Significance
On encountering antigens, antibody’s variable domains evolve and mature through multiple rounds of somatic mutation. By surveying the effects of all possible single mutations of an antibody’s variable domains on folding stability and antigen binding, we showed that even for a high-affinity antibody such as the one used in our study, beneficial mutations can be found both near and far from the antigen-binding site. Furthermore, our mutational scan revealed an antigen-distal framework position working as a structural switch whereby a mutation can allow the variable domains to sample different conformational spaces and thus potentially different binding functions. This understanding of the mechanism of somatic mutation in human antibodies illustrates how antibodies efficiently use somatic mutation for evolving diversity in immune recognition.
Keywords: antibody, affinity maturation, immunology, deep mutational scanning, conformational dynamics
Abstract
Somatic mutations within the antibody variable domains are critical to the immense capacity of the immune repertoire. Here, via a deep mutational scan, we dissect how mutations at all positions of the variable domains of a high-affinity anti-VEGF antibody G6.31 impact its antigen-binding function. The resulting mutational landscape demonstrates that large portions of antibody variable domain positions are open to mutation, and that beneficial mutations can be found throughout the variable domains. We determine the role of one antigen-distal light chain position 83, demonstrating that mutation at this site optimizes both antigen affinity and thermostability by modulating the interdomain conformational dynamics of the antigen-binding fragment. Furthermore, by analyzing a large number of human antibody sequences and structures, we demonstrate that somatic mutations occur frequently at position 83, with corresponding domain conformations observed for G6.31. Therefore, the modulation of interdomain dynamics represents an important mechanism during antibody maturation in vivo.
Unlike T-cell receptors, B-cell–derived antibodies accumulate somatic mutations in the antigen-binding variable domains of heavy chain (HC) and light chain (LC), in both the complementarity-determining regions (CDRs) and the framework regions (FWRs) that scaffold the CDRs (1, 2). Together with the V(D)J gene recombination, somatic hypermutation (SHM) adds to the gene diversity of antibody variable domains (VH and VL) necessary for efficient immune recognition (3).
Deciphering the molecular mechanism of SHMs in improving antibodies is critical for understanding the evolution of the immune repertoire. It is well recognized that mutations at the CDRs or FWR structurally adjacent to the CDRs can modulate and optimize the antigen-binding interface (4–7), and several studies have used saturated mutagenesis of CDR position to explore the effect of various mutations on affinity and specificity (8–11).
The role of SHM in antigen-distal FWR is less well understood, however. Previous studies have suggested that the antigen-distal FWR contributes primarily to the variable domain structural integrity; thus, mutations at these positions would be either detrimental to or neutral for antigen-binding function (12–14). Other authors have characterized the potential of antigen-distal framework mutation as beneficial for antigen-binding function. For example, framework mutations can compensate for the destabilizing effect of mutations at CDRs needed for antigen binding (15). In other cases, non-CDR SHMs have been shown to be required for the neutralization activity of broadly neutralizing anti-HIV antibodies (16).
Thus far, the roles of SHM have been studied primarily by examining the functional consequences of reverting the somatic mutation of selected antibodies back to the germline sequences (15–21). Although these studies have demonstrated that antibodies depend on somatic mutations to achieve high-affinity antigen binding, the approach is limited to the small set of mutations present in a given antibody.
Here we took a systematic approach to assessing the mutability of the whole variable domain for maintaining or improving folding stability and antigen binding. We used a well-optimized anti-vascular endothelial cell growth factor (VEGF), G6.31, with high affinity (Kd = 0.4 nM) and excellent thermostability [fragment antigen-binding (Fab) melting temperature (Tm) = 84 °C]. G6.31 derives its HC and LC variable domains from the frequently used human germline V families VH3 and VK1, respectively, and exhibits the canonical variable domain backbone structure (22–24), and thus is a good representative of human antibodies.
The present study had two aims: first, to map the mutational landscape of the two variable domains to delineate the potential effects of all possible single mutations, and second, to identify novel molecular mechanisms of antigen distal framework mutations for improving antibody function. A high-affinity antibody such as G6.31 likely has well-optimized CDRs, thus providing an opportunity to identify beneficial antigen-distal mutations.
We generated a phage display library of G6.31 Fab variants with all possible single mutations throughout the variable HC (VH library) and LC (VL library) domains. We compared the frequency of each mutation in the library before and after selection for binding to its various binding partners, including protein A, protein L, and VEGF, using deep sequencing (25, 26). We found that a large number of single mutations in the framework outside the structural core and the VH–VL interface are well tolerated. We identified mutations at positions distal to CDRs that can further improve the antigen-binding affinity and/or thermostability of this well-optimized antibody, and examined the molecular mechanism of mutations at LC-83 in modulating Fab stability and antigen binding. We found that approximately 25 Å away from the antigen-binding site, the mutation at LC-83 can alter the predominant orientation of the constant domains (CL and CH1) to the variable domains VL and CL (the Fab elbow angle), as well as the orientations of the two variable domains toward each other (the torsion angle between VL and VH). We also found that somatic mutations at LC-83 are common among human antibodies of the VKappa1 germline origin. Our work suggests that antigen-distal framework SHMs occurring during antibody maturation can improve antibody properties by optimizing interdomain structural dynamics.
Results and Discussion
High Tolerance to Single Mutations in VL and VH for Stable Fab Assembly.
We displayed G6.31 Fab on the surface of M13 phage by coexpressing the LC that is C-terminally fused to a 14-mer peptide expression tag gD and the HC up to the CH1 domain, which is then fused to the C-terminal domain of M13 p3 minor coat protein (Fig. 1 and Fig. S1) (24). We generated a single-site saturated mutagenesis library of both variable domains. The VL library and VH library mutated VL positions 2–107 and VH positions 2–113 (Kabat numbering), respectively, one residue at a time, to all 20 amino acids. The first position was omitted to avoid the effect of mutation on signal peptide processing. To detect the impact of mutation on functional expression and fold stability, we panned both libraries against an anti-gD antibody, protein A, or protein L, which binds the LC tag, the side of VH, the side of VL, respectively, to enrich for well-expressed Fab variants (Fig. 1A and Figs. S1 and S2). We previously showed that detection of the gD tag on the LC indicates stably assembled Fab with properly folded VL and VH, because only the HC is physically linked to the phage coat (27). The sequences from the selected and unselected library pools were determined using Illumina MiSeq pair-end reads spanning ∼300 base pairs in each direction, as described previously (26).
Fig. 1.
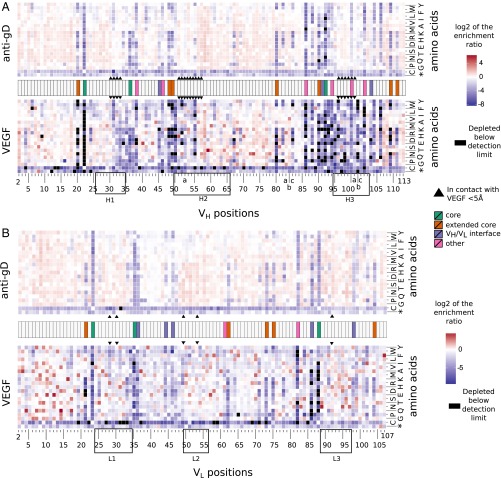
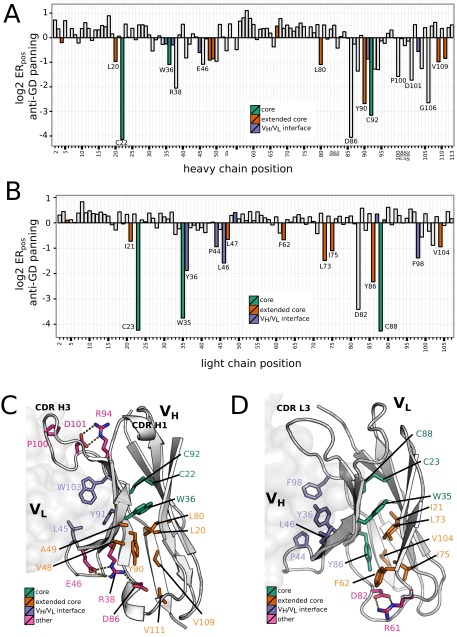
Mutational landscape of VH and VL. The change of the abundance of each mutation in the VH (A) and VL (B) during selection for stable expression using anti-gD tag or for binding antigen VEGF (Fig. S1A) is calculated as log2ER and depicted in the heat maps with a scale of red (enriched) to blue (depleted). All mutations were accounted for in the preselection libraries. Mutations in black are those that are absent after selection indicating strong depletion. Highly conserved positions of VH or VL are colored based on their structural role, including hydrophobic core (colored green), extended core (orange), VH–VL interface (purple), or other (pink; hydrogen-bonded cluster) (Figs. S1–S4). CDR positions are boxed. Residues that contact VEGF (≤5 Å in distance based on the G6/VEGF complex structure; PDB ID code 2FJG) are marked with arrowheads.
Fig. S1.
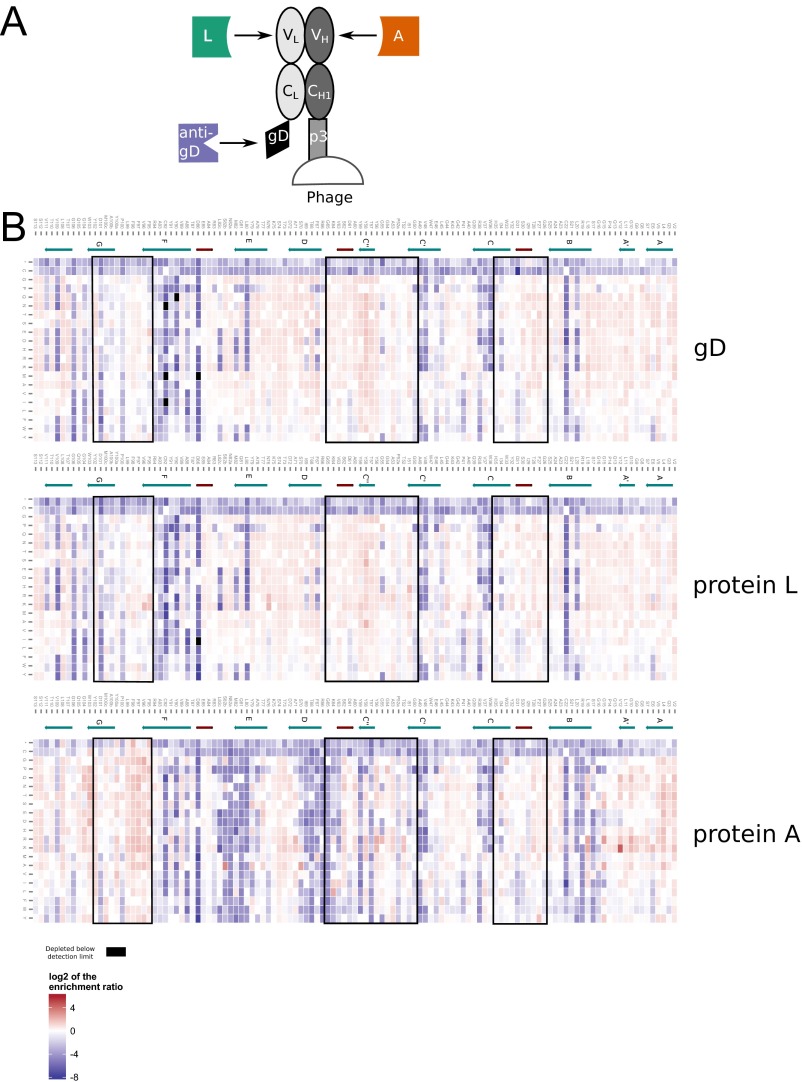
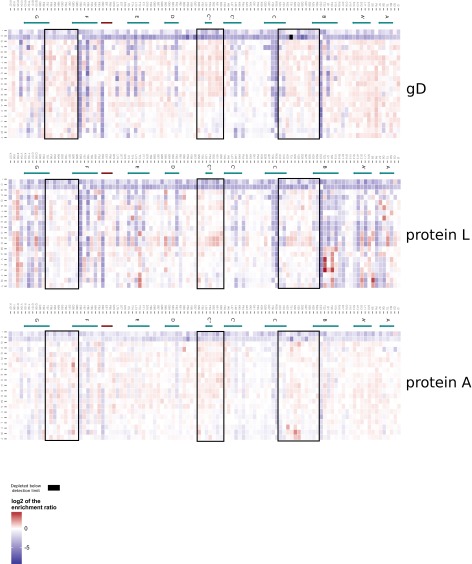
Mutational landscape of the G6.31 VH obtained from stability selection. (A) Schematic representation of the Fab-displaying phage and the location where the three selection ligands for stable expression bind. (B) Enrichment (red) or depletion (blue) of all VH mutations from panning against anti-gD, protein L, and protein A were calculated as log2ER and are displayed as a heat map. CDR positions are boxed (Kabat definition). Secondary structure elements as observed in the structure of G6 are depicted by green arrows (β-strand) and red tubes (α-helix).
Fig. S2.
Mutational landscape of the G6.31 VL obtained from stability selection. Enrichment (red) or depletion (blue) of all VL mutations from panning against anti-gD, protein L, and protein A were calculated as log2ER and are displayed as a heat map. CDR positions are boxed (Kabat definition). Secondary structure elements as observed in the structure of G6 are depicted by green arrows (β-strand) and red tubes (α-helix).
From the 105–106 reads per sample, we verified that all expected mutations were found in the unselected pools, and further filtered the sequences to reduce biases from PCR or sequencing (Methods and Table S1). We then calculated the enrichment ratio (ER) for each mutation by dividing the frequency in the selected pools with the frequency in the unselected pools, as described previously (25, 26). The ER of a particular mutation likely aggregates several effects, which include the impact of the mutation on functional expression, the stability of the fold as well as the binding affinity of the ligands in selection.
Table S1.
Read count overview of VH and VL library Miseq sequencing before and after sorting
| Panning | Library | No. of raw reads | No. of reads after filtering and quality control |
| Presort | VH | 3,307,273 | 1,683,830 |
| Protein A | VH | 3,013,411 | 1,796,501 |
| Protein L | VH | 3,135,884 | 1,904,145 |
| Anti-gD | VH | 2,115,752 | 1,281,856 |
| Presort | VH 1–13, 26–113# | 1,045,661 | 545,825 |
| VEGF | VH 1–13, 26–113# | 774,397 | 515,928 |
| Presort | VH 14–25* | 1,210,000 | 532,861 |
| VEGF | VH 14–25* | 770,000 | 548,901 |
| Presort | Vl | 2,554,326 | 1,507,703 |
| Protein A | VL | 4,487,694 | 2,798,116 |
| Protein L | VL | 5,889,742 | 3,661,983 |
| Anti-gD | VL | 868,034 | 651,100 |
| VEGF | VL | 3,070,495 | 2,106,826 |
Sorting of two VH libraries was performed. The original VH library (#) contained a segment (Kabat positions 14–25) that had some error and did not display well on phage. A second library was generated with correction in this segment. VEGF selection sequence data from the first library minus this 14–25 segment (#) were combined with the data of the 14–25 segment from the second library (*) for analysis. For data analysis purposes, the reads obtained from the second library were reduced by randomly selecting a similar number of reads to match that obtained in the first VH library.
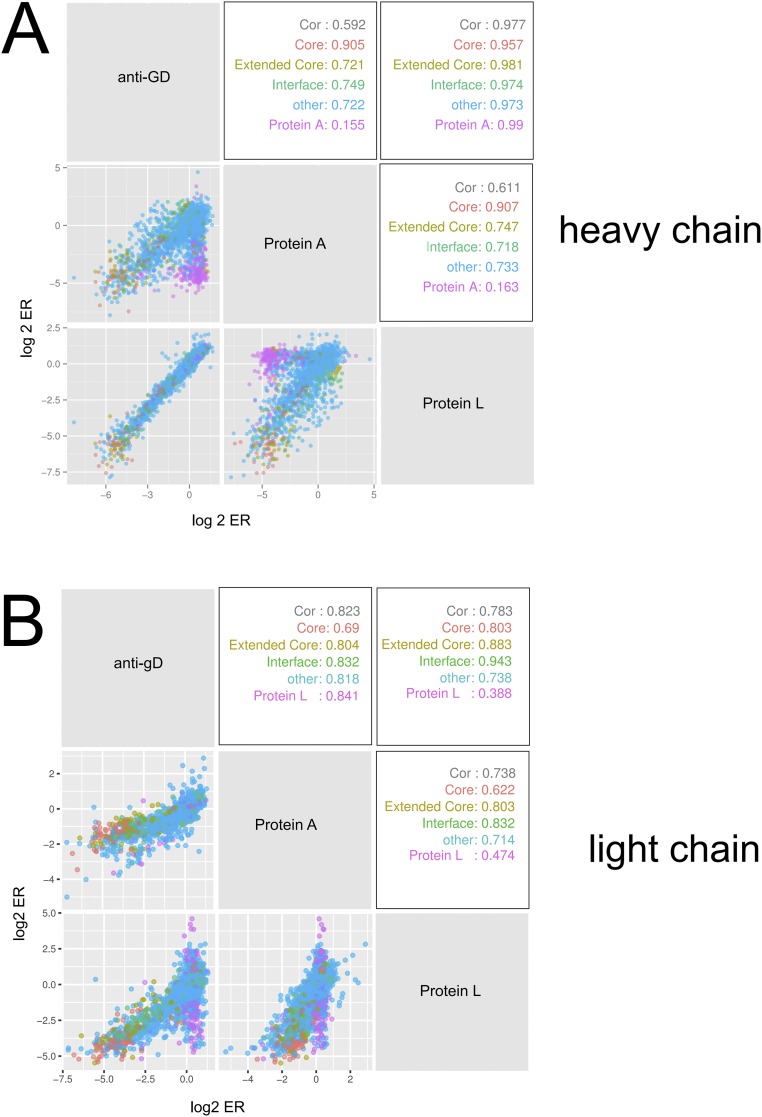
The ERs of all single mutations in VH library and VL library from panning with three different ligands (anti-gD, protein A, and protein L) yielded similar and consistent results demonstrating the validity of the approach (Figs. S1–S3). The ERs of mutations in VH library from anti-gD and protein L panning were closely correlated (r2 = 0.98). The ERs from the protein A and protein l-panned VH were also correlated (r2 = 0.611) with the differential enrichment attributed to the subset of VH positions binding to protein A (28). A similar pattern was seen for the VL library panning data.
Fig. S3.
Correlation of ERs from panning of the VH or VL libraries against anti-gD antibody, protein A, and protein L. Log2ERs obtained from panning against two of the three ligands are plotted together to examine correlation (Cor). Each dot represents a given mutation in the VH (A) or VL (B) library. The dots are color-coded according to the position in the structural subsets to which they belong: hydrophobic core (Core), extended hydrophobic core (Extended core), VH–VL interface (Interface), protein A-binding site (protein A), or two protein L-binding sites (protein L1 and protein L2). The Pearson correlation coefficient is shown for each of the different structural subsets for two given pannings.
We chose the ER dataset from anti-gD panning to represent folding selection, and used a heat map to display the patterns of enrichment (red) or depletion (blue) for each mutation (Fig. 1 and Figs. S1 and S2). Enrichment indicates that the mutation is likely beneficial for the selected function (stable expression), whereas depletion indicates unfavorable mutations. Overall, the vast majority of positions appear to be tolerant to either the full range of mutations or a set of conserved mutations, because mutations at these sites do not exhibit overt depletion (Fig. 1 and Figs. S1, S2, and S4).
Fig. S4.
Identification of conserved residues important for Fab stability. (A and B) The mean log2ER of all mutations at a given position (log2ERpos) in the VH (A) and VL (B) domains, plotted on bar graphs. Positions are color-coded according to location in the hydrophobic core (green), in the extended hydrophobic core (red), or in the VH–VL interface (violet). Positions that are conserved and do not tolerate mutations (log2ERpos <−0.5) are labeled. (C and D) Using the same color-coding scheme, selected mutations mapped on the VL (C) and VH (D) of the G6 structure (in the G6/VEGF complex; PDB ID code 2FJG).
Consistent with what is known about the folding of the antibody variable domain (7, 13, 14, 29, 30), we found a set of mainly structurally buried framework positions resisting mutation (Fig. S4). These “conserved” framework positions include the hydrophobic core and VH–VL interface residues, as well as the residues involved in a hydrogen bond network at the bottom half of the variable domains. Mutations at some of the hydrogen bond network positions, like LC-R61 and LC-D82, have been shown to induce aggregation and fibril formation in LCs (31). Furthermore, a few CDR positions (e.g., R94, P100, D101 in the CDR-H3) also exhibited low tolerance to mutation, suggesting their role in overall stability. R94 and D101 are known to be important for the “kinked” structure of CDR-H3 (32), whereas HC-P100 may be structurally important for the VH–VL interface as it packs against LC-Y49 in the G6 structure.
Antigen-Distal Framework Mutations That Improve G6.31 Affinity.
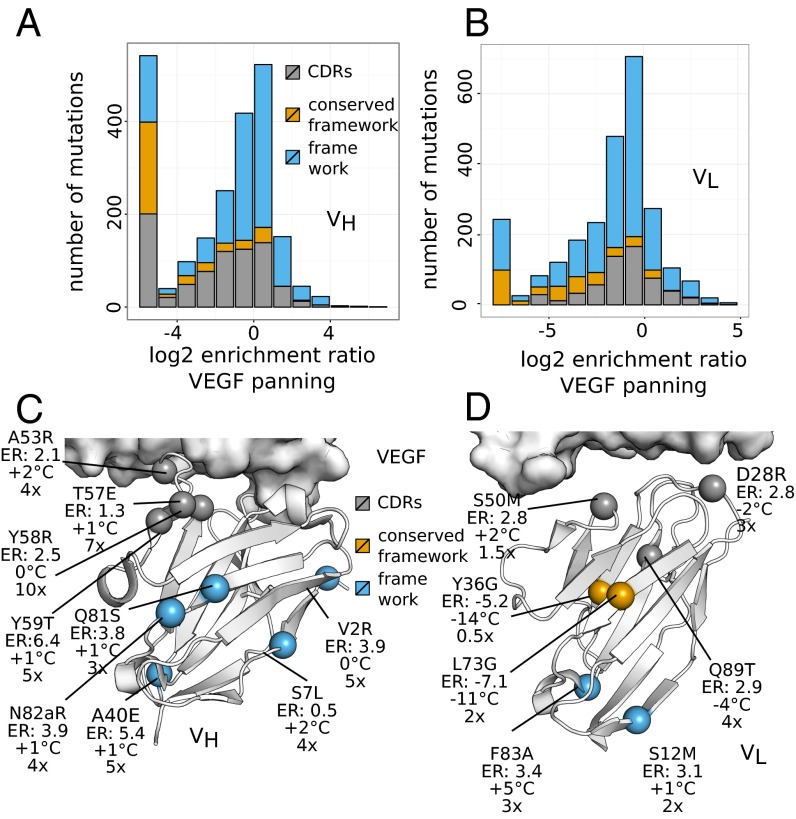
The VL and VH libraries were also selected against VEGF to obtain an overview of the effects of single substitutions on antigen binding (Fig. 1). Compared with the selection for stable expression using anti-gD, protein A, or protein L, greater stringency selection was used to enrich clones that not only bind VEGF, but bind with high affinity. Thus, the extents of enrichment/depletion obtained were more prominent. We first note that except for the CDR positions expected to be important for VEGF interaction, the ER profile was strikingly similar to that observed from the selection for functional folding and expression (Fig. 1). This is consistent with the fact that stable expression is a prerequisite for antigen binding. Along with confirming the mutational landscape for stable expression, the prominent ERs obtained from VEGF panning showed that VEGF binding for G6.31 is a HC CDR-centric event (Fig. 1), consistent with the finding that HC CDRs account for most of the structural contact with VEGF (22). Collectively, the ERs showed a bimodal distribution (Fig. 2). Whereas most mutations appear to have a neutral or negative impact on binding or folding, many potentially beneficial mutations are located in CDRs as well as in nonconserved regions of the framework. Similar bimodal distributions have been observed in deep mutagenesis scanning experiments, suggesting that this is a frequent fitness landscape for protein interactions (33).
Fig. 2.
Identification of VEGF affinity-improving mutations in G6.31. Shown is the distribution of log2ERs obtained from VEGF panning of the VH library (A) and the VL library (B). Mutations that are depleted beyond the detection limit are set to the maximum observed depletion. Mutations are color-coded according to their location in the CDRs (gray), in the conserved framework as identified in Fig. 1 and Fig. S4 (yellow), or in the rest of the framework (blue). (C and D) Using the same color scheme, selected mutations are mapped on the G6/VEGF complex structure (PDB ID code 2FJG), and the change in Tm (°C) and the fold improvement in antigen-binding affinity (Kd) compared with G6.31 Fab are shown.
We next expressed and purified several Fab variants based on the ERs obtained, which should reasonably classify the mutation as favorable or unfavorable for ligand binding and/or thermostability (Fig. S5 A and B), and measured their affinity for VEGF using Biacore and their thermostability using differential scanning fluorimetry (DSF). As expected, two examples of highly depleted mutations in the conserved framework (LC-Y36G and LC-L73G) exhibited greatly reduced thermostability (with a drop in Tm of 14 °C and 11 °C, respectively) but with intact antigen-binding affinity (0.5- and 2-fold improvement). Thus, depletion of such mutants is an effect of unstable folding, which clearly dominates over the selection for VEGF binding.
Fig. S5.
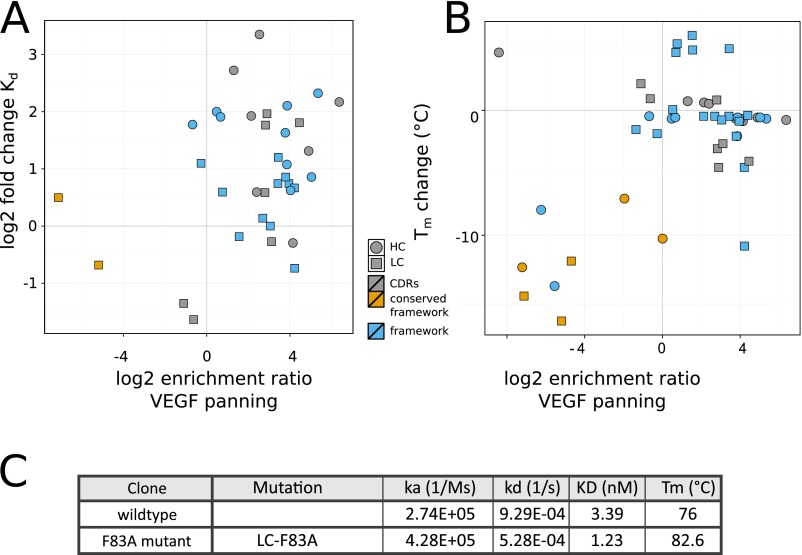
VEGF binding affinity and thermostability of selected Fab variants compared with log2ER obtained from selection against VEGF. (A and B) Relationship of the log2ER from VEGF panning of selected VH and VL mutations with log2 of the fold improvement in Kd as Fab (A) or the change in Tm from the G6.31 parent Fab (B) shown as scatterplots. Mutations were selected mainly to verify their beneficial or neutral effect on antigen binding and stability. A few detrimental mutations were selected to verify and demonstrate the range of effect. (C) The LC-F83A mutation was introduced into an anti–BACE-1 antibody, and the affinity of the WT and the mutant for BACE-1 was determined using Biacore. The thermostability of the antibodies was measured by DSF.
To identify antigen-distal mutations that improve the antibody, we mapped 16 mainly beneficial mutations (9 in VH and 7 in VL) on the G6/VEGF complex structure (Fig. 2 C and D). Whereas two CDR mutations (HC-T57E and HC-Y58R) resulted in the largest increase in VEGF binding affinity (7- to 10-fold), many antigen-distal framework mutations also produced a significant increase in affinity (2- to 5-fold). We found LC-F83 particularly interesting, because it locates 25 Å away from the antigen near the VL–CL interface. The F83A mutant exhibited not only the greatest increase in thermostability of all examined mutations (Tm +5 °C), but also a threefold increase in VEGF-binding affinity, predominantly due to a ∼twofold improvement in the rate of dissociation (off-rate), which suggests an optimized interface. Transferring the F83A mutation to another antibody (34) of the same framework as G6.31 similarly improved thermostability (Tm +6.6 °C) and binding affinity (2.8-fold, to KD of 1.23 nM), demonstrating that this result is not specific to G6.31 (Fig. S5C).
LC-F83 Conformations Correlate with the Fab Elbow Angle and the VL–CL Interface in G6 Structures.
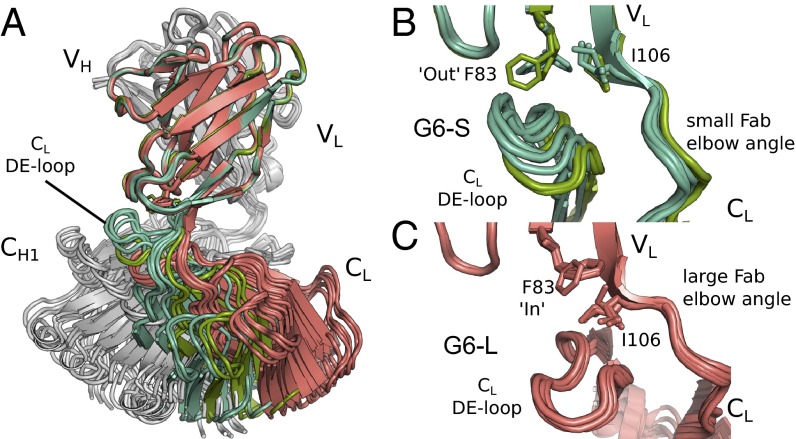
To understand how the antigen-distal mutation LC-F83A can have such a strong impact on antigen binding and stability, we first looked at the potential structural role of the LC-F83A mutation. Because G6.31 is a variant of G6 by four substitutions in CDR-L3, we used structures of G6 in the VEGF bound (G6VEGF) and free form (G6free) that we previously reported as a model for G6.31 (22). Notably, the G6free crystal structure contains 12 Fab molecules in the asymmetric unit, which can be clustered into two groups based on the Fab elbow angle (Fig. 3A), that is, the orientation of the constant domains to the variable domains (35, 36).
Fig. 3.
Association of G6 Fab elbow angles with LC-F83 conformation. (A) Fab elbow angles of the12 molecules present in the G6 Fab crystal structure asymmetric unit (PDB ID code 2FJF). LCs of the molecules exhibiting a small elbow angle (G6-S) are colored in green, and those with a large elbow angle (G6-L) are in red. HCs are in gray. (B and C) Detailed views of the VL–CL interface and the side chain conformation of LC-F83 and LC-I106 in G6-S molecules (green) or G6-L (red). The β-strand E, the helix α-1, and the loop connecting both elements are removed for clarity.
The particular class of LC in a given antibody is known to exert a dominant influence on distribution of Fab elbow angles (35). Lambda LC antibodies adopt a wide range of angles, whereas kappa LCs show a bimodal distribution peaking at ∼140° and ∼175° (35). G6 behaves like a typical human kappa LC antibody. Six G6free molecules (from the asymmetric unit) have small elbow angles (143–155°) which, interestingly, are associated with a tightly packed VL–CL interface (comprising the DE loop of the CL domain packed against α1 of VL; 304 ± 48 Å2), whereas the other six G6free molecules have a large elbow angle (170–187°) with a less tightly packed VL–CL interface (234 ± 29 Å2) (Fig. 3 B and C). Moreover, the side chain conformation of LC-F83 correlates with the G6 Fab elbow angle. In the large elbow angle structures, the LC-F83 side chain is tightly held in a hydrophobic pocket (the “in” conformation) in the distal capping region of the VL domain (Fig. 3C). In the small elbow angle structures, the LC-F83 side chain is not held in the hydrophobic pocket (the “out” conformation) and adopts rotamers that are partly solvent-exposed (Fig. 3B). Although LC-F83 is not part of the Fab elbow, its rotamers are associated with changes in the elbow residue LC-I106. In structures where LC-F83 is in the “out” conformation, the side chain of LC-I106 moves “up” to occupy the hydrophobic pocket, whereas when LC-F83 is in the “in” conformation, the side chain of LC-I106 moves “down.” In contrast to LC-F83, the movement of LC-I106 is not limited to the side chain, but also involves changes in the local protein backbone (LC-105 Φ-angle, LC-106 ψ-angle) that determine the elbow angles as observed. In summary, the crystal structures of G6 indicate that change in the G6 Fab elbow angle is associated with the conformational states of LC-F83.
Molecular Dynamics Simulations Indicate the Effect of LC-83 Mutation on the G6 Elbow Conformation.
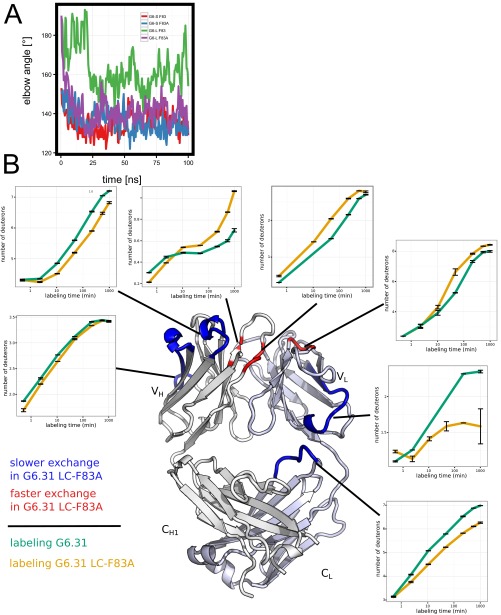
To gain further insight into the effects of the LC-F83A mutation found on our structural analysis, we turned to molecular dynamics (MD) simulations. The effect of the LC-F83A mutation was simulated in two different structural backgrounds, a G6 Fab crystal structure with a large elbow angle, G6-L [G6 HC V and LC U in G6Free; Protein Data Bank (PDB) ID code 2FJF], or a G6 Fab crystal structure with a small elbow angle, G6-S (G6 chains B and A; PDB ID code 2FJF). Both structures, together with the two structures modeled with an LC-F83A mutation (G6-L-83A and G6-S-83A), were simulated in water for 100 ns. All molecules except G6-L-83A showed a slight reduction of the elbow angle of the respective crystal structure in the first 25 ns of the simulation; however, after the initial 25 ns, the elbow angles of G6-L, G6-S, and G6-S-83A remained stable. For G6-L, the F83 stayed in the “in” conformation, and the elbow angle remained large (∼161°), whereas G6-S maintained the “out” F83 conformation and a small elbow angle (∼135°). G6-S-83A maintained the small elbow angle distribution (Fig. 4 A and B and Fig. S6A). In contrast to the other three structures, G6-L-83A showed a quick transition from a large elbow angle to a smaller elbow angle (∼140°) and a larger VL–CL interface (Fig. 4 A and B and Fig. S6A), a conformation that is maintained during the remainder of the simulation.
Fig. 4.
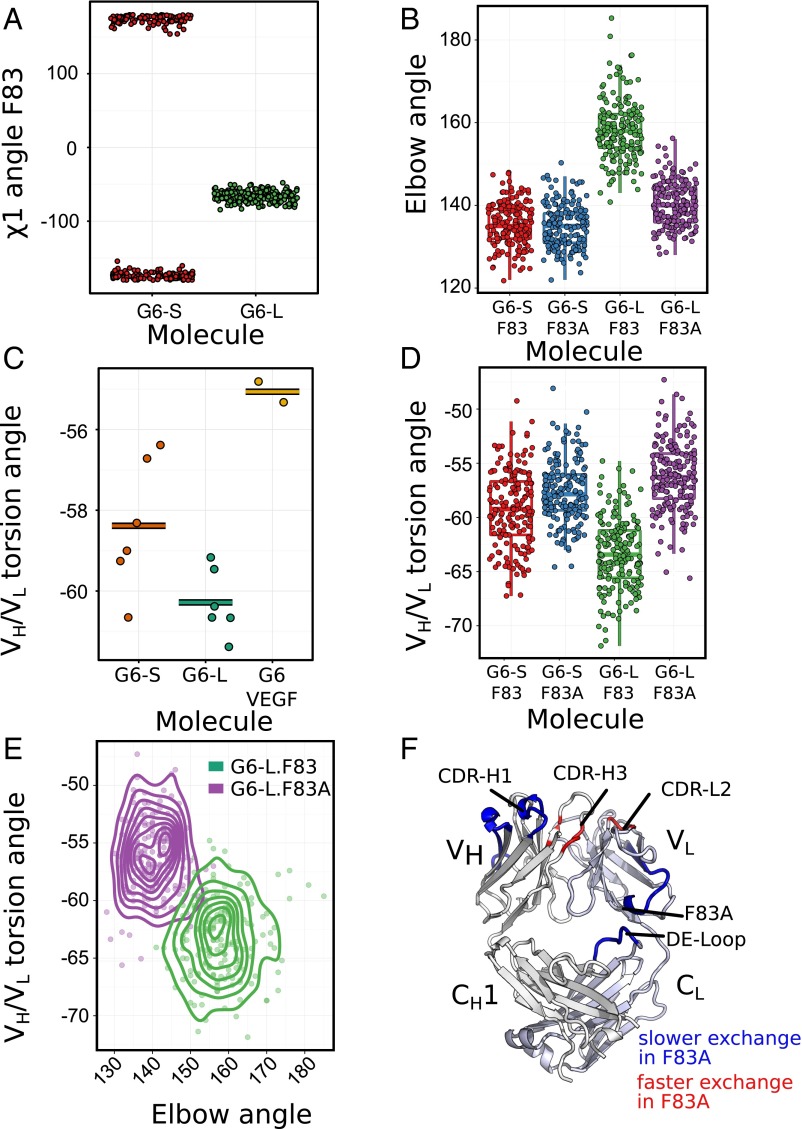
Mutations at LC-F83 regulate G6 conformation and domain dynamics. (A) Distribution of the F83 side chain conformation (χ1 angle) of G6-S (small elbow angle) or G6-L (large elbow angle) of the G6free (PDB ID code 2FJF) during a 100-ns MD simulation, showing little transition. The χ1 angle of the G6-S F83 was artificially split because it was plotted in a range of −180° to +180°. (B, D, and E) Data from the last 75 ns of the 100-ns MD simulations. (B) Distribution of the Fab elbow angle for the molecules G6-S, G6-S-83A, G6-L, and G6-L-83A. All samples are significantly different (P < 0.001) except G6-S and G6-S-83A, as determined by ANOVA with Tukey’s honest significance difference (HSD) test. (C) VH–VL torsion angle for the six molecules of G6free structures with F83 in an “out” conformation (orange) and for the six molecules with G6free with F83 in an “in” conformation (green), as well as the VH–VL torsion angle of the VEGF-bound G6 structures (yellow). (D) VH–VL torsion angle distribution of the same molecules obtained from the same MD simulation. All samples except G6-S-83A and G6-L-83A are significantly different (P < 0.001) by the HSD test. (E) Contour plot showing the overall Fab interdomain dynamics. The elbow angle vs. the VH–VL torsion angle is plotted for selected time points of the MD simulation using a G6 structure with LC-F83 in an “in” conformation (G6-L; red) and the same structure with an LC-F83A mutation (G6-L-83A; green) as starting points. Two structures occupy distinct regions in the conformational landscape. (F) In HX-MS experiments comparing G6.31 F83A and G6.31 Fab, the positions that exchange faster (red) or slower (blue) are depicted on the cartoon representation of the G6free Fab structure (PDB ID code 2FJF) (Fig. S6).
Fig. S6.
G6 Fab elbow angle movement during the MD simulation and effect of F83A in HX-MS. (A) G6 Fab elbow angle movement during a 100-ns MD simulation using either G6-S (chain AB; PDB ID code 2FJF) or G6-L (chain VU; PDB ID code 2FJF) or respective molecules with a LC-F83A mutation (G6-S-83A and G6-L-83A) of the antigen-free form of G6. (B) Cartoon representation of the G6 Fab (PDB ID code 2FJF), with regions that exchange faster (red) or slower (blue) in G6.31 F83A Fab compared with G6.31 Fab WT in HDX-MS experiments highlighted. Representative titration curves of hydrogen-deuterium exchange for the regions with changes in the exchange rate are shown. The line of the titration curves passes through the mean of two independent experiments; the upper limit and the lower limit of the bars represent the two data points obtained from the two experiments.
Meta-Analysis of Human Antibody Structures Showing a Correlation Between LC-83 Conformation and VL–CL Packing.
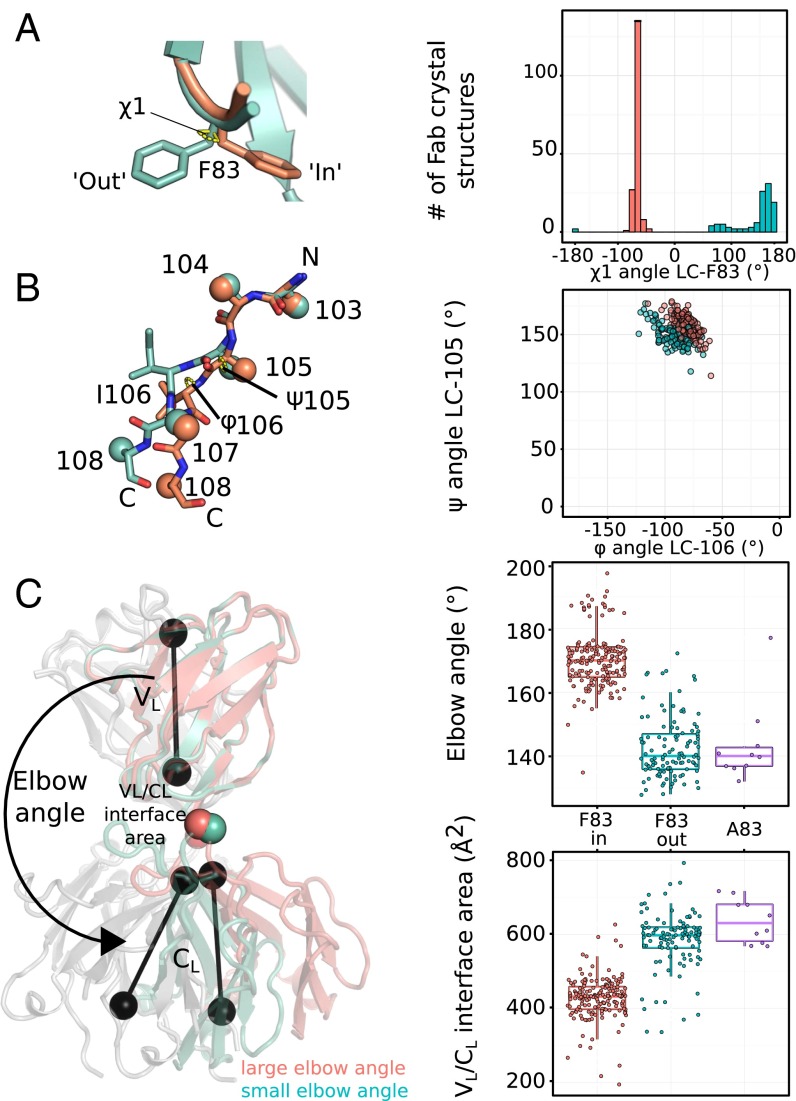
We next wished to determine whether the coupling of the side chain conformation of LC-83 with the Fab elbow conformation is a common feature in human antibodies. The most frequently used human kappa germline families, IGKV1 and IGKV3 (IMGT database) (37), carry phenylalanine at position 83, whereas the other, less frequently used germlines carry mostly valine (IGKV2 and IGKV4) or alanine (IGKV5 and IGKV6). The isoleucine at position LC-106 is conserved in all five human IGKJ germline genes. We examined the conformation of LC-F83 of 319 Vkappa-containing human Fab crystal structures in the PDB and divided these structures into two groups: LC-F83 in an “in” conformation (dihedral χ1 angle between −50° and −100°) and LC-F83 in an “out” conformation (dihedral χ1 angle mostly between 50° and 180°) (Fig. 5A). Indeed, the two groups of LC-F83 rotamers correlate with changes in the Fab elbow region at position LC-106 (LC-105 Φ-angle, LC-106 ψ-angle) in the vast majority of structures (Fig. 5B). Furthermore, the structures with a large elbow angle have a smaller, less tightly packed VL–CL interface, whereas structures with a small elbow angle have a large VL–CL interface (Fig. 5C).
Fig. 5.
LC-83 conformation correlates with the elbow angles in human antibody structures. (A) The bimodal distribution of F83 side chain χ1 angle rotamers of 319 Vkappa-containing human antibody structures from PDB with Phe at the position 83 in an “in” (red) or an “out” (green) conformation. (B) The 319 structures categorized according to their F83 conformation as in A exhibit a distinct distribution of elbow angle, as represented by the backbone conformation at position 105 (ψ angle) and position 106 (φ angle). (C, Left) Illustration of how elbow angles are determined for G6 Fab structures (in cartoon representations) with LC-F83 in an “in” (orange) or an “out” (green) conformation. The black dumbbells represent the pseudo-twofold symmetry axis through the variable domains or the constant domains used to calculate Fab elbow angles. The red and green spheres represent the border residue (LC-107) between the VL and CL domains. (C, Right) Elbow angle or VL–CL interface area of the Vkappa containing human Fab crystal structures from the PDB with LC-83 with an “in” or “out” conformation, with nine Vkappa Fab structures that harbor LC-A83 singled out for comparison.
Given that the LC-F83A mutation in G6.31 resulted in greater thermostability and greater affinity toward the antigen, we examined the elbow angle of nine human Vkappa-containing Fab structures carrying LC-A83 and LC-I106 available in the PDB. We showed that those structures mirror the properties of the LC-F83 “out” confirmation structures with a small elbow angle and a large VL–CL interface (Fig. 5 B and C). We determined that for eight of the nine antibodies, LC-A83 is a result of somatic mutations. The remaining antibody LC is from IGKV6, which carries an alanine as a germline at position LC-83. In summary, we have demonstrated that the bimodal distribution in the Fab elbow angle observed in human kappa LC antibody structures (35) is associated with conformations of the phenylalanine side chain at position LC-83. Furthermore, the type of amino acid present at position LC-83 indeed influences the range of the elbow angle and, consequently, the VL–CL interface area.
Therefore, our meta-analysis of human antibody structures, along with the MD simulations, strongly suggest that the greater thermostability of G6.31LC-F83A compared with G6.31 can be explained by an increase in the VL–CL interface interaction area through the decrease in the Fab elbow angle, which likely further stabilizes the fold of the four domains composing the Fab.
Elbow Angle Dynamics of the G6 Fab Influence the Antigen-Binding Interface by Modulating the VH–VL Torsion Angle.
Whereas the change in Fab elbow angle explains the increase in thermostability of the LC-F83A variant, the effect of the mutation on antigen-binding affinity cannot be explained. We hypothesize that the LC-F83A mutation may influence the orientation of the VH and VL domains toward each other (i.e., the VH–VL torsion angle) either indirectly via the change in Fab elbow angle or directly because LC-83 sits close to the VH–VL interface. The VH–VL interface has been shown to have a strong influence on antigen-binding affinity (38, 39). Furthermore, the VH–VL torsion angle and flexibility of the VH–VL interface have been reported to vary substantially between the antigen-free and the antigen-bound forms (40, 41).
Indeed, the crystal structures of G6 show significant differences in the VH–VL torsion angle between the G6free and the G6VEGF. The mean VH–VL angle was −60° for the six G6free molecules with a large elbow angle and –58° for the six G6free molecules with a small elbow. The VH–VL torsion angle of G6VEGF (PDB ID code 2FJG) was −55°, closer to that of the G6free with small elbow angle (Fig. 4C).
To build confidence that the observed torsion angles are not an artifact of crystallization, we turned again to MD simulation. We found that the median VH–VL torsion angle was −62° for G6-L (with a large elbow angle) and −59° for G6-S (with a small elbow angle), consistent with that seen in G6free crystal structures (Fig. 4D). Moreover, the two mutant Fabs (G6-L-83A and G6-S-83A) exhibited an even smaller VH–VL angle (both −57°) (Fig. 4D). The dynamic distribution of the VH–VL torsion angle and Fab elbow angle for G6-L and G6-L-83A (Fig. 4E) clearly indicates that the LC-F83A mutation modulates the interdomain conformations. The analysis suggests that, through its influence on the VH–VL torsion angle, the LC-83 mutation improves G6-VEGF binding affinity likely by optimizing the interface complementarity with VEGF or reducing the entropy cost for VEGF binding.
Hydrogen-Deuterium Exchange Mass Spectrometry Confirms the Effect of F83A Mutation in the VL–CL Interface and the Antigen-Binding Regions.
To assess how the LC-F83A mutation affects interdomain dynamics of the G6.31 Fab, we used hydrogen-deuterium exchange mass spectrometry (HX-MS) to measure the exchange rate of backbone amide hydrogen atoms in D2O by following the increase in mass as backbone protons exchange with deuterium over time. The exchange rate of each backbone amide proton reflects the aggregate structural energetics of all H-bonding interactions with that specific exchange site (42). Increased structural stability or reduced conformational fluctuation generally causes a relative reduction in the observed exchange rate of nearby residues, and the converse is true as well.
Comparing exchange rates between G6.31 and G6.31F83A Fab revealed both short-range and long-range consequences of the mutation on structural energetics (Fig. 4F and Fig. S6B). The significant reduction in the exchange rate near position 83 in G6.31F83A is consistent with the prediction that VL–CL interface packing increases on mutation. Long-range allosteric effects of the mutation are manifested near the antigen-binding site. The CDR loop H1 exchanges more slowly in G6.31F83A, whereas the sections of the CDR-H3 and CDR-L2 known to be a part of the VH–VL interface (7, 43, 44) exchange more rapidly.
The results obtained by the MD simulation and HX-MS are in good agreement, and thus both methods independently confirm the impact of this antigen-distal mutation at LC-83 on Fab domain orientation and dynamics.
LC-83 and LC-106 Are Frequently Mutated in Human Antibodies.
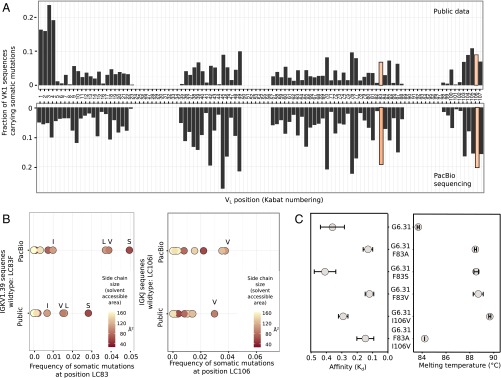
We next investigated whether mutation at the position LC-83 occurs frequently during affinity maturation in vivo among human antibodies of the same germline family. We used two approaches to examine the distribution of SHMs in LC sequences originating from IGKV1 (Vkappa1) germlines. In the first approach, we collated the SHMs of 4,623 unique human IGKV1 LC sequences available in several publicly available databases (Fig. 6A, Upper). In the second approach, we amplified IGKV1 sequences from RNA originating from more than 1,000 individual human lymphoid tissues containing naïve and antigen-experienced B cells, followed by high-fidelity circular consensus single-molecule real-time sequencing (SMRT; Methods) and SHM collating (Fig. 6A, Lower). Despite the differing origins of the two datasets, the distribution of somatic mutations was very similar. Interestingly, LC-83 is one of the most frequently mutated positions in the VL segment; 7% and 19% of all IGKV1 sequences in the two datasets, respectively, carry a mutation at LC83. Interestingly, position LC-106 also frequently mutated in IGKV1 sequences (9% and 20%, respectively), with valine the most common mutation (Fig. 6C).
Fig. 6.
Somatic mutations in human antibody sequences in IGKV1 germline-derived antibodies. (A) Histogram showing the fraction somatically mutated for the indicated VL positions of antibody sequences originating from IGKV1 germlines. The mutational distribution in the upper panel was obtained using publicly available human antibody sequences from the GenBank, PDB, Kabat, Abysis, and IMGT databases (Public). The lower panel was obtained using SMRT sequencing (PacBio) of cDNA obtained from more than 1,000 individual human lymphoid tissues. The high mutation rates at the N terminus of the public sequences likely come from cloning artifacts. (B) The most common mutations at position LC-83 for IGKV1-39 sequences (Left) or at position LC-106 (Right) from the Public dataset or the PacBio dataset. IGKV1-39 carries a phenylalanine at position LC-83. Points are color-coded by the respective amino acid size as noted. (C) The affinity (Kd) (Left) and Tm (Right) of selected mutations of G6.31 as measured by Biacore and DSF, respectively. Values represent the mean from three replicates, and error bars show the SD.
The closest germline gene segments for the G6 VL are IGKV1-39 and IGJ1. Thus, we examined the antibodies of the IGKV1-39 germline source, and found that somatic mutations at F83 primarily result in smaller side chains (serine, valine, and leucine) (Fig. 6B). We did not observe any alanine in antibodies of IGKV1-39, possibly due to inadequate sampling, because an alanine mutation would require a two-base substitution in the codon at F83 and thus occur only infrequently.
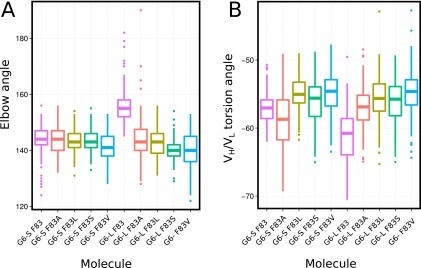
We next ran MD simulations of the three most frequent LC-F83 mutations (F83S, F83V, and F83L) in the context of the G6 structure (Fig. S7). The simulations show a similar effect of the LC-F83S, LC-F83V, and LC-F83L mutations on the elbow and the HL angle as that produced by LC-F83A (Fig. 4 B and D). In addition, G6.31 Fab variants carrying the most frequent somatic mutations for positions 83 and 106 (F83A, F83S, F83V, and I106V) were generated and tested for their effect on antigen binding and thermostability (Fig. 6C). All single-mutation variants increased the Tm by 4.8–6 °C, presumably by decreasing the Fab elbow angle and increasing the VL–CL interface area. Consistently, the double mutation LC-F83A/I106V did not result in a significant increase in Tm, because simultaneously reducing the size of both side chains left a void in the hydrophobic cavity typically occupied by F83 or I106. The effect of the LC-83 mutation on VEGF affinity was mixed. F83V led to a threefold increase in affinity, very similar to LC-F83A, whereas the F83S and I106V mutations showed nearly no increase in affinity. The effect of the LC-83 mutation on antigen affinity may be context-dependent. We note that the F83S mutation reportedly increased the affinity of a Vkappa1-containing anti-estradiol antibody (45).
Fig. S7.
MD simulation study of the G6 Fab F83 mutants. The most frequent somatic mutations at the LC-83 position identified in human kappa-containing LC were modeled in G6-S or G6-L, and the distributions of the Fab elbow angle (A) and the VH–VL torsion angle (B) obtained during the last 75 ns of the 100-ns MD simulation are plotted.
In summary, LC-83 and LC-106 are frequently mutated in human antibodies. Given the influence of these two positions on affinity and stability, optimization of the overall Fab domain dynamics may be a widely used molecular mechanism during antibody affinity maturation in vivo.
Conclusions
In this work, we mapped the entire mutational landscape of the VH and VL domains of a human antibody by single-site saturated mutagenesis, showing the impact of mutation at each position on antibody stability and antigen binding. Using a similar approach, Traxylmeyr et al. (46) recently reported that residues of the HC constant domains CH2 and CH3 are generally conserved and resist mutation for proper folding, in stark contrast to the high mutational tolerance of the numerous variable domain positions of G6.31 (Figs 1 and 2 and Figs. S1 and S3). This suggests that antibody constant domains are highly evolved and optimized for folding and stability, whereas variable domains can be mutated with excellent potential to further improve stability and optimize binding function. The comprehensive mutational landscape of the variable domain suggests a road map for the somatic mutation process to enhance antibody function and also guide the engineering and optimization of antibodies needed for therapeutic development.
To search for novel mechanisms of somatic mutation that affect antigen-binding function, we focused on beneficial mutations at framework positions distal to the antigen. We report how mutation at residue 83 of G6.31 LC can modulate interdomain structural dynamics of Fab molecules through interaction with elbow residue LC-I106. The interplay of these two residues is associated not only with the dynamics of Fab elbow angle, but also with the interaction between VL and CL and between VL and VH (Figs. 3–5). We further demonstrate the high frequency of mutations at LC-F83 in human antibodies of the IGKV1 germline source, often to smaller residues (Fig. 6), and show that these mutations are associated with similar changes in the interdomain conformations as for G6.31.
Our analysis of anti-VEGF antibody G6.31 indicates several specific roles for the residue in LC position 83 during in vivo affinity maturation. First, LC-F83 mutations have an impact on antigen binding by influencing the orientation of VH and VL (Fig. 6), and thus the topography of the CDRs. Second, F83 mutation to the smaller side chain (A, V, or S) induces a more compact Fab conformation and increases structural stability. Third, LC-F83 mutation can affect the biological activity of an antibody–antigen interaction through its modification of the Fab elbow angle and interdomain dynamics (47–49). Furthermore, B-cell signaling is sensitive to small conformational changes in the B-cell receptor (50). Modulation of the Fab elbow angle dynamics may have a favorable effect on B-cell receptor signaling (51).
Our study provides insight into the active role of framework positions in modulating antibody affinity and stability. The CDRs typically provide the main direct contacts with antigens, but—as highlighted by our analysis—affinity also depends on framework residues, even when the FWR positions are structurally far away from the antigen. Our investigation into the role of mutations at LC position 83 has unraveled the complex relationship between a framework mutation and the conformational dynamics of a Fab molecule, emphasizing the importance of conformational dynamics in antibody–antigen interaction.
Materials and Methods
Full Variable Domain NNK-Walk Library Design, Generation, and Phage Panning.
VH residues 2–113 and VL residues 2–107 of G6.31 Fab in a phagemid construct, as described previously (26) (Fig. S1), were subjected to randomization by oligonucleotide-directed mutagenesis (52). Between 10 and 12 sequential residues were mutagenized in a single reaction using a mixture of oligonucleotides, each with an NNK codon at one position targeted for randomization. NNK encodes for all 20 amino acids (n = G, A, T, and C; K= G and T, in equal proportions). The template for each mutagenesis reaction was generated by introducing stop codons (TAA) at all of the positions targeted for randomization in each reaction. The distribution of NNK vs. wild-type (WT) codons at a given position depended on the size of the randomized segment; for example, for the VL domain, 106 positions (amino acid position 2–107) were randomized. Thus, for a given position in the library, 99.1% of reads carried the WT codon, whereas only 0.9% of the reads carried mutations resulting from the NNK randomization. The VH and VL libraries were generated separately by electroporating the pool of DNA products from mutagenesis reactions into Escherichia coli XL1 cells, yielding ∼109 transformants as described previously (24).
Phage Panning Selection.
To select for stably expressed clones, the VH and VL libraries were incubated with protein A, protein L, or anti-gD immobilized on ELISA wells for 2 h, followed by washing away of the unbound phage. Bound phage was eluted in 100 mM HCl for 20 min, then neutralized with 1/10 volume of 1 M Tris pH 11.0 and used to infect E. coli for amplification, followed by one round or two rounds of plate-based selection. VEGF-binding clones were selected by incubating the phage display libraries with 5, 0.5, and 0.1 nM biotinylated VEGF in solution in the successive rounds of selection with increasing stringency, as described previously (53). Bound clones were captured on ELISA wells coated with neutravidin, washed, eluted, and amplified as above.
Illumina Sequencing of G6.31 Deep Mutational Scanning Libraries.
For deep sequencing, phagemid DNA was isolated from E. coli XLI cells from either the unselected or the selected G6.31 NNK library and used for a limited-cycle PCR-based amplification of VL and VH regions using Phusion polymerase (New England BioLabs). PCR products were purified to generate libraries using the TruSeq Nano DNA library preparation kit (Illumina). Multiplexed adapter-ligated libraries with unique barcodes were sequenced on an Illumina MiSeq sequencing system for 300-bp paired-end sequencing.
Deep Mutational Scanning Data Analysis.
After merging of paired-end reads using FLASh (54), data analysis was performed using the statistical programming language R (55) and the ShortRead package (56) (Table S1). The first step in quality control was to filter out sequences that did not carry the respective VH and VL barcodes. In a second step, the flanking regions of the VH and VL domains were identified for each merged read and checked for the correct lengths to remove the reads with indels (insertion and deletion). To further correct for sequencing errors, non-NNK mutations were converted back to the WT base, and reads that contained two or more NNK mutations were filtered out. Calculating the frequency of all mutations at each randomized position generated position weight matrices. ERs for all mutations were calculated by dividing the frequency of a given mutation at a given position in the sorted sample by the frequency of the same mutation in the unsorted sample, as described previously (25, 26). Mutagenesis of one segment of VH (position 14–25) in the original VH library resulted in errors which led to low expression of the respective segment. This result was likely related to a mutational error in the original stop template used in the mutagenesis reaction for this VH segment 14–25, which affected expression of the variants. This error was discovered only after sequencing and data analysis. Therefore, a second VH library was generated with correction in the 14–25 segment (by making a new stop template vector), and phage selection was repeated. Outside of the 14–25 regions, data from the two VH libraries were generally correlated with each other, and data from the second library were used in all of the selections for folding and expression (anti-gD antibody, protein A, and protein L). The VEGF selection data of the corrected segment from the second library were merged with the sequence data of the original library after calculation of ERs. To adjust for different sequencing depths, the dataset of the second VH library was adjusted by obtaining a random sample of sequences of sizes matched to the dataset obtained from the first VH library (Table S1).
Antibody Expression and Characterization.
VH and VL sequences of selected variants were cloned into a mammalian Fab vector for expression and purification with a protein G column. Surface plasmon resonance measurements with a BIAcore T200 instrument was used for affinity determination as described previously (24, 26). In brief, single cycle injection was used for kinetic measurement. VEGF (8–109) was coupled to Series S sensor chip CM5 (GE Healthcare) to achieve an Rmax value of ∼50 response units. Serial dilutions of Fab in HBS-P buffer (0.01 M Hepes pH 7.4, 0.15 M NaCl, and 0.005% Tween 20) (0.08–50 nM) were injected. The VEGF-binding responses were corrected by subtracting responses from a blank flow cell. Association rates (kon), dissociation rates (koff), and Kd (koff/kon) were calculated using a simple one-to-one Langmuir binding model. For Tm, we used DSF, which monitors thermal unfolding of proteins in the presence of fluorescent SYPRO orange dye (Invitrogen). The diluted dye (1:20; 1 µL) was added into 24 µL of Fab protein (∼100 µg/mL). The fluorescence intensity during the temperature increase from 20 °C to 100 °C was plotted, and Tm, the inflection point of the transition curve, was calculated using the Boltzmann equation (57).
Structural Meta-Analysis.
Human antibody structures were obtained from the structural antibody database SAbDab (58). Additional structure filtering and manipulation were performed using Bio3D (59). The interface area between constant and variable domains was calculated using a local instance of CCP4-Pisa (60). ABangle (40) was used to characterize the VH–VL interaction. Pymol and a publicly available script (www.pymolwiki.org/index.php/Elbow_angle) were used to calculate the elbow angle.
MD Simulations.
Before the MD simulations were performed, segments in the constant domains of G6 (HC130-145, HC220-223, and LC211-213) that had not been resolved in the G6 crystal structure were merged from a high-resolution crystal structure (4hh9.DC) into the G6 structure. Mutations at the LC-83 position were introduced using Pymol. The AMBER12 simulation package (61) with the ff99SB force field was used in all minimization, equilibration, and production stages. All structures were solvated in a truncated octahedron box using the TIP3P water mode with a 10-Å buffer of solvent between the solute's furthest dimensions in each direction to avoid interaction with self-images. Each system was neutralized by adding Cl counterions using a Coulomb potential on a 1-Å grid. (All initial systems were of net positive charge.) The system (solute and solvent) was minimized in six stages with a harmonic restraint on the solute slowly decreasing from 40 to 0 kcal/mol/Å2, with each stage over 100 steps of steepest descent. Following the energy minimization, the system temperature was increased from 150 K to 300 K over 50 psec at 2-fsec integration steps. Bonds involving hydrogens were constrained by the SHAKE algorithm. Production data of 100 ns were collected at the NPT ensemble at 3-fsec time steps and recorded every 15 psec. The particle mesh Ewald method was used to calculate the full electrostatic energy of a periodic box. The obtained simulated structures were analyzed using VMD (62), Bio3d, CCP4-Pisa, Pymol, and ABangle.
HX-MS.
G6.31 WT and F83A mutant Fab (45 μM) were diluted 15-fold into a 20 mM histidine-acetate buffer at pH 7.0 and 50 mM NaCl with >90% D2O content to begin the labeling reaction at 20 °C. At six logarithmically spaced time intervals spanning 30 s to 1,000 min, the labeling reaction was quenched by pH reduction (pH 2.5) and the addition of 2 M guanidinium chloride and 0.25 M Tris(2-carboxyethyl)phosphine before injection onto a cold online system (63). In brief, quenched samples were first passed through an immobilized pepsin column (2.1 × 30 mm; Applied Biosystems) at 0 °C for proteolysis and then bound to a trap column (Acquity Vanguard C8) for desalting before being separated by reverse-phase chromatography (Acquity UPLC BEH C18; 1.7 μm particle size; 1.0 × 50 mm) and introduced into the mass spectrometer (Thermo Orbitrap Elite; 120 kHz resolution at m/z 400) for measurement of deuterium content. Chromatographic mobile phases were prepared as described previously (64) with a pH of 2.25 to maximize deuterium recovery, which averaged 82% by measurement of fully deuterated controls. Mutant and WT experiments were interleaved, and randomized time points were collected in triplicate.
This process resulted in 152 unique peptides, consistently identified by the ExMS program (65) for both WT and mutant samples, providing sequence coverage of roughly 95%. The analysis of deuterium content involved the use of previously described custom Python scripts (64, 66, 67). Significant differences in deuterium uptake levels between WT and mutant were identified by Student’s t test as having a P value < 0.05, as described previously (68).
Somatic Mutations in Human Antibodies.
This analysis was performed for antibody sequences obtained from publicly available sources. Duplicated sequences were removed, and the closest germline genes were assigned for the respective V-segments using IGBlast (69). Sequences to which a mouse or rat germline was assigned were removed from the dataset. Sequences were Kabat-numbered (70) and somatic mutations in the framework of the V-segment were identified. Mutations in the CDR regions (as defined by the Kabat numbering scheme) were not considered in our analysis.
In a second experiment, the RNA from peripheral blood lymphocytes, tonsil, spleen, and bone marrow of more than 1,000 individuals was obtained from commercial source (Clontech, Amsbio, and Biochain) either as total RNA or poly(A)-positive RNA. The RNA was first transcribed into cDNA using the SMARTer RACE cDNA Amplification Kit (Clontech). IGKV-specific DNA was amplified using 5′ RACE (Advantage 2 PCR Kit; Clontech) with a primer annealing the 5′ region of the IGKC region (5′ CATCAGATGGCGGGAAGATGAAGACAGATGGTGC 3′), whereas IGKV1 segments were enriched in a second PCR step using the following primers: forward, 5′-GCCATCCAGATGACCCAGTCTCC-3′; reverse, 5′-GGCTGCACCATCTGTCTTC-3′. The PCR product was circularly (and hence repeatedly) sequenced using Pacific Bioscience RSII (EA Genomic Services). A total of 994.8 Mbp were obtained. A consensus of each circular sequencing read was created to improve the fidelity of sequencing using Pacific Bioscience’s SMRT Analysis 2.3 software package. The germline annotation of the consensus sequences was performed using VDJFasta (71). Sequences were Kabat-numbered, and somatic mutations in the framework of the V-segment were identified. A total of 19,034 sequences were annotated as IGKV1; among these, 3,796 contained at least three FWRs and two CDRs and were included to generate the SHM distributions.
Acknowledgments
We thank the Genentech Oligonucleotide Synthesis group, Sequencing group, Bioinformatics group, Early-Stage Cell Culture group, Antibody Engineering Protein group, and Outsourcing group for technical assistance, and Isidro Hötzel, Wenwu Zhai, and Greg Lazar for insightful discussions.
Footnotes
Conflict of interest statement: All authors are or were once paid employees of Genentech Inc., a member of the Roche Group, and are inventors of a patent application based on the work described herein.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1613231114/-/DCSupplemental.
References
- 1.Kim S, Davis M, Sinn E, Patten P, Hood L. Antibody diversity: Somatic hypermutation of rearranged VH genes. Cell. 1981;27(3 Pt 2):573–581. doi: 10.1016/0092-8674(81)90399-8. [DOI] [PubMed] [Google Scholar]
- 2.Neuberger MS, Milstein C. Somatic hypermutation. Curr Opin Immunol. 1995;7(2):248–254. doi: 10.1016/0952-7915(95)80010-7. [DOI] [PubMed] [Google Scholar]
- 3.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302(5909):575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 4.Foote J, Winter G. Antibody framework residues affecting the conformation of the hypervariable loops. J Mol Biol. 1992;224(2):487–499. doi: 10.1016/0022-2836(92)91010-m. [DOI] [PubMed] [Google Scholar]
- 5.Tomlinson IM, et al. The imprint of somatic hypermutation on the repertoire of human germline V genes. J Mol Biol. 1996;256(5):813–817. doi: 10.1006/jmbi.1996.0127. [DOI] [PubMed] [Google Scholar]
- 6.Burkovitz A, Sela-Culang I, Ofran Y. Large-scale analysis of somatic hypermutations in antibodies reveals which structural regions, positions and amino acids are modified to improve affinity. FEBS J. 2014;281(1):306–319. doi: 10.1111/febs.12597. [DOI] [PubMed] [Google Scholar]
- 7.Padlan EA. Anatomy of the antibody molecule. Mol Immunol. 1994;31(3):169–217. doi: 10.1016/0161-5890(94)90001-9. [DOI] [PubMed] [Google Scholar]
- 8.Brummell DA, et al. Probing the combining site of an anti-carbohydrate antibody by saturation-mutagenesis: Role of the heavy-chain CDR3 residues. Biochemistry. 1993;32(4):1180–1187. doi: 10.1021/bi00055a024. [DOI] [PubMed] [Google Scholar]
- 9.Burks EA, Chen G, Georgiou G, Iverson BL. In vitro scanning saturation mutagenesis of an antibody binding pocket. Proc Natl Acad Sci USA. 1997;94(2):412–417. doi: 10.1073/pnas.94.2.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chowdhury PS, Pastan I. Improving antibody affinity by mimicking somatic hypermutation in vitro. Nat Biotechnol. 1999;17(6):568–572. doi: 10.1038/9872. [DOI] [PubMed] [Google Scholar]
- 11.Yang WP, et al. CDR walking mutagenesis for the affinity maturation of a potent human anti–HIV-1 antibody into the picomolar range. J Mol Biol. 1995;254(3):392–403. doi: 10.1006/jmbi.1995.0626. [DOI] [PubMed] [Google Scholar]
- 12.Wagner SD, Milstein C, Neuberger MS. Codon bias targets mutation. Nature. 1995;376(6543):732. doi: 10.1038/376732a0. [DOI] [PubMed] [Google Scholar]
- 13.Hsu HJ, et al. Antibody variable domain interface and framework sequence requirements for stability and function by high-throughput experiments. Structure. 2014;22(1):22–34. doi: 10.1016/j.str.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 14.Chang HJ, et al. Loop-sequence features and stability determinants in antibody variable domains by high-throughput experiments. Structure. 2014;22(1):9–21. doi: 10.1016/j.str.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 15.Wang F, et al. Somatic hypermutation maintains antibody thermodynamic stability during affinity maturation. Proc Natl Acad Sci USA. 2013;110(11):4261–4266. doi: 10.1073/pnas.1301810110. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Klein F, et al. Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell. 2013;153(1):126–138. doi: 10.1016/j.cell.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu X, et al. NISC Comparative Sequencing Program Focused evolution of HIV-1–neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333(6049):1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doria-Rose NA, et al. NISC Comparative Sequencing Program Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature. 2014;509(7498):55–62. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Banfield MJ, King DJ, Mountain A, Brady RL. VL:VH domain rotations in engineered antibodies: Crystal structures of the Fab fragments from two murine antitumor antibodies and their engineered human constructs. Proteins. 1997;29(2):161–171. doi: 10.1002/(sici)1097-0134(199710)29:2<161::aid-prot4>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 20.Wedemayer GJ, Patten PA, Wang LH, Schultz PG, Stevens RC. Structural insights into the evolution of an antibody combining site. Science. 1997;276(5319):1665–1669. doi: 10.1126/science.276.5319.1665. [DOI] [PubMed] [Google Scholar]
- 21.Xiao X, et al. Germline-like predecessors of broadly neutralizing antibodies lack measurable binding to HIV-1 envelope glycoproteins: Implications for evasion of immune responses and design of vaccine immunogens. Biochem Biophys Res Commun. 2009;390(3):404–409. doi: 10.1016/j.bbrc.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuh G, et al. Structure-function studies of two synthetic anti-vascular endothelial growth factor Fabs and comparison with the Avastin Fab. J Biol Chem. 2006;281(10):6625–6631. doi: 10.1074/jbc.M507783200. [DOI] [PubMed] [Google Scholar]
- 23.Liang WC, et al. Cross-species vascular endothelial growth factor (VEGF)-blocking antibodies completely inhibit the growth of human tumor xenografts and measure the contribution of stromal VEGF. J Biol Chem. 2006;281(2):951–961. doi: 10.1074/jbc.M508199200. [DOI] [PubMed] [Google Scholar]
- 24.Lee CV, et al. High-affinity human antibodies from phage-displayed synthetic Fab libraries with a single framework scaffold. J Mol Biol. 2004;340(5):1073–1093. doi: 10.1016/j.jmb.2004.05.051. [DOI] [PubMed] [Google Scholar]
- 25.Fowler DM, et al. High-resolution mapping of protein sequence-function relationships. Nat Methods. 2010;7(9):741–746. doi: 10.1038/nmeth.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koenig P, et al. Deep sequencing-guided design of a high-affinity dual-specificity antibody to target two angiogenic factors in neovascular age-related macular degeneration. J Biol Chem. 2015;290(36):21773–21786. doi: 10.1074/jbc.M115.662783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee CV, et al. Synthetic anti-BR3 antibodies that mimic BAFF binding and target both human and murine B cells. Blood. 2006;108(9):3103–3111. doi: 10.1182/blood-2006-03-011031. [DOI] [PubMed] [Google Scholar]
- 28.Donaldson JM, et al. Identification and grafting of a unique peptide-binding site in the Fab framework of monoclonal antibodies. Proc Natl Acad Sci USA. 2013;110(43):17456–17461. doi: 10.1073/pnas.1307309110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bork P, Holm L, Sander C. The immunoglobulin fold: Structural classification, sequence patterns and common core. J Mol Biol. 1994;242(4):309–320. doi: 10.1006/jmbi.1994.1582. [DOI] [PubMed] [Google Scholar]
- 30.Hamill SJ, Cota E, Chothia C, Clarke J. Conservation of folding and stability within a protein family: The tyrosine corner as an evolutionary cul-de-sac. J Mol Biol. 2000;295(3):641–649. doi: 10.1006/jmbi.1999.3360. [DOI] [PubMed] [Google Scholar]
- 31.Helms LR, Wetzel R. Specificity of abnormal assembly in immunoglobulin light chain deposition disease and amyloidosis. J Mol Biol. 1996;257(1):77–86. doi: 10.1006/jmbi.1996.0148. [DOI] [PubMed] [Google Scholar]
- 32.Morea V, Tramontano A, Rustici M, Chothia C, Lesk AM. Conformations of the third hypervariable region in the VH domain of immunoglobulins. J Mol Biol. 1998;275(2):269–294. doi: 10.1006/jmbi.1997.1442. [DOI] [PubMed] [Google Scholar]
- 33.Hietpas RT, Jensen JD, Bolon DN. Experimental illumination of a fitness landscape. Proc Natl Acad Sci USA. 2011;108(19):7896–7901. doi: 10.1073/pnas.1016024108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Atwal JK, et al. A therapeutic antibody targeting BACE1 inhibits amyloid-β production in vivo. Sci Transl Med. 2011;3(84):84ra43. doi: 10.1126/scitranslmed.3002254. [DOI] [PubMed] [Google Scholar]
- 35.Stanfield RL, Zemla A, Wilson IA, Rupp B. Antibody elbow angles are influenced by their light chain class. J Mol Biol. 2006;357(5):1566–1574. doi: 10.1016/j.jmb.2006.01.023. [DOI] [PubMed] [Google Scholar]
- 36.Lesk AM, Chothia C. Elbow motion in the immunoglobulins involves a molecular ball-and-socket joint. Nature. 1988;335(6186):188–190. doi: 10.1038/335188a0. [DOI] [PubMed] [Google Scholar]
- 37.Lefranc MP, et al. IMGT-choreography for immunogenetics and immunoinformatics. In Silico Biol. 2005;5(1):45–60. [PubMed] [Google Scholar]
- 38.Masuda K, et al. The role of interface framework residues in determining antibody V(H)/V(L) interaction strength and antigen-binding affinity. FEBS J. 2006;273(10):2184–2194. doi: 10.1111/j.1742-4658.2006.05232.x. [DOI] [PubMed] [Google Scholar]
- 39.Khalifa MB, et al. Effects on interaction kinetics of mutations at the VH-VL interface of Fabs depend on the structural context. J Mol Recognit. 2000;13(3):127–139. doi: 10.1002/1099-1352(200005/06)13:3<127::AID-JMR495>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 40.Dunbar J, Fuchs A, Shi J, Deane CM. ABangle: Characterising the VH-VL orientation in antibodies. Protein Eng Des Sel. 2013;26(10):611–620. doi: 10.1093/protein/gzt020. [DOI] [PubMed] [Google Scholar]
- 41.Stanfield RL, Takimoto-Kamimura M, Rini JM, Profy AT, Wilson IA. Major antigen-induced domain rearrangements in an antibody. Structure. 1993;1(2):83–93. doi: 10.1016/0969-2126(93)90024-b. [DOI] [PubMed] [Google Scholar]
- 42.Skinner JJ, Lim WK, Bédard S, Black BE, Englander SW. Protein hydrogen exchange: Testing current models. Protein Sci. 2012;21(7):987–995. doi: 10.1002/pro.2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vargas-Madrazo E, Paz-García E. An improved model of association for VH-VL immunoglobulin domains: Asymmetries between VH and VL in the packing of some interface residues. J Mol Recognit. 2003;16(3):113–120. doi: 10.1002/jmr.613. [DOI] [PubMed] [Google Scholar]
- 44.Aburatani T, Ueda H, Nagamune T. Importance of a CDR H3 basal residue in V(H)/V(L) interaction of human antibodies. J Biochem. 2002;132(5):775–782. doi: 10.1093/oxfordjournals.jbchem.a003286. [DOI] [PubMed] [Google Scholar]
- 45.Lamminmäki U, Westerlund-Karlsson A, Toivola M, Saviranta P. Modulating the binding properties of an anti-17beta-estradiol antibody by systematic mutation combinations. Protein Sci. 2003;12(11):2549–2558. doi: 10.1110/ps.0353903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Traxlmayr MW, et al. Construction of a stability landscape of the CH3 domain of human IgG1 by combining directed evolution with high-throughput sequencing. J Mol Biol. 2012;423(3):397–412. doi: 10.1016/j.jmb.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Landolfi NF, et al. The integrity of the ball-and-socket joint between V and C domains is essential for complete activity of a humanized antibody. J Immunol. 2001;166(3):1748–1754. doi: 10.4049/jimmunol.166.3.1748. [DOI] [PubMed] [Google Scholar]
- 48.Sampei Z, et al. Non–antigen-contacting region of an asymmetric bispecific antibody to factors IXa/X significantly affects factor VIII-mimetic activity. MAbs. 2015;7(1):120–128. doi: 10.4161/19420862.2015.989028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Niederfellner G, et al. Epitope characterization and crystal structure of GA101 provide insights into the molecular basis for type I/II distinction of CD20 antibodies. Blood. 2011;118(2):358–367. doi: 10.1182/blood-2010-09-305847. [DOI] [PubMed] [Google Scholar]
- 50.Pierce SK, Liu W. The tipping points in the initiation of B cell signalling: How small changes make big differences. Nat Rev Immunol. 2010;10(11):767–777. doi: 10.1038/nri2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaplinsky J, et al. Antibody repertoire deep sequencing reveals antigen-independent selection in maturing B cells. Proc Natl Acad Sci USA. 2014;111(25):E2622–E2629. doi: 10.1073/pnas.1403278111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kunkel TA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bostrom J, Lee CV, Haber L, Fuh G. Improving antibody binding affinity and specificity for therapeutic development. Methods Mol Biol. 2009;525:353–376, xiii. doi: 10.1007/978-1-59745-554-1_19. [DOI] [PubMed] [Google Scholar]
- 54.Magoč T, Salzberg SL. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27(21):2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Team RC. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2014. [Google Scholar]
- 56.Morgan M, et al. ShortRead: A bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics. 2009;25(19):2607–2608. doi: 10.1093/bioinformatics/btp450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc. 2007;2(9):2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 58.Dunbar J, et al. SAbDab: The structural antibody database. Nucleic Acids Res. 2014;42(Database issue):D1140–D1146. doi: 10.1093/nar/gkt1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grant BJ, Rodrigues AP, ElSawy KM, McCammon JA, Caves LS. Bio3d: An R package for the comparative analysis of protein structures. Bioinformatics. 2006;22(21):2695–2696. doi: 10.1093/bioinformatics/btl461. [DOI] [PubMed] [Google Scholar]
- 60.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 61.Case DA, et al. The Amber biomolecular simulation programs. J Comput Chem. 2005;26(16):1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14(1):33–38, 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 63.Mayne L, et al. Many overlapping peptides for protein hydrogen exchange experiments by the fragment separation-mass spectrometry method. J Am Soc Mass Spectrom. 2011;22(11):1898–1905. doi: 10.1007/s13361-011-0235-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Walters BT, Ricciuti A, Mayne L, Englander SW. Minimizing back exchange in the hydrogen exchange-mass spectrometry experiment. J Am Soc Mass Spectrom. 2012;23(12):2132–2139. doi: 10.1007/s13361-012-0476-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kan ZY, Mayne L, Chetty PS, Englander SW. ExMS: Data analysis for HX-MS experiments. J Am Soc Mass Spectrom. 2011;22(11):1906–1915. doi: 10.1007/s13361-011-0236-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Walters BT, Mayne L, Hinshaw JR, Sosnick TR, Englander SW. Folding of a large protein at high structural resolution. Proc Natl Acad Sci USA. 2013;110(47):18898–18903. doi: 10.1073/pnas.1319482110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hu W, et al. Stepwise protein folding at near amino acid resolution by hydrogen exchange and mass spectrometry. Proc Natl Acad Sci USA. 2013;110(19):7684–7689. doi: 10.1073/pnas.1305887110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Leurs U, et al. Dissecting the binding mode of low-affinity phage display peptide ligands to protein targets by hydrogen/deuterium exchange coupled to mass spectrometry. Anal Chem. 2014;86(23):11734–11741. doi: 10.1021/ac503137u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ye J, Ma N, Madden TL, Ostell JM. IgBLAST: An immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013;41(Web Server issue):W34–40. doi: 10.1093/nar/gkt382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu TT, Kabat EA. An analysis of the sequences of the variable regions of Bence Jones proteins and myeloma light chains and their implications for antibody complementarity. J Exp Med. 1970;132(2):211–250. doi: 10.1084/jem.132.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Glanville J, et al. Precise determination of the diversity of a combinatorial antibody library gives insight into the human immunoglobulin repertoire. Proc Natl Acad Sci USA. 2009;106(48):20216–20221. doi: 10.1073/pnas.0909775106. [DOI] [PMC free article] [PubMed] [Google Scholar]