Significance
The evolution of photosynthetic cyanobacteria under 24-h cycles of light and darkness selected for a robust circadian clock. Understanding how cyanobacteria integrate circadian clock signals with natural light–dark cycles to control metabolism is critical, because these organisms are central to global carbon cycling and hold promise for development of renewable energy. Here we assess how the circadian transcription factor regulator of phycobilisome association A (RpaA) influences metabolism as a cyanobacterium goes through a light-to-dark transition. The data show that RpaA plays a key role in maintaining metabolic stability during the night period. Additionally, RpaA is important in controlling redox balance, which in turn is very important for regulating metabolism at night.
Keywords: cyanobacteria, metabolism, circadian clock, metabolomics, diurnal
Abstract
Cyanobacteria evolved a robust circadian clock, which has a profound influence on fitness and metabolism under daily light–dark (LD) cycles. In the model cyanobacterium Synechococcus elongatus PCC 7942, a functional clock is not required for diurnal growth, but mutants defective for the response regulator that mediates transcriptional rhythms in the wild-type, regulator of phycobilisome association A (RpaA), cannot be cultured under LD conditions. We found that rpaA-null mutants are inviable after several hours in the dark and compared the metabolomes of wild-type and rpaA-null strains to identify the source of lethality. Here, we show that the wild-type metabolome is very stable throughout the night, and this stability is lost in the absence of RpaA. Additionally, an rpaA mutant accumulates excessive reactive oxygen species (ROS) during the day and is unable to clear it during the night. The rpaA-null metabolome indicates that these cells are reductant-starved in the dark, likely because enzymes of the primary nighttime NADPH-producing pathway are direct targets of RpaA. Because NADPH is required for processes that detoxify ROS, conditional LD lethality likely results from inability of the mutant to activate reductant-requiring pathways that detoxify ROS when photosynthesis is not active. We identified second-site mutations and growth conditions that suppress LD lethality in the mutant background that support these conclusions. These results provide a mechanistic explanation as to why rpaA-null mutants die in the dark, further connect the clock to metabolism under diurnal growth, and indicate that RpaA likely has important unidentified functions during the day.
Cyanobacteria are both key agents of global carbon and nitrogen cycles and promising platforms for renewable chemicals, fuels, and nutraceuticals (1–3). Understanding the control mechanisms that govern the flow of carbon and nitrogen through these organisms is crucial for predicting their behavior in natural environments as well as for improving engineering strategies. Although the basic pathways for carbon and nitrogen metabolism, and their regulation, are well understood in heterotrophic bacteria, cyanobacteria exhibit important deviations in these core metabolic pathways (4–7). Additionally, metabolic control mechanisms in cyanobacteria evolved to be compatible with photoautotrophic metabolism and the dramatic shifts that are imposed on those pathways by predictable daily light–dark (LD) cycles. Examples include enzymatic activity that responds to light-dependent cellular redox changes (8–11); the preference for NADPH, the reductant produced by the photochemical reactions, over NADH by many biosynthetic enzymes (12, 13); and a circadian clock that drives 24-h transcriptional rhythms in most genes (14–16).
A daily LD cycle presents a strong metabolic driver for the photosynthetic cyanobacteria, but a circadian clock also imposes daily cycles in transcription and redox regulatory systems (17, 18). Circadian measurements historically have been performed in constant light (LL) conditions to distinguish internal circadian regulation from that which is environmentally driven (16, 17). However, diurnal physiology in a natural environment must integrate the two sources of regulation. We recently showed that the circadian clock regulates carbon metabolism in Synechococcus elongatus PCC 7942 as cells transition from the dark into the light during diurnal growth (19). Specifically, in the morning the clock represses the activity of the conserved circadian transcriptional regulator regulator of phycobilisome association A (RpaA), which normally activates nighttime metabolic processes (19, 20). This action suppresses primary metabolic processes in the morning, allowing carbon to flow toward secondary metabolic processes when light energy is not limiting (19).
The circadian clock in S. elongatus comprises a core oscillator formed by the proteins KaiA, KaiB, and KaiC (16). The oscillator relays timing information to the SasA–RpaA two-component output pathway, in which RpaA is a transcription factor that binds 170 known downstream gene targets (20, 21). RpaA was first identified in another cyanobacterium, Synechocystis sp. strain PCC 6803, as an OmpR-type response regulator that influences the ratio of light energy transfer from light-harvesting phycobilisomes to photosystem I (PSI) vs. PSII (22). Thus, it is not surprising that RpaA affects core energy-producing pathways when cells are exposed to light. RpaA protein activity is directly controlled by the circadian oscillator, which represses the activity of RpaA in the late night/early morning, and subsequently relieves repression throughout the day such that RpaA reaches its peak activity at dusk (19, 20, 23). This temporal activity pattern, and the fact that RpaA directly binds and activates nighttime metabolism genes (20, 24), suggests that it also plays an important role in metabolic control at night. Although the transcriptional targets of RpaA have been identified, and it is clear that LD conditions are deleterious to rpaA-null mutants (25), the metabolic and physiological changes that attenuate growth under LD conditions have not been explored.
Under LD conditions, S. elongatus performs photosynthesis and carbon fixation during the day via the Calvin–Benson cycle, with excess fixed carbon stored as the branched chain glucose polymer glycogen (19). As cells enter a dark period, glycogen is rapidly degraded via the oxidative pentose phosphate pathway (OPPP), which serves as the primary source of energy and reducing power (NADPH) at night (26, 27). The OPPP shares many reactions with the Calvin–Benson cycle, and the transition from photosynthetic to oxidative metabolism occurs through both transcriptional and redox-regulated steps (28–30). Strict control of cellular redox via the NADPH/NADP+ ratio is a common and important mechanism across plant and cyanobacterial species (28). Additionally, RpaA transcriptionally activates genes that code for sugar catabolic and OPPP enzymes at the end of the day, before entering the dark, including glgP (glycogen phosphorylase), gap1 (glyceraldehyde-3-phosphate dehydrogenase 1), opcA (OxPP cycle protein A), and the OPPP rate-limiting enzyme zwf (glucose-6-phosphate dehydrogenase) (20). In rpaA mutants, glycogen degradation is strongly attenuated, which reflects an inability to activate these sugar catabolic pathways (19).
In this study, we investigated whether the LD growth defect in a rpaA-null mutant is attributable to specific misregulation of metabolism and physiology as S. elongatus transitions into darkness and over the night period. We initially addressed the viability of an rpaA-null mutant (hereafter ΔrpaA) over a 12-h dark period. Subsequently, we used untargeted metabolic profiling to investigate how loss of RpaA affects the abundance of primary metabolites at time points after cells enter the dark. Finally, we identified both second-site mutations and physiological growth conditions that suppress LD lethality in the ΔrpaA mutant and correlated these data with metabolomics, gene expression, and measurements of global oxidative stress. We present a model in which RpaA acts as a critical transcriptional activator of reductant-producing pathways and show that its activity is important to maintain strict metabolic stability at night. This work shows that, even in cyanobacteria that do not carry out obvious nighttime programs such as nitrogen fixation, carbon catabolism and reductant production at night are crucial for homeostasis. Thus, metabolic quiescence is not sustainable under diurnal growth conditions.
Results
Darkness Initiates Pigmentation Changes and Rapid Cell Death in the ΔrpaA Mutant.
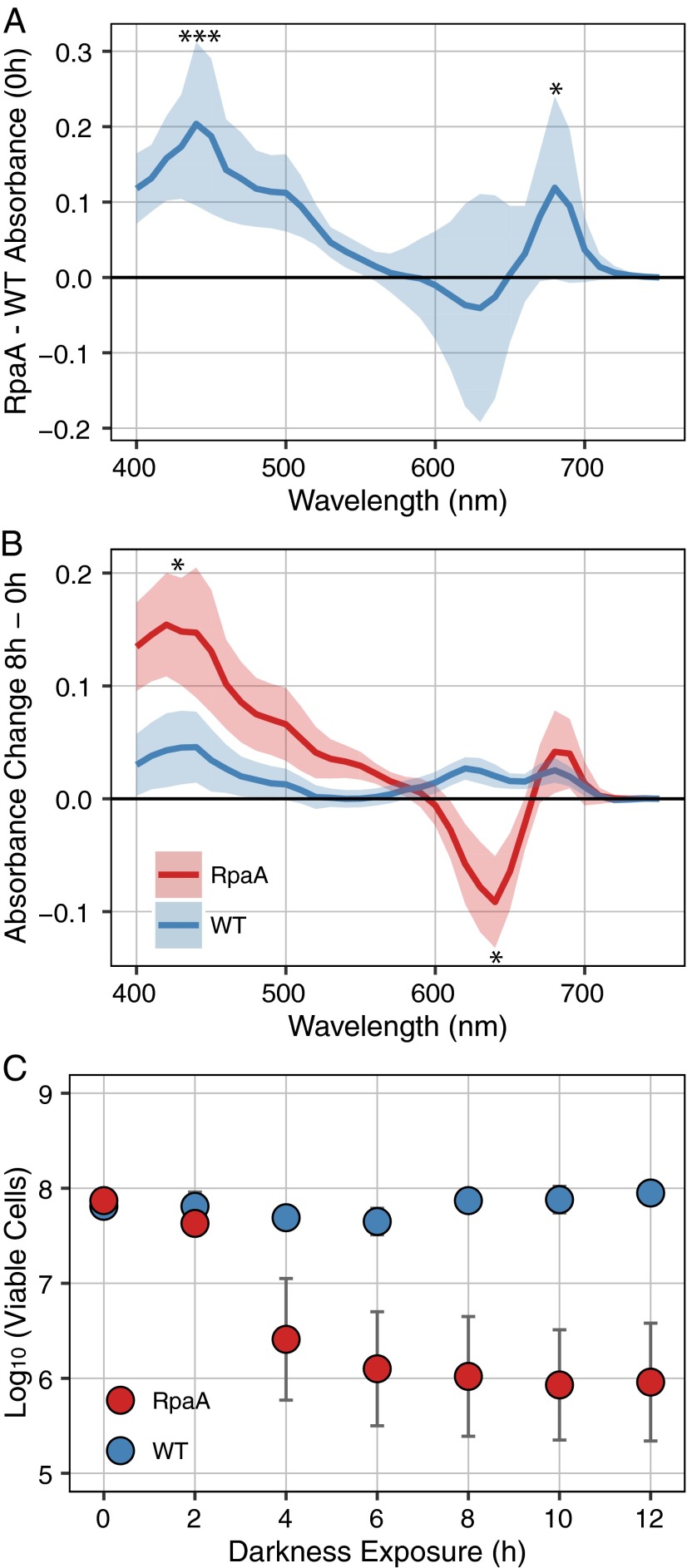
Although it is known that ΔrpaA strains do not grow under LD conditions (19, 25), the nature of the defect has not been characterized. We initially examined changes in cell viability and whole-cell absorbance as cultures entered the dark. WT and ΔrpaA cultures were sampled immediately before entry into the dark (0 h) and at intervals thereafter. Before dark exposure, the ΔrpaA strain had significantly elevated absorbance at 440 and 680 nm relative to WT (Fig. 1A), indicating an increase in chlorophyll absorbance. Although this study did not directly address changes in photosynthesis, a similar relative increase in chlorophyll was observed in Synechocystis sp. strain PCC 6803 ΔrpaA mutants (22). The broad differences in pigmentation at 0 h between WT and ΔrpaA (Fig. 1A) may indicate that the S. elongatus ΔrpaA mutant has altered energy-transfer kinetics between phycobilisomes, PSI, and PSII, as is true for Synechocystis sp. strain PCC 6803. Beginning 1–2 h after entering the dark and reaching a maximal change at 8 h after dark exposure, ΔrpaA had a significant decrease in absorbance at 630 nm and a further increase in absorbance at 440 nm, whereas WT showed no significant change in its absorbance spectrum (Fig. 1B and Fig. S1A). The decrease in absorbance at 630 nm indicates a loss of phycobilisome-specific pigmentation, which is a well-characterized response to stress and macronutrient deprivation (31).
Fig. 1.
Absorbance and viability data from WT and the ΔrpaA mutant. (A) Mean absorbance of WT subtracted from the ΔrpaA mutant at 0 h, immediately before entering darkness. Shaded area indicates SD of mean, significance of difference between WT and ΔrpaA calculated by Student’s t test (n = 8). *P < 0.05; ***P < 0.001. (B) Change in absorbance of WT and ΔrpaA from 0 h immediately before entering darkness to 8 h of dark exposure. Shaded area indicates SD of mean, significance of difference between 0 and 8 h for each strain calculated by Student’s t test (n = 8). *P < 0.05. (C) Mean viable cells counted at time points after WT and the ΔrpaA mutant entered the dark. Error bars indicate SEM. Significance was calculated by using Student’s t test (n = 4).
Fig. S1.
Summary of phenotypic effects of darkness on WT and the ΔrpaA mutant. (A) Representative photographs of WT and ΔrpaA before and after 8 h of dark exposure. Chlorotic bleaching is evident in the ΔrpaA strain after incubation in darkness (A, Lower). (B) Representative data collected from the photobioreactor optical density sensor (900 nm) over the course of an experiment where WT and the ΔrpaA mutant were exposed to darkness. Time is given from the inoculation of photobioreactors, and gray bars indicate 12-h periods of darkness. Turbidostatic growth of both cultures can be observed at ∼72–84 h from the start of the experiment. Both cultures show a decrease in optical density in the final dark period; however, the WT culture resumes growth in the following light period (B, Upper, black arrow), whereas the optical density of the ΔrpaA mutant continues to decrease despite being in a light period (B, Lower, black arrow).
In parallel, separate samples were removed and plated under LL conditions to assess viable cell counts (colony-forming units) at each time point. Samples taken immediately before dark exposure (0 h) showed similar numbers of viable cells in the two strains (Fig. 1C). After 4 h of dark exposure, a large reproducible decrease in cell number was evident for the ΔrpaA strain, with no corresponding decrease for WT (Fig. 1C); by 8 h of darkness, only ∼1% of ΔrpaA cells were viable (Fig. 1C). Optical density measurements of the sampled cultures also showed that the ΔrpaA strain did not resume growth during a following light period, even when transitioned back to LL growth conditions (Fig. S1B). These data indicate that ΔrpaA cells die soon after entering the dark. Together, the speed of the cell death response, the rapid changes in pigment absorbance, and an inability to regain viability in LL support an active mechanism that drives cell death in LD, as opposed to simple failure to thrive under LD conditions.
Temporal Metabolic Changes in the ΔrpaA Mutant.
Previous data showing attenuated glycogen degradation in the ΔrpaA mutant (19) and RpaA transcriptional regulation of carbon catabolic pathways (20), and our observation of active nutrient deprivation-like bleaching (Fig. 1B), suggest that broad changes in central carbon metabolism likely occur in ΔrpaA cells after a light-to-dark transition. To characterize metabolic changes, we applied untargeted gas chromatography time-of-flight mass spectrometry (GC-TOF-MS) to samples collected from photobioreactors under LD (12-h light:12-h dark) growth conditions. Samples were collected directly before the dark onset (0 h) and at 1, 2, 4, and 6 h thereafter. A total of 114 known compounds were identified and measured (Dataset S1). Dramatic differences in primary carbon metabolites and metabolites that require NADPH for their biosynthesis were found between WT and ΔrpaA samples, suggesting that RpaA-mediated reductant production via the OPPP at night is critically important to keep metabolite levels stable. Additionally, the detection of stress-associated metabolites before dark exposure suggests that RpaA also plays a role in mitigating cellular stress during the day.
Elevated polyamines in ΔrpaA before entering dark indicate stress.
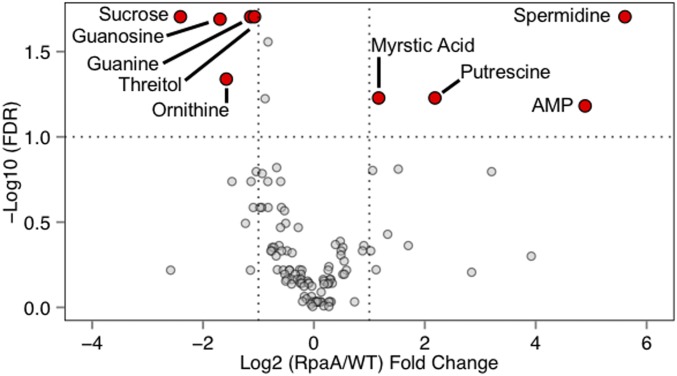
Before entering the dark (0 h), polyamines were highly elevated in ΔrpaA with spermidine and putrescine showing a 48.8- and 4.5-fold increase relative to WT, respectively (Fig. 2A and Fig. S2). Correspondingly, ornithine, which is a known precursor for polyamines (32), was one-third less abundant in ΔrpaA, suggesting mobilization of carbon toward polyamines. Accumulation of polyamines is a known general stress response in cyanobacteria (33), and the observed differences indicate that ΔrpaA cells may be stressed even before they enter the dark. Thus, in addition to nighttime functions, RpaA may have other, less understood, functions during the day.
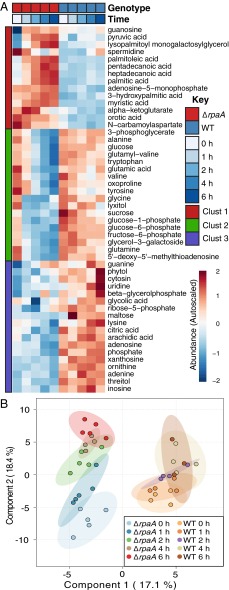
Fig. 2.
Summary of metabolic changes in WT and ΔrpaA. (A) Heatmap showing the autoscaled abundances of all metabolites where a significant difference was detected between WT and ΔrpaA over the time course as analyzed by two-way ANOVA and Tukey’s honest significant difference (n = 4 for WT; n = 5 for ΔrpaA; P < 0.05). Autoscaling represents a Z-score difference from the mean value of the metabolite across all time points. (B) Plot of PLS-DA components 1 and 2 for all metabolomics samples. Components 1 and 2 account for 35.5% of the variance in the dataset and are significant predictors of class membership (Materials and Methods). Ellipses indicate the 95% confidence interval (CI) for each sample grouping (n = 4 for WT; n = 5 for ΔrpaA; 114 metabolites per sample).
Fig. S2.
Comparison of metabolite abundance between WT and ΔrpaA before entering the dark. This volcano plot of metabolites highlights metabolites that show a significant difference in abundance between WT and ΔrpaA at 0 h (named red points) vs. those without a detectable difference (gray points). Dotted lines indicate required thresholds for significance. Metabolites on the right and left sides of the plot were elevated and decreased in ΔrpaA relative to WT, respectively. Significance was calculated using Student’s t test (n = 4 for WT and n = 5 for ΔrpaA), and correction for multiple testing used the method of Benjamini–Hochberg.
WT maintains strict metabolic stability at night, which is lost in ΔrpaA.
Over the first half of the dark period, a large number of additional metabolite differences rapidly formed between WT and the ΔrpaA mutant. Using two-way ANOVA, we identified 50 compounds with significant differences in abundance patterns over the time course (Fig. 2A and Dataset S1). We also applied a multivariate modeling method, partial least-squares discriminate analysis (PLS-DA), to visualize and statistically test the overall similarity of sample groups, as well as determine which metabolites were more associated with differences in genotype or time (Fig. 2B).
One of the most striking observations from both analysis methods was the overall stability of metabolite levels in WT relative to ΔrpaA (Fig. 2). We had expected that WT would show significant metabolic changes downstream of the OPPP, because cyanobacteria have significant flux through glycogen degradation and the OPPP at night (19, 34, 35). However, WT maintained a stable metabolic profile, whereas ΔrpaA exhibited broad metabolic changes. Metabolites in the first and second clusters of the heatmap (Fig. 2A) showed large increases and decreases, respectively, over time in ΔrpaA. In WT, the corresponding metabolites showed very gradual or no change in abundance (Fig. 2A). This effect was also pronounced in the plot of PLS-DA components 1 and 2 (Fig. 2B). Component 1 discriminated well between the two sample genotypes, and component 2 discriminated based on sampling time (Fig. 2B). The ΔrpaA mutant shows a clear separation across component 2, with temporally close samples more similar to each other than temporally distant samples (Fig. 2B). This pattern was absent in WT samples, indicating that these samples were globally similar over the time course (Fig. 2B). Thus, ANOVA and PLS-DA both indicated that WT cells maintain a high level of metabolic stability in the early night period, which is lost in ΔrpaA mutants.
Metabolic changes in ΔrpaA indicate OPPP depression and NADPH deficit.
Metabolites connected to the OPPP and those that require NADPH for biosynthesis rapidly decrease in ΔrpaA after dark exposure. The OPPP-connected compounds sucrose, glucose-1-phosphate (G1P), glucose-6-phosphate (G6P), and fructose-6-phosphate (F6P) all showed a precipitous drop in abundance as soon as ΔrpaA entered the dark (Figs. 2A and 3A). WT also showed decreases in these metabolites over time, which was expected as glycogen stores are used, but the decrease was much more gradual (Fig. 2A). However, WT, unlike ΔrpaA, showed an increase in F6P at the 1-h time point after dark onset. F6P is a known indicator of OPPP activation in cyanobacteria (28), and its stark decrease in ΔrpaA cells is consistent with highly attenuated OPPP activity (Fig. 2A).
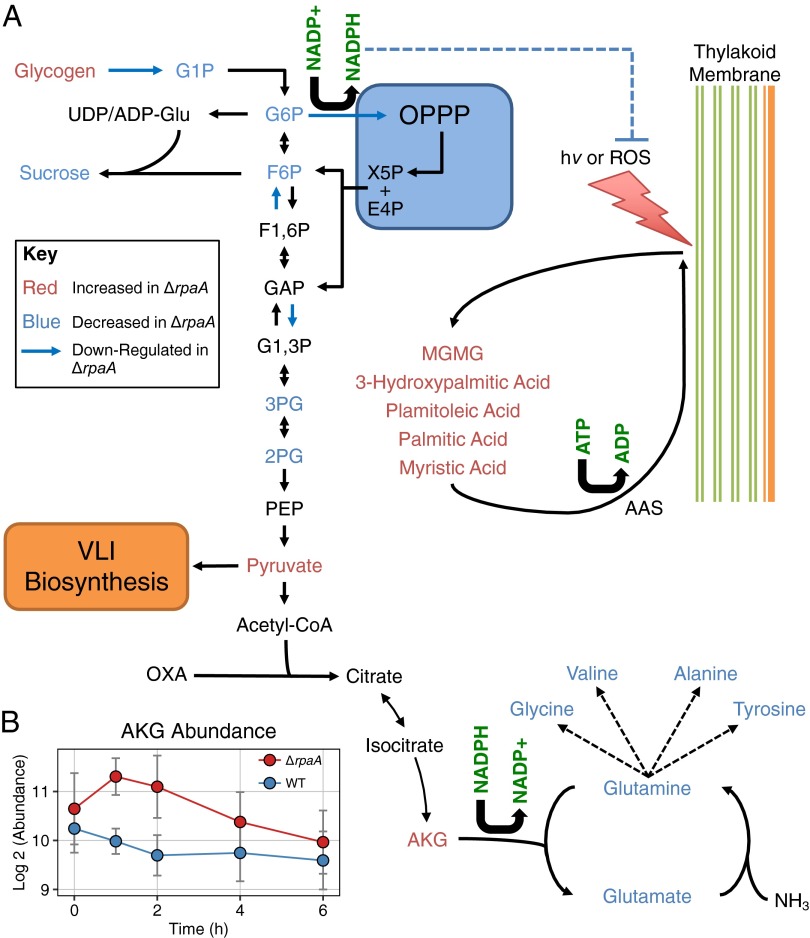
Fig. 3.
Metabolic changes in the context of central carbon and nitrogen metabolism. (A) Diagram of relevant reactions in central carbon and nitrogen metabolism. Also included is a model of lipid recycling from photosynthetic membranes (right side). Metabolites are colored based on whether they were elevated (red text) or decreased (blue text) in ΔrpaA relative to WT at some point during the 6-h time course. Data for RpaA gene regulation were taken from Markson et al. (20). Dotted lines indicate that metabolites are linked, but details are not displayed. (B) Plot of AKG abundance in ΔrpaA and WT across the metabolic time course. Error bars indicate SD (n = 4 for WT; n = 5 for ΔrpaA).
A primary sink of NADPH in cyanobacteria is amino acid biosynthesis, and many amino acids showed strong decreases in the ΔrpaA mutant (Fig. 2A). The primary nitrogen donors to amino acid synthesis, glutamine and glutamate, dropped in abundance rapidly when ΔrpaA cells entered the dark (Figs. 2A and 3A). In turn, ΔrpaA showed a corresponding increase in α-ketoglutarate (AKG), the precursor metabolite for nitrogen assimilation (Fig. 3B). The conversion of AKG into glutamate in cyanobacteria is catalyzed by ferredoxin-dependent enzymes that use NADPH exclusively in their oxidation/reduction cycle (12, 13). A decrease in amino acid pools with a corresponding increase in AKG is consistent with an NADPH deficit in ΔrpaA cells. Additionally, elevated AKG can activate a nitrogen-starvation transcriptional response in cyanobacteria (36, 37), and although these cells were not nitrogen-starved, we found that elevated AKG levels in ΔrpaA were accompanied by this transcriptional response (Fig. S3 and SI Text).
Fig. S3.
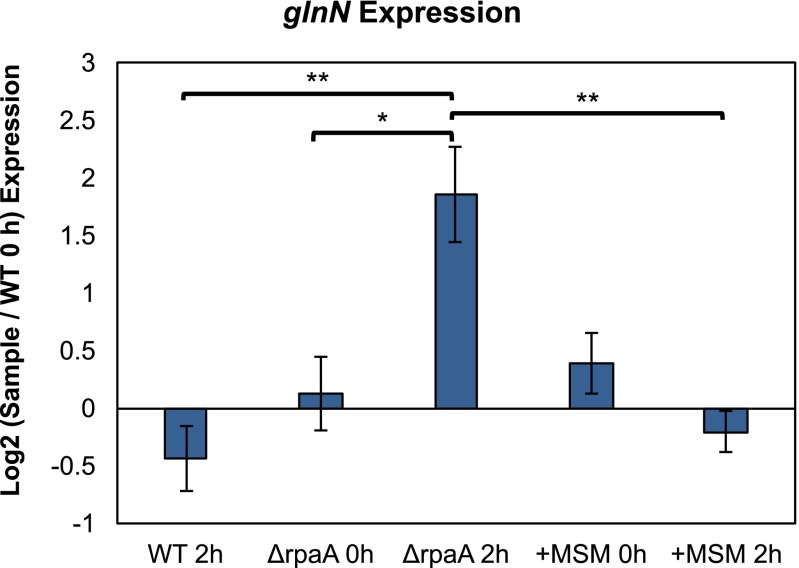
Changes in expression of glnN. Relative expression levels of the glnN transcript at 0 h (before entering darkness) and 2 h after exposure to darkness as measured by qRT-PCR. Log2 values were calculated relative to WT at 0 h. +MSM sample indicates ∆rpaA treated with 25 µM MSM for a 12-h period in the light before entering the dark. Error bars indicate SEM. Significance calculated using one-way ANOVA and Tukey’s HSD (n = 3). *P < 0.05; **P < 0.01. Additionally, there was no significant difference between ∆rpaA treated with MSM, a WT control treated with MSM, and WT untreated with MSM at any time point, indicating that transcript abundance of glnN was not generally affected by MSM treatment.
Fatty acid accumulation in ΔrpaA likely causes damage and may result from redox imbalance.
Intracellular accumulation of free fatty acids (FFAs) is uncommon in cyanobacteria, because they are activated and recycled into membrane lipids in an ATP-dependent reaction catalyzed by acyl-ACP synthetase (AAS) (38). However, in ΔrpaA, we observed a large increase in the abundance of palmitic acid, palmitoleic acid, and myristic acid at the 4- to 6-h time points after dark exposure (Figs. 2A and 3A). The accumulation of intracellular FFAs in S. elongatus is generally toxic and directly causes damage to photosystem complexes, potentially exacerbating redox stress (39, 40). We saw that the accumulation of FFAs in ΔrpaA temporally coincided with the observed decrease in ΔrpaA cell viability (Figs. 1C and 2A), which is consistent with accumulation of these compounds contributing to the LD lethality phenotype.
Fatty acid recycling in S. elongatus increases dramatically under high light, and functional AAS is important to maintain cell viability under these conditions (40). In the ΔrpaA mutant, the presence of elevated lysopalmitoyl monogalactosylglycerol and 3-hydroxypalmiticacid indicated that active membrane remodeling and fatty acid recycling are taking place (Figs. 2A and 3A). Although the mechanism that drives membrane remodeling and subsequent lipid accumulation in cyanobacteria is still unclear, de novo synthesis of lipids would be unlikely in ΔrpaA cells that are reductant-poor. We hypothesize that regulation of membrane lipid turnover and FFA accumulation may be responsive to a change in cellular redox state. This hypothesis is consistent with membrane remodeling occurring under both high light (40) and in ΔrpaA cells that lack sufficient reductant (NADPH) to control cellular redox state in the dark. We posit that, although both WT and ΔrpaA may respond to oxidative stress by activation of lipid recycling, only the mutant reaches a triggering threshold of activation under moderate-light growth and does so during the night period, when AAS activity may be limited by ATP availability.
Interventions That Suppress the ΔrpaA LD Lethality Phenotype Support Reductant Imbalance as a Cause of Cell Death.
Suppression of ΔrpaA LD lethality by second-site mutagenesis.
Older cultures of ΔrpaA mutants accumulate cells with the ability to grow under normally restrictive LD conditions (Fig. S4A). These clones still maintain fully segregated deletions at the rpaA locus; thus, it was surmised that they have accumulated compensatory changes at secondary genetic loci. To investigate the types of mutations that could suppress ΔrpaA LD lethality, we mutagenized freshly constructed, and still LD-sensitive, ΔrpaA mutant cells with ethyl methanesulfonate (EMS). Both EMS-exposed and unexposed ΔrpaA samples were then incubated under a restrictive LD growth condition. Hundreds of colonies with a wide degree of coloration and morphology appeared exclusively on the plate containing EMS-exposed ΔrpaA cells (Fig. S4B). We isolated 20 colonies, confirmed that all maintained fully segregated deletions at the rpaA locus (Fig. S4C), and performed full genome resequencing on each. Comparison of the mutagenized genomes to both a WT control and the ΔrpaA parent strain revealed a total of 63 single nucleotide changes across all strains with an average of 3.15 ± 1.2 new mutations per strain. Subsequently, we filtered the mutations (Materials and Methods) and identified a subset of 56 that we categorized as “high confidence for biological effect” (Dataset S2).
Fig. S4.
Supporting data for EMS mutagenesis of the ΔrpaA mutant. (A) A representative photo of an older ΔrpaA culture that was plated as a serial dilution and grown in an LD cycle. Black arrows indicate ΔrpaA colonies showing robust growth even under normally restrictive LD conditions. (B) Photographs of plates of unmutagenized and EMS-mutagenized ΔrpaA cultures after incubation in an LD cycle for 15 d. The photographs show that hundreds of colonies form on the EMS-mutagenized ΔrpaA plate (B, Right), but not on the plate that received unmutagenized cells (B, Left). (C) PCR amplification of the rpaA locus from ΔrpaA cells carrying second-site mutations and from WT controls. The recombination at the rpaA locus with the pAM4420 vector to produce the ΔrpaA strain results in an expected amplified fragment of 1.8 kb. This region is larger than the 1.2-kb amplicon expected from WT cells. Comparison with amplification from WT cells (white arrows) shows that all ΔrpaA strains tested in this study produce the expected increased amplicon size for a strain that carries the ΔrpaA mutation. Additionally, no WT-size bands are present in the ΔrpaA strains, indicating that the mutation is fully segregated.
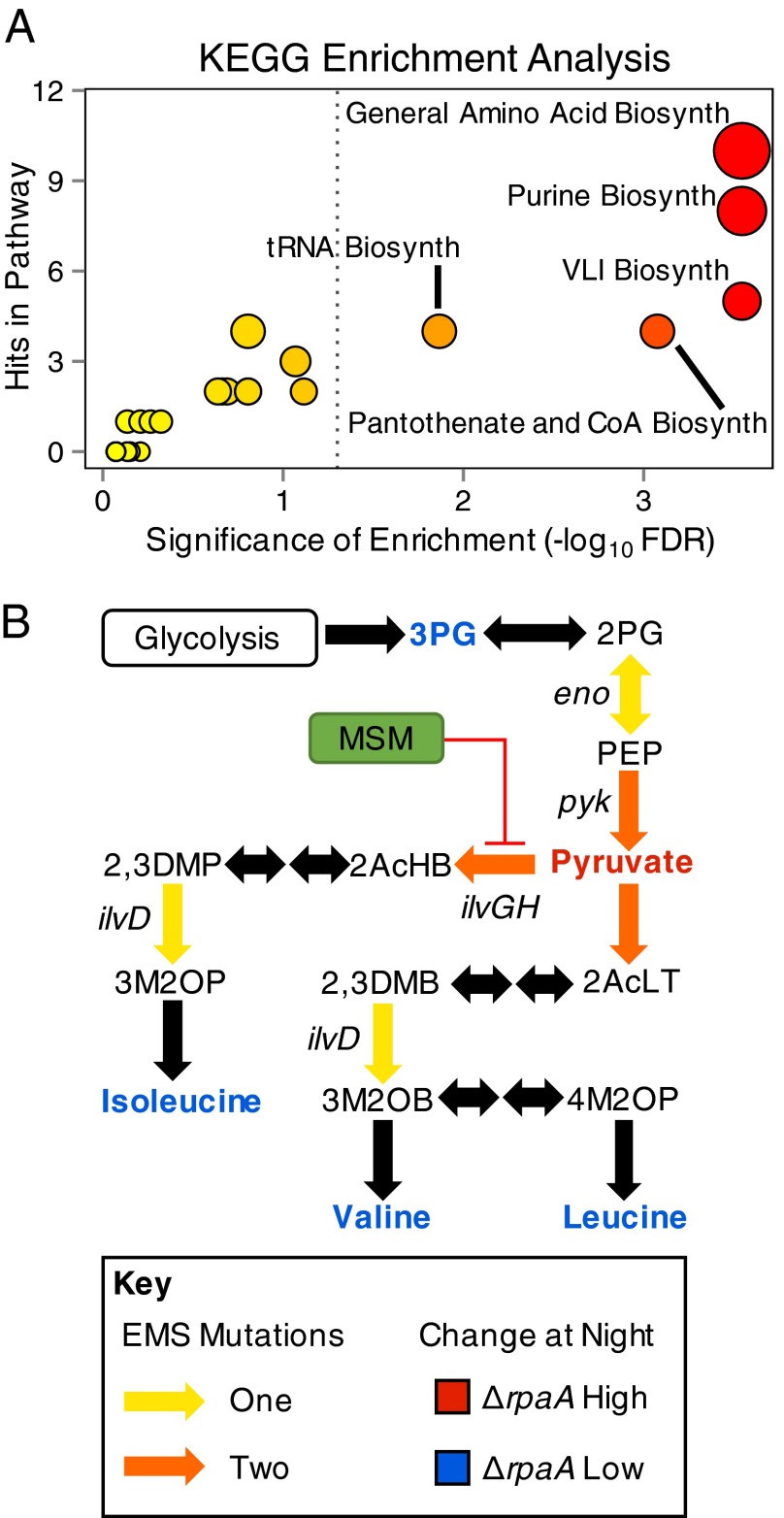
Of the mutations, 47% occurred in genes that code for metabolic enzymes (Dataset S2). Pathways that synthesize and use amino acids were significantly enriched in these mutations, including valine, leucine, and isoleucine (VLI) biosynthesis [false discovery rate (FDR) = 2.8e-4]; aminoacyl–tRNA biosynthesis (FDR = 0.01); and global amino acid biosynthesis (FDR = 2.8e-4) (Fig. 4A and Dataset S2). Additionally, multiple independent strains carried mutations affecting the same biochemical step in some of these pathways (Fig. 4B). These mutations affecting the same biochemical step included independent point mutations in ilvG and ilvH (acetolactate synthase catalytic and regulatory subunits), the rate-limiting enzyme complex in VLI biosynthesis (41), and two in pyk (pyruvate kinase), which produces pyruvate, the substrate of ilvGH (Fig. 4B) (42). Although mutations were found that could affect a number of metabolic pathways (Dataset S2), the high concentration of mutations in pathways that produce and consume amino acids was interesting, considering the large decreases observed for these compounds in ΔrpaA at night (Figs. 2A and 4B).
Fig. 4.
Summary of enriched KEGG functional categories identified by suppressor mutations and metabolic pathway topology represented by mutated genes. (A) Plot of KEGG metabolic categories that were enriched in the gene set of suppressor mutations. The x axis indicates the number of times a specific KEGG pathway was matched to genes in the set; dots scale from small to large with increasing number of matches, and color of dots scales from yellow to red with increasing significance. Significance was calculated by using the binomial distribution, corrected for multiple testing using the method of Benjamini–Hochberg, and significance cutoff is indicated with a gray dotted line (FDR < 0.05). (B) Subpathway diagram of VLI biosynthesis indicating locations of ΔrpaA suppressor mutations and average abundance of compounds in the ΔrpaA strain relative to WT over the metabolomics time course. Genes were named for reactions where a suppressing mutation was identified, and colors are detailed in the key.
Blocking VLI biosynthesis suppresses LD lethality in ΔrpaA.
Because each suppressed ΔrpaA strain carries multiple EMS generated mutations, we targeted the VLI biosynthetic pathway to test whether manipulation of a single pathway identified through EMS mutagenesis is sufficient for the suppression of LD lethality in ΔrpaA. VLI biosynthesis was chosen because a known herbicide, metsulfuron methyl (MSM), specifically and potently inhibits IlvGH (6, 43), and two suppressor mutations mapped to genes coding for this complex (Fig. 4B). Suppression of LD sensitivity with MSM, if successful, would also imply that the point mutations identified have a negative impact on complex activity.
Treatment of ΔrpaA with MSM under LD growth suppressed cell death of ΔrpaA on solid (100 nM MSM) and in liquid (25 μM MSM) media (Fig. 5A and Fig. S5A). Additionally, although all MSM-treated cultures showed a change in pigmentation (Fig. 5A), we confirmed that when ΔrpaA entered the dark, treated cells no longer showed phycobilisome-specific pigment bleaching or the activation of a nitrogen-deprivation transcriptional response (Fig. 5B and Fig. S3). Therefore, inhibition of the VLI biosynthetic pathway is sufficient to rescue ΔrpaA cells from LD lethality, as well as inhibit specific phenotypes associated with cell death in the dark. Additionally, the fact that MSM exerts its suppressive effect by ilvGH inhibition suggests that the point mutations identified in ilvG and ilvH reduce native enzyme complex activity.
Fig. 5.
Summary of data on MSM- and light intensity-mediated suppression of the ΔrpaA LD lethality phenotype. (A) Representative photo of dilution series of WT and ΔrpaA cells treated (Left) and not treated (Right) with 100 nM MSM (n = 6 biological replications of experiment). Pictured samples were grown in an LD cycle with a light intensity of 120 μE⋅m−2⋅s−1. (B) Difference in absorbance of ΔrpaA cells treated with 25 μM MSM between 0 and 8 h after dark exposure. Shaded region indicates SD of mean, and black arrow points to absorbance at 630 nm highlighting no significant change. Significance was calculated by using Student’s t test for absorbance at 440, 630, and 680 nm, with no significant change observed (n = 3). (C) Mean absorbance values of WT and ΔrpaA untreated (solid line) and treated (dotted line) with 25 μM MSM after 12 h in the light. Shaded area indicates SEM. MSM-exposed cells show significantly lower absorption values at 440, 630, and 680 nm, as calculated by a one-sided Student’s t test (n = 3 for MSM treated samples and n = 4 for untreated samples). *P < 0.05. (D) Representative photo of dilution series of WT and ΔrpaA cells grown in an LD cycle with decreasing daytime light intensity (indicated below each image; n = 2).
Fig. S5.
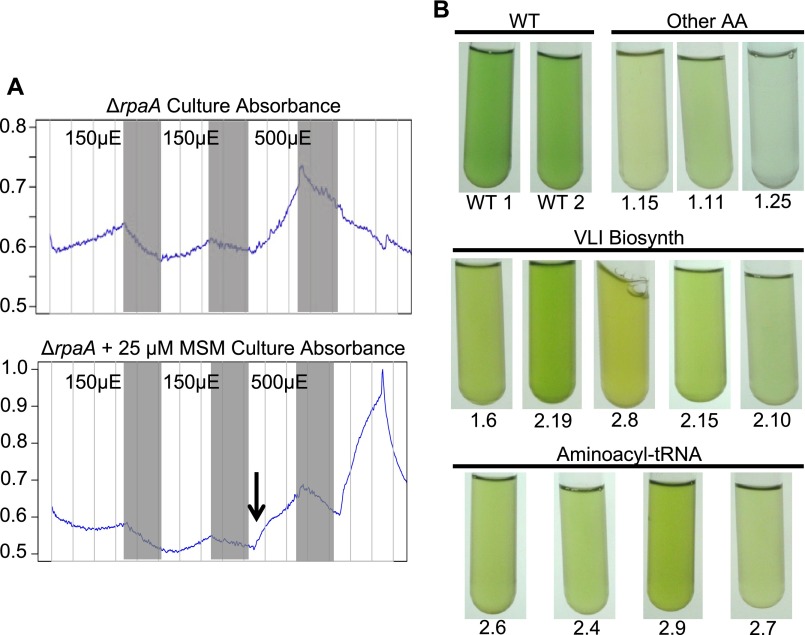
Supporting data for potential mechanisms that suppress LD lethality in the ΔrpaA mutant. (A) Representative data collected from the photobioreactor optical density sensor (900 nm; y axis) over the course of an experiment where ΔrpaA mutants were exposed to darkness and not treated (Upper) or treated (Lower) with MSM. Time (x axis) starts at inoculation of photobioreactors, gray bars indicate 12-h periods of darkness, and light intensity during the light periods is noted with black text. The black arrow in the bottom panel indicates when 25 μM MSM was added to one culture. The ΔrpaA mutant receiving MSM was able to continue growth even after an LD cycle (Lower) that was lethal to the ΔrpaA mutant not receiving MSM (Upper). (B) Photographs of EMS-mutagenized ΔrpaA strains before genomic DNA extraction. Strains are organized by the type of mutation they were found to carry, and only strains with mutations that affected amino acid metabolism in some way are included. The general metabolic pathway affected is indicated above each photograph panel and the specific EMS mutant number is indicated below each test tube (Dataset S2). The photos highlight the altered pigmentation that is present in these strains relative to WT cells.
Suppression of VLI biosynthesis lowers phycobilisome content during the daytime.
ΔrpaA strains treated with MSM and those that carry VLI pathway mutations had a strong yellow color (Fig. 5A and Fig. S5B). Absorbance scans taken before genomic DNA extraction for sequencing revealed that these mutants had a decreased phycobilisome-to-chlorophyll ratio (630/680 nm) relative to WT (Table S1). To investigate whether pigmentation changes occur during repression of VLI biosynthesis, we incubated WT and ΔrpaA with 25 μM MSM in the light for 12 h and compared whole-cell absorbance spectra of treated and untreated cells. Cultures exposed to MSM visibly appeared more yellow. WT treated with MSM had decreases in all three pigment absorption maxima: 440, 630, and 680 nm, with the phycobilisome peak at 630 nm showing the largest difference (Fig. 5C). MSM-treated ΔrpaA samples also had a significant reduction in absorbance at the 630-nm phycobilisome absorbance peak (Fig. 5C). These results show that treatment with MSM reduces phycobilisome content in both WT and ΔrpaA. Because phycobilisomes are the primary light-collecting pigment proteins in S. elongatus, the MSM-mediated decrease at 630 nm represents a significant change in the ability of cells to collect light and likely alters both photosynthetic output and cellular redox state (44, 45).
Table S1.
Absorbance ratio (630/680 nm) of select ΔrpaA suppressor mutants
| EMS mutant | Gene affected* | Color | Ratio | Significant† |
| WT controls | No | |||
| WT_1 | None | Green | 0.939 | |
| WT_2 | None | Green | 0.944 | |
| WT_3 | None | Green | 0.970 | |
| Purine biosynthesis | No | |||
| 1_9 | guaA (0189) | Light green | 0.914 | |
| 1_10 | guaA (0189) | Light green | 1.004 | |
| 1_3 | guaB (1831) | Very light green | 1.056 | |
| 1_1 | guaB (1831) | Light green | 0.979 | |
| 2_3 | guaA (0189) | Yellow green | 0.929 | |
| BCAA biosynthesis | Yes (P < 0.01) | |||
| 1_6 | pyk (0098) | Yellow green | 0.847 | |
| 2_19 | AHAS (0139) | Yellow green | 0.853 | |
| 2_8 | ilvD (0626) | Strong yellow | 0.915 | |
| 2_10 | eno (0639) | Light green | 0.874 | |
| 2_15 | ilvH (2434) | Yellow green | 0.892 | |
| tRNA-related | Yes (P < 0.05) | |||
| 2_4 | ileS (2437) | Yellow green | 0.807 | |
| 2_6 | glyQ (2457) | Yellow green | 0.889 | |
| 2_7 | tRNA-Arg (R0011) | Pale yellow green | 0.890 | |
| 2_9 | leuS (1920) | Strong yellow green | 0.854 | |
| Other amino acid biosynthetic | No | |||
| 1_11 | trpB (2143) | Pale green | 0.931 | |
| 1_15 | thrA (2090) | Pale yellow | 0.861 | |
| 1_25 | hisD (1519) | Pale yellow/clear | 1.049 |
SynPCC7942_# of gene given in parentheses next to gene name.
Average of ratio from a functional gene group was different from WT average by Student’s t test.
Decreasing light intensity suppresses ΔrpaA LD lethality.
Given that reducing the light-collecting ability of cells is one effect of MSM treatment, we tested whether modulating growth light intensity has an effect on the ΔrpaA LD lethality phenotype. WT and an ΔrpaA mutant were serially diluted, plated, and incubated under three light intensities in LL and LD. At the highest light intensity tested (120 μE·M−2.s−1), we observed the expected ΔrpaA lethality phenotype under LD conditions (Fig. 5D). However, at an intermediate light intensity (33 μE·M−2.s−1), the ΔrpaA mutant had slightly improved growth in LD (Fig. 5D), and at the lowest light intensity (17 μE·M−2.s−1), ΔrpaA cells grew to almost WT levels (Fig. 5D). Thus, a low-light LD cycle creates a permissive condition where ΔrpaA cells can survive. These results show that light intensity during the day period is at least one factor that contributes to the ΔrpaA LD lethality phenotype. Thus, the reduction of phycobilisome pigment associated with MSM treatment may partially contribute to its mechanism for suppressing LD lethality via reducing absorbed light energy during the day.
Redox Stress Is Associated with Cell Death in the ΔrpaA Mutant.
Although ΔrpaA strains can tolerate high light intensity under LL conditions (Fig. S6A), we observed that ΔrpaA cells had high levels of metabolites, indicating cellular stress before entering the dark (Fig. 2A and Fig. S2). Additionally, the protective effects of MSM treatment (Fig. 5A) and decreased light intensity (Fig. 5D) suggest that photosynthetically generated reactive oxygen species (ROS) may act as a destructive agent that drives ΔrpaA LD lethality. S. elongatus maintains a strict cellular redox balance and controls ROS using multiple systems, including modulation of phycobilisome abundance, glutathione redox control, and enzymatic ROS scavenging (46, 47). Glutathione biosynthesis is particularly important for modulating redox state and counteracting ROS in cyanobacteria (46, 48, 49). Regeneration of reduced glutathione requires NADPH, and our data suggest that ΔrpaA cells are NADPH-limited at night (Figs. 2A and 3A). Additionally, metabolites of the glutathione biosynthetic pathway, including oxoproline, glycine, glutamate, and glutamyl–valine all showed large decreases in ΔrpaA after dark transition (Fig. 2A). To determine the influence of redox stress on ΔrpaA LD phenotypes, we tracked total ROS in WT and ΔrpaA over a 24-h LD cycle (at high light intensity) using the fluorescent marker 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA) (50, 51). Samples were taken every 2 h during the 12-h day period and every hour during the 12-h night period. Additionally, we assessed whether MSM addition impacted the ROS detected in WT and ΔrpaA cells over the 24-h LD cycle.
Fig. S6.
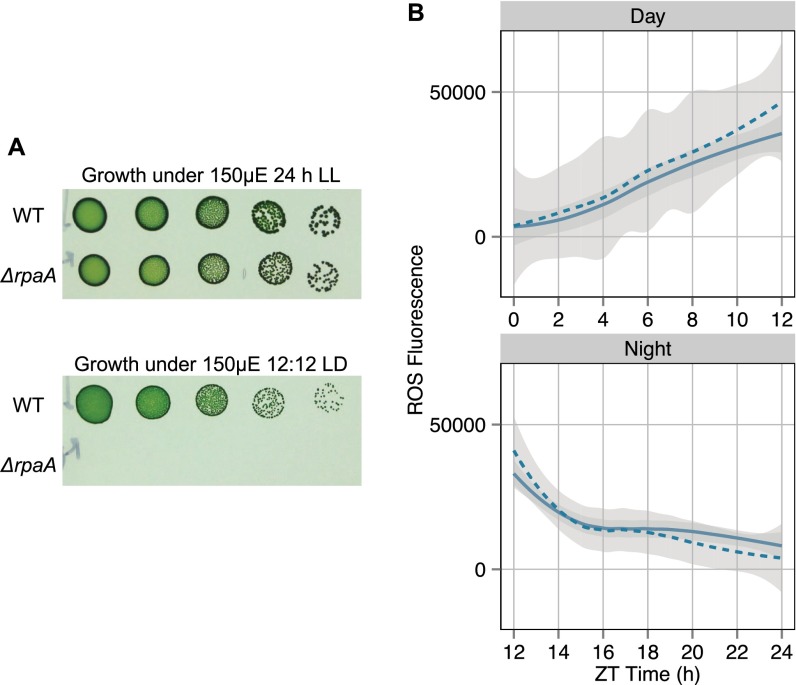
Controls for LD plating and ROS experiments. (A) Serial dilutions of WT and ΔrpaA grown at a light intensity of 150 μE·m−2·s−1 using 24-h LL (Upper) and 12 h:12 h LD (Lower) light regimes for 7 d. These data show the ability of ΔrpaA to tolerate high light conditions as long as a 24-h constant light regime is used. (B) Plot of H2DCFDA fluorescence over a 24-h LD cycle indicating total cellular ROS in WT untreated (solid line) and treated (dashed line) with 25 μM MSM. Curves are best-fit lines calculated by using LOESS regression to all data points in a given sample; the gray-shaded area indicates the 95% CI of the regression line (n = 21 data points for day samples; n = 42 data points for night samples). The data show that treatment of WT with 25 μM MSM does not significantly affect levels of ROS over the 24-h LD cycle.
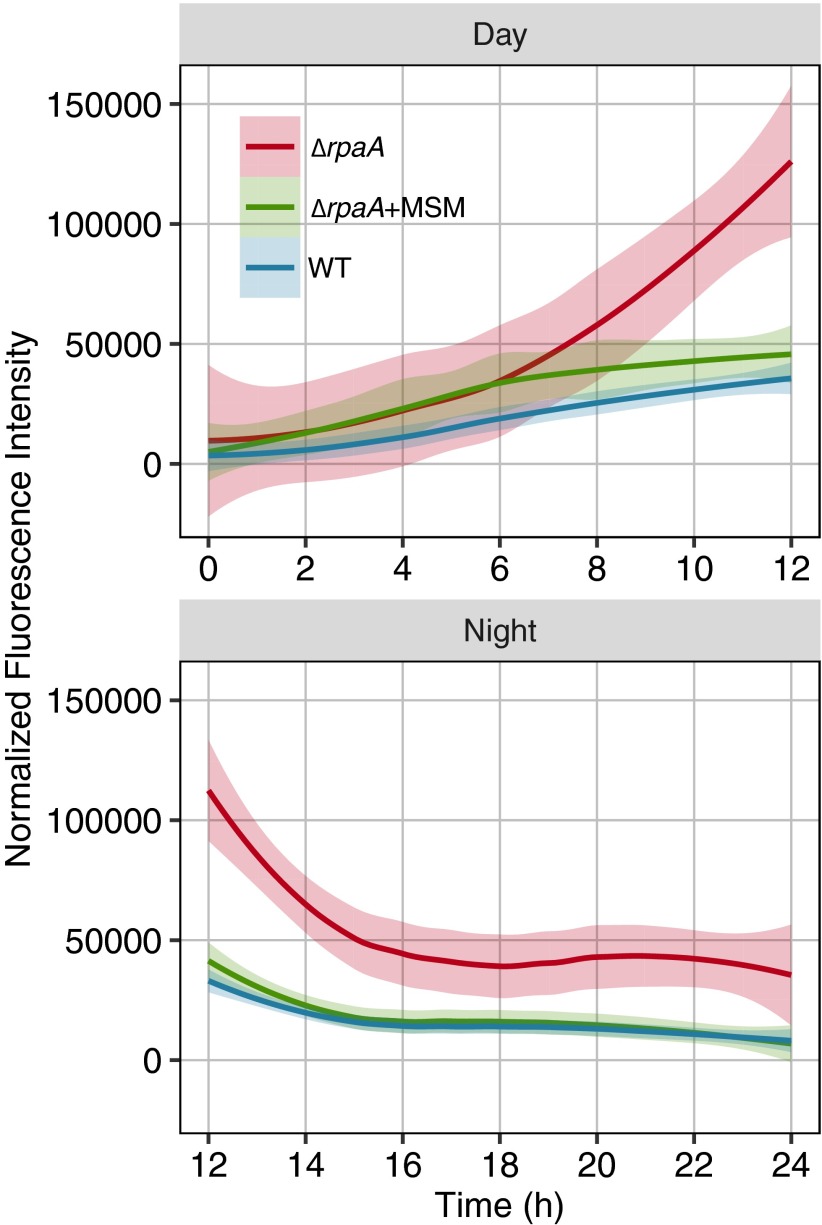
ROS levels were similar for all strains at the start of the experiment (Fig. 6). ROS increased gradually in all strains through the first 6 h of the day, then rapidly increased in the ΔrpaA mutant exclusively, and by the end of the light period (12 h), were 3.5-fold higher than in the WT or MSM-treated ΔrpaA cultures (Fig. 6). Upon entering the dark, all strains showed a rapid drop in ROS within the first 2 h (Fig. 6). ROS continued to drop in WT and the MSM-treated ΔrpaA mutant throughout the night, reaching a level similar to the start of the experiment. In contrast, the untreated ΔrpaA mutant maintained high static ROS levels after the first 2–3 h of darkness (Fig. 6). MSM treatment had no significant effect on WT ROS levels (Fig. S6B).
Fig. 6.
Plot of H2DCFDA fluorescence over a 24-h LD cycle indicating total cellular ROS in WT, ΔrpaA, and ΔrpaA treated with 25 μM MSM. Cells were grown at a light intensity in bioreactors empirically determined to support ΔrpaA growth in LD, and the experiment began after a 12-h dark period for all cells. Curves shown are best-fit lines calculated using LOESS regression to all data points in a given sample; the gray shaded area indicates the 95% CI of the regression line (n = 21 data points for day samples; n = 42 data points for night samples). Day and night from the same experiment were split to more effectively fit regressions. Places where the CI does not overlap indicate a statistically significant difference in the model.
Comparison of ROS levels with cell viability (Fig. 1C) showed that cell death begins in ΔrpaA cells around the time ROS levels stabilize. Overall, the ΔrpaA mutant accumulates high levels of ROS under the restrictive light condition and has trouble clearing ROS over the night period. However, treatment with MSM alleviates the elevated ROS phenotype. We propose that the rapid lethal effect of darkness on a ΔrpaA mutant results from a failure to clear ROS that accumulates late in the day period. This hypothesis is consistent with the observation that cell death begins in ΔrpaA cells when ROS levels stop decreasing at night and that the ΔrpaA metabolome indicates an NADPH deficit. Detoxification of ROS at night places an additional strain on the reductant pool, which likely exacerbates the metabolic imbalances we observe in this strain.
Discussion
This study highlights three functions of RpaA in WT cells that are critical for survival under LD growth conditions: (i) RpaA has daytime functions that are important for limiting ROS buildup; (ii) RpaA activates genes that encode the enzymes of the OPPP, which are critical for NADPH production in the absence of photosynthesis, and inability to generate sufficient reductant at night results in failure to detoxify ROS accumulated during the day and cell death; and (iii) overall, RpaA exerts a strong influence over the control of redox balance, which seems to be critical for maintaining the inherent metabolic stability of WT cells at night. These results are consistent with recent findings in Synechocystis PCC 6803 that show proper NADPH balance is important for appropriate diurnal regulation of metabolic processes (52), as well as data indicating that the fitness advantage conferred by the S. elongatus circadian clock is more likely due to the ability to anticipate a coming dark period rather than a morning period (53).
One of the surprising findings is that events during the day ultimately affect death of the ΔrpaA mutant in the dark. This outcome was not anticipated because the level of light that is lethal under LD growth was well tolerated by ΔrpaA cells when they are grown under LL (Fig. S6A). Thus, it was originally hypothesized that events exclusively occurring at night were driving cell death. However, the original identification of RpaA derived from its effect on the association of phycobilisomes with photosystem reaction centers in Synechocystis sp. strain PCC 6803. In that strain, rpaA-null mutants have an increased efficiency of energy transfer to PSII relative to PSI (22). Our pigment absorbance data collected at 0 h support the hypothesis that RpaA also affects photosynthetic parameters in S. elongatus (Fig. 1A). If energy transfer to PSII relative to PSI is also increased in ΔrpaA S. elongatus cells, this alteration could both serve as a source of redox stress via excess excitation energy in PSII as well as a decrease the cells’ ability to produce NADPH via PSI-driven reduction. Regardless of the source, the ROS and metabolomics data both indicate that once cells reach the light-to-dark transition, the ΔrpaA mutant is already under a great deal of cellular redox stress (Figs. 1A, 2A, and 6 and Fig. S2) (33, 46).
We propose that high redox stress generated from photosynthetic activity is a critical component of the ΔrpaA LD lethality phenotype. Manipulations that reduce light energy absorbed by cells, including reduction of growth light intensity (Fig. 5D) and treatment with MSM (Fig. 5A), rescue ΔrpaA from death under LD conditions. These results are consistent with overstimulation of photosynthetic pathways as a source of ROS. Although we focused on the EMS suppressor mutations that affect the VLI biosynthetic pathway, via treatment with MSM, strains that carry other mutations in amino acid and aminoacyl–tRNA biosynthesis exhibited similarly depressed phycobilisome absorbance (Fig. S5B and Table S1). As some of the most abundant proteins in cyanobacteria (54), phycobilisome levels are affected significantly by amino acid limitation. Cells with reduced phycobilisome content would collect less light energy and generate less ROS during the day period. Additionally, amino acid biosynthetic pathways consume large amounts of NADPH, and partially blocking these pathways may serve to preserve the limited NADPH pools present in ΔrpaA at night, which are needed for ROS-scavenging pathways.
The viability of the ΔrpaA mutant in LL (Fig. S6A), at a light intensity that generates high ROS, suggests that NADPH produced via photosynthesis can drive the ROS detoxification mechanisms necessary to maintain viability while ΔrpaA cells are in the light; however, when the ΔrpaA mutant enters the dark under redox stress, it lacks a source of NADPH because of an attenuated ability to degrade glycogen and activate the OPPP (19, 20). Consistent with an inability to activate the OPPP in the dark, ΔrpaA cells rapidly deplete soluble sugars connected to this pathway (Fig. 2A). Because glycolysis is still active in ΔrpaA and the OPPP functions as a cycle at night, recycling its inputs (34, 35), we saw a significant increase only in metabolites downstream of glycolysis in the ΔrpaA mutant, including pyruvate and AKG (Figs. 2A and 3A). Although glycolysis can produce some NADH as reducing power, NADH is a poor electron source for ROS-detoxifying processes in S. elongatus (46). AKG buildup normally results in its conversion to glutamine and glutamate via nitrogen assimilation (12, 13). The rapid depletion of glutamine, glutamate, and many other amino acid species in the ΔrpaA mutant, concurrent with AKG elevation (Figs. 2A and 3B), is consistent with an NADPH deficit that precludes cells from performing AKG to amino acid biosynthesis. Additionally, the activation of a nitrogen-deprivation transcriptional response (Fig. S3) and phycobilisome degradation (Fig. 1B) indicate that ΔrpaA cells accumulate AKG to a level that is perceived as C/N imbalance. The EMS mutations in amino acid biosynthetic and utilization pathways slow the mobilization of carbon toward amino acids and lower NADPH consumption, allowing ΔrpaA to more easily achieve homeostasis with limited NADPH pools. This hypothesis is consistent with the fact that ΔrpaA cells treated with MSM no longer show nitrogen-deprivation transcriptional or bleaching responses at night (Fig. 5B and Fig. S3).
Protein redox modifications drive important metabolic shifts in cyanobacteria and plant chloroplasts (10, 28, 55, 56). Recent work has shown that redox modifications are pervasive across all metabolic pathways in cyanobacteria and that LD transitions drive global changes in the oxidation state of redox-modified proteins (8, 9, 49). Because de novo transcription is limited to the early night period in S. elongatus (57), redox modifications on metabolic enzymes likely play a major role in modulating enzymatic activity and dictating metabolic flux over the dark period. Some of the metabolic changes in the ΔrpaA mutant after the termination of ROS detoxification may be driven by an inability to further modulate the protein redox state. Specifically, there is evidence that accumulation of lipids can be driven by redox changes and may contribute directly to LD lethality (40, 58).
Lipids are particularly sensitive to oxidative stress, and their turnover is important because oxidized lipid species can further perpetuate oxidative damage (59). Activation of lipid recycling by high light in S. elongatus is consistent with a redox-driven mechanism to signal this process. The increase of lysopalmitoyl monogalactosylglycerol and 3-hydroxypalmitic acid in ΔrpaA indicates that a lipid-recycling response has been activated at night (43). We propose that ROS detoxification terminates in the ΔrpaA mutant when the limited NADPH pool is exhausted, and subsequent redox stress activates the membrane recycling process (46). We propose that the ΔrpaA mutant, but not WT, has reached an ROS threshold that would trigger lipid recycling by the end of the day. Although both WT and ΔrpaA cells should properly express the AAS protein required to activate lipids for recycling (20), AAS is ATP-dependent, and its functionality at night in cyanobacteria would likely be impaired (39). Thus, the activation of lipid recycling in ΔrpaA occurs when ATP levels are dropping in the dark, and AAS may be unable to deal with FAA load. Indeed, we observed bleaching (Fig. 1B), AKG elevation (Figs. 2A and 3B), fatty acid accumulation (Fig. 2A), and the start of cell death (Fig. 1C) around the time that ROS detoxification terminated (Fig. 6). Thus, the primary mechanisms directly driving cell death likely occur between the 2- to 6-h period after entering the dark, and these mechanisms are likely the result of a strong redox imbalance.
The daytime functions of RpaA are unexpected and should be further explored. In particular, the pigmentation changes during LL growth of the ΔrpaA mutant (Fig. 1A) suggest that core photosynthetic parameters are altered. Metrics such as photosystem efficiency, capacity, and oxygen evolution in S. elongatus have not been explored in the context of circadian rhythms. Additionally, it is interesting that ROS levels in the ΔrpaA mutant consistently increase at ∼6 h after a transition from darkness into light (Fig. 6), because this mutant lacks a clock output mechanism and does not exhibit transcriptional rhythms (20, 21). This timing may represent a point where ROS scavenging resources are naturally exhausted, or it may indicate that other known rhythmic processes, such as 24-h peroxiredoxin rhythms, may be important for modulating ROS (18, 60). The participation of peroxiredoxins in ROS modulation would be consistent with their roles in oxidative stress-mediated signaling (61). This observation hints that Kai-mediated and other rhythmic processes may interact to control the cellular redox state. Overall, the integration of light conditions, circadian rhythms, and the cellular redox state in the control of cyanobacterial metabolism will be of crucial importance to advance the engineering and understanding of cyanobacteria growing under natural diurnal conditions.
Materials and Methods
The full description of experimental techniques is provided in SI Materials and Methods.
Cyanobacterial Strains, Media, and Culture Conditions.
All ΔrpaA mutants were constructed by transformation in a WT S. elongatus background with plasmid pAM4420 (25) and were validated by PCR. For all experiments, precultures were first prepared in 100 mL of fresh BG-11 medium as in Diamond et al. (19).
For metabolomics experiments, precultures were used to inoculate Phenometrics ePBR photobioreactors (Version 1.1; Phenometrics Inc.) at an initial density of OD750 = 0.1 in 400 mL of BG-11 medium without antibiotics. Temperature was maintained at 30 °C; filtered (0.2 μm) air was sparged at a rate of 50 mL/min; and light intensity was either 150 or 500 μE⋅m−2⋅s−1 provided from the top of the culture while lights were on. After inoculation, cultures were grown at a constant light intensity of 150 μE⋅m−2⋅s−1 until OD750 = 0.3, then maintained turbidostatically at this density for the duration of the experiment. In the metabolomics experiment, WT circadian rhythms were entrained by growth in a 12:12 LD at a light intensity of 150 μE⋅m−2⋅s−1 cycle for 1 d and subsequently at a light intensity of 500 μE⋅m−2⋅s−1 for 2 d before release into experimental conditions and sampling. The ΔrpaA strains were maintained in constant light at the same intensities as for the WT strain before the sampling procedure (Fig. S7).
Fig. S7.
Diagram of the growth and sampling scheme used for experiments conducted in photobioreactors. Each box indicates a 12-h period with white boxes corresponding to periods of light and black boxes corresponding to periods of darkness. Where indicated, only the WT strain was exposed to darkness. Light intensity during each light period is indicated in black text within each white box. Times when samples were taken during the metabolomics experiment are indicated by red arrows. Red arrows correspond to time points ZT12 (0 h), ZT13 (1 h), ZT14 (2 h), ZT16 (4 h), and ZT18 (6 h), from left to right.
For absorbance scanning, viable cell counts, quantitative reverse transcription-polymerase chain reaction (qRT-PCR), MSM-treatment absorbance measurements, and oxidative stress measurements precultures were used to inoculate the photobioreactors at an initial density of OD750 = 0.2 in 400 mL of BG-11 medium without antibiotics. Temperature, airflow rate, and light intensity settings were the same as above. For these experiments, both WT and ΔrpaA mutants were maintained at a constant light intensity of 150 μE⋅m−2⋅s−1 for 1 d. Subsequently, both strains were subjected to growth in a 12:12 LD cycle at a light intensity of 150 μE⋅m−2⋅s−1 for 2 d. Light intensity was then increased to 500 μE⋅m−2⋅s−1 over the final 12:12 LD period, during which sampling took place.
For viable cell plating, 200 μL of the indicated sample was serially diluted 1:5 in fresh BG-11 medium without antibiotics five times. For LD sensitivity testing, samples were first all diluted to an OD750 = 0.2, and the same dilution scheme was then followed. Subsequently, 4 μL of each sample was spotted onto solid BG-11 plates without antibiotics. Plates were incubated at 30 °C and 150 μE⋅m−2⋅s−1 constant light for 5–6 d. For LD sensitivity testing, samples were plated in duplicate, with one set incubated in constant light for 5–6 d, and a second set under a 12:12 LD cycle for 6–8 d.
Metabolomics and Data Analysis.
Strains for metabolomics were grown in Photobioreactors and sampled (n = 5 at each time point and for each genotype) as described above (Fig. S7). Metabolite extraction and GC-TOF-MS were conducted by the West Coast Metabolomics Center (WCMC) at the University of California, Davis identically to the methods used in Diamond et al. (19) and Fiehn et al. (62, 63).
Raw metabolite abundance data for known metabolites (Dataset S1) were analyzed by using a combination of the online analysis platform MetaboAnalyst (Version 3.0) (64) and the statistical package R (65). Principal component analysis (PCA) was applied to log2-normalized and autoscaled data to detect outlying samples. Based on PCA, replicate A of the WT sample was removed from the dataset before statistical analysis (Fig. S8A). In addition, WT replicate C time T6 and ΔrpaA replicate C time T4 were removed before analysis because of problems during sample extraction reported by the WCMC. Statistical analysis of metabolomics data are detailed in SI Materials and Methods. Further details and statistical methods are provided in SI Text.
Fig. S8.
Supporting data for metabolomics statistical analysis. (A) Plot of PCA components 1 and 2 for log2 autoscaled metabolite abundance data for all WT samples. Ellipses indicate the 95% CI for each sample group. The black arrow indicates all time points collected for WT biological replicate A. The statistically significant separation of the WT_A replicate indicates that this sample is an outlier relative to the other WT samples collected. (B) Plot showing result from LOOCV performed on the PLS-DA model. The red star indicates the accuracy of the model is highest when it includes only two components. Due to a high degree of variability in the data, which is typical of metabolomics datasets, we chose prediction accuracy as a metric to select a number of components over other metrics such as model fit (Q2). (C) Plot showing the results of accuracy permutation testing on the PLS-DA model. The red arrow indicates the test statistic. The data indicate that our PLS-DA model is significantly better at predicting class membership than a random model (P < 0.01; n = 1,000 permutations).
Mutagenesis and Identification of ΔrpaA Suppressing Mutations.
EMS mutagenesis of fresh ΔrpaA mutant cyanobacterial strains was carried out as in Kondo et al. (66). Absorbance scans were taken of all cultures, as detailed in SI Materials and Methods, and genomic DNA was extracted by using standard methods (67). Before sequencing genomic DNA, the disruption of rpaA in all strains was verified by PCR (Fig. S4C). Genomic library preparation for Illumina short-read sequencing was performed by using the NEBNext DNA library preparation kit (NEB, catalog no. E6040S/L) with NEXTflex barcoded cDNA adaptors (BIOO Scientific, catalog no. 514104). Samples were run on an Illumina HiSeq 2500 DNA sequencer at the University of California, Berkeley QB3 Genomics Sequencing Laboratory. Sequencing runs resulted in 50-bp reads with a median coverage depth of 45.7× per sample over the S. elongatus genome. Reads from all sequencing methods were mapped against the S. elongatus genome (GenBank accession no. NC_007604) and the large plasmid pANL (GenBank accession no. AF441790), and polymorphisms were called by using the program breseq (68). Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of mutated genes was conducted by using a custom-written R script and the metabolic categories in Dataset S2. Statistical overrepresentation was determined by using the binomial test, and P values were corrected by using the method of Benjamini and Hochberg (69).
SI Text
The ΔrpaA Mutant Activates a Nitrogen-Starvation Transcriptional Response
In cyanobacteria, elevated AKG is the primary signal for nitrogen deprivation, and activation of this system results in a coordinated transcriptional response with the subsequent degradation of phycobilisome proteins (36, 37). The result is a bleaching process termed chlorosis. The observed AKG elevation in ΔrpaA mutants temporally precedes a rapid decrease in phycobilisome absorbance at 630 nm in ΔrpaA mutants that is evident after 8 h in the dark (Figs. 1B and 3B and Fig. S1). However, a variety of stress conditions can cause chlorosis (31). To determine whether elevated AKG is accompanied by a transcriptional response linked to nitrogen deprivation, we tracked transcript levels of glnN (glutamine synthase), a primary transcriptional target of this transcriptional response, before and 2 h after WT and the ΔrpaA mutant entered the dark (37). WT showed a slight decrease in glnN transcript levels after the dark transition, whereas the ΔrpaA mutant showed a statistically significant approximately fourfold increase in glnN transcripts (Fig. S3). Combined, these results are consistent with a transcriptionally activated nitrogen starvation response and indicate that AKG reaches high enough levels in ΔrpaA cells to activate this response in the dark. The activation of this response is normally repressed at night, and its activation may squander already limited cellular resources.
SI Materials and Methods
Cyanobacterial Strains, Media, and Culture Conditions.
All ΔrpaA mutants were constructed by transformation in a WT S. elongatus background with plasmid pAM4420 (25) and were validated by PCR. For all experiments, precultures were first prepared in 100 mL of fresh BG-11 medium as described in Diamond et al. (19).
For metabolomics experiments, precultures were used to inoculate Phenometrics ePBR photobioreactors (Version 1.1; Phenometrics Inc.) at an initial density of OD750 = 0.1 in 400 mL of BG-11 medium without antibiotics. Temperature was maintained at 30 °C; filtered (0.2 μm) air was sparged at a rate of 50 mL/min, and light intensity was either 150 or 500 μE·m−2·s−1 provided from the top of the culture while lights were on. After inoculation, cultures were grown at a constant light intensity of 150 μE·m−2·s−1 until OD750 = 0.3, then maintained turbidostatically at this density for the duration of the experiment. In the metabolomics experiment, WT circadian rhythms were entrained by growth in a 12:12 LD cycle with the light portion at an intensity of 150 μE·m−2·s−1 for 1 d and subsequently at 500 μE·m−2·s−1 for 2 d before release into experimental conditions and sampling. The ΔrpaA strains were maintained in constant light at the same intensities as for the WT strain before the sampling procedure (Fig. S7).
For absorbance scanning, viable cell counts, qRT-PCR, MSM-treatment absorbance measurements, and oxidative stress measurements, precultures were used to inoculate the photobioreactors at an initial density of OD750 = 0.2 in 400 mL of BG-11 medium without antibiotics. Temperature, airflow rate, and light intensity settings were the same as above. For these experiments, both WT and ΔrpaA mutants were maintained at a constant light intensity of 150 μE·m−2·s−1 for 1 d. Subsequently, both strains were subjected to growth in a LD cycle with the light portion at an intensity of 150 μE·m−2·s−1 for 2 d, and then increased to 500 μE·m−2·s−1 over the final LD period during which sampling took place.
For viable cell plating, 200 μL of the indicated sample was serially diluted 1:5 in fresh BG-11 medium without antibiotics five times. For LD sensitivity testing, samples were first all diluted to an OD750 = 0.2, and the same dilution scheme was then followed. Subsequently, 4 μL of each sample was spotted onto solid BG-11 plates without antibiotics. Plates were incubated at 30 °C and 150 μE·m−2·s−1 constant light for 5–6 d. For LD sensitivity testing, samples were plated in duplicate, with one set incubated in constant light for 5–6 d and a second set under a LD cycle for 6–8 d.
Whole-Cell Absorbance Spectra Analysis.
For all reported absorbance spectra, the absorbance between 400 and 750 nm was determined for 200 μL of the indicated sample by using a Tecan Infinite M200 plate reader. Raw absorbance values were normalized to OD750 of each sample. Statistical analyses used the Student’s t test, with a P value < 0.05 considered as significant (n ≥ 3).
Metabolomics and Data Analysis.
Strains for metabolomics sampling were grown in photobioreactors and sampled (n = 5 at each time point and for each genotype) as described above (Fig. S7). Metabolite extraction and GC-TOF-MS were conducted by the WCMC at the University of California, Davis identically to the methods used in Diamond et al. (19) and Fiehn et al. (62, 63).
Raw metabolite abundance data for known metabolites (Dataset S1) were analyzed by using a combination of the online analysis platform MetaboAnalyst (Version 3.0) (64) and the statistical package R (65). PCA was applied to log2-normalized and autoscaled data to detect outlying samples. Based on PCA, replicate A of the WT sample was removed from the dataset before statistical analysis (Fig. S8A). Also, WT replicate C time T6 and ΔrpaA replicate C time T4 were removed before analysis because of problems during sample extraction reported by the WCMC.
To detect metabolites that changed between WT and ΔrpaA at the initial sampling time point (0 h), we used a t test on log2-normalized data (FDR corrected P value < 0.05) (69) and required significant metabolites to show a greater than twofold change between genotypes. To detect metabolites that changed between WT and ΔrpaA over the entire time course, we applied two separate statistical methods to log2-normalized abundance data: two-way ANOVA analysis [Tukey’s honest significant difference (HSD); P < 0.05] using the MetaboAnalyst platform (Version 3.0) and a linear mixed-effect model via the lme4 package in R (65, 70). For mixed-effect linear models, genotype and time were set as fixed effects, while biological replicate and time of sampling were allowed to contribute random effects. Models produced using genotype and an interaction of genotype and time as fixed effects were compared with a base model without genotype or the interaction effect using ANOVA. Final P values generated for each metabolite were corrected by using the method of Benjamini and Hochberg (69) (FDR < 0.05 was considered significant). Both the ANOVA and lme4 methods produced almost identical lists of significant metabolites (Dataset S1); we chose to use the output from two-way ANOVA in our analysis, because of the broader understanding of this method and for simplification of downstream analysis. Hierarchical clustering of significant metabolites for heatmap ordering (Fig. 2A) was performed by using the R package pheatmap, with Euclidian distance and complete linkage for leaf ordering.
PLS-DA modeling was carried out on log2-normalized and autoscaled data with genotype and time as class factors. Using leave-one-out cross-validation indicated that the PLS-DA model providing the most prediction accuracy used two components (Fig. S8B). Permutation testing was used to confirm that our PLS-DA model had a statistically significant ability to correctly predict class membership of samples based on the top 25 discriminating metabolites relative to a random permuted model (P = 0.008; n = 1,000) (Fig. S8C).
Metabolite KEGG enrichment analysis was conducted by using a custom-written R script and the metabolic categories in Dataset S2. Statistical overrepresentation was determined by using Fisher’s exact test. Because of the difficulty in achieving high statistical significance with the low numbers of tested metabolites, we did not apply a multiple-testing correction to P values, but required more than three metabolites to be present in a pathway with a P value < 0.05 to be considered significant.
qRT-PCR Analysis.
For each cyanobacterial sample, 10 mL of culture at an OD750 of 0.2–0.4 was collected and immediately placed on ice. The cultures were then centrifuged for 10 min at 4,000 × g and −10 °C. Pellets were then frozen at −80 °C until extraction. Total RNA was extracted by using the TriZol reagent (Life Technologies) and the Direct-zol RNA MiniPrep Kit (Zymo Research). Briefly, the frozen pellets were thawed on ice and resuspended thoroughly in 1 mL of TriZol reagent. Cell suspensions were then transferred to 1.5-mL microcentrifuge tubes on ice, and cells were lysed by 5–10 cycles of vortexing for 30 s at room temperature and then allowed to sit on ice for 30 s. Cell debris was pelleted by centrifuging at 16,000 × g at room temperature for 5 min. After transferring the supernatant fraction to an RNase-free 2-mL tube, 1 volume of 100% ethanol for every volume of TriZol (typically 1 mL) was added and mixed by pipetting up and down. Total RNA was isolated from the TriZol–ethanol mixture following the manual of the Direct-zol RNA MiniPrep Kit. The RNA quality was checked with agarose gel imaging (1% agarose, 0.5× TBE, 75 V/60 min), using 10,000× SYBR green II RNA gel stain (Lonza). The extracted RNA samples were treated with DNaseI (Thermo Scientific) to remove contaminating genomic DNA. cDNA was synthesized with the SuperScript III First-Strand Synthesis System for RT-PCR (Life Technologies) following the kit manual. For qRT-PCR experiments, standard reactions in triplicate were set up with the Power SYBR Green PCR Master Mix (Life Technologies) and run on a StepOnePlus Real-Time PCR System (Life Technologies) following the instructions of the manufacturer. Significance for a change in glnN expression was calculated by using one-way ANOVA and Tukey’s HSD (n = 3).
Mutagenesis and Identification of ΔrpaA Suppressor Mutations.
EMS mutagenesis of fresh ΔrpaA mutant cyanobacterial strains was carried out as described in Kondo et al. (66). Mutagenized cultures were resuspended in 5 mL of BG-11 medium with appropriate antibiotics and incubated at 30 °C, using 150 rpm shaking, under 30 μE·m−2·s−1 constant light for 2 d. Subsequently 300 μL of mutagenized ΔrpaA cells (as well as untreated controls) were plated on BG-11 plates with appropriate antibiotics. Plates were prepared in duplicate, and one set was incubated at 30 °C and 150 μE·m−2·s−1 constant light for 15 d, with the second set being incubated at 30 °C and a LD cycle with a light intensity of 150 μE·m−2·s−1 for 15 d. Colonies (40) that formed on the LD-grown plate that contained EMS-mutagenized ΔrpaA cells were picked and patched onto BG-11 plates with appropriate antibiotics and grown for a further 10 d at 30 °C and a LD cycle with a light intensity of 150 μE·m−2·s−1 to confirm suppression of the LD death phenotype. Patches of surviving ΔrpaA EMS mutants were then transferred to 10 mL BG-11 medium and incubated at 30 °C, using 150 rpm shaking, with a LD cycle and light intensity of 120 μE·m−2·s−1 for between 11 and 16 d. Absorbance scans were taken of all cultures as described above, and genomic DNA was extracted by using standard methods (67). Before sequencing genomic DNA, the disruption of rpaA in all strains was verified by PCR (Fig. S4C).
Genomic library preparation for Illumina short-read sequencing was performed by using the NEBNext DNA library preparation kit (NEB, catalog no. E6040S/L) with NEXTflex barcoded cDNA adaptors (BIOO Scientific, catalog no. 514104). Samples were run on an Illumina HiSeq2500 DNA sequencer at the University of California, Berkeley QB3 Genomics Sequencing Laboratory. Sequencing runs resulted in 50-bp reads with a median coverage depth of 45.7× per sample over the S. elongatus genome. However, for samples 1.1, 1.25, and 2.3, (Dataset S2), genome resequencing was performed at Bio Applied Technologies Joint, Inc., by using the Ion PGM system (Life Technologies) following the standard workflow illustrated in the manuals of the Life Technologies kits. Sequencing libraries were prepared with the Ion Xpress Plus Fragment Library Kit. In brief, 1 µg of total gDNA was sheared into desired fragment size (200–400 bp) via enzymatic digestion and then ligated to specified sequencing adapters and/or barcode adapters. The sequencing templates were amplified from constructed libraries by using the Ion OneTouch 2 System and the Ion PGM Hi-Q OT2 Kit. Amplified templates were processed with the Ion PGM Hi-Q Sequencing Kit and loaded on an Ion 316 Chip Kit (Version 2) for sequencing on a Ion PGM system with 500 sequencing flows (200 bp reads). Reads from all sequencing methods were mapped against the S. elongatus genome (GenBank accession no. NC_007604) and the large plasmid pANL (GenBank accession no. AF441790), and polymorphisms were called by using the program breseq (68). Polymorphisims were filtered for “high confidence” mutations by removing all mutations that resulted in a high-use codon coding for the same amino acid. All mutations in noncoding regions were included in the high-confidence set.
KEGG enrichment analysis of mutated genes was conducted by using a custom-written R script and the metabolic categories in Dataset S2. Statistical overrepresentation was determined by using the binomial test, and P values were corrected by using the method of Benjamini and Hochberg (69).
MSM Treatment.
Initially by testing decreasing concentrations of MSM (Sigma-Aldrich catalog no. N12482) on WT and ΔrpaA cells, we were able to determine that concentrations of 100 nM and 25 µM, in solid and liquid media, respectively, were nonlethal to WT and ΔrpaA, yet resulted in slightly suppressed growth and suppression of LD lethality. For all experiments using MSM, a stock solution was filter-sterilized (0.2 μm), and subsequently added to sterile medium to reach the desired working concentration. For experiments on solid medium, MSM was directly added to plate agar before plating and growing cells as described above. For experiments in photobioreactors, cells were grown as described above, and sterile MSM was added to a concentration of 25 µM at the beginning of a 12-h light period that preceded the transition into darkness for ΔrpaA cultures.
Quantification of ROS.
ROS were quantified by using the fluorescent marker H2DCFDA (Life Technologies catalog no. D399). Briefly, 2 mL of photobioreactor-grown (see above) culture was collected and split into 1-mL aliquots. H2DCFDA was added to one sample at a final concentration of 5 μM. Tubes were protected from light and shaken at 30 °C for 30 min. After incubation, 200 μL of each tube was added to a separate well in a 96-well plate. Fluorescence was quantified at an excitation of 480 nm and an emission of 520 nm on a Tecan Infinite M200 plate reader with the gain manually set to 120. Fluorescence data were normalized to OD750 of each sample, and untreated-sample background fluorescence was then subtracted from treated-sample fluorescence values.
Supplementary Material
Acknowledgments
We thank the members of the Ralph Greenspan Laboratory and the Berkeley QB3 Genomics Sequencing Laboratory for their assistance with library preparation and sequencing; BATJ, Inc., for their collaboration in sequencing additional suppressor mutants; Profs. Stephen Mayfield and Terry Hwa for particularly useful guidance on metabolomics and microbial growth kinetics, respectively; Dr. Mark Paddock for helpful discussion that improved the manuscript; and Anish Pal for assistance with sample collection and strain maintenance. This work was supported by National Science Foundation Grant MCB1244108. S.D. and B.E.R. were supported in part by National Institutes of Health and Molecular Genetics Training Grant T32GM007240.
Footnotes
The authors declare no conflict of interest.
Dedicated to the memory of Dr. David B. Knaff.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1613078114/-/DCSupplemental.
References
- 1.Oliver JW, Atsumi S. Metabolic design for cyanobacterial chemical synthesis. Photosynth Res. 2014;120(3):249–261. doi: 10.1007/s11120-014-9997-4. [DOI] [PubMed] [Google Scholar]
- 2.Ducat DC, Way JC, Silver PA. Engineering cyanobacteria to generate high-value products. Trends Biotechnol. 2011;29(2):95–103. doi: 10.1016/j.tibtech.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Bryant DA. The beauty in small things revealed. Proc Natl Acad Sci USA. 2003;100(17):9647–9649. doi: 10.1073/pnas.1834558100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang S, Bryant DA. Biochemical validation of the glyoxylate cycle in the cyanobacterium Chlorogloeopsis fritschii strain PCC 9212. J Biol Chem. 2015;290(22):14019–14030. doi: 10.1074/jbc.M115.648170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang S, Bryant DA. The tricarboxylic acid cycle in cyanobacteria. Science. 2011;334(6062):1551–1553. doi: 10.1126/science.1210858. [DOI] [PubMed] [Google Scholar]
- 6.Kouhen OM-E, Joset F. Biosynthesis of the branched-chain amino acids in the cyanobacterium Synechocystis PCC6803: Existence of compensatory pathways. Curr Microbiol. 2002;45(2):94–98. doi: 10.1007/s00284-001-0097-2. [DOI] [PubMed] [Google Scholar]
- 7.Rubin BE, et al. The essential gene set of a photosynthetic organism. Proc Natl Acad Sci USA. 2015;112(48):E6634–E6643. doi: 10.1073/pnas.1519220112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo J, et al. Proteome-wide light/dark modulation of thiol oxidation in cyanobacteria revealed by quantitative site-specific redox proteomics. Mol Cell Proteomics. 2014;13(12):3270–3285. doi: 10.1074/mcp.M114.041160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ansong C, Sadler NC, Hill EA, Lewis MP. Characterization of protein redox dynamics induced during light-to-dark transitions and nutrient limitation in cyanobacteria. Front Microbiol. 2014;5:325. doi: 10.3389/fmicb.2014.00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindahl M, Kieselbach T. Disulphide proteomes and interactions with thioredoxin on the track towards understanding redox regulation in chloroplasts and cyanobacteria. J Proteomics. 2009;72(3):416–438. doi: 10.1016/j.jprot.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 11.Schürmann P. Redox signaling in the chloroplast: The ferredoxin/thioredoxin system. Antioxid Redox Signal. 2003;5(1):69–78. doi: 10.1089/152308603321223559. [DOI] [PubMed] [Google Scholar]
- 12.Ohashi Y, et al. Regulation of nitrate assimilation in cyanobacteria. J Exp Bot. 2011;62(4):1411–1424. doi: 10.1093/jxb/erq427. [DOI] [PubMed] [Google Scholar]
- 13.Muro-Pastor MI, Reyes JC, Florencio FJ. Ammonium assimilation in cyanobacteria. Photosynth Res. 2005;83(2):135–150. doi: 10.1007/s11120-004-2082-7. [DOI] [PubMed] [Google Scholar]
- 14.Vijayan V, Zuzow R, O’Shea EK. Oscillations in supercoiling drive circadian gene expression in cyanobacteria. Proc Natl Acad Sci USA. 2009;106(52):22564–22568. doi: 10.1073/pnas.0912673106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mackey SR, Golden SS, Ditty JL. 2011. The Itty-Bitty Time Machine, Advances in Genetics (Elsevier, New York) pp 13–53.
- 16.Cohen SE, Golden SS. Circadian rhythms in cyanobacteria. Microbiol Mol Biol Rev. 2015;79(4):373–385. doi: 10.1128/MMBR.00036-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shultzaberger RK, Boyd JS, Diamond S, Greenspan RJ, Golden SS. Giving time purpose: The Synechococcus elongatus Clock in a broader network context. Annu Rev Genet. 2015;49:485–505. doi: 10.1146/annurev-genet-111212-133227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edgar RS, et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature. 2012;485(7399):459–464. doi: 10.1038/nature11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diamond S, Jun D, Rubin BE, Golden SS. The circadian oscillator in Synechococcus elongatus controls metabolite partitioning during diurnal growth. Proc Natl Acad Sci USA. 2015;112(15):E1916–E1925. doi: 10.1073/pnas.1504576112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Markson JS, Piechura JR, Puszynska AM, O’Shea EK. Circadian control of global gene expression by the cyanobacterial master regulator RpaA. Cell. 2013;155(6):1396–1408. doi: 10.1016/j.cell.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takai N, et al. A KaiC-associating SasA-RpaA two-component regulatory system as a major circadian timing mediator in cyanobacteria. Proc Natl Acad Sci USA. 2006;103(32):12109–12114. doi: 10.1073/pnas.0602955103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashby MK, Mullineaux CW. Cyanobacterial ycf27 gene products regulate energy transfer from phycobilisomes to photosystems I and II. FEMS Microbiol Lett. 1999;181(2):253–260. doi: 10.1111/j.1574-6968.1999.tb08852.x. [DOI] [PubMed] [Google Scholar]
- 23.Paddock ML, Boyd JS, Adin DM, Golden SS. Active output state of the Synechococcus Kai circadian oscillator. Proc Natl Acad Sci USA. 2013;110(40):E3849–E3857. doi: 10.1073/pnas.1315170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iijima H, et al. Changes in primary metabolism under light and dark conditions in response to overproduction of a response regulator RpaA in the unicellular cyanobacterium Synechocystis sp. PCC 6803. Front Microbiol. 2015;6:888. doi: 10.3389/fmicb.2015.00888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boyd JS, Bordowitz JR, Bree AC, Golden SS. An allele of the crm gene blocks cyanobacterial circadian rhythms. Proc Natl Acad Sci USA. 2013;110(34):13950–13955. doi: 10.1073/pnas.1312793110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang C, Hua Q, Shimizu K. Integration of the information from gene expression and metabolic fluxes for the analysis of the regulatory mechanisms in Synechocystis. Appl Microbiol Biotechnol. 2002;58(6):813–822. doi: 10.1007/s00253-002-0949-0. [DOI] [PubMed] [Google Scholar]
- 27.Osanai T, Azuma M, Tanaka K. Sugar catabolism regulated by light- and nitrogen-status in the cyanobacterium Synechocystis sp. PCC 6803. Photochem Photobiol Sci. 2007;6(5):508–514. doi: 10.1039/b616219n. [DOI] [PubMed] [Google Scholar]
- 28.Tamoi M, Miyazaki T, Fukamizo T, Shigeoka S. The Calvin cycle in cyanobacteria is regulated by CP12 via the NAD(H)/NADP(H) ratio under light/dark conditions. Plant J. 2005;42(4):504–513. doi: 10.1111/j.1365-313X.2005.02391.x. [DOI] [PubMed] [Google Scholar]
- 29.Wedel N, Soll J.1998Evolutionary conserved light regulation of Calvin cycle activity by NADPH-mediated reversible phosphoribulokinase/CP12/ glyceraldehyde-3-phosphate dehydrogenase complex dissociation Proc Natl Acad Sci USA 95169699.- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waldbauer JR, Rodrigue S, Coleman ML, Chisholm SW. Transcriptome and proteome dynamics of a light-dark synchronized bacterial cell cycle. PLoS One. 2012;7(8):e43432. doi: 10.1371/journal.pone.0043432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collier JL, Grossman AR. Chlorosis induced by nutrient deprivation in Synechococcus sp. strain PCC 7942: Not all bleaching is the same. J Bacteriol. 1992;174(14):4718–4726. doi: 10.1128/jb.174.14.4718-4726.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu X, Li Q, Yin C, Fang X, Xu X. Role of spermidine in overwintering of cyanobacteria. J Bacteriol. 2015;197(14):2325–2334. doi: 10.1128/JB.00153-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Incharoensakdi A, Jantaro S, Raksajit W, Mäenpää P. Polyamines in cyanobacteria: Biosynthesis, transport and abiotic stress response. In: Méndez-Vilas A, editor. Current Research Topics in Applied Microbiology and Microbial Biotechnology. Formatex; Badajoz, Spain: 2010. pp. 23–32. [Google Scholar]
- 34.Young JD, Shastri AA, Stephanopoulos G, Morgan JA. Mapping photoautotrophic metabolism with isotopically nonstationary (13)C flux analysis. Metab Eng. 2011;13(6):656–665. doi: 10.1016/j.ymben.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakajima T, et al. Integrated metabolic flux and omics analysis of Synechocystis sp. PCC 6803 under mixotrophic and photoheterotrophic conditions. Plant Cell Physiol. 2014;55(9):1605–1612. doi: 10.1093/pcp/pcu091. [DOI] [PubMed] [Google Scholar]
- 36.Klotz A, Reinhold E, Doello S, Forchhammer K. Nitrogen starvation acclimation in Synechococcus elongatus: Redox-control and the role of nitrate reduction as an electron sink. Life (Basel) 2015;5(1):888–904. doi: 10.3390/life5010888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flores E, Herrero A. Nitrogen assimilation and nitrogen control in cyanobacteria. Biochem Soc Trans. 2005;33(Pt 1):164–167. doi: 10.1042/BST0330164. [DOI] [PubMed] [Google Scholar]
- 38.Kaczmarzyk D, Fulda M. Fatty acid activation in cyanobacteria mediated by acyl-acyl carrier protein synthetase enables fatty acid recycling. Plant Physiol. 2010;152(3):1598–1610. doi: 10.1104/pp.109.148007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kato A, et al. Modulation of the balance of fatty acid production and secretion is crucial for enhancement of growth and productivity of the engineered mutant of the cyanobacterium Synechococcus elongatus. Biotechnol Biofuels. 2016;9:91. doi: 10.1186/s13068-016-0506-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takatani N, et al. Essential role of acyl-ACP synthetase in acclimation of the cyanobacterium Synechococcus elongatus strain PCC 7942 to high-light conditions. Plant Cell Physiol. 2015;56(8):1608–1615. doi: 10.1093/pcp/pcv086. [DOI] [PubMed] [Google Scholar]
- 41.Shen J, DiTommaso A, Shen M, Lu W, Li Z. Molecular basis for differential metabolic responses to monosulfuron in three nitrogen-fixing cyanobacteria. Weed Sci. 2009;57(March 2009):133–141. [Google Scholar]
- 42.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Friedberg D, Seijffers J. Sulfonylurea-resistant mutants and natural tolerance of cyanobacteria. Arch Microbiol. 1988;150(3):278–281. [Google Scholar]
- 44.Page LE, Liberton M, Pakrasi HB. Phycobilisome antenna truncation reduces photoautotrophic productivity in Synechocystis sp. PCC 6803, a cyanobacterium. Appl Environ Microbiol. 2012;165(2):705–714. doi: 10.1128/AEM.00499-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grossman AR, Schaefer MR, Chiang GG, Collier JL. The phycobilisome, a light-harvesting complex responsive to environmental conditions. Microbiol Rev. 1993;57(3):725–749. doi: 10.1128/mr.57.3.725-749.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Latifi A, Ruiz M, Zhang C-C. Oxidative stress in cyanobacteria. FEMS Microbiol Rev. 2009;33(2):258–278. doi: 10.1111/j.1574-6976.2008.00134.x. [DOI] [PubMed] [Google Scholar]
- 47.Perelman A, Uzan A, Hacohen D, Schwarz R. Oxidative stress in Synechococcus sp. strain PCC 7942: Various mechanisms for H2O2 detoxification with different physiological roles. J Bacteriol. 2003;185(12):3654–3660. doi: 10.1128/JB.185.12.3654-3660.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cameron JC, Pakrasi HB. Essential role of glutathione in acclimation to environmental and redox perturbations in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 2010;154(4):1672–1685. doi: 10.1104/pp.110.162990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chardonnet S, et al. First proteomic study of S-glutathionylation in cyanobacteria. J Proteome Res. 2015;14(1):59–71. doi: 10.1021/pr500625a. [DOI] [PubMed] [Google Scholar]
- 50.Rastogi RP, Singh SP, Häder D-P, Sinha RP. Detection of reactive oxygen species (ROS) by the oxidant-sensing probe 2′,7′-dichlorodihydrofluorescein diacetate in the cyanobacterium Anabaena variabilis PCC 7937. Biochem Biophys Res Commun. 2010;397(3):603–607. doi: 10.1016/j.bbrc.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 51.Lea-Smith DJ, et al. Thylakoid terminal oxidases are essential for the cyanobacterium Synechocystis sp. PCC 6803 to survive rapidly changing light intensities. Plant Physiol. 2013;162(1):484–495. doi: 10.1104/pp.112.210260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saha R, et al. Diurnal regulation of cellular processes in the cyanobacterium Synechocystis sp. Strain PCC 6803: Insights from transcriptomic, fluxomic, and physiological analyses. mBio. 2016;7(3):e00464–16. doi: 10.1128/mBio.00464-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lambert G, Chew J, Rust MJ. Costs of clock-environment misalignment in individual cyanobacterial cells. Biophys J. 2016;111(4):883–891. doi: 10.1016/j.bpj.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guerreiro ACL, et al. Daily rhythms in the cyanobacterium synechococcus elongatus probed by high-resolution mass spectrometry-based proteomics reveals a small defined set of cyclic proteins. Mol Cell Proteomics. 2014;13(8):2042–2055. doi: 10.1074/mcp.M113.035840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nikkanen L, Rintamäki E. Thioredoxin-dependent regulatory networks in chloroplasts under fluctuating light conditions. Philos Trans R Soc Lond B Biol Sci. 2014;369(1640):20130224. doi: 10.1098/rstb.2013.0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Díaz-Troya S, López-Maury L, Sánchez-Riego AM, Roldán M, Florencio FJ. Redox regulation of glycogen biosynthesis in the cyanobacterium Synechocystis sp. PCC 6803: Analysis of the AGP and glycogen synthases. Mol Plant. 2014;7(1):87–100. doi: 10.1093/mp/sst137. [DOI] [PubMed] [Google Scholar]
- 57.Hosokawa N, et al. Circadian transcriptional regulation by the posttranslational oscillator without de novo clock gene expression in Synechococcus. Proc Natl Acad Sci USA. 2011;108(37):15396–15401. doi: 10.1073/pnas.1019612108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singh SC, Sinha RP, Häder D-P. Role of lipids and fatty acids in stress tolerance in cyanobacteria. Acta Protozool. 2002;41(4):297–308. [Google Scholar]
- 59.Girotti AW. Lipid hydroperoxide generation, turnover, and effector action in biological systems. J Lipid Res. 1998;39(8):1529–1542. [PubMed] [Google Scholar]
- 60.Hoyle NP, O’Neill JS. Oxidation-reduction cycles of peroxiredoxin proteins and nontranscriptional aspects of timekeeping. Biochemistry. 2015;54(2):184–193. doi: 10.1021/bi5008386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dietz K-J. Peroxiredoxins in plants and cyanobacteria. Antioxid Redox Signal. 2011;15(4):1129–1159. doi: 10.1089/ars.2010.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fiehn O, et al. Plasma metabolomic profiles reflective of glucose homeostasis in non-diabetic and type 2 diabetic obese African-American women. PLoS One. 2010;5(12):e15234. doi: 10.1371/journal.pone.0015234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fiehn O, et al. Quality control for plant metabolomics: Reporting MSI-compliant studies. Plant J. 2008;53(4):691–704. doi: 10.1111/j.1365-313X.2007.03387.x. [DOI] [PubMed] [Google Scholar]
- 64.Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0-making metabolomics more meaningful. Nucleic Acids Res. 2015;43(W1):W252–7. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.R Core Team . R: A Language and Environment for Statistical Computing. R Core Team; Vienna: 2014. [Google Scholar]
- 66.Kondo T, et al. Circadian clock mutants of cyanobacteria. Science. 1994;266(5188):1233–1236. doi: 10.1126/science.7973706. [DOI] [PubMed] [Google Scholar]
- 67.Clerico EM, Ditty JL, Golden SS. Specialized techniques for site-directed mutagenesis in cyanobacteria. Methods Mol Biol. 2007;362:155–171. doi: 10.1007/978-1-59745-257-1_11. [DOI] [PubMed] [Google Scholar]
- 68.Deatherage DE, Barrick JE. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol. 2014;1151:165–188. doi: 10.1007/978-1-4939-0554-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 70.Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015;67(1):1–48. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.