Significance
Aminoacyl-tRNA synthetases (AARSs) comprise a family of 20 enzymes that catalyze the first step of protein synthesis by esterifying specific amino acids to the 3′ ends of their cognate tRNAs. Protein acetylation, an evolutionarily conserved posttranslational modification, has recently been extensively investigated to regulate diverse cellular processes. This work provides the molecular evidence that one of the aminoacyl-tRNA synthetases is modulated by reversible acetylation. This study not only reveals a regulatory mechanism for the tRNA synthetase tyrosyl-tRNA synthetase (TyrRS) due to a posttranslational modification but also enriches knowledge of the regulatory pathway between p300/CBP-associated factor (PCAF)/sirtuin 1 (SIRT1) and the DNA damage/repair response.
Keywords: tRNA synthetases, acetylation, sirtuins, oxidative stress, DNA damage repair
Abstract
Tyrosyl-tRNA synthetase (TyrRS) is well known for its essential aminoacylation function in protein synthesis. Recently, TyrRS has been shown to translocate to the nucleus and protect against DNA damage due to oxidative stress. However, the mechanism of TyrRS nuclear localization has not yet been determined. Herein, we report that TyrRS becomes highly acetylated in response to oxidative stress, which promotes nuclear translocation. Moreover, p300/CBP-associated factor (PCAF), an acetyltransferase, and sirtuin 1 (SIRT1), a NAD+-dependent deacetylase, regulate the nuclear localization of TyrRS in an acetylation-dependent manner. Oxidative stress increases the level of PCAF and decreases the level of SIRT1 and deacetylase activity, all of which promote the nuclear translocation of hyperacetylated TyrRS. Furthermore, TyrRS is primarily acetylated on the K244 residue near the nuclear localization signal (NLS), and acetylation inhibits the aminoacylation activity of TyrRS. Molecular dynamics simulations have shown that the in silico acetylation of K244 induces conformational changes in TyrRS near the NLS, which may promote the nuclear translocation of acetylated TyrRS. Herein, we show that the acetylated K244 residue of TyrRS protects against DNA damage in mammalian cells and zebrafish by activating DNA repair genes downstream of transcription factor E2F1. Our study reveals a previously unknown mechanism by which acetylation regulates an aminoacyl-tRNA synthetase, thus affecting the repair pathways for damaged DNA.
Aminoacyl-tRNA synthetases (AARSs) are a family of 20 enzymes that catalyze the first step of protein synthesis by esterifying specific amino acids on the 3′ ends of their cognate tRNAs (1). Recently, AARSs have been implicated in specific physiological responses, such as apoptosis (2), cellular growth (3), and angiogenesis (4). Tyrosyl-tRNA synthetase (TyrRS or YARS) is conserved in eukaryotes ranging from insects to humans, and multiple mutations have been identified in its catalytic domain. These mutations are associated with dominant intermediate Charcot-Marie-Tooth (CMT) disease, a common peripheral nervous system disorder caused by axonal degeneration and demyelination (5).
In response to oxidative damage or serum starvation stress, TyrRS translocates to the nucleus, where it protects against DNA damage by activating the transcription factor E2F1 and subsequent downstream DNA repair genes (6, 7). Notably, resveratrol directly binds to the active site of TyrRS, thereby directing TyrRS to the nucleus, and stimulates the activation of NAD+-dependent auto-poly-ADP ribosylation of poly(ADP ribose) polymerase 1 (PARP1) (6, 7). Interestingly, a hexapeptide motif in the anticodon recognition domain of TyrRS has been identified as a nuclear localization signal (NLS) (8). However, the mechanism by which TyrRS is redistributed between the cytoplasmic and nuclear compartments during stress and the effect of TyrRS localization on its aminoacylation and activity toward DNA damage protection remain unknown.
Protein acetylation, a posttranslational modification process that has been conserved through evolution, has been extensively investigated in recent years (9–12). Acetylation regulates diverse cellular processes, including gene silencing (13), oxidative stress (13, 14), DNA repair (15), cell survival and migration (16, 17), and metabolism (9, 18, 19). Most acetylated proteins act as transcription factors in the nucleus and as metabolic enzymes outside the nucleus (9). Strikingly, the acetylation of multiple aminoacyl-tRNA synthetases, including tyrosyl-tRNA synthetase, has been reported in a number of proteomic studies (10, 12). However, the link between acetylation and AARS remains to be established.
Herein, we propose a model in which acetylation of the NLS in TyrRS enhances its nuclear transport under oxidative stress.
Results
Nicotinamide and Oxidative Stress Enhance the Acetylation Level of TyrRS.
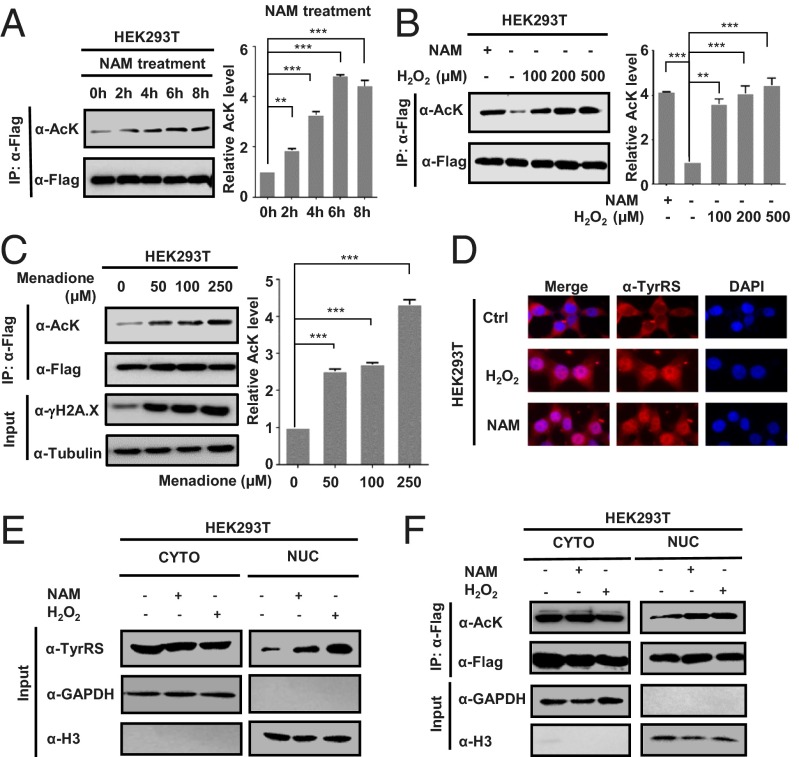
Several recent proteomic studies have shown that lysine residues on TyrRS are often acetylated (10, 12). To confirm that TyrRS was acetylated, human full-length TyrRS was overexpressed in HEK293T cells, which were subsequently treated with nicotinamide (NAM), a common deacetylase inhibitor that is effective against the sirtuin family of proteins (20). The results indicated that TyrRS was indeed acetylated and that the acetylation level of TyrRS was approximately fourfold higher after treatment with 5 mM NAM for 4–6 h (Fig. 1A), indicating that an NAD+-dependent deacetylase of the sirtuin family, most likely cytosolic sirtuin 1 (SIRT1) or SIRT2 (21), was the deacetylase of TyrRS.
Fig. 1.
NAM and oxidative stress promote TyrRS nuclear translocation through acetylation. (A) TyrRS is acetylated. HEK293T cells were transfected with TyrRS-Flag and then treated with 5 mM nicotinamide (NAM) for the indicated times. The TyrRS acetylation and protein levels were analyzed by Western blotting with the indicated antibodies. Relative TyrRS acetylation over protein level was quantified by ImageJ. Error bars represent ± SD for triplicate experiments. **P < 0.01; ***P < 0.001. (B and C) HEK293T cells were transfected with TyrRS-Flag and then treated with hydrogen peroxide (H2O2) (B) or menadione (C) at the indicated concentrations for 1 h. Error bars represent ± SD for triplicate experiments. **P < 0.01; ***P < 0.001. (D) NAM and H2O2 treatments promote TyrRS nuclear translocation. The subcellular localization of endogenous TyrRS was examined by immunofluorescence microscopy in human HEK293T cells subjected to the indicated treatments (NAM, 5 mM for 4 h; H2O2, 200 μM for 1 h). (E) HEK293T cells were harvested, and the cytosolic and nuclear fractions were separated after the indicated treatments. Results were analyzed by Western blotting. GAPDH, marker for cytosolic fraction; H3, marker for nuclear fraction. (F) Nuclear TyrRS is highly acetylated. TyrRS-Flag was transfected into HEK293T cells before the indicated treatments. Cytosolic and nuclear fractions were separated. Results were determined by Western blotting.
Because TyrRS has been reported to be involved in the oxidative stress response (7), its acetylation level was assessed after inducing oxidative stress. Strikingly, upon H2O2 treatment, the acetylation level of TyrRS significantly increased in a concentration-dependent manner (Fig. 1B). To further validate this result, cells overexpressing TyrRS were treated with the oxidative stressor menadione, a polycyclic aromatic ketone that generates intracellular reactive oxygen species (ROS) at multiple cellular sites via futile redox cycling (14). Menadione treatment also significantly increased the acetylation level of TyrRS (Fig. 1C), indicating that TyrRS becomes highly acetylated in response to oxidative stress.
NAM and Oxidative Stress Promote TyrRS Nuclear Localization.
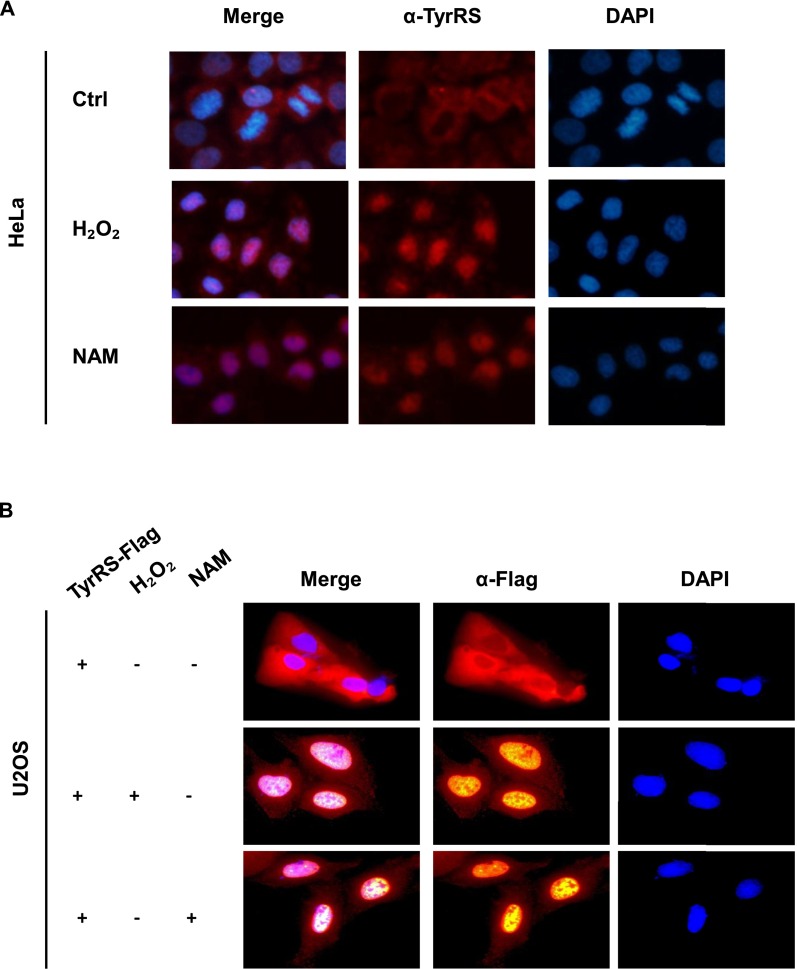
Despite its central role in the translational machinery of the cytoplasm, TyrRS rapidly translocates to the nucleus in response to oxidative stress (7). Having shown that the acetylation levels of TyrRS significantly increased in response to stress (H2O2 and NAM), we determined whether acetylation induced nuclear translocation, by treating HEK293T, HeLa, and human osteosarcoma U2OS cells with H2O2 and NAM, and assessed the distribution of endogenous TyrRS after treatment by cell fractionation and immunofluorescence microscopy. In all three different types of cells, NAM and H2O2 dramatically enhanced the nuclear import of TyrRS (Fig. 1 D and E and Fig. S1A), and cells overexpressing exogenous TyrRS displayed the same phenotypes as cells expressing endogenous TyrRS (Fig. S1B and Fig. 1E). We expressed TyrRS-Flag in HEK293T cells and fractionated the cells after treatment with NAM or H2O2. The acetylation level of overexpressed TyrRS in the nucleus was strongly elevated after treatment, indicating that increased acetylation may play an important role in the nuclear localization of TyrRS due to NAM addition or oxidative stress (Fig. 1F). The observations described above indicated that NAM and oxidative stress promoted the nuclear translocation of TyrRS in an acetylation-dependent manner.
Fig. S1.
(A) NAM and H2O2 treatments promote TyrRS nuclear translocation in HELA cells. Subcellular localization of endogenous TyrRS was examined by immunofluorescence microscopy with indicated treatments. (B) NAM and H2O2 treatments also promote ectopic TyrRS-Flag nuclear translocation. Subcellular localization of exogenous Flag-TyrRS was examined by immunofluorescence microscopy with indicated treatments.
Oxidative Stress-Mediated SIRT1 and PCAF Regulate the Nuclear Localization of TyrRS in an Acetylation-Dependent Manner.
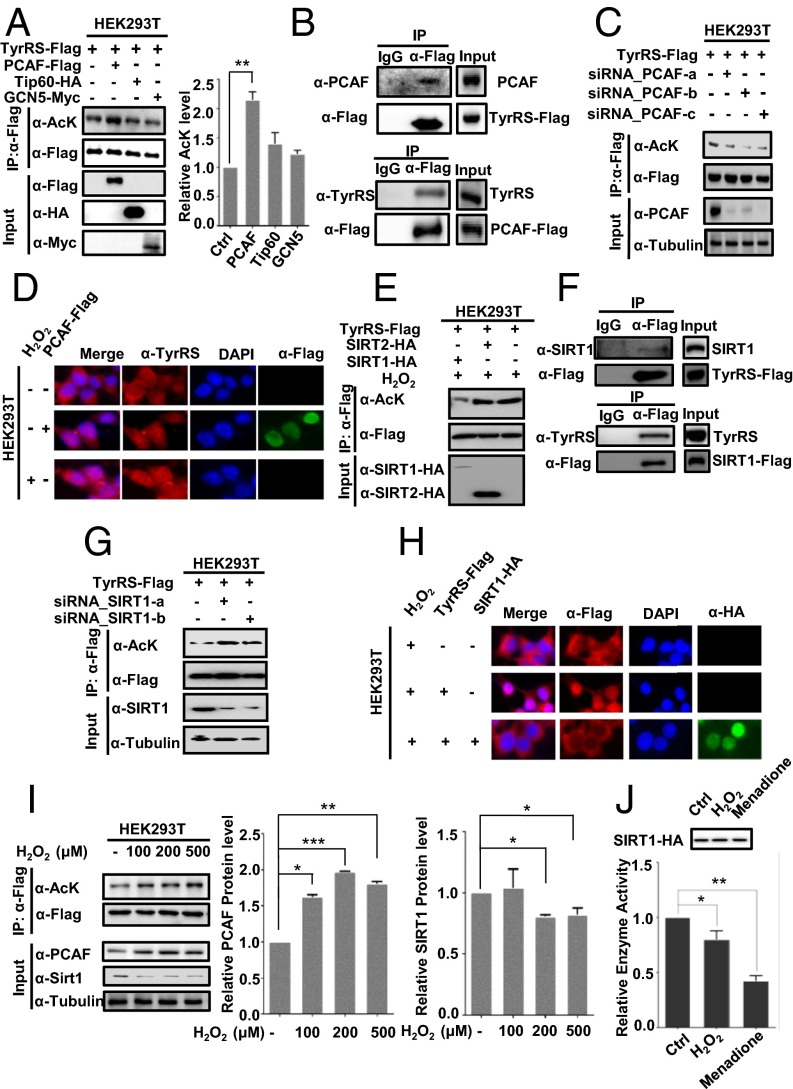
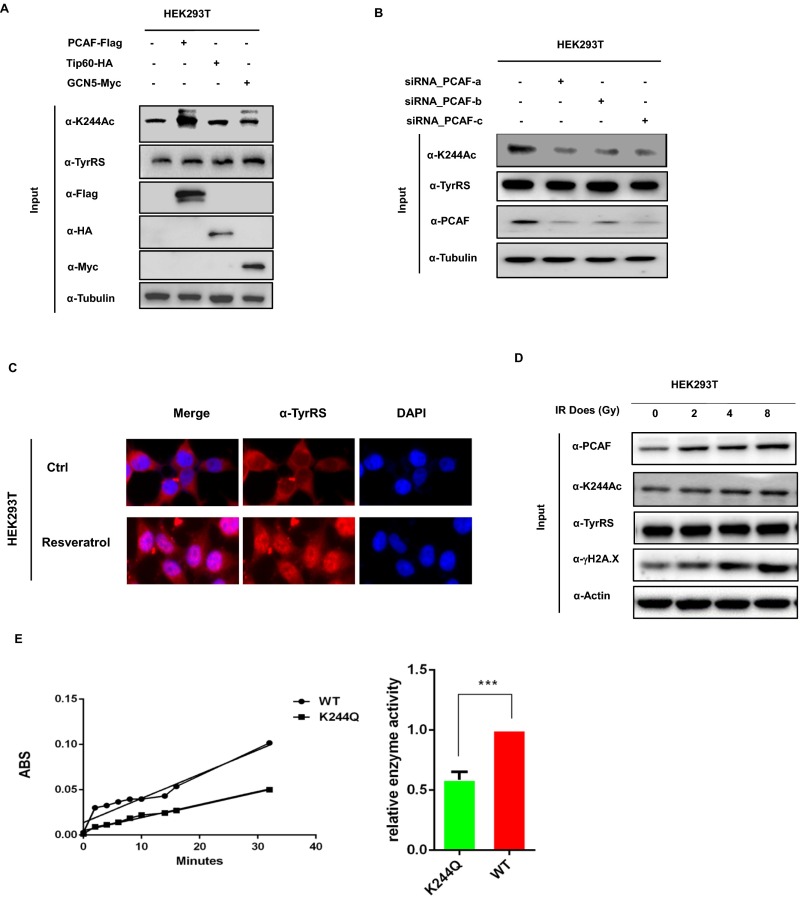
Lysine acetylation is a reversible process catalyzed by acetyltransferases and deacetylases (9). To identify the acetyltransferase responsible for TyrRS acetylation, we overexpressed p300/CBP-associated factor (PCAF), Tip60, and GCN5 into HEK293T cells, because PCAF, Tip60, and GCN5 have been shown to respond to oxidative stress (22–25), and found that the acetylation of TyrRS was elevated only after the ectopic expression of PCAF, not the other acetyltransferases (Fig. 2A). We also detected a specific interaction between TyrRS and PCAF (Fig. 2B). Furthermore, PCAF knockdown using siRNA significantly reduced the acetylation of TyrRS (Fig. 2C). To determine whether PCAF influenced the nuclear localization of TyrRS, the effect of ectopic PCAF expression on TyrRS localization was analyzed. As expected, ectopic PCAF expression promoted the nuclear localization of TyrRS (Fig. 2D). Together, these results demonstrated that PCAF acetylates TyrRS, thus facilitating nuclear translocation.
Fig. 2.
Oxidative stress-mediated PCAF and SIRT1 regulate the nuclear localization of TyrRS. (A) PCAF acetylates TyrRS. TyrRS-Flag was coexpressed with different acetyltransferases and purified by Flag beads. **P < 0.01. (B) Coimmunoprecipitation assay detecting TyrRS-PCAF binding in HEK293 cells transiently transfected with Flag-tagged PCAF or TyrRS. (C) PCAF knockdown decreases TyrRS acetylation. Acetylation levels of TyrRS-Flag expressed and purified from 293T cells with or without PCAF knocked down by siRNA were detected by Western blotting. (D) PCAF overexpression promotes TyrRS translocation. The subcellular localization of TyrRS was examined in human HEK293T cells expressing the indicated plasmids or subjected to the indicated treatments. (E) SIRT1 overexpression decreases TyrRS acetylation. The acetylation levels of TyrRS in HEK293T cells expressing the indicated plasmids and then treated with H2O2 were analyzed by Western blotting. (F) Coimmunoprecipitation assay detecting TyrRS-SIRT1 binding in HEK293 cells transiently transfected with Flag-tagged SIRT1 or TyrRS. (G) SIRT1 knockdown promotes TyrRS acetylation. Acetylation levels of TyrRS-Flag expressed and purified from 293T cells with or without SIRT1 knocked down by siRNA were detected by Western blotting. (H) SIRT1 overexpression prevents oxidative stress-mediated TyrRS translocation. The subcellular localization of TyrRS was examined in human HEK293T cells expressing the indicated plasmids or subjected to the indicated treatments. (I) H2O2 treatments decrease SIRT1 level and increase both PCAF level and TyrRS acetylation. TyrRS-Flag was transfected into HEK293T cells followed by indicated treatments. Average quantified relative protein abundance from all three repeats is shown with SD. *P < 0.05; **P < 0.01; ***P < 0.001. (J) SIRT1 deacetylase activity is significantly decreased under oxidative stress. HEK293T cells transfected with HA-tagged SIRT1 were performed with indicated treatments. Then HA-tagged SIRT1 was immunoprecipitated from cells and used in the in vitro deacetylation assay. The mean value of triplicate experiments and SD are presented. *P < 0.05; **P < 0.01.
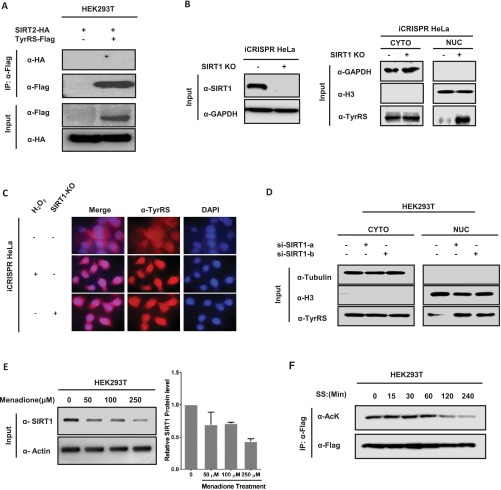
Next, the deacetylase responsible for TyrRS regulation was investigated. We confirmed that NAM (a sirtuin inhibitor) and oxidative stress treatment significantly increased the acetylation level of TyrRS (Fig. 1A). Previous studies have shown that oxidative stress reduces the protein expression of SIRT1 (26, 27). To determine whether SIRT1 interacted with TyrRS as its deacetylase, we coexpressed TyrRS with either SIRT1 or SIRT2 in HEK293T cells and then measured the acetylation level of TyrRS. We found that the ectopic expression of SIRT1, but not SIRT2, decreased TyrRS acetylation (Fig. 2E). We further verified the SIRT1–TyrRS interaction in HEK293T cells by coimmunoprecipitation. However, SIRT2, a sirtuin deacetylase that is abundant in the cytoplasm, did not interact with TyrRS (Fig. 2F and Fig. S2A). Moreover, SIRT1 knockdown via transfection of two siRNAs significantly increased the acetylation level of TyrRS (Fig. 2G). To further confirm that SIRT1 regulated the acetylation and subsequent subcellular relocalization of TyrRS, we coexpressed SIRT1 with TyrRS in HEK293T cells and found that ectopic SIRT1 inhibited the nuclear translocation of TyrRS in response to H2O2 treatment (Fig. 2H). We also constructed a SIRT1 knockout (KO) HeLa cell line using the iCRISPR method (28) to further verify the regulation of TyrRS by SIRT1 (Fig. S2B). Through cell fractionation and immunofluorescence staining experiments, we discovered that the loss of SIRT1 promoted the nuclear translocation of TyrRS (Fig. S2 B and C). The knockdown of SIRT1 in HEK293T with two siRNAs further confirmed this result (Fig. S2D). Collectively, these results demonstrated that SIRT1 prevented TyrRS localization to the nucleus via deacetylation.
Fig. S2.
(A) SIRT2 doesn’t interact with TyrRS. HEK293T cells were cotransfected with SIRT2-HA and TyrRS-Flag. Interaction between SIRT2 and TyrRS cannot be detected by immunoprecipitation and Western blot analysis. (B) SIRT1-KO promotes TyrRS nuclear translocation in subcellular isolation assay. SIRT1 protein level was detected by Western blotting analysis in SIRT1-KO HeLa cells by iCRISPR method (Left). Subcellular localization of endogenous TyrRS of SIRT1-KO iCRISPR HeLa cells was analyzed by Western blotting (Right). (C) Knocking out SIRT1 promotes TyrRS nuclear translocation in Immunocytochemistry. Nuclear TyrRS was detected by immunofluorescence microscopy in iCRISPR HeLa cells or SIRT1-KO iCRISPR HeLa cells with indicated treatments. (D) SIRT1 knockdown promotes TyrRS nuclear translocation. HEK293T cells were transfected with two designed siRNAs targeting SIRT1. Cells were harvested, and cytosolic and nuclear fractions were separated. Subcellular localization of TyrRS was examined by Western blotting. (E) Menadione treatment decreases SIRT1 level. The HEK293T cell lysates were prepared after treatment with menadione at the doses shown. Endogenous SIRT1 were determined by Western blotting with indicated antibodies. (F) TyrRS acetylation decreases a lot after long-time serum starvation. HEK293T cells were transfected with Flag-TyrRS followed by indicated treatment. TyrRS acetylation and protein levels were analyzed by Western blotting with indicated antibody.
To understand whether oxidative stress regulated SIRT1 and PCAF to affect TyrRS acetylation levels, we investigated the SIRT1 expression levels and its deacetylase activity using an assay under oxidative stress. Consistent with previous reports, H2O2 and menadione treatment decreased SIRT1 protein levels (Fig. 2I and Fig. S2E) and reduced its deacetylase activity (Fig. 2J), resulting in significantly increased TyrRS acetylation (Fig. 2A). Moreover, we found that PCAF levels were elevated under H2O2 treatment, enhancing the acetylation of TyrRS. Taken together, these results suggest that oxidative stress-mediated PCAF and SIRT1 regulate TyrRS acetylation and its subsequent translocation to the nucleus.
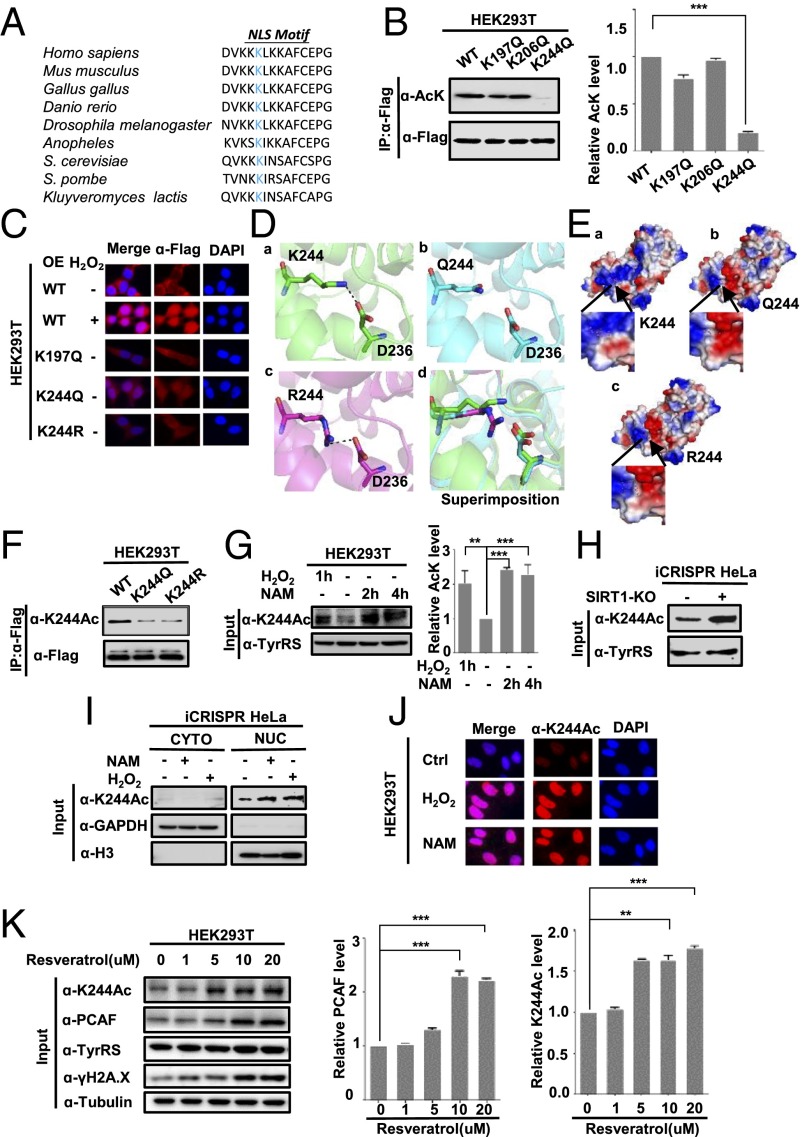
TyrRS K244 Is the Major Acetylation Site Regulated by PCAF and SIRT1.
Large-scale proteomic analyses of acetylation (10–12) have identified K197, K206, and other lysine residues in TyrRS as putative acetylation sites. Because the NLS of TyrRS plays an important role in determining its subcellular localization (8), we also considered the role of K244, a lysine residue in the NLS motif that is the most conserved lysine residue across different TyrRS proteins, as a putative acetylation site (Fig. 3A). To identify the primary acetylation site of TyrRS, we replaced several lysine residues with glutamine (Q), an acetyl-mimetic amino acid (18), and transfected 293T cells with individual mutants. We found that substitution of the K244 residue significantly decreased the overall acetylation of TyrRS compared with that of the WT protein, suggesting that K244, but not K197 or K206, was the primary acetylation site (Fig. 3B). The acetylation of TyrRS was not completely abolished in the K244 mutant, indicating that other sites may be acetylated in TyrRS.
Fig. 3.
K244 acetylation affects TyrRS nuclear translocation under the regulation of PCAF and SIRT1. (A) Alignment of the TyrRS sequence surrounding K244 from various species, including human [Homo sapiens, National Center for Biotechnology Information (NCBI) reference number NP_003671.1], mouse (Mus musculus, NP_598912.4), chicken (Gallus gallus, NP_001006314.1), zebrafish (Danio rerio, NP_958473.1), fruit fly (Drosophila melanogaster, NP_648895.1), budding yeast (Saccharomyces cerevisiae, NP_011701.3), and fission yeast (Schizosaccharomyces pombe, NP_587876.1). Bold blue text indicates lysine 244. (B) K244 is the primary acetylation site of TyrRS. The indicated plasmids were transfected into HEK293T cells, and the proteins were immunoprecipitated before being subjected to Western blotting for acetylation analysis. ***P < 0.001. (C) Exogenous K244Q mutant locates in nucleus. Subcellular localization of exogenous TyrRS-Flag and Flag-K244 mutants was examined in HEK293T cells with indicated treatments or expressed plasmids. (D and E) Molecular modeling of acetylation of K244 in TyrRS. TyrRS K244R caused a hydrogen-bond disruption between K244 and D236 (D), as well as a change on the electronic potential surface of TyrRS (E). (F) Characterization of acetyl-TyrRS (K244) antibody. The acetylation level of immunoprecipitated TyrRS-Flag was measured by the site-specific K244 acetylation antibody. (G) Endogenous TyrRS is acetylated at K244. HEK293T cell lysates were prepared after H2O2 and NAM treatment. Results were determined by Western blotting with the indicated antibodies. Shown are representative Western blot results of three replicates with SD. **P < 0.01; ***P < 0.001. (H) K244 acetylation level is elevated in SIRT1-KO iCRISPR HeLa cells. K244 acetylation in WT and SIRT1-KO iCRISPR HeLa cells was determined by Western blotting. (I) K244-acetylated TyrRS accumulates in the nucleus. The subcellular localization of endogenous K244-acetylated TyrRS was analyzed in iCRISPR HeLa cells by Western blotting. (J) Subcellular localization of K244-acetylated TyrRS was examined in HEK293T cells after the indicated treatments. (K) Resveratrol promotes the expression of PCAF and increases K244 acetylation of TyrRS. Endogenous PCAF and TyrRS K244 acetylation levels of HEK293T cells were detected by Western blotting after treatment with resveratrol at the doses shown. Shown are representative Western blot results of three replicates with SD. **P < 0.01; ***P < 0.001.
To test the hypothesis that K244 acetylation promoted TyrRS nuclear translocation in response to oxidative stress, we transfected HEK293T cells with either WT TyrRS or mutant versions (K244Q, acetylated mimic, and K244R, deacetylated mimic) of TyrRS and detected their subcellular localization. The acetylation mimic mutant K244Q showed greater nuclear accumulation than both the WT and K244R mutant, demonstrating the importance of acetylation for this change in localization (Fig. 3C). We also found that the phenotype of the K244Q mutant was identical to that of the H2O2-treated WT, showing that substantial TyrRS accumulation occurred in the nucleus. Moreover, the phenotype of the K244R mutant was identical to that of the untreated WT (Fig. 3C). To further investigate how the acetylation of K244 modulates the nuclear import of TyrRS, we performed molecular dynamics simulations on the deacetylation mimetic mutation K244R and the acetylation mimetic mutation K244Q in human TyrRS (PDB ID code 4BQT). First, acetylation of K244 affected the conformation of TyrRS around the NLS (Fig. 3D) by abrogating the interaction with an aspartate residue (D236) because acetylation eliminates the positive charge on the lysine side chain. Moreover, the acetylation-induced changes on K244 affected the electrostatic surface potential of the NLS in human TyrRS (Fig. 3E). Taken together, these simulations suggested that acetylation of K244 induced changes in TyrRS that affected its conformation and the electrostatic field around the NLS, which may have promoted TyrRS nuclear translocation. Therefore, we decided to generate an antibody specific to acetylated K244 and verify its specificity to further investigate the function of K244 acetylation. The proposed antibody readily detected K244-acetylated TyrRS (Fig. 3F). Furthermore, Western blotting analysis of a whole-cell extract from HEK293T cells with the anti–acetyl-K244 antibody detected a band with a substantial increase in intensity after treatment with either NAM or H2O2, thereby demonstrating endogenous acetylation of TyrRS at K244 in cultured cells (Fig. 3G).
Given that SIRT1 regulates TyrRS acetylation, we measured the endogenous TyrRS K244 acetylation level in SIRT1-KO iCRISPR HeLa cells and found that K244 acetylation was markedly enhanced (Fig. 3H), indicating that K244 was the major site of TyrRS acetylation and that SIRT1 was responsible for the deacetylation of TyrRS. Moreover, we confirmed that the ectopic expression of PCAF increased K244 acetylation and that the knockdown of PCAF using siRNA significantly reduced K244 acetylation (Fig. S3 A and B). These results demonstrated that PCAF was the major acetyltransferase for TyrRS at lysine 244 and that TyrRS K244 was the major acetylation site regulated by SIRT1 and PCAF.
Fig. S3.
(A) Overexpression of PCAF increases endogenous TyrRS K244 acetylation level. Endogenous TyrRS K244 acetylation levels of 293T cells overexpressing different acetyltransferases were determined by Western blotting. (B) PCAF knockdown decreases endogenous TyrRS K244 acetylation. Endogenous TyrRS K244 acetylation levels of 293T cells with or without PCAF knocked down by siRNA were detected by Western blotting. (C) Resveratrol treatments promote TyrRS nuclear translocation in HEK293T cells. Subcellular localization of endogenous TyrRS was examined by immunofluorescence microscopy with indicated treatments. (D) The γ-irradiation promotes the expression of PCAF and increases K244 acetylation of TyrRS. Endogenous PCAF and TyrRS K244 acetylation levels of HEK293T cells were detected by Western blotting after treatment with irradiation at the doses shown. (E) K244Q mutation decreases TyrRS enzyme activity. TyrRS enzyme activity was detected by pyrophosphate release assay. Recombinant WT TyrRS and K244Q TyrRS were used to assay the step of aminoacylation reaction. ABS, absorbency. The enzyme activity was measured and normalized against protein level. Error bars ± represent SD for triplicate experiments. ***P < 0.001.
Acetylation at K244 Promotes TyrRS Nuclear Translocation but Inhibits Enzyme Activity.
To further show that the nuclear translocation of TyrRS was directly regulated by K244 acetylation, we conducted cell fractionation and immunofluorescence experiments and discovered that K244-acetylated TyrRS proteins were highly enriched in the nucleus after NAM and H2O2 treatment (Fig. 3 I and J). In addition to promoting the nuclear translocation of TyrRS (6), resveratrol also enhanced the protein level of PCAF and the acetylation level of K244, thus facilitating the nuclear localization of TyrRS (Fig. 3K and Fig. S3C). Using classical DNA-damaging agents, we also discovered that exposure to IR resulted in a dose-dependent increase in the content of PCAF and the acetylation of K244 (Fig. S3D). Therefore, we confirmed that K244 acetylation of TyrRS promoted nuclear translocation.
We also tested the effect of K244 acetylation on the canonical function of TyrRS: i.e., to catalyze esterification reactions that conjugate amino acids with their cognate tRNAs in the protein translation process (1). We immunopurified the WT and K244Q mutants of TyrRS from transfected HEK293T cells and then measured the TyrRS enzymatic activity using optimized methods (5, 29). We found that the acetylation mimic mutant K244Q showed a nearly 50% decrease in TyrRS catalytic activity (Fig. S3E), indicating that K244 acetylation inhibits TyrRS activity.
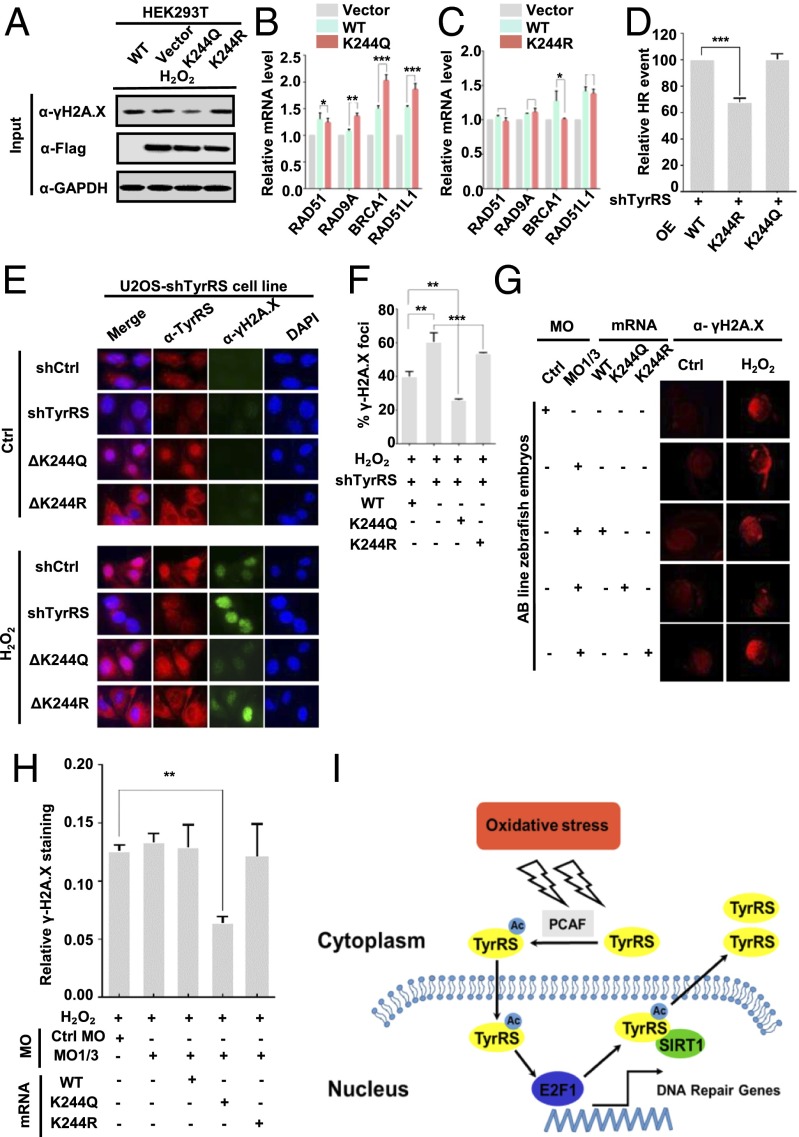
The Acetylation-Mimic K244Q Mutant TyrRS Protects Against DNA Damage Under Oxidative Stress.
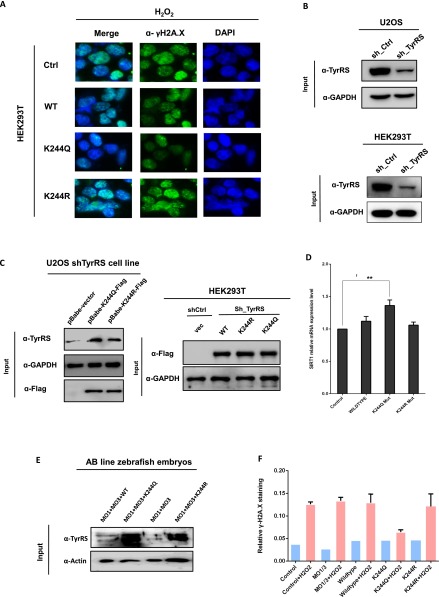
Nucleus-localized TyrRS has previously been shown to protect cells from DNA damage under certain conditions (7). We hypothesized that the acetylation mimic K244Q mutant TyrRS would be able to perform the same function by accumulating in the nucleus at the cost of decreasing its enzymatic activity. To test this hypothesis, we transfected HEK293T cells and U2OS cells with WT, K244Q, and K244R TyrRS and detected γ-H2A.X levels by both Western blotting and immunofluorescence after treatment with 200 μM H2O2 for 1 h. The K244Q mutant, but not the K244R mutant, showed significantly less DNA damage under oxidative stress conditions (Fig. 4A and Fig. S4A). To further test these results, we transfected HEK293T cells with WT, K244Q, and K244R TyrRS and used quantitative real-time PCR (qRT-PCR) to measure the expression level of the DNA damage-response gene cluster, which is activated by the transcription factor E2F1. In support of our hypothesis, we found that the K244Q-mutant TyrRS significantly increased the DNA damage repair and sensing functions of BRCA1 and RAD51L1, respectively (30, 31), in the absence of stress (Fig. 4B). In contrast, the phenotypes of the K244R mutant and vector-transfected cells were identical. Specifically, the transcription of DNA repair genes was not up-regulated (Fig. 4C). We next investigated whether mutants of TyrRS would affect homologous recombination (HR) during the DNA damage repair process. Using a reporter assay for HR (32), we found that reconstituting cells with shRNA-resistant K244R TyrRS mutants resulted in compromised HR in TyrRS-depleted HEK293T cells compared with that in cells reconstituted with WT or K244Q TyrRS mutants (Fig. 4D and Fig. S4 B and C). These results support the idea that K244Q TyrRS has a stronger effect on the promotion of DNA damage repair.
Fig. 4.
Acetylation-mimic K244Q mutant TyrRS protects against DNA damage under oxidative stress. (A) K244Q TyrRS protects cells from DNA damage caused by oxidative stress. HEK293T cells were transfected with the indicated plasmids and then subjected to the indicated treatments. Results were detected by Western blotting. (B) Overexpression of K244Q TyrRS in HEK293T cells up-regulates DNA damage response genes relative to the WT, as detected by qRT-PCR. Error bars represent the error for triplicate experiments. *P < 0.05; **P < 0.01; ***P < 0.001. (C) Overexpressing K244R TyrRS in HEK293T cells did not activate DNA damage response gene clusters. *P < 0.05. (D) K244R TyrRS mutant resulted in compromised HR. The HR assay is well-described in SI Materials and Methods. Data are presented as mean ± SD of three independent experiments. ***P < 0.001. (E) K244 acetylation promotes TyrRS nuclear translocation, thereby protecting against DNA damage. The subcellular localization of TyrRS was examined in shTyrRS cells and in K244Q and K244R cell lines with indicated treatments. The γ-H2A.X level was also measured. (F) Cells positive for γ-H2A.X foci were defined as cells with one nucleus containing at least 10 foci. γ-H2A.X focus-positive cells were randomly counted from N total cells subjected to the indicated treatments (n = 50 to 100). Error bars represent the error for triplicate experiments. **P < 0.01; ***P < 0.001. (G and H) K244Q protects zebrafish embryos from oxidative stress-mediated DNA damage. The embryos of zebrafish were injected with the indicated mRNA and morpholinos. The γ-H2A.X levels of 28-h postfertilization fish were examined by immunofluorescence microscopy. The images (G) and fluorescence intensities (H) were analyzed and quantified with ImageJ. Error bars represent the error for N independent fish embryos (n = 3 to 6). **P < 0.01. (I) Schematic illustration of how acetylation regulates TyrRS relocalization in response to oxidative stress.
Fig. S4.
(A) Exogenous K244Q mutants protect cells from oxidative damage. HEK293T cells were transfected with Flag-TyrRS and Flag-K244 mutants with indicated treatment. γH2A.X level was examined by immunofluorescence microscopy. (B and C) TyrRS was knocked down in WT U2OS and HEK293T cells (B), and then shRNA-resistant WT K244R or K244Q mutant was reintroduced into these cells (C). (D) Overexpression of K244Q mutant protein enhances SIRT1 mRNA level. Error bars ± represent SD for triplicate experiments. **P < 0.01. (E) Proteins were extracted from 28 hours postfertilization (hpf) AB line zebrafish embryos with indicated morpholinos (MOs) and mRNA injections. Human-TyrRS (WT and K244R, K244Q mutant) rescue was detected by Western blotting. Actin served as a loading control. (F) Zebrafish embryos’ DNA damage under H2O2 treatment was measured through γH2A.X staining. Images and fluorescence intensities were analyzed and quantified by using ImageJ. Error bars ± represent SD for independent N fish embryos (n = 3–6).
TyrRS is an essential component of the translation machinery (1). However, severe knockdown of TyrRS expression in human cells does not result in clear cytotoxicity (7). To eliminate the possibility that the overexpression of exogenous TyrRS did not mimic authentic intracellular conditions, we stably transfected U2OS cells with TyrRS shRNA (Fig. S4B) and reconstituted the cells with shRNA-resistant K244Q or K244R mutants using a lentivirus (Fig. S4C). We detected the subcellular localization of TyrRS in these cell lines and found that the K244Q-mutant TyrRS accumulated in the nucleus, even in the absence of stress (Fig. 4E). In contrast, the K244R mutant accumulated in the cytoplasm and did not translocate to the nucleus in response to H2O2 treatment (Fig. 4E), which is consistent with the results shown in Fig. 3. We also found that the K244Q mutant promoted translocation and protected against DNA damage after H2O2 treatment, as detected by γ-H2A.X staining (Fig. 4E). Specifically, γ-H2A.X foci in positive nuclei were counted and quantified, and the counts were normalized to the total number of analyzed nuclei (Fig. 4F). These results further indicated that TyrRS K244 acetylation protects cells against DNA damage due to oxidative stress through TyrRS nuclear translocation.
Finally, we explored the protection conferred by the acetylation-mimic K244Q TyrRS against DNA damage in vivo using the zebrafish as a model organism and the well-established method described by Wei et al (7). Briefly, 0 to 1 h postfertilization, we injected zebrafish embryos with two TyrRS-targeting antisense morpholinos to suppress the endogenous fish TyrRS and reconstituted the embryos with WT, K244Q, and K244R TyrRS mRNAs to induce the expression of exogenous human TyrRS (Fig. S4E). We then examined the number of γ-H2A.X foci by immunofluorescence after treatment with 20 mM H2O2 for 2 h. H2O2 treatment significantly increased DNA damage in noninjected, WT-injected, and K244R mutant-injected fish (Fig. 4G and Fig. S4F). In comparison, DNA damage was much less extensive in K244Q mutant-injected fish after H2O2 treatment (Fig. 4 G and H), suggesting that K244Q TyrRS was strongly protective against DNA damage in vivo. These results suggested that TyrRS-mediated protection was strongly correlated with subcellular localization, which is regulated by acetylation at K244 (Fig. 4I).
Discussion
TyrRS has been recently shown to translocate to the nucleus, exhibiting protective effects against DNA damage due to oxidative stress (7) A potential mechanism of the aforementioned protective effects was analyzed in the present study. Specifically, we identified factors that promote the nuclear translocation of acetylated TyrRS and established a physiological role for acetylation in the prevention of DNA damage (Fig. 4I). Although acetylation plays an important role in regulating the function of several metabolic enzymes (14, 18, 19), acetylation has been shown to modify several proteins, such as PKM2 and FOXO1, enhancing their translocation into the nucleus (18, 33). Our study provided further insights into the mechanism of acetylation-regulated TyrRS nuclear localization. TyrRS is a member of the aminoacyl-tRNA synthetase family, whose members are among the most abundant proteins in cells and are known for their essential aminoacylation roles in protein synthesis. In the present study, we discovered that oxidative stress markedly increased the acetylation level of TyrRS, which promoted the nuclear translocation of TyrRS. Moreover, acetylation at K244 in TyrRS regulated both its cytoplasmic aminoacylation activity and nuclear translocation, thereby increasing protection against DNA damage. Indeed, K244 acetylation strongly protected cell cultures and zebrafish against DNA damage by activating DNA repair genes located downstream of E2F1. More importantly, we demonstrated that PCAF and SIRT1 were the acetylation enzymes that acted on TyrRS and regulated its nuclear translocation in response to oxidative stress. Along with previous findings, our data were indicative of a unique mechanism in which acetylation regulates TyrRS, thus protecting cells from DNA damage (Fig. 4I). These findings suggest that targeting the K244 residue of TyrRS may be a therapeutic strategy for physiological diseases characterized by DNA damage.
SIRT1, an NAD+-dependent deacetylase, is the best-studied member of the sirtuin family and has been implicated in signaling pathways in various diseases, including cancer (34), cardiovascular diseases (35), diabetes (36), and neurodegeneration (37). DNA damage due to oxidative stress is an important hallmark of many diseases, and SIRT1 plays several critical roles in the DNA damage response, as previously discussed (15, 38). Notably, PARP1, another protein that responds to DNA damage, is part of an intricate network that includes SIRT1 (16, 38). Specifically, SIRT1 and PARP1 share a common cofactor, NAD+, as well as several common substrates (39). More strikingly, Sajish and Schimmel recently found that serum starvation or resveratrol addition promotes the nuclear translocation of TyrRS, which enhances the interaction of TyrRS with PARP1 and increases the PARP1 activity (6). However, serum starvation increases SIRT1 protein expression, thereby inhibiting the function of PARP1 (16, 40). Thus, the function of this regulatory network remains controversial. Because resveratrol has been shown to promote the localization of TyrRS in the nucleus, we verified herein that resveratrol increased the amount of acetylated K244 of TyrRS in a dose-dependent manner by up-regulating the protein expression levels of PCAF (Fig. 3K). We also found that SIRT1 deacetylated TyrRS and prevented nuclear localization. Interestingly, the acetylation level of TyrRS significantly decreased after 2 h of serum starvation (Fig. S2F), similar to the effects of PARP1 activation (6). Moreover, the mRNA level of SIRT1 increased when the K244Q mutant of TyrRS was overexpressed in cells (Fig. S4D). Collectively, these results suggested that regulation occurred due to negative feedback among TyrRS, PARP1, and SIRT1 under oxidative stress.
In addition to their aminoacylation functions in protein synthesis, many aminoacyl-tRNA synthetases, including TyrRS, have been shown to take on multiple roles (41–43). Specifically, TyrRS was found to act as a sensor for oxidative stress after translocating to the nucleus, where it activates DNA damage repair genes that are downstream of E2F1 (7). Our study suggests that acetylation of K244 in the NLS of TyrRS results in the relocalization of this protein to the nucleus and prevents DNA damage by significantly activating DNA repair genes located downstream of E2F1, as observed in our in vivo models. Therefore, the present study not only reveals a regulatory mechanism for the tRNA synthetase TyrRS due to posttranslational modification but also enriches our knowledge of the regulatory pathway between SIRT1 and the repair response to damaged DNA. These findings provide a basis for the development of drugs that target the K244 residue of TyrRS to treat diseases characterized by DNA damage.
Materials and Methods
All experiments were approved by the Animal Care Committee at the Fudan University, China. HeLa cells were cultured in DMEM and transfected with a total of 9 µg of AAVS1-TALEN-L, AAVS1-TALEN-R, Puro-Cas9, and Neo-M2rtTA donor (1:1:8:8). After transfection, cells were treated with G418 and puromycin. After antibiotic selection, 12 to 24 colonies were randomly picked based on HeLa morphology, mechanically disaggregated, and replated into individual wells of 96-well plates. iCRISPR HeLa cells were treated with doxycycline before and during transfection. The DNA cassettes targeting SIRT1 were used to generate guide RNA (gRNA). After the gRNA transfection, iCRISPR HeLa cells were dissociated into single cells and replated at 2,000 cells per 10-cm dish. Cells were allowed to grow until colonies from single cells became visible (∼10 d). Then single colonies were randomly picked mechanically to amplify in a 24-well dish. Detailed descriptions of all materials and methods are provided in SI Materials and Methods.
SI Materials and Methods
Plasmid Construction.
Full-length cDNA of TyrRS (TyrRS) was amplified by PCR and cloned into indicated vectors including pRK7-N-FLAG. SIRT1 and SIRT2 were cloned to pCDNA3-HA vector. Point mutations for TyrRS were generated by site-directed mutagenesis (Toyobo KOD Mut Kit).
Cell Lines and Culture Conditions.
HEK293T (human embryonic kidney), HeLa (human cervical cancer), and U2OS (human osteosarcoma) cell lines were from the Fudan Molecular and Cell Biology (MCB) research laboratory. HEK293T, HeLa, and U2OS cells were grown in DMEM (Gibco) supplemented with 10% (vol/vol) heat-inactivated FBS (Gibco). Cultures were maintained at 37 °C in a humidified atmosphere containing 5% (vol/vol) CO2.
Cell Treatment and Transfection.
Cells were grown to 50 to 70% confluency before treatments. For H2O2, nicotinamide (Beyotime), and menadione (Sigma Aldrich) treatments, the reagents were diluted in DMEM (Gibco) serum-free and added into the cell media with different final concentrations and incubated for indicated time at 37 °C. Cell transfection except for siRNA was carried out by polyethylenimine (PEI) according to the manufacturer's protocol. Cell transfection for siRNA was carried out by Lipofectamine 2000 according to the manufacturer's protocol.
The sequences of siRNAs were as follows: siRNA_SIRT1a, 5′-AAATGTCTCCACGAACAGC-3′; siRNA_SIRT1b, 5′-GCTGCATCCAAGGGCCATG-3′; siRNA_PCAFa, 5′-GCAGATACCAAACAAGTTT-3′; siRNA_PCAFb, 5′-GCATCCAAACAGTTATCAA-3′; siRNA_PCAFc, 5′-CCGTATGTTCCCATCTCAA-3′.
Cytoplasmic and Nuclear Fractionation.
One 10-cm-diameter plate of HEK293T cells was lysed in1 mL of Harvest buffer (10 mM Hepes, pH 7.9, 50 mM NaCl, 0.5 M sucrose, 0.1 mM EDTA, 0.1% Triton X-100, and freshly added protease and phosphatase inhibitors) at 4 °C for 10 min, and then centrifuged at 500 × g for 10 min to pellet the nucleus. The pellet was then washed three times with washing buffer (10 mM Hepes, pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, and freshly added protease and phosphatase inhibitors). The supernatant was subjected to 17,000 × g centrifugation for another 10 min to remove any nuclear contamination and transferred to a new tube. Both the pellet and supernatant were boiled separately in SDS sample buffer.
Immunofluorescence Staining.
Cells were seeded in a 20-mm glass bottom cell culture dish (Nest) and treated with the reagents indicated. Cells were washed in PBS, fixed in 4% (wt/vol) formaldehyde, immunostained overnight with a TyrRS antibody (1/200; Abcam), Flag antibody (1/250; Sigma Aldrich), anti–acetyl-TyrRS-K244 (1/200; Customized), or γ-H2A.X antibody (1/200; Abcam) with 1% BSA (Sigma Aldrich) and washed three times with cold PBS followed by a 1-h exposure to an FITC-conjugated secondary antibody (Jackson ImmunoResearch). Immunofluorescence was visualized by an Olympus IX81.
Immunoprecipitated Assay.
TyrRS-Flag was transfected into HEK293T cells, and cell lysates were immunoprecipitated with Flag beads (Sigma Aldrich) overnight at 4 °C, and then boiled with SDS loading buffer and subjected to Western blotting. TyrRS-Flag was detected as indicated.
Coimmunoprecipitated Assay.
SIRT1-HA was transfected into HEK293T cells, and cell lysates were immunoprecipitated with HA antibody (SinoBiology) overnight at 4 °C, immunoprecipitated with Protein A/G beads (Invitrogen), and then boiled with SDS loading buffer and subjected to Western blotting. SIRT1-HA and endogenous TyrRS were detected as indicated.
Cell Lysis and Western Blot Analysis.
For cell-based experiments, cells were washed twice in PBS, scraped into PBS, pelleted, and resuspended in 1% Nonidet P-40 lysis buffer containing 50 mM Tris, pH 7.5, 150 mM NaCl, 1% Nonidet P-40, 1 mg/mL aprotinin, 1 mg/mL leupeptin, 1 mg/mL pepstatin, 1 mM Na3VO4, and 1 mM phenylmethylsulfonylfluoride (PMSF) (additional 25 mM NAM and trichostatin A (TSA) are needed for acetylation assay) for 30 min and centrifuged for 15 min at 12,000 × g; the insoluble fraction was discarded. The lysates were fractionated by SDS/PAGE and transferred to nitrocellulose filter (NC) membranes. The membranes were blocked for 1 h with Tris-buffered saline with Tween 20 (TBST) containing 5% (mass/vol) nonfat dry milk. Antibodies for Flag were purchased from Sigma Aldrich, and antibodies for acetylated lysine, GAPDH were purchased from Cell Signaling. Antibodies for SIRT1, TyrRS, γ-H2A.X (phosphor S139), α-tubulin, and histone 3 were purchased from Abcam. Anti–acetyl-TyrRS-K244 was customized from Shanghai Youke Biotechnology Co. Ltd. After incubation with primary antibodies, the membranes were washed and incubated with HRP-conjugated anti-mouse or anti-rabbit secondary antibodies, followed by detection using ECL Western blotting substrate (Bio-Rad).
Quantitative Real-time PCR.
Total RNAs were extracted from cells by using TRIzol reagents and then reverse-transcribed by using the first strand cDNA synthesis kit (Thermo Scientific). The products were then used as templates for real-time PCR using the SYBR Green real-time PCR master mixes (Invitrogen). The primers used for different target genes were as follows: RAD51L1 (forward), 5′-GCA TTC TTA TCT ACT ACC CTT TC-3′; RAD51L1 (reverse), 5′-CTG CTA TTT CAA CCA GTC TTT CA-3′; BRCA1 (forward), 5′- GAA ACC AGT CTC AGT GTC CAA CTC TCT A-3′; BRCA1 (reverse), 5′-GGT GAT TTG TAA CAA TTC TTG ATC TCC C-3′; RAD51 (forward), 5′- GAG TTT GGT GTA GCA GTG GTA AT-3′; RAD51 (reverse), 5′- GAC AGG GAG AGT CGT AGA TTT TG-3′; RAD9A (forward), 5′-TGA GAT GTG CCT TGG AGA GGA GG-3′; RAD9A (reverse), 5′-GCG AGT CGG TGT CTG AGA GTG TG-3′; GAPDH (forward), 5′- GAC CTG CCG TCT AGA AAA ACC TGC CAA ATA TGA-3′; GAPDH (reverse), 5′-GTG GAG GAG TGG GTG TCG CTG TTG AAG TCA GAG-3′; SIRT1 (forward), 5′-CAG GTT GCG GGA ATC CAA AG- 3′; SIRT1 (reverse), 5′-GCT GGG CAC CTA GGA CAT CG- 3′. The real-time PCR was performed using the Bio-Rad system. The relative gene expression levels were normalized to GAPDH as an internal control.
shTyrRS Cell Line Construction.
U2OS cells with TyrRS shRNA knockdown were generated using a lenti-viral–mediated delivery system. The DNA cassettes used to generate shRNA against different regions of TyrRS mRNA in this experiment were as follows: shRNA-1, 5′CCGGCGAGTCAGTTACTATGAGAATCTCGAGATTCTCATAGTAACTGACTCGTTTTTG-3′ and shRNA-2, 5′CCGGGCTCCATCCATCACCCATTTACTCGAGTAAATGGGTGATGGATGGAGCTTTTTG-3′.
The 21-nt sense and reverse complementary targeting sequences were underlined. Briefly, double-stranded oligos were inserted into the EcoRI/AgeI site of pMKO vector (gift from Shimin Zhao, Fudan University, Shanghai, China). pMKO-shRNA lenti-viral vectors were transfected into HEK293T cells together with three packaging plasmids: pMDLg /pRRE, CMV-VSVG, and RSV-Rev. The recombinant lenti-viruses produced from the transfected HEK293T cells were used to infect U2OS cells.
SIRT1-KO iCRISPR HeLa Cell Line Construction and Culture.
HeLa cells were cultured in DMEM (Life Technologies) supplemented with 10% (vol/vol) FBS (Life Technologies), 100 U/mL penicillin/100 µg/mL streptomycin (Shenggong), and 0.11 g/L sodium pyruvate (Shenggong). Cultures were passaged at 1:5 to 1:10 split ratios every 3 to 5 d using 0.05% trypsin/EDTA. Then, ∼80 to 90% confluent HeLa cells in 12-well dishes were transfected using Lipofectamine 2000 reagent (Life Technologies) following the manufacturer’s instructions. For each well, a total of 9 µg of AAVS1-TALEN-L, AAVS1-TALEN-L, Puro-Cas9, and Neo-M2rtTA donor (1:1:8:8) was used. Two days after transfection, cells were treated with 100 µg/mL G418 (Life Technologies) for 4 d, followed by treatment with 2 µg/mL puromycin (Life Technologies) for another 3 d. After antibiotic selection, 12 to 24 colonies were randomly picked based on HeLa morphology, mechanically disaggregated, and replated into individual wells of 96-well plates. Colonies were allowed to grow to near confluence and split and replica-plated in another 96-well plate. Once confluent, one replica was used for genomic DNA extraction, and the other was expanded for frozen stocks.
iCRISPR HeLa cells were treated with doxycycline (2 μg/mL) for 1 or 2 d before and during transfection. The DNA cassettes used to generate gRNA targeting SIRT1 were as follows: (Forward), 5′CACCGCTCCCCGGCGGGGGACGACG-3′; (Reverse), 5′AAACCGTCGTCCCCCGCCGGGGAGC-3′. Two days after the gRNA transfection, iCRISPR HeLa cells were dissociated into single cells and replated at 2,000 cells per 10-cm dish. Cells were allowed to grow until colonies from single cells became visible (∼10 d). Then single colonies were randomly picked mechanically to amplify in a 24-well dish.
TyrRS Enzyme Activity Assay.
Pyrophosphate release was assayed at 37 °C in 100 μL of reaction buffer containing 100 mM Tris⋅HCl (pH 7.6), 10 mM MgCl2, 40 mM KCl, 1 mM DTT, 50 U/mL yeast tRNA (Sigma), 5 U/mL yeast inorganic pyrophosphatase (Sigma), 1 mM ATP, and 0.2 mM l-tyrosine. Flag-tagged WT and mutant TyrRS proteins were expressed in HEK293T cells and purified by immunoprecipitation. Purified proteins were assayed for 0 to 40 min. Stop reaction buffer [4.2% (mass/vol) (NH4)2MoO4 in 4 M HCl diluted in 3 volumes 0.045% Malachite Green in 1.5% (vol/vol) polyvinyl alcohol (PVA)] was added to the mixture, and absorbency at 620 nm was measured using a BioTek Epoch2 Microplate reader. Spontaneous hydrolysis of ATP was negligible in the assay conditions used.
In Vitro SIRT1 Deacetylation Assay.
SIRT1 activity in vitro was determined with a SIRT1 Fluorometric kit (Biomol International) according to the manufacturer’s instructions. This assay uses a small lysine-acetylated peptide, corresponding to K382 of human p53, as a substrate. The lysine residue is deacetylated by SIRT1, and this process is dependent on the addition of exogenous NAD+. Cultured cells were transfected with the indicated constructs. After 2 d, cells were untreated or treated with H2O2 (500 μM) or menadione (250 μM). One hour later, cells were lysed in Nonidet P-40 buffer as described above. HA-tagged SIRT1 was immunoprecipitated from cells and eluted with biotin (Sigma). An equal amount of SIRT1 was incubated with 50 mM Fluor de Lys–SIRT1 substrate and 500 mM NAD+ in 50 mL of reaction buffer (BML-KI286; Biomol International). The mixture was incubated for 60 min at 37 °C, and the reaction was terminated by adding a solution containing Fluor de Lys Developer (Enzo Life Sciences) and 2 mM nicotinamide. Plates were incubated for 1 h at 37 °C. Values were determined by reading fluorescence on a fluorometric plate reader (Spectramax GeminiXPS; Molecular Devices) with an excitation wavelength of 360 nm and an emission wavelength of 460 nm. Calculation of net fluorescence included the subtraction of a blank consisting of buffer containing no NAD+.
In Vivo Zebrafish Experiments.
The AB-line zebrafish were maintained at 28.5 °C under continuous water flow and filtration with automatic control for a 14:10 h light/dark cycle. The following antisense morpholinos (MOs) targeting TyrRS were purchased from Gene Tools: MO1, 5′-GCT GCT CTC CCA TGA TGT CTG CTG C-3′; MO3, 5′-AGT GCT GAA TAC AAA CCT CAC AGC C-3′; and control-MO, 5′-CCT CGT CAC AAA AGA TCC AGG TAA A-3′, and have been verified by Wei et al. (7). The synthetic WT, K244Q, and K244R mutants mRNAs (500 pg per embryo) (made by an Ambion mMessage Machine SP6) and MOs (4∼5 ng per embryo) were injected into the yolk of 1- to 2-cell-stage embryos. After injection, embryos were incubated in E3 embryo medium; 24 h after injection, 20 mM H2O2 was added to the medium. Then, 3 to 4 h later, all of the fish were anesthetized with 0.168 mg/mL tricaine (Sigma-Aldrich), fixed in 4% (mass/vol) paraformaldehyde (PFA) overnight at 4 °C, blocked with 10% (vol/vol) goat serum in PBST, and then incubated with an 1:200 diluted anti–γ-H2A.X antibody (Genetex) overnight at 4 °C followed by a 1 h exposure to an FITC-conjugated secondary antibody (Jackson ImmunoResearch). Immunofluorescence was visualized by Olympus IX81.
HR Assay.
The in vivo HR reporter (DR-GFP) was a gift from Jiaxue Wu, Fudan University, Shanghai, China. HEK293T cells stably expressing shRNA targeting TyrRS were cotransfected with shRNA-resistant WT TyrRS-Flag, TyrRS-Flag K244Q, or TyrRS-Flag K244R, and an I-SceI expression vector (pCBA-I-SceI). Cells were harvested 36 h after transfection and subjected to flow cytometric analysis to determine recombination induced by I-SceI digestion. The parallel transfection with pApple-C1 was used to normalize for transfection efficiency.
Statistical Analysis.
The statistical data are from three independent experiments. Statistical analysis was performed by the Student’s t test for two groups and by ANOVA for multiple groups.
Analysis of Molecular Dynamics Simulations.
The structure models of mutations (K244Q and K244R) are predicted based on the human tyrosyl tRNA synthetase (TyrRS) (PDB ID code 4QBT) by Phyre2 (49). After several rounds of simulations, the rank 1 model was chosen with 100% confidence. The figures were depicted by PyMOL software.
Acknowledgments
We thank Dr. Kunliang Guan and Dr. Yue Xiong for engaging in helpful discussions throughout this study and providing PCAF, GCN5, and Tip60 constructs; Dr. Jiaxue Wu for providing plasmids for the HR assay; and Dr. Chunlin Shao for assistance with the γ-irradiation experiments. This work was supported by National Key Research and Development Program of China Grant 2016YFA0500600, Shanghai Committee of Science and Technology Grants KBH1322367 and 16JC1404000, and National Natural Science Foundation of China Grant 31571492. W.Y. was supported by China “Thousand Youth Talents” Program Grant KHH1322026 and the Program for Professors of Special Appointment (Eastern Scholar) at the Shanghai Institutions of Higher Learning (Grant SHH1322013).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. W.G. is a Guest Editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1608488114/-/DCSupplemental.
References
- 1.Carter CW., Jr Cognition, mechanism, and evolutionary relationships in aminoacyl-tRNA synthetases. Annu Rev Biochem. 1993;62(1):715–748. doi: 10.1146/annurev.bi.62.070193.003435. [DOI] [PubMed] [Google Scholar]
- 2.Ko YG, et al. Glutamine-dependent antiapoptotic interaction of human glutaminyl-tRNA synthetase with apoptosis signal-regulating kinase 1. J Biol Chem. 2001;276(8):6030–6036. doi: 10.1074/jbc.M006189200. [DOI] [PubMed] [Google Scholar]
- 3.Han JM, et al. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell. 2012;149(2):410–424. doi: 10.1016/j.cell.2012.02.044. [DOI] [PubMed] [Google Scholar]
- 4.Shi Y, et al. tRNA synthetase counteracts c-Myc to develop functional vasculature. eLife. 2014;3:e02349. doi: 10.7554/eLife.02349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jordanova A, et al. Disrupted function and axonal distribution of mutant tyrosyl-tRNA synthetase in dominant intermediate Charcot-Marie-Tooth neuropathy. Nat Genet. 2006;38(2):197–202. doi: 10.1038/ng1727. [DOI] [PubMed] [Google Scholar]
- 6.Sajish M, Schimmel P. A human tRNA synthetase is a potent PARP1-activating effector target for resveratrol. Nature. 2015;519(7543):370–373. doi: 10.1038/nature14028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei N, et al. Oxidative stress diverts tRNA synthetase to nucleus for protection against DNA damage. Mol Cell. 2014;56(2):323–332. doi: 10.1016/j.molcel.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu G, Xu T, Shi Y, Wei N, Yang XL. tRNA-controlled nuclear import of a human tRNA synthetase. J Biol Chem. 2012;287(12):9330–9334. doi: 10.1074/jbc.C111.325902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choudhary C, Weinert BT, Nishida Y, Verdin E, Mann M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat Rev Mol Cell Biol. 2014;15(8):536–550. doi: 10.1038/nrm3841. [DOI] [PubMed] [Google Scholar]
- 10.Zhao S, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327(5968):1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim SC, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23(4):607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 12.Choudhary C, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325(5942):834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 13.Peng L, et al. SIRT1 deacetylates the DNA methyltransferase 1 (DNMT1) protein and alters its activities. Mol Cell Biol. 2011;31(23):4720–4734. doi: 10.1128/MCB.06147-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Y, et al. Oxidative stress activates SIRT2 to deacetylate and stimulate phosphoglycerate mutase. Cancer Res. 2014;74(13):3630–3642. doi: 10.1158/0008-5472.CAN-13-3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang C, et al. Interactions between E2F1 and SirT1 regulate apoptotic response to DNA damage. Nat Cell Biol. 2006;8(9):1025–1031. doi: 10.1038/ncb1468. [DOI] [PubMed] [Google Scholar]
- 16.Rajamohan SB, et al. SIRT1 promotes cell survival under stress by deacetylation-dependent deactivation of poly(ADP-ribose) polymerase 1. Mol Cell Biol. 2009;29(15):4116–4129. doi: 10.1128/MCB.00121-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inuzuka H, et al. Acetylation-dependent regulation of Skp2 function. Cell. 2012;150(1):179–193. doi: 10.1016/j.cell.2012.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lv L, et al. Mitogenic and oncogenic stimulation of K433 acetylation promotes PKM2 protein kinase activity and nuclear localization. Mol Cell. 2013;52(3):340–352. doi: 10.1016/j.molcel.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang W, et al. Acetylation regulates gluconeogenesis by promoting PEPCK1 degradation via recruiting the UBR5 ubiquitin ligase. Mol Cell. 2011;43(1):33–44. doi: 10.1016/j.molcel.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Avalos JL, Bever KM, Wolberger C. Mechanism of sirtuin inhibition by nicotinamide: Altering the NAD(+) cosubstrate specificity of a Sir2 enzyme. Mol Cell. 2005;17(6):855–868. doi: 10.1016/j.molcel.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 21.Milne JC, Denu JM. The Sirtuin family: Therapeutic targets to treat diseases of aging. Curr Opin Chem Biol. 2008;12(1):11–17. doi: 10.1016/j.cbpa.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 22.Kim MY, et al. Tip60 histone acetyltransferase acts as a negative regulator of Notch1 signaling by means of acetylation. Mol Cell Biol. 2007;27(18):6506–6519. doi: 10.1128/MCB.01515-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Love IM, Sekaric P, Shi D, Grossman SR, Androphy EJ. The histone acetyltransferase PCAF regulates p21 transcription through stress-induced acetylation of histone H3. Cell Cycle. 2012;11(13):2458–2466. doi: 10.4161/cc.20864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bakker WJ, Harris IS, Mak TW. FOXO3a is activated in response to hypoxic stress and inhibits HIF1-induced apoptosis via regulation of CITED2. Mol Cell. 2007;28(6):941–953. doi: 10.1016/j.molcel.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 25.Gaupel AC, Begley TJ, Tenniswood M. Gcn5 modulates the cellular response to oxidative stress and histone deacetylase inhibition. J Cell Biochem. 2015;116(9):1982–1992. doi: 10.1002/jcb.25153. [DOI] [PubMed] [Google Scholar]
- 26.Caito S, et al. SIRT1 is a redox-sensitive deacetylase that is post-translationally modified by oxidants and carbonyl stress. FASEB J. 2010;24(9):3145–3159. doi: 10.1096/fj.09-151308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abdelmohsen K, et al. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol Cell. 2007;25(4):543–557. doi: 10.1016/j.molcel.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.González F, et al. An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell. 2014;15(2):215–226. doi: 10.1016/j.stem.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cestari I, Stuart K. A spectrophotometric assay for quantitative measurement of aminoacyl-tRNA synthetase activity. J Biomol Screen. 2013;18(4):490–497. doi: 10.1177/1087057112465980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhattacharyya A, Ear US, Koller BH, Weichselbaum RR, Bishop DK. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J Biol Chem. 2000;275(31):23899–23903. doi: 10.1074/jbc.C000276200. [DOI] [PubMed] [Google Scholar]
- 31.Deng CX. BRCA1: Cell cycle checkpoint, genetic instability, DNA damage response and cancer evolution. Nucleic Acids Res. 2006;34(5):1416–1426. doi: 10.1093/nar/gkl010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell. 2001;7(2):263–72. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 33.Qiang L, Banks AS, Accili D. Uncoupling of acetylation from phosphorylation regulates FoxO1 function independent of its subcellular localization. J Biol Chem. 2010;285(35):27396–27401. doi: 10.1074/jbc.M110.140228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu T, Liu PY, Marshall GM. The critical role of the class III histone deacetylase SIRT1 in cancer. Cancer Res. 2009;69(5):1702–1705. doi: 10.1158/0008-5472.CAN-08-3365. [DOI] [PubMed] [Google Scholar]
- 35.Alcendor RR, et al. Sirt1 regulates aging and resistance to oxidative stress in the heart. Circ Res. 2007;100(10):1512–1521. doi: 10.1161/01.RES.0000267723.65696.4a. [DOI] [PubMed] [Google Scholar]
- 36.Banks AS, et al. SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab. 2008;8(4):333–341. doi: 10.1016/j.cmet.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gan L, Mucke L. Paths of convergence: Sirtuins in aging and neurodegeneration. Neuron. 2008;58(1):10–14. doi: 10.1016/j.neuron.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salminen A, Kaarniranta K, Kauppinen A. Crosstalk between oxidative stress and SIRT1: Impact on the aging process. Int J Mol Sci. 2013;14(2):3834–3859. doi: 10.3390/ijms14023834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bai P, Cantó C. The role of PARP-1 and PARP-2 enzymes in metabolic regulation and disease. Cell Metab. 2012;16(3):290–295. doi: 10.1016/j.cmet.2012.06.016. [DOI] [PubMed] [Google Scholar]
- 40.Kanfi Y, et al. Regulation of SIRT1 protein levels by nutrient availability. FEBS Lett. 2008;582(16):2417–2423. doi: 10.1016/j.febslet.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 41.Yannay-Cohen N, et al. LysRS serves as a key signaling molecule in the immune response by regulating gene expression. Mol Cell. 2009;34(5):603–611. doi: 10.1016/j.molcel.2009.05.019. [DOI] [PubMed] [Google Scholar]
- 42.Sajish M, et al. Trp-tRNA synthetase bridges DNA-PKcs to PARP-1 to link IFN-γ and p53 signaling. Nat Chem Biol. 2012;8(6):547–554. doi: 10.1038/nchembio.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He W, et al. CMT2D neuropathy is linked to the neomorphic binding activity of glycyl-tRNA synthetase. Nature. 2015;526(7575):710–714. doi: 10.1038/nature15510. [DOI] [PMC free article] [PubMed] [Google Scholar]