGA) of plasma vRNA, which precludes PCR artifacts such as Taq polymerase-mediated recombination (15–18), revealed that in the great majority (∼80%) of sexual transmission cases, a single TF virus establishes the new infection (9, 12, 13, 19–21).
The ability to infer and molecularly clone the genomes of TF viruses has permitted their biological characterization. Initial studies showed that TF viruses use the cell-surface molecules CD4 and CCR5 as their receptor and coreceptor, and replicate efficiently in activated CD4+ T cells but not macrophages (14, 22–25). Moreover, analysis of a comprehensive panel of infectious molecular clones (IMCs) showed that TF viruses packaged more envelope glycoprotein (Env), exhibited greater infectivity, bound to monocyte-derived dendritic cells more efficiently, and replicated to higher titers in CD4+ T cells in the presence of the type 1 interferon IFNα2 than chronic control (CC) viruses (26). However, a potential confounder of these studies was the fact that TF and CC viruses were not derived from epidemiologically linked transmission pairs. To compare transmitted and nontransmitted viruses close to the time of transmission, two recent studies characterized the phenotype of viruses from matched donor and recipient pairs (27, 28). Examining various biological properties, including the sensitivity of donor and recipient viruses to IFNα2, both studies failed to identify viral traits that were indicative of enhanced transmission fitness (27, 28).
Innate immune responses, in particular type 1 IFNs, represent a potent first-line defense against many pathogens, including primate lentiviruses (29–33). Consistent with these data, treatment of rhesus macaques with pegylated IFNα2 increased the number of intrarectal challenges required to achieve systemic simian immunodeficiency virus (SIV) infection and decreased the number of transmitted founder viruses (34). Similarly, mucosal application of IFNβ protected macaques from repeated intrarectal and intravaginal challenges with a simian–human immunodeficiency virus (SHIV) (35). Because type 1 IFNs are rapidly up-regulated at mucosal sites of virus replication in SIV-infected macaques (36), and bioactive IFN levels are highly elevated during acute HIV-1 infection (37), we hypothesized that IFN-mediated antiviral activity contributes to the HIV-1 transmission bottleneck. To test this hypothesis, we generated a large panel of limiting dilution-derived isolates from the plasma and genital secretions of chronically infected donors and their matched recipients. Analyzing 300 such isolates, we identified a number of biological properties that are associated with increased transmission fitness, all of which serve to enhance HIV-1 replication and spread in the face of a vigorous innate immune response.
Results
Generation of Limiting Dilution HIV-1 Isolates from Sexual Transmission Pairs.
Molecular cloning of HIV-1 genomes is labor intensive and thus limits the number of IMCs that can reasonably be characterized. Moreover, predicting which viral genomes are functional in chronically infected individuals is challenging, because immune escape mutations frequently incur fitness costs (38–45). Virus isolation represents an alternative to cloning, but bulk cultures cannot account for the biological variation of individual quasispecies members. Here, we used limiting dilution virus isolation to generate single virion-derived HIV-1 strains from eight sexual transmission pairs. These included four female-to-male (FTM) transmissions (subtype C) from southern Africa as well as from one male-to-female (MTF) and three men-who-have-sex-with-men (MSM) transmissions (subtype B) from the United States (SI Appendix, Table S1). In all but one case, the newly infected recipient was identified first as part of an acute infection cohort, whereas the transmitting partner was identified retrospectively. Phylogenetic analysis of SGA-derived plasma viral sequences confirmed that all transmission pairs were epidemiologically linked (SI Appendix, Fig. S1) and showed that two recipients (CH378 and CH831) had acquired their infection from the same donor (CH742). Seven of the eight recipients were infected with a single TF virus, whereas the remaining subject (CH378) acquired at least two TF viruses (SI Appendix, Fig. S2). All subjects remained treatment naïve throughout the study.
To generate limiting dilution-derived viral isolates, plasma as well as cell-free fractions of cervicovaginal lavage (CVL) and semen (SEM) samples were end-point diluted and used to infect activated normal donor CD4+ T cells in 24-well plates. According to a Poisson distribution, a dilution that yields positive cultures in no more than 30% of wells should contain a single infectious unit more than 80% of the time. Cultures were maintained for 20 d, tested for p24 antigen production, and virus positive wells were expanded further in normal donor CD4+ T cells for an additional 10 d. The resulting viral stocks were used for all subsequent genetic and biological analyses.
To ensure that the limiting dilution isolates were indeed single virion derived, we sequenced all stocks before biological characterization. Briefly, 5′ and 3′ half genomes were PCR amplified, MiSeq sequenced, and the resulting reads assembled to generate an isolate-specific consensus sequence. Viral reads were then mapped to this consensus sequence and the extent of genetic diversity was examined for each position along the genome. Isolates that exhibited more than 15% diversity at any one position in the alignment were considered to contain more than one variant and thus removed from further analysis. To control for the emergence of phenotypically distinct variants in the culture, we also generated limiting dilution isolates from all acutely infected subjects even though TF IMCs were available for two recipients (14, 26). Using plasma samples collected closest to the time of transmission, we generated 95 donor and 61 recipient isolates (SI Appendix, Table S1). Virus isolation from CVL and SEM samples was more challenging, because of lower viral loads, frequent bacterial and yeast contaminations, and the fact that many genital secretions were inherently cytotoxic for CD4+ T cells (46). Nonetheless, we were able to generate limiting dilution isolates from the CVL or semen samples of three transmitting donors (SI Appendix, Table S1).
Limiting Dilution HIV-1 Isolates Are Representative of the Donor Quasispecies.
To determine whether the limiting dilution isolates were representative of the viral quasispecies present in both donors and recipients, we compared all isolate-derived sequences to SGA-derived vRNA sequences amplified directly from the blood of the same individual. In phylogenetic trees of 3′ half genome sequences, isolate and plasma vRNA sequences were completely interspersed (Fig. 1A and SI Appendix, Fig. S3). To assess whether isolate and plasma viral sequences from chronically infected donors were segregated, we calculated their genealogical sorting index (gsi) (47). Two donor samples yielded gsi values that were higher than expected from random segregation (SI Appendix, Table S2A). In one case (CH212), available isolates represented only two of three diverse viral lineages present in this donor’s quasispecies, indicating limited sampling (SI Appendix, Fig. S3F). In the other case (CH728), two pairs of near identical isolate sequences indicated repeat culture of the same virus (SI Appendix, Fig. S3C). Collapsing one of these to a single sequence rendered the gsi value nonsignificant. For all other subjects, there was no evidence for segregation (SI Appendix, Table S2A), indicating that the isolates were fully representative of the viral diversity present in the plasma. As expected, plasma isolates from single TF infections were very closely related, differing from each other by fewer than 8 (range 2–7) and from the inferred TF genome by fewer than 12 (range 2–11) nucleotides across the entire genome (SI Appendix, Fig. S2). Plasma isolates from subject CH378 exhibited greater diversity, because they represented the progeny of two TF viruses as well as their recombinants (SI Appendix, Fig. S2A). Unlike in some previous studies (8), there was no evidence of compartmentalization of plasma and genital secretion isolates from donors CH492 and CH742 (SI Appendix, Fig. S3 and Table S2B).
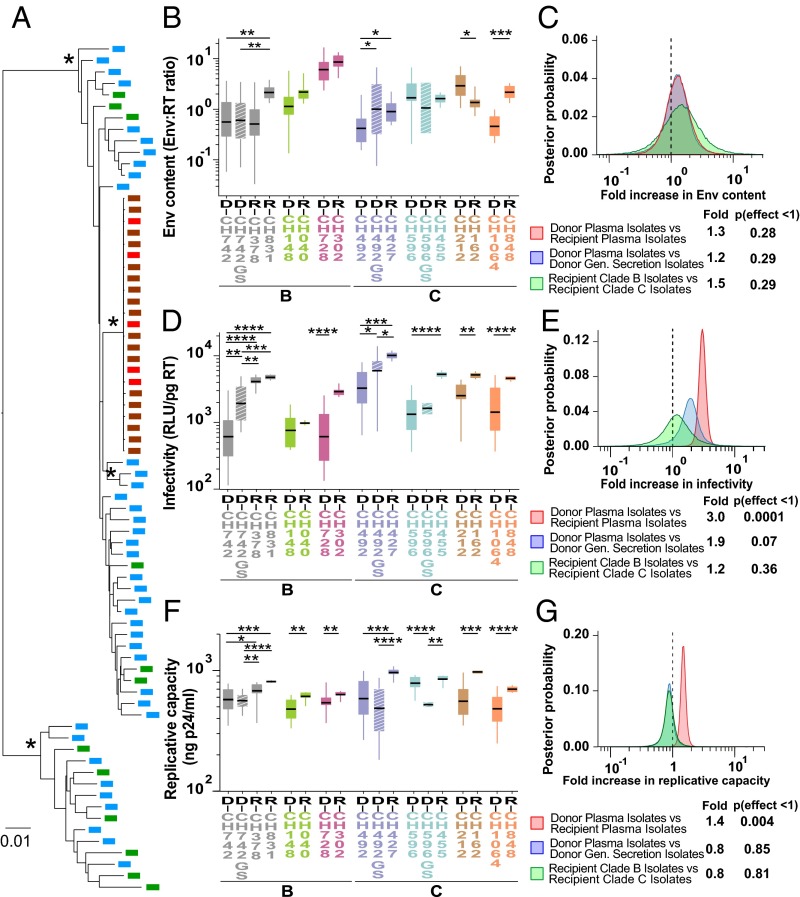
Fig. 1.
Genetic and biological characterization of matched donor and recipient limiting dilution-derived isolates. (A) The phylogenetic relationships of donor (green) and recipient (brown) isolate sequences to donor (blue) and recipient (red) SGA-derived plasma viral sequences are shown for the CH596–CH455 transmission pair (maximum likelihood trees for all other transmission pairs are shown in SI Appendix, Fig. S3). Asterisks denote nodes with 100% bootstrap support (the scale bar indicates 0.01 substitutions per site). (B, D, and F) Viral Env content (mass ratio of gp120 and RT), particle infectivity (RLU in TZM-bl cells per picogram of RT), and replicative capacity (p24 antigen levels in CD4+ T-cell culture supernatants 7 d postinfection) of plasma isolates from matched donor (D) and recipient (R) pairs (color coded) are shown, with HIV-1 subtype classification indicated below. Data are grouped for each transmission pair, with genital secretion isolates (GS) shown as hashed boxes. Donor D-CH472 transmitted to two recipients R-CH378 and R-CH831. Boxes show the interquartile range, a black bar within each box indicates the geometric mean, and whiskers span the range of the data, respectively. Asterisks indicate significant differences (determined by unpaired t test) between groups (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001). (C, E, and G) Hierarchical Bayesian regression models were used to estimate the population-wide fold change of Env content (C), particle infectivity (E), and replicative capacity (G) across all transmission pairs between donor and recipient plasma isolates (red), donor plasma and genital (Gen) secretion isolates (blue), and clade B and C recipient isolates (green). A dashed line indicates a fold change of 1, indicating no effect. The estimated posterior probability distribution for each parameter is shown along with a table summarizing the expected fold change and the probability that the effect is <1 (analogous to a one-sided P value).
Increased Env Content Is Not a Characteristic Feature of Transmitted Viruses.
Comparing viruses from unrelated subjects, we previously reported that TF IMCs package on average 1.9-fold more Env than viruses circulating in the plasma of chronically infected individuals (26). To examine the Env content of matched donor and recipient isolates, we generated viral stocks in normal donor CD4+ T cells, depleted these of microvesicles, purified virions using antibody-coated magnetic beads, and quantified Env by ELISA per unit of RT activity. We found that plasma isolates varied widely in the amounts of Env that they packaged, but failed to identify consistent differences between donor and recipient isolates. Recipient isolates packaged either significantly more, less, or similar amounts of Env compared with their corresponding donor viruses (Fig. 1B and SI Appendix, Fig. S4A). For one donor (CH492), genital tract isolates had a 2.4-fold higher mean Env content than the corresponding plasma isolates, but this was not the case for the other two donors (Fig. 1B and SI Appendix, Fig. S4A). When data from all pairs were combined, no significant differences in Env content were observed between donor and recipient isolates, plasma and genital secretion isolates, and subtype B and C recipient isolates (Fig. 1C). These data indicate that mucosal transmission does not select for viruses with an increased Env content.
Transmitted Viruses Exhibit Increased Particle Infectivity.
We previously reported that TF viruses were 2-fold more infectious than chronic viruses from unrelated subjects (26), but two subsequent studies failed to identify virus infectivity as a determinant of transmission fitness (27, 28). Here, we used TZM-bl cells, which express luciferase under the control of an HIV-1 promoter (48, 49), to determine the per-particle infectivity of CD4+ T-cell–derived viral stocks. To limit virus infection to a single round, we added the fusion inhibitor T1249 (50) to all cultures 12–15 h following infection. Plotting relative light units (RLUs) per amount of input virus (picograms of RT), we found that donor plasma isolates exhibited a wide range of particle infectivity both within and between individuals, whereas the infectivity of recipient isolates was much less variable. Moreover, for seven transmission pairs, recipient viruses were significantly (2- to 8-fold) more infectious than the corresponding donor viruses, with a trend observed for the eighth pair (Fig. 1D and SI Appendix, Fig. S4B). Higher particle infectivity relative to plasma viruses was also observed for CVL and SEM isolates from two donors (1.8- and 3.2-fold, respectively), but not for the third donor, although in the latter case only two CVL isolates were available for comparison (Fig. 1D and SI Appendix, Fig. S4B). When data from all transmission pairs were combined, recipient isolates were on average 3-fold more infectious (P = 0.0001) than the corresponding donor isolates irrespective of their subtype (Fig. 1E). Donor genital secretion isolates tended to be more infectious than the corresponding plasma isolates, but these did not reach statistical significance. Thus, mucosal transmission selects for viruses with increased particle infectivity, some of which are present in genital secretions.
Transmitted Viruses Replicate to Higher Titers.
The replicative capacity of viruses can influence their reproductive ratio (R0) and thus their ability to expand an initial infection (51). Comparing IMCs from unrelated subjects, we previously failed to detect differences in the growth potential of TF and chronic HIV-1 strains (26), and similar results were reported for donor and recipient viruses from transmission pairs (27, 28). Here, we compared the replicative capacity of limiting dilution-derived isolates in normal donor CD4+ T cells. Using equal numbers of particles for viral input (1 ng of RT activity), we monitored the growth kinetics of a subset of isolates (n = 25) for 9 d by measuring p24 antigen in culture supernatants every 48 h. We then determined the area under the curve (AUC) and compared it with p24 values measured at individual time points. This analysis revealed a strong correlation between the AUC and p24 production at day 7 (r = 0.99, P < 0.0001). We thus used the latter as a measure of viral replicative capacity for all remaining isolates.
Transmitting donor isolates varied widely in their replicative capacity, and this was also true for some recipient isolates. However, recipient isolates replicated on average between 1.2- and 1.7-fold more efficiently than viruses isolated from the corresponding donors (Fig. 1F and SI Appendix, Fig. S4C). These differences were significant for seven transmission pairs, with a trend observed for the eighth pair (Fig. 1F and SI Appendix, Fig. S4C). In contrast, genital secretion isolates did not exhibit an increased replicative capacity. Combining results from all transmission pairs, we found that on average recipient isolates grew to 1.4-fold higher titers than their corresponding donor isolates (P = 0.004), whereas no significant differences were observed between plasma and genital secretion isolates, or between recipient isolates representing subtype B and C infections (Fig. 1G). These data indicate that mucosal transmission selects for viruses with enhanced replicative capacity.
Transmitted Viruses Are Uniformly Resistant to Type I Interferons.
We previously reported that TF viruses are more resistant to IFNα2 than viruses from chronically infected individuals (26, 52). However, two subsequent studies of linked transmission pairs failed to confirm this phenotype, with one study finding no differences in IFNα2 resistance between transmitted and nontransmitted viruses (27), and the other reporting transmitted viruses being more IFNα2 sensitive (28). To resolve these differences, we tested the IFN sensitivity of the limiting dilution-derived isolates, but with some experimental modifications. First, instead of testing only IFNα2, we measured the antiviral effect of a second potent inhibitor of HIV-1, IFNβ (35, 52). Second, rather than examining the effect of only a single IFN inhibitory dose (26–28, 53), we determined the half-maximal inhibitory concentration (IC50) of both IFNα2 and IFNβ for every single isolate. This was done by treating normal donor CD4+ T cells with increasing quantities of IFN, infecting them with equal amounts of virus, and culturing the cells for 7 d while replenishing IFN-containing medium. Virus replication was then measured for each IFN concentration as the amount of p24 produced at day 7 and plotted as the percentage of viral growth in the absence of IFN, which was set to 100% (Fig. 2 A and B). As an independent measure of IFN resistance, we also measured viral replication at the highest IFN dose and expressed this residual replication capacity (Vres) as the percentage of viral growth in the absence of IFN (Fig. 2 A and B and SI Appendix, Fig. S5 B and D).
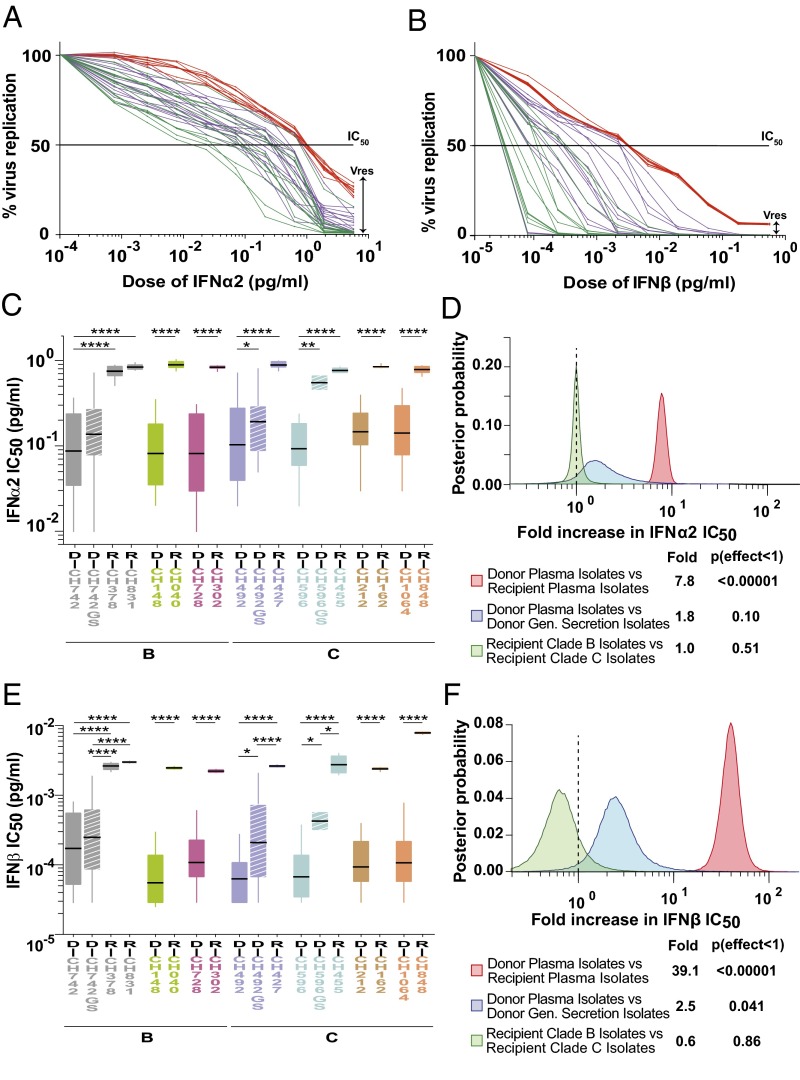
Fig. 2.
IFN resistance of matched donor and recipient isolates. (A and B) Dose–response curves for IFNα2 (A) and IFNβ (B) are shown for plasma (green) and genital secretion (magenta) isolates of one chronically infected donor as well as plasma isolates of the corresponding acutely infected recipient (red) of a representative transmission pair (CH492–CH427). A black line indicates the half-maximal inhibitory concentration (IC50) and a double arrow, the residual viral replication (Vres) capacity at the highest IFN dose. (C and E) IFNα2 (C) and IFNβ (E) concentrations (picograms per milliliter), which resulted in 50% viral inhibition, are shown for plasma isolates from matched donor (D) and recipient (R) pairs (color coded as in Fig. 1), with HIV-1 subtype classification indicated below. Data are grouped for each transmission pair, with genital secretion isolates (GS) shown as hashed boxes. Donor D-CH472 transmitted to two recipients R-CH378 and R-CH831. Boxes show the interquartile range, a black bar within each box indicates the geometric mean, and whiskers span the range of the data, respectively. Asterisks indicate significant differences (determined by unpaired t test) between groups (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001). IFN IC50 values were determined in pooled CD4+ T cells from multiple donors. (D and F) Hierarchical Bayesian regression models were used to estimate the population-wide fold change of IFNα2 (D) and IFNβ (F) IC50 values across all transmission pairs between donor and recipient plasma isolates (red), donor plasma and genital (Gen) secretion isolates (blue), and clade B and C recipient isolates (green). A dashed vertical line marks a fold change of 1, indicating no effect. The estimated posterior probability distribution for each parameter is shown along with a table summarizing the expected fold change and the probability that the effect is <1 (analogous to a one-sided P value).
For each transmission pair, plasma isolates from donors exhibited a wide range of sensitivities to both IFNα2 and IFNβ, whereas recipient isolates were much less variable as well as uniformly more resistant to both IFNα2 and IFNβ (Fig. 2 C and E). Compared with the respective donor isolates, recipient isolates exhibited on average 6- to 11-fold higher IFNα2, and 15- to 71-fold higher IFNβ IC50 values (SI Appendix, Fig. S5 A and C). Analysis of the residual replicative capacity, Vres, yielded similar results, although the differences between donor and recipient isolates were much more pronounced. At the highest IFNα2 dose (5.5 pg/mL), recipient isolates retained on average 15–26% of their replicative capacity, whereas the corresponding donor viruses reached only 0.8–2% (SI Appendix, Fig. S5B). At the highest IFNβ dose (0.44 pg/mL), recipient viruses retained on average 4–13% of their replicative capacity, whereas the corresponding donor isolates were either suppressed below the limits of p24 detection or reached Vres values of 0.01–0.1% (SI Appendix, Fig. S5D). Thus, the ability of recipient isolates to replicate at the highest IFN dose was 13- to 51-fold higher for IFNα2, and 123- to 541-fold higher for IFNβ, compared with the corresponding donor viruses (SI Appendix, Fig. S5 B and D).
Examining genital tract isolates, we found that they also exhibited higher IFNα2 and IFNβ IC50 values than the corresponding plasma isolates, although significance was reached only for IFNβ (P = 0.04) (Fig. 2F). In addition, genital tract isolates exhibited higher Vres values, but in this case, significance was reached only for IFNα2 (P = 0.008) (SI Appendix, Fig. S6B). Comparing IC50 and Vres, we found that these values correlated strongly for donor plasma isolates (IFNα2: r = 0.89, P < 0.0001; IFNβ: r = 0.57, P < 0.0001), but only weakly for donor genital secretion isolates (IFNα2: r = 0.34, P < 0.05; IFNβ: r = 0.40, P < 0.01), indicating that IC50 and Vres provide different measures of the antiviral effect of IFNs in these compartments. Similarly, IC50 values for IFNα2 and IFNβ correlated only weakly (r = 0.33, P = 0.048), indicating only a partial overlap in the effects of the two IFN subtypes on the activation state, survival, and IFN stimulated gene (ISG) expression levels of CD4+ T cells.
Combining data from all transmission pairs, we found that recipient isolates were on average significantly more resistant to both IFNα2 and IFNβ than the corresponding donor isolates, exhibiting 7.8-fold (P < 0.00001) and 39-fold (P < 0.00001) higher IC50 values, respectively (Fig. 2 D and F). Moreover, recipient isolates had 35-fold (P < 0.00001) and 250-fold (P < 0.00001) greater odds of replicating at the highest IFNα2 (SI Appendix, Fig. S6B) and IFNβ (SI Appendix, Fig. S6D) doses than the great majority of donor viruses, respectively. These differences were not dependent on the viral subtype (Fig. 2 D and F). Thus, resistance to type 1 IFNs is a characteristic feature of transmitted viruses.
Selection with IFNβ, but Not IFNα2, Yields Donor Isolates with a Transmitted Virus-Like Phenotype.
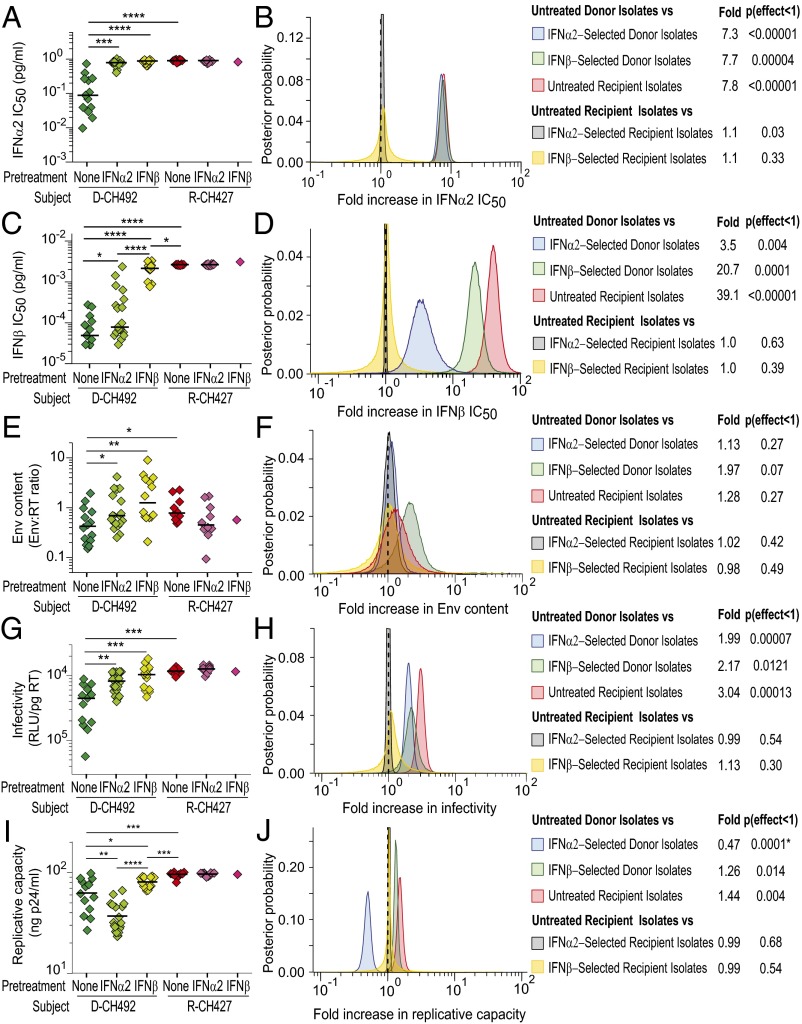
To search for IFN-resistant viruses in donor plasma, we treated CD4+ T cells with high doses of IFNα2 (4.0 pg/mL) or IFNβ (0.44 ng/mL) 24 h before virus isolation. The rationale was to maximally up-regulate antiviral ISGs in these target cells (without causing cell toxicity), thereby simulating, at least in part, conditions during the earliest stages of HIV-1 infection. As a control, the same approach was used to isolate viruses from recipient plasma. As expected, the number of viral isolates recovered from pretreated CD4+ T cells was lower than from untreated CD4+ T cells, especially when IFNβ was used for selection (SI Appendix, Table S1). Thus, whereas IFNα2 pretreatment yielded plasma isolates for all donors and recipients, only three donors and two recipients yielded IFNβ-preselected plasma isolates. This was as expected because the selection dose of IFNβ was six orders of magnitude higher than the average IFNβ IC50 value of all isolates (IFNα2 doses higher than 5.5 pg/mL caused cell toxicity). Phylogenetic analyses of full-length genome sequences revealed no evidence of compartmentalization of selected and nonselected isolates (SI Appendix, Fig. S7 and Table S2C).
IC50 determinations confirmed that donor isolates from IFN-pretreated cells were indeed more IFN resistant than those derived from untreated CD4+ T cells, whereas no changes were observed for recipient isolates (Fig. 3). For example, IFNα2- and IFNβ-selected plasma isolates from donor CH492 had mean IFNα2 and IFNβ IC50 values that were 7.6-fold and 31-fold higher than those of untreated plasma isolates (Fig. 3 A and C). However, resistance to one IFN subtype did not always predict resistance to the other. For donor CH492, IFNβ pretreatment generated isolates that were also highly resistant to IFNα2 (Fig. 3A), whereas IFNα2 pretreatment generated isolates with a wide range of IFNβ IC50 values, including some as low as untreated isolates (Fig. 3C). When results from all donors were combined, both IFNα2- and IFNβ-selected isolates were as resistant to IFNα2 as were untreated recipient isolates (Fig. 3B). In contrast, IFNα2-selected isolates were 7-fold less resistant to IFNβ than IFNβ-selected isolates, and these in turn were 2-fold less resistant than untreated recipient isolates (Fig. 3D). Similar results were obtained for Vres, which showed that IFNα2 selection did not generally yield IFNβ-resistant isolates, and that IFNβ-selected isolates were less resistant to IFNβ than untreated recipient isolates (SI Appendix, Fig. S6). Interestingly, IFN selection did not increase the IC50 or Vres values of recipient isolates, suggesting that transmitted viruses are already maximally resistant to both of these IFN subtypes (Fig. 3 B and D and SI Appendix, Fig. S6 B and D).
Fig. 3.
Biological characterization of IFNα2- and IFNβ-selected donor and recipient isolates. (A, C, E, G, and I) IFNα2 IC50 (picograms per milliliter) (A), IFNβ IC50 (picograms per milliliter) (C), viral Env content (mass ratio of gp120 and RT) (E), particle infectivity (RLU per picogram of RT) (G), and replicative capacity in CD4+ T cells (nanograms of p24 antigen per milliliter) (I) values are shown for limiting dilution-derived viral isolates from one representative matched donor (D-CH492) and recipient (R-CH427) pair. In each panel, untreated (dark green), IFNα2-selected (light green), and IFNβ-selected (yellow) isolates from the donor (D-492) are compared with untreated (red), IFNα2-selected (dark pink), and IFNβ-selected (light pink) isolates from the corresponding recipient R-CH427. Boxes show the interquartile range, a black bar within each box indicates the geometric mean, and whiskers span the range of the data, respectively. Asterisks indicate significant differences (determined by unpaired t test) between groups (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001). Because IFN selection did not alter the phenotype of recipient isolates, only statistical comparisons of donor isolates to untreated recipient isolates are shown. (B, D, F, H, and J) Hierarchical Bayesian regression models were used to estimate the population-wide fold change of IFNα2 IC50 (B), IFNβ IC50 (D), Env content (F), particle infectivity (H), and replicative capacity in CD4+ T cells (J) across all transmission pairs between untreated and IFNα2-selected donor isolates (blue), untreated and IFNβ-selected donor isolates (green), untreated and IFNα2-selected recipient isolates (gray), and untreated and IFNβ-selected recipient isolates (yellow). The fold change between untreated donor and recipient plasma isolates (red), as in Figs. 1 and 2, is also shown for comparison. A dashed vertical line marks a fold change of 1, indicating no effect. The estimated posterior probability distribution for each parameter is shown along with a table summarizing the expected fold change and the probability that the effect is <1 [or where indicated by an asterisk (*) the probability that the effect is >1].
Having generated IFNα2 or IFNβ preselected isolates, we next examined their biological properties. For donor CH492, IFNα2 and IFNβ pretreatment resulted in isolates that packaged 2.0- and 3.3-fold more Env than untreated isolates, respectively (Fig. 3E). However, no significant differences in Env content were detected between treated and untreated isolates when data from all subjects were combined (Fig. 3F). However, pretreatment with IFNα2 and IFNβ resulted in donor isolates that exhibited increased infectivity. This was observed for donor CH492 (Fig. 3G) as well as all donor isolates combined (Fig. 3H). IFNα2 and IFNβ pretreatment yielded plasma isolates that were on average 2- and 2.2-fold more infectious, respectively, than isolates obtained without selection, although neither pretreated group was as infectious as the recipient isolates. Interestingly, IFN pretreatment had no effect on the infectivity of recipient isolates (Fig. 3H).
Reasoning that IFN pretreatment may favor the outgrowth of viruses that replicated to higher titers, we compared the replicative capacity of IFN-selected and -unselected donor and recipient isolates. Indeed, pretreatment of CD4+ target cells with IFNβ resulted in donor isolates that replicated more efficiently than untreated viruses, both for CH492 (1.3-fold; Fig. 3I) and all donor isolates combined (1.3-fold; Fig. 3J). However, this was not observed when CD4+ T cells were pretreated with IFNα2. Surprisingly, IFNα2-selected isolates replicated significantly less well, both for donor CH492 (1.7-fold; Fig. 3I) and all donor isolates combined (2.1-fold; Fig. 3J). For each of the seven donors, IFNα2 treatment selected isolates whose replicative capacity was much reduced compared with untreated isolates despite higher infectivity and in some cases greater amounts of packaged Env (e.g., CH492). These data indicate that IFNα2 and IFNβ selection can have opposing effects on some viral properties, and that in contrast to previous suggestions (27), IFN resistance is not simply a consequence of a higher replicative fitness. As expected, IFNα2 and IFNβ selection did not increase the growth potential of recipient isolates (Fig. 3J). Taken together, these results indicate that both IFNα2- and IFNβ-resistant viruses are present, albeit at low levels, in the plasma of chronically infected individuals, and that in vitro treatment of CD4+ T cells with IFNβ, but not IFNα2, selects isolates that approach the phenotype of transmitted viruses (Fig. 3 and SI Appendix, Fig. S6).
Transmitted Viruses Are More Efficiently Released from Infected Cells.
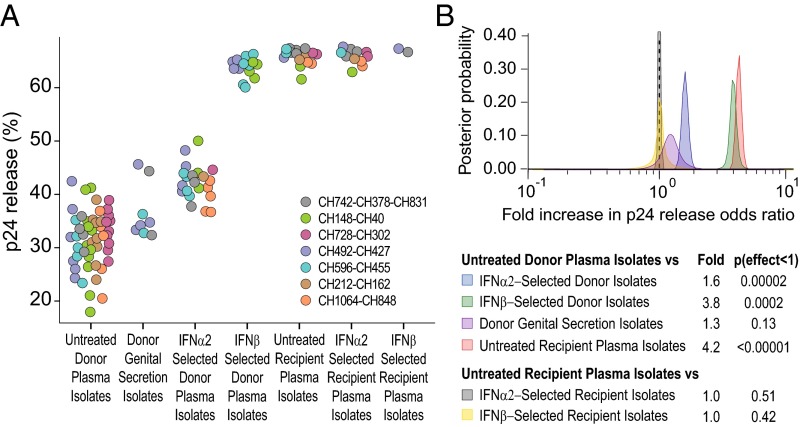
We previously reported that CD4+ T cells infected with TF viruses released larger quantities of cell-free virions than cultures infected with CC viruses (54). However, because only two TF and two CC IMCs were studied, we examined this property in a much larger number (n = 127) of matched donor and recipient isolates. To quantify particle release from infected CD4+ T cells, we measured the amounts of cell-free and cell-associated p24 antigen 7 d postinfection, and used these values to calculate the percentage of p24 that was released into the supernatant. Consistent with our previous observations (54), we found that donor isolates produced on average much less cell-free virus than recipient isolates (Fig. 4), although the total amount of p24 in these cultures was comparable. Plasma and genital secretion isolates from chronically infected donors released on average 31% and 38% of their total p24, respectively, whereas recipient isolates released 65%. In addition, IFN-selected isolates released more p24 than unselected donor isolates, although this effect was less pronounced for IFNα2 (42%) than for IFNβ (64%). Combining results from all isolates, the odds of p24 antigen being released from CD4+ T-cell cultures infected with IFNα2- and IFNβ-selected donor isolates were 1.6-fold and 3.8-fold higher, respectively, than from cultures infected with untreated donor isolates, and the odds of release were even higher (4.2-fold) for untreated recipient isolates (Fig. 4B). In contrast, no differences were observed for donor genital secretion isolates as well as for IFN-treated and untreated recipient isolates (Fig. 4B). Thus, mucosal transmission selects for viruses with a significantly enhanced particle release capacity, suggesting that the production of cell-free virions is important in the transmission process.
Fig. 4.
Particle release capacity of matched donor and recipient isolates. (A) Donor and recipient isolates were tested for their ability to be released from infected CD4+ T cells. The percent of viral release was determined as the ratio of cell-free p24 divided by the total amount (cell associated plus cell free) of p24 7 d postinfection. Only a subset of isolates (n = 132) was tested. Values are color coded by transmission pair. (B) A hierarchical Bayesian regression model was used to estimate the population-wide fold change in the odds of release (the probability of release divided by the probability of retention) of p24 between untreated and IFNα2-selected donor plasma isolates (blue), untreated and IFNβ-selected donor plasma isolates (green), untreated donor plasma and genital secretion isolates (purple), untreated donor and recipient plasma isolates (red), untreated and IFNα2-selected recipient isolates (gray), and untreated and IFNβ-selected recipient isolates (yellow). A dashed vertical line marks a fold change of 1, indicating no effect. The estimated posterior probability distribution for each parameter is shown along with a table summarizing the expected fold change and the probability that the effect is <1 (analogous to a one-sided P value).
Discussion
An effective AIDS vaccine will need to prevent acquisition of HIV-1 at mucosal surfaces (5). In this context, it is critical to know whether transmitted viruses possess unique biological properties that predispose them to establish new infections more efficiently. This is a controversial topic, because some studies have reported TF-specific traits (22, 24, 26, 52, 55–57), whereas others have failed to confirm these results (27, 28, 53, 58, 59). Some of these discrepancies are likely due to the fact that most previous analyses did not compare HIV-1 strains from transmission pairs. Using a more rigorous approach, two recent studies characterized viruses from matched donors and recipients, but failed to identify viral properties that were indicative of enhanced transmission fitness (27, 28). These findings led to the prevailing view that HIV-1 transmission is a stochastic process in which any reasonably fit virus has the potential of crossing the mucosa.
Both transmission pair studies characterized only very few donor and recipient viruses, using either infectious molecular clones (27) or peripheral blood mononuclear cell (PBMC)-derived bulk cultures (28). Reasoning that this approach had likely led to erroneous conclusions, we used limiting dilution isolation to generate a much larger number of donor and recipient viruses for phenotypic comparisons. We also used plasma rather than PBMCs for virus isolation to preclude the characterization of archived HIV-1 strains, generated genital secretion isolates for a subset of donors, and examined viral properties, such as virion release and resistance to IFNβ, which have not been previously characterized. Finally, we rendered the CD4+ T cells used for virus isolation more resistant to infection by treating them with high doses of type 1 IFNs to simulate host innate defenses that may be operative during the earliest stages of infection. We found that both recipient and in vitro IFN-selected donor isolates were more infectious, replicated to higher titers, were released from infected cells more efficiently, and were much more resistant to both IFNα2 and IFNβ than the great majority of unselected donor isolates (Figs. 1–4). Thus, it seems clear that these viral properties collectively contribute to transmission fitness.
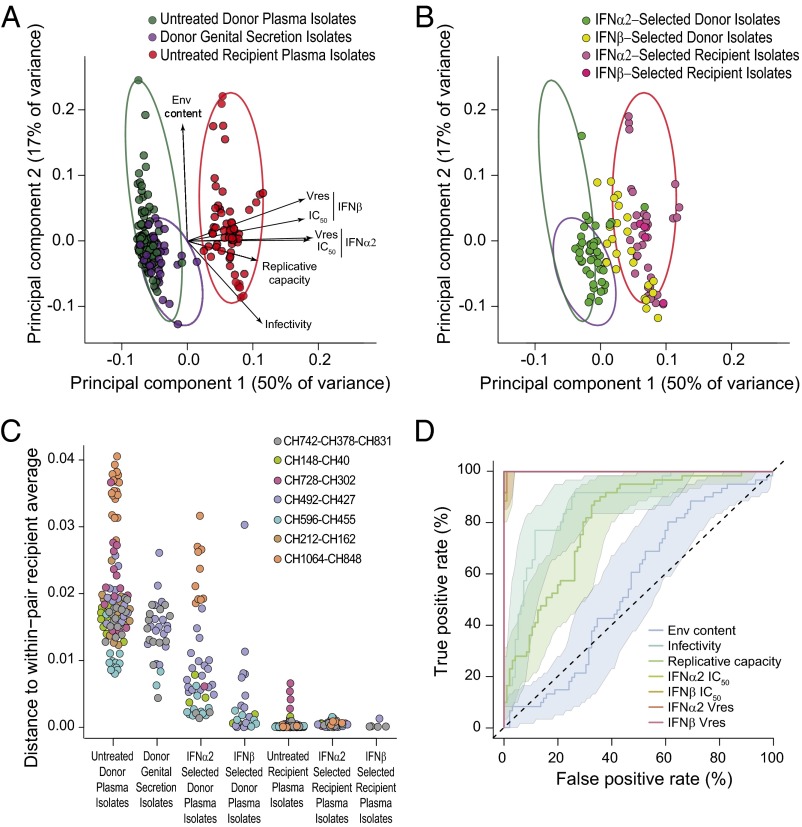
To visualize the biological properties examined for all virus isolates (particle Env content, infectivity, replicative capacity, IFN IC50, and Vres values) in combination, we conducted a principal component analysis (Fig. 5 A and B). This approach revealed two major groups, one that contained all plasma and genital secretion isolates from chronically infected donors, and another that included all plasma isolates from acutely infected recipients (Fig. 5A). The fact that there was no overlap between these groups indicates that transmitted viruses are phenotypically distinct. This conclusion was confirmed when IFN-treated isolates were plotted on the same principal components (Fig. 5B). Whereas most IFNα2-selected donor isolates grouped within the untreated donor cluster, most IFNβ-selected donor isolates overlapped the cluster of recipient viruses (Fig. 5B).
Fig. 5.
Phenotypic properties distinguishing donor and recipient isolates. (A) Principal component analysis was used to visualize properties that were determined for all viral isolates (Env content, particle infectivity, replicative capacity, IFNα2 IC50, IFNβ IC50, IFNα2 Vres, and IFNβ Vres) in combination. The positions of untreated donor plasma (green), donor genital secretion (purple), and recipient plasma (red) isolates are shown on the first two components. Length and direction of arrows show how each variable contributes to the two axes. The minimum spanning ellipses that contain all data points for each group are shown in corresponding colors. (B) To visualize the effect of IFN selection, IFNα2-selected (green), and IFNβ-selected (yellow) donor isolates are plotted together with IFNα2-selected (dark pink) and IFNβ-selected (light pink) recipient isolates on the same principal components as in A. Minimum spanning ellipses encompassing the untreated donor plasma isolates (green), donor genital secretion isolates (purple), and untreated recipient plasma isolates (red) as shown in A were retained. (C) To quantify the groupings apparent in A and B, we calculated the distance of the first two principal components for each isolate to the average position of the corresponding untreated recipient isolates for that transmission pair. Isolates are color coded by transmission pairs and grouped as in A and B. (D) The accuracy with which the seven viral properties predicted whether an isolate came from a donor or recipient was measured using receiver operating characteristic curves. Each line indicates the trade-off between true and false positive rates as a threshold is moved through the range of the data. Shading indicates the 95% confidence interval of the true positive rate. The dashed line indicates the expected performance of a predictor with no relationship to donor–recipient status. A line that reaches a true positive rate of 100% with a 0% false positive rate indicates that there is perfect separation between donor and recipient isolates.
To quantify these relationships, we calculated the distance between each virus and its pair-matched recipient average of the first two principal components (Fig. 5C). As expected, untreated and IFN-treated recipient isolates were the closest to the recipient average, exhibiting only minimal variation. In contrast, untreated donor plasma and genital secretion isolates as well as IFNα2-selected donor isolates were most distant from the average position of their respective recipient isolates and exhibited a wide distribution of distances. Interestingly, IFNβ-selected donor isolates were much closer to their recipient isolate average, consistent with IFNβ selection yielding a transmitted virus-like phenotype. We also examined the accuracy with which an isolate could be predicted to be derived from either a donor or a recipient on the basis of the seven biological properties examined (Fig. 5D). This analysis showed that IFN IC50 and Vres values predicted donor and recipient isolates with near 100% accuracy, indicating that resistance to type 1 IFNs is the most distinguishing characteristic of transmitted viruses.
If IFN resistance represents such a discerning feature, why did previous transmission pair studies miss this property? As shown in Fig. 2, chronic viruses exhibit a wide range of IFN IC50 values, indicating that random selection of just two such viruses per transmitting donor as reported by Deymier et al. (27) may not reveal donor/recipient differences. Moreover, measuring viral inhibition in response to a single IFN dose (26–28, 53) is likely less accurate than a formal IC50 determination. It should also be noted that the resistance of HIV-1 to IFNs is not constant during the course of infection. IFN resistance declines rapidly within the first 6 mo (52, 57), but then increases again when subjects progress toward AIDS (52, 60, 61). Thus, depending on when during the course of infection a virus is transmitted to another person, donor viruses may be more or less IFN resistant. For example, viruses from donors who transmit during acute HIV-1 infection or immediately following treatment interruption as described by Oberle et al. (28) would be expected to exhibit much higher levels of IFN resistance than viruses from subjects who transmit during asymptomatic chronic infection. In addition, PBMC cultures may reactivate latent viruses, which would be expected to exhibit IFN resistance levels consistent with their entry into the latent pool.
None of the previous transmission pair studies analyzed viral resistance to IFNβ, which produced the most pronounced donor/recipient differences. Indeed, the 39-fold higher IFNβ IC50 values of recipient isolates (Fig. 2F) is likely a gross underestimate, because many donor viruses were already more than 50% inhibited at the lowest IFNβ dose (Fig. 2B). This finding explains why the donor/recipient differences for IFNβ Vres values are so much higher than the corresponding IC50 values and why these differences are not observed for IFNα2 (Fig. 2F and SI Appendix, Fig. S5D). Whereas both IC50 and Vres values provide an indicator of IFN resistance, they seem to describe only partially overlapping biological effects. For example, the strong correlation of both IFNα2 and IFNβ IC50 and Vres values for donor plasma isolates likely indicates restriction by an IFN dose-driven increase in ISG activity. In contrast, the lack of a similarly strong correlation for donor genital secretion isolates suggests that some of these viruses are restricted by ISGs whose inhibitory activity is not IFN dose dependent. In addition, Vres may be a more relevant indicator of IFN resistance during the acute phase of infection when IFN levels are particularly high in the mucosa, whereas IC50 may be a more appropriate measure of systemic immune activation during later stages of infection. Future studies will need to determine the full range of IFNα2 and IFNβ IC50 and Vres values in HIV-1–infected subjects over time.
Not all viral properties studied contributed, or contributed equally, to HIV-1 transmission fitness. For example, virion-associated Env content, although previously identified as a characteristic feature of TF viruses (26), did not differentiate donor and recipient isolates (Figs. 1C and 5D). Nonetheless, in half of the transmission pairs studied, recipient isolates packaged significantly more Env than the respective donor viruses (Fig. 1B), suggesting that increased Env content may increase transmission fitness under certain circumstances. Similarly, particle infectivity and replicative capacity were significantly increased in most, but not all, recipient isolates. The successful transmission of viruses lacking these properties suggests that they are not absolutely required and/or that their absence can be compensated by other factors. In contrast, enhanced resistance to type I IFNs was observed for every single recipient isolate, indicating that the ability to counteract these innate immune responses is essential for successful mucosal transmission.
The need to overcome innate defenses is also exemplified by the fact that recipient and IFNβ-selected donor isolates produced much higher levels of cell-free virus than the corresponding untreated donor isolates (Fig. 4). Type 1 IFNs induce tetherin, which prevents the release of virus particles from the plasma membrane of infected cells. HIV-1 counteracts tetherin using its Vpu protein, which binds tetherin and prevents its expression on the cell surface (62–64). However, TF Vpu proteins do not seem to counteract tetherin more effectively than the Vpu protein of chronic viruses (65). Moreover, TF-infected CD4+ T cells were shown to produce more cell-free virions even in the absence of Vpu (54). Although we have not mapped the genomic region(s) responsible for the significantly enhanced virion release capacity of recipient isolates, it is unlikely that Vpu alone is responsible. In fact, several isolates that differed significantly in their particle release function encoded identical vpu genes (SI Appendix, Fig. S7). Thus, it seems clear that other as-yet-unknown factors must be responsible for the increased particle release function of recipient (and IFNβ-selected donor) isolates, which may be critical to enhance virus spread in the mucosa during the first rounds of replication when extracellular IFN levels are particularly high.
In summary, we have identified resistance to type 1 IFNs, in particular IFNβ, as a key determinant of HIV-1 transmission fitness. This observation is consistent with previous studies showing that innate responses in the mucosa immediately following infection are inducing a potent antiviral state through the up-regulation of ISGs, many of which have anti–HIV-1 activity (63, 64, 66–73). All IFN subtypes signal through the same heterodimeric receptor (30), but differences in receptor binding and/or downstream signal transduction pathways are thought to be responsible for IFN subtype-specific biological effects (74–77). IFNβ has been reported to bind the IFN receptor (IFNAR) with the highest affinity (76) and ligates the IFNAR1 chain in an IFNAR2-independent manner, resulting in the expression of a distinct set of genes (78). Either of these properties could explain its greater potency in placing selection on the transmitted virus pool. Nonetheless, IFNβ selection did not fully recapitulate the biological properties of recipient isolates despite the extremely high dose that was used to treat the target cells (Figs. 3D and 5 B and C and SI Appendix, Fig. S6D). These results indicate that additional factors, possibly including IFNα2 and/or other IFN subtypes, shape the transmitted founder phenotype. Because there are a total of 13 IFNα subtypes as well as other type 1 IFNs such as IFNω, some of which inhibit HIV-1 even more potently in vitro and in animal models (79–81), it will be critical to evaluate to what extent they contribute alone, or in combination, to the HIV-1 transmission bottleneck.
Materials and Methods
Study Subjects.
Transmission pairs were identified in the context of the Center for HIV/AIDS Vaccine Immunology (CHAVI-001) acute infection cohort (82). Plasma samples were obtained from eight acutely infected subjects (recipients) and their transmission partners (donors), with epidemiological linkage confirmed by viral sequence analysis (SI Appendix, Fig. S1). For a subset of donors, cervicovaginal lavage and semen samples were also available. Relevant epidemiological information is listed in SI Appendix, Table S1. Written informed consent was obtained from each subject and the study was approved by the institutional review boards of the University of Pennsylvania and Duke University.
Generation of Limiting Dilution HIV-1 Isolates.
Plasma samples as well as cell-depleted genital secretions were end-point diluted and used to infect 1 × 106 positively selected, activated CD4+ T cells (pooled from multiple donors) in 24-well plates. Cultures were maintained for 20 d, tested for p24 antigen (26), and virus positive wells were expanded in normal donor CD4+ T cells for an additional 10 d. The resulting (one time expanded) viral stocks were used for all genetic and biological analyses.
Isolate Sequencing.
Viral RNA was extracted from isolate stocks, reverse transcribed, and the resulting cDNA was used to amplify overlapping 5′ and 3′ genome halves in separate PCR reactions (14, 26). Amplicons were sequenced using an Illumina NGS platform, and paired-end reads were assembled to generate a sample-specific reference sequence. Viral reads were mapped to this reference, and the extent of genetic diversity was examined for each position along the alignment. Isolates that exhibited more than 15% diversity at any one position were judged to contain more than one variant and removed from further analysis.
Phylogenetic Analyses.
Nucleotide sequences were aligned using CLUSTALW v2 (83), with ambiguous regions removed. Maximum likelihood trees with bootstrap support (1,000 replicates) were constructed using PhyML v3.1 (84) with evolutionary models selected using jModelTest v2.1.4 (85) or for larger datasets, RAxML using a GTRGAMMA model (86). The gsi was used to calculate the degree of phylogenetic association of sequences (47).
Env Content and Particle Infectivity.
Particle-associated Env content was measured as described (26), with minor modifications (SI Appendix). Virion infectivity was measured using the TZM-bl reporter cell line as described (26), except for adding the fusion inhibitor T1249 (0.01 mg/mL) to prevent multiple rounds of infection.
Replicative Capacity and IFN Resistance.
To determine the IFNα2 and IFNβ concentrations required to inhibit virus replication by 50% (IC50), activated CD4+ T cells were left untreated or cultured in the presence of increasing amounts of IFNα2 (0.00074 pg/mL–5.5 pg/mL) or IFNβ (0.000067 pg/mL–0.44 pg/mL) for 24 h, infected with equal amounts of virus (1 ng of RT), and cultured for 7 d, while replenishing IFN-containing medium every 48 h. Virus replication was measured as the amount of p24 produced at day 7 and plotted for each IFN concentration as the percentage of viral growth in the absence of IFN. Residual virus replication (Vres) was measured at the highest IFNα and IFNβ concentrations and expressed as the percentage of replication in the absence of IFN. Replication in the untreated cells was used to determine the replicative capacity of viral isolates. Experiments were conducted in pooled CD4+ T cells from multiple normal subjects (SI Appendix provides details).
Isolation of IFN-Resistant Viruses.
To generate IFN-resistant viruses, pooled CD4+ T cells were treated with high (but nontoxic) doses of IFNα2 (4.0 pg/mL) or IFNβ (44 ng/mL) for 24 h before the addition of end-point diluted donor and recipient plasma. Limiting dilution isolates were generated and maintained as described above.
Virion Release.
Pooled CD4+ T cells were infected with equal amounts of virus and maintained for 7 d. To quantify particle release, both cell-free and cell-associated amounts of p24 antigen levels were measured, and the percentage of p24 that was released into the supernatant was calculated.
Statistical Analyses.
For intrapair comparisons, P values were derived using Welch’s unequal variance t test to compare log-transformed values. Population-wide fold changes of viral properties were estimated using hierarchical Bayesian regression models (87), which accounted for (i) nested measurements within a transmission pair, (ii) multiple transmissions from a single donor, (iii) heteroscedasticity between groups, and (iv) censored data where measurements were less than a given value. These models assumed that observations were normally distributed, with mean and variance drawn from population-level distributions (SI Appendix, SI Materials and Methods). Posterior probability distributions were estimated using Markov chain Monte Carlo sampling, implemented in Stan v2.12.0 (88) and R v3.3.1 (89). The models were used to estimate the fold change of log-transformed functional data (or logit-transformed proportional data) between donor and recipient plasma isolates as well as donor plasma and genital secretion isolates, along with effects of HIV-1 subtype and IFNα2 or IFNβ selection. Fold change for viral properties was based on the estimated posterior mean, and probability values were calculated from the estimated cumulative posterior probability for a fold change of <1 (or >1 in the case of a posterior mean of <1) for the population-level parameters. Principal component and receiver operating characteristic analysis (90) were performed using R v3.3.1.
Supplementary Material
Acknowledgments
We thank Shivani Sethi and Michelle Krysztofiak for artwork and manuscript preparation and the University of Pennsylvania’s Center for AIDS Research Human Immunology, Flow Cytometry, and Viral and Molecular Core facilities for reagents and protocols. This work was supported by the National Institutes of Health (Grants R01 AI114266, R01 AI111789, and P30 AI45008), the Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery (Grant UM1 AI100645), and the BEAT-HIV Delaney Collaboratory (Grant UM1 AI 126620). S.S.I., S.S.-M., H.J.B., and R.M.R. were supported by Training Grants T32 AI007632 and T32 AI055400. P.B. is a Jenner Institute Investigator.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KY111920–KY112584 and KY364886). Biological data and analysis code are archived on Zenodo, https://zenodo.org (doi: 10.5281/zenodo.21645).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1620144114/-/DCSupplemental.
References
- 1.UNAIDS 2016. Global AIDS Update 2016. Available at www.unaids.org/sites/default/files/media_asset/global-AIDS-update-2016_en.pdf. Accessed December 21, 2016.
- 2.Shaw GM, Hunter E. HIV transmission. Cold Spring Harb Perspect Med. 2012;2(11):a006965. doi: 10.1101/cshperspect.a006965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Passmore JA, Jaspan HB, Masson L. Genital inflammation, immune activation and risk of sexual HIV acquisition. Curr Opin HIV AIDS. 2016;11(2):156–162. doi: 10.1097/COH.0000000000000232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burgener A, McGowan I, Klatt NR. HIV and mucosal barrier interactions: consequences for transmission and pathogenesis. Curr Opin Immunol. 2015;36:22–30. doi: 10.1016/j.coi.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 5.Haynes BF, et al. HIV-host interactions: Implications for vaccine design. Cell Host Microbe. 2016;19(3):292–303. doi: 10.1016/j.chom.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hladik F, McElrath MJ. Setting the stage: Host invasion by HIV. Nat Rev Immunol. 2008;8(6):447–457. doi: 10.1038/nri2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keele BF, Estes JD. Barriers to mucosal transmission of immunodeficiency viruses. Blood. 2011;118(4):839–846. doi: 10.1182/blood-2010-12-325860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joseph SB, Swanstrom R, Kashuba AD, Cohen MS. Bottlenecks in HIV-1 transmission: Insights from the study of founder viruses. Nat Rev Microbiol. 2015;13(7):414–425. doi: 10.1038/nrmicro3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keele BF, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci USA. 2008;105(21):7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boeras DI, et al. Role of donor genital tract HIV-1 diversity in the transmission bottleneck. Proc Natl Acad Sci USA. 2011;108(46):E1156–E1163. doi: 10.1073/pnas.1103764108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlson JM, et al. HIV transmission. Selection bias at the heterosexual HIV-1 transmission bottleneck. Science. 2014;345(6193):1254031. doi: 10.1126/science.1254031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bar KJ, et al. Wide variation in the multiplicity of HIV-1 infection among injection drug users. J Virol. 2010;84(12):6241–6247. doi: 10.1128/JVI.00077-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, et al. High multiplicity infection by HIV-1 in men who have sex with men. PLoS Pathog. 2010;6(5):e1000890. doi: 10.1371/journal.ppat.1000890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salazar-Gonzalez JF, et al. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J Exp Med. 2009;206(6):1273–1289. doi: 10.1084/jem.20090378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salazar-Gonzalez JF, et al. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J Virol. 2008;82(8):3952–3970. doi: 10.1128/JVI.02660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simmonds P, Balfe P, Ludlam CA, Bishop JO, Brown AJ. Analysis of sequence diversity in hypervariable regions of the external glycoprotein of human immunodeficiency virus type 1. J Virol. 1990;64(12):5840–5850. doi: 10.1128/jvi.64.12.5840-5850.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palmer S, et al. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J Clin Microbiol. 2005;43(1):406–413. doi: 10.1128/JCM.43.1.406-413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu SL, et al. HIV quasispecies and resampling. Science. 1996;273(5274):415–416. doi: 10.1126/science.273.5274.415. [DOI] [PubMed] [Google Scholar]
- 19.Abrahams MR, et al. CAPRISA Acute Infection Study Team; Center for HIV-AIDS Vaccine Immunology Consortium Quantitating the multiplicity of infection with human immunodeficiency virus type 1 subtype C reveals a non-poisson distribution of transmitted variants. J Virol. 2009;83(8):3556–3567. doi: 10.1128/JVI.02132-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haaland RE, et al. Inflammatory genital infections mitigate a severe genetic bottleneck in heterosexual transmission of subtype A and C HIV-1. PLoS Pathog. 2009;5(1):e1000274. doi: 10.1371/journal.ppat.1000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tully DC, et al. Differences in the selection bottleneck between modes of sexual transmission influence the genetic composition of the HIV-1 founder virus. PLoS Pathog. 2016;12(5):e1005619. doi: 10.1371/journal.ppat.1005619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilen CB, et al. Phenotypic and immunologic comparison of clade B transmitted/founder and chronic HIV-1 envelope glycoproteins. J Virol. 2011;85(17):8514–8527. doi: 10.1128/JVI.00736-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parrish NF, et al. Transmitted/founder and chronic subtype C HIV-1 use CD4 and CCR5 receptors with equal efficiency and are not inhibited by blocking the integrin α4β7. PLoS Pathog. 2012;8(5):e1002686. doi: 10.1371/journal.ppat.1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ping LH, et al. CAPRISA Acute Infection Study and the Center for HIV-AIDS Vaccine Immunology Consortium Comparison of viral Env proteins from acute and chronic infections with subtype C human immunodeficiency virus type 1 identifies differences in glycosylation and CCR5 utilization and suggests a new strategy for immunogen design. J Virol. 2013;87(13):7218–7233. doi: 10.1128/JVI.03577-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ochsenbauer C, et al. Generation of transmitted/founder HIV-1 infectious molecular clones and characterization of their replication capacity in CD4 T lymphocytes and monocyte-derived macrophages. J Virol. 2012;86(5):2715–2728. doi: 10.1128/JVI.06157-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parrish NF, et al. Phenotypic properties of transmitted founder HIV-1. Proc Natl Acad Sci USA. 2013;110(17):6626–6633. doi: 10.1073/pnas.1304288110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deymier MJ, et al. Heterosexual transmission of subtype C HIV-1 selects consensus-like variants without increased replicative capacity or interferon-α resistance. PLoS Pathog. 2015;11(9):e1005154. doi: 10.1371/journal.ppat.1005154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oberle CS, et al. Swiss HIV Cohort Study Tracing HIV-1 transmission: Envelope traits of HIV-1 transmitter and recipient pairs. Retrovirology. 2016;13(1):62. doi: 10.1186/s12977-016-0299-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altfeld M, Gale M., Jr Innate immunity against HIV-1 infection. Nat Immunol. 2015;16(6):554–562. doi: 10.1038/ni.3157. [DOI] [PubMed] [Google Scholar]
- 30.Stetson DB, Medzhitov R. Type I interferons in host defense. Immunity. 2006;25(3):373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 31.Schoggins JW. Interferon-stimulated genes: Roles in viral pathogenesis. Curr Opin Virol. 2014;6:40–46. doi: 10.1016/j.coviro.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doyle T, Goujon C, Malim MH. HIV-1 and interferons: Who’s interfering with whom? Nat Rev Microbiol. 2015;13(7):403–413. doi: 10.1038/nrmicro3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Towers GJ, Noursadeghi M. Interactions between HIV-1 and the cell-autonomous innate immune system. Cell Host Microbe. 2014;16(1):10–18. doi: 10.1016/j.chom.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sandler NG, et al. Type I interferon responses in rhesus macaques prevent SIV infection and slow disease progression. Nature. 2014;511(7511):601–605. doi: 10.1038/nature13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Veazey RS, et al. Prevention of SHIV transmission by topical IFN-β treatment. Mucosal Immunol. 2016;9(6):1528–1536. doi: 10.1038/mi.2015.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abel K, Rocke DM, Chohan B, Fritts L, Miller CJ. Temporal and anatomic relationship between virus replication and cytokine gene expression after vaginal simian immunodeficiency virus infection. J Virol. 2005;79(19):12164–12172. doi: 10.1128/JVI.79.19.12164-12172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stacey AR, et al. Induction of a striking systemic cytokine cascade prior to peak viremia in acute human immunodeficiency virus type 1 infection, in contrast to more modest and delayed responses in acute hepatitis B and C virus infections. J Virol. 2009;83(8):3719–3733. doi: 10.1128/JVI.01844-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frater AJ, et al. Effective T-cell responses select human immunodeficiency virus mutants and slow disease progression. J Virol. 2007;81(12):6742–6751. doi: 10.1128/JVI.00022-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crawford H, et al. Evolution of HLA-B*5703 HIV-1 escape mutations in HLA-B*5703-positive individuals and their transmission recipients. J Exp Med. 2009;206(4):909–921. doi: 10.1084/jem.20081984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ganusov VV, et al. Fitness costs and diversity of the cytotoxic T lymphocyte (CTL) response determine the rate of CTL escape during acute and chronic phases of HIV infection. J Virol. 2011;85(20):10518–10528. doi: 10.1128/JVI.00655-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bar KJ, et al. Early low-titer neutralizing antibodies impede HIV-1 replication and select for virus escape. PLoS Pathog. 2012;8(5):e1002721. doi: 10.1371/journal.ppat.1002721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song H, et al. Impact of immune escape mutations on HIV-1 fitness in the context of the cognate transmitted/founder genome. Retrovirology. 2012;9:89. doi: 10.1186/1742-4690-9-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sather DN, et al. Broadly neutralizing antibodies developed by an HIV-positive elite neutralizer exact a replication fitness cost on the contemporaneous virus. J Virol. 2012;86(23):12676–12685. doi: 10.1128/JVI.01893-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yue L, et al. Transmitted virus fitness and host T cell responses collectively define divergent infection outcomes in two HIV-1 recipients. PLoS Pathog. 2015;11(1):e1004565. doi: 10.1371/journal.ppat.1004565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lynch RM, et al. HIV-1 fitness cost associated with escape from the VRC01 class of CD4 binding site neutralizing antibodies. J Virol. 2015;89(8):4201–4213. doi: 10.1128/JVI.03608-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Politch JA, Marathe J, Anderson DJ. Characteristics and quantities of HIV host cells in human genital tract secretions. J Infect Dis. 2014;210(Suppl 3):S609–S615. doi: 10.1093/infdis/jiu390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cummings MP, Neel MC, Shaw KL. A genealogical approach to quantifying lineage divergence. Evolution. 2008;62(9):2411–2422. doi: 10.1111/j.1558-5646.2008.00442.x. [DOI] [PubMed] [Google Scholar]
- 48.Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72(4):2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wei X, et al. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother. 2002;46(6):1896–1905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eron JJ, et al. Short-term safety and antiretroviral activity of T-1249, a second-generation fusion inhibitor of HIV. J Infect Dis. 2004;189(6):1075–1083. doi: 10.1086/381707. [DOI] [PubMed] [Google Scholar]
- 51.Ribeiro RM, et al. Estimation of the initial viral growth rate and basic reproductive number during acute HIV-1 infection. J Virol. 2010;84(12):6096–6102. doi: 10.1128/JVI.00127-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fenton-May AE, et al. Relative resistance of HIV-1 founder viruses to control by interferon-alpha. Retrovirology. 2013;10:146. doi: 10.1186/1742-4690-10-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Song H, et al. Transmission of multiple HIV-1 subtype C transmitted/founder viruses into the same recipients was not determined by modest phenotypic differences. Sci Rep. 2016;6:38130. doi: 10.1038/srep38130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kmiec D, et al. Vpu-mediated counteraction of tetherin is a major determinant of HIV-1 interferon resistance. MBio. 2016;7(4):e00934-16. doi: 10.1128/mBio.00934-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gnanakaran S, et al. Recurrent signature patterns in HIV-1 B clade envelope glycoproteins associated with either early or chronic infections. PLoS Pathog. 2011;7(9):e1002209. doi: 10.1371/journal.ppat.1002209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parker ZF, et al. Transmitted/founder and chronic HIV-1 envelope proteins are distinguished by differential utilization of CCR5. J Virol. 2013;87(5):2401–2411. doi: 10.1128/JVI.02964-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Foster TL, et al. Resistance of transmitted founder HIV-1 to IFITM-mediated restriction. Cell Host Microbe. 2016;20(4):429–442. doi: 10.1016/j.chom.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Etemad B, et al. Characterization of HIV-1 envelopes in acutely and chronically infected injection drug users. Retrovirology. 2014;11:106. doi: 10.1186/s12977-014-0106-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Isaacman-Beck J, et al. Heterosexual transmission of human immunodeficiency virus type 1 subtype C: Macrophage tropism, alternative coreceptor use, and the molecular anatomy of CCR5 utilization. J Virol. 2009;83(16):8208–8220. doi: 10.1128/JVI.00296-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Künzi MS, Farzadegan H, Margolick JB, Vlahov D, Pitha PM. Identification of human immunodeficiency virus primary isolates resistant to interferon-alpha and correlation of prevalence to disease progression. J Infect Dis. 1995;171(4):822–828. doi: 10.1093/infdis/171.4.822. [DOI] [PubMed] [Google Scholar]
- 61.Edlin BR, et al. In-vitro resistance to zidovudine and alpha-interferon in HIV-1 isolates from patients: Correlations with treatment duration and response. Ann Intern Med. 1992;117(6):457–460. doi: 10.7326/0003-4819-117-6-457. [DOI] [PubMed] [Google Scholar]
- 62.Neil SJ, Sandrin V, Sundquist WI, Bieniasz PD. An interferon-alpha-induced tethering mechanism inhibits HIV-1 and Ebola virus particle release but is counteracted by the HIV-1 Vpu protein. Cell Host Microbe. 2007;2(3):193–203. doi: 10.1016/j.chom.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Van Damme N, et al. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe. 2008;3(4):245–252. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451(7177):425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 65.Jafari M, Guatelli J, Lewinski MK. Activities of transmitted/founder and chronic clade B HIV-1 Vpu and a C-terminal polymorphism specifically affecting virion release. J Virol. 2014;88(9):5062–5078. doi: 10.1128/JVI.03472-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418(6898):646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 67.Stremlau M, et al. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427(6977):848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 68.Ribeiro C, et al. 2016. Receptor usage dictates HIV-1 restriction by human TRIM5α in dendritic cell subsets. Nature 540(7633):448–452.
- 69.Laguette N, et al. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474(7353):654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hrecka K, et al. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature. 2011;474(7353):658–661. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Goujon C, et al. Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature. 2013;502(7472):559–562. doi: 10.1038/nature12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu Z, et al. The interferon-inducible MxB protein inhibits HIV-1 infection. Cell Host Microbe. 2013;14(4):398–410. doi: 10.1016/j.chom.2013.08.015. [DOI] [PubMed] [Google Scholar]
- 73.Li M, et al. Codon-usage-based inhibition of HIV protein synthesis by human schlafen 11. Nature. 2012;491(7422):125–128. doi: 10.1038/nature11433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lavoie TB, et al. Binding and activity of all human alpha interferon subtypes. Cytokine. 2011;56(2):282–289. doi: 10.1016/j.cyto.2011.07.019. [DOI] [PubMed] [Google Scholar]
- 75.Gibbert K, Schlaak JF, Yang D, Dittmer U. IFN-α subtypes: Distinct biological activities in anti-viral therapy. Br J Pharmacol. 2013;168(5):1048–1058. doi: 10.1111/bph.12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jaks E, Gavutis M, Uzé G, Martal J, Piehler J. Differential receptor subunit affinities of type I interferons govern differential signal activation. J Mol Biol. 2007;366(2):525–539. doi: 10.1016/j.jmb.2006.11.053. [DOI] [PubMed] [Google Scholar]
- 77.Schreiber G, Piehler J. The molecular basis for functional plasticity in type I interferon signaling. Trends Immunol. 2015;36(3):139–149. doi: 10.1016/j.it.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 78.de Weerd NA, et al. Structural basis of a unique interferon-β signaling axis mediated via the receptor IFNAR1. Nat Immunol. 2013;14(9):901–907. doi: 10.1038/ni.2667. [DOI] [PubMed] [Google Scholar]
- 79.Lavender KJ, et al. Interferon alpha subtype-specific suppression of HIV-1 infection in vivo. J Virol. 2016;90(13):6001–6013. doi: 10.1128/JVI.00451-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Harper MS, et al. Interferon-α subtypes in an ex vivo model of acute HIV-1 infection: Expression, potency and effector mechanisms. PLoS Pathog. 2015;11(11):e1005254. doi: 10.1371/journal.ppat.1005254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Künzi MS, Pitha PM. Role of interferon-stimulated gene ISG-15 in the interferon-omega-mediated inhibition of human immunodeficiency virus replication. J Interferon Cytokine Res. 1996;16(11):919–927. doi: 10.1089/jir.1996.16.919. [DOI] [PubMed] [Google Scholar]
- 82.Cope AB, et al. Ongoing HIV transmission and the HIV care continuum in North Carolina. PLoS One. 2015;10(6):e0127950. doi: 10.1371/journal.pone.0127950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 84.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 85.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: More models, new heuristics and parallel computing. Nat Methods. 2012;9(8):772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gelman A, Carlin J, Stern H, Rubin D. Bayesian Data Analysis. 2nd Ed Chapman & Hall/CRC; New York, NY: 2003. [Google Scholar]
- 88.Carpenter B, et al. Stan: A probabilistic programming language. J Stat Softw, in press. [DOI] [PMC free article] [PubMed]
- 89.R Development Core Team 2016. A language and environment for statistical computing (R Foundation for Statistical Computing, Vienna). Available at https://www.R-project.org/. Accessed December 21, 2016.
- 90.Robin X, et al. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.