Significance
The role of properdin in stabilization of the alternative pathway C3 convertase is indisputable, whereas its role as pattern recognition molecule remains controversial. Properdin lacks the structural homology shared by other pattern recognition molecules of the complement system, and has its major function in stabilizing the C3bBb convertase. We found that properdin binding was completely abolished by C3 inhibition after the exposure of human serum to myeloperoxidase, human umbilical vein endothelial cells, and Neisseria meningitidis, showing that properdin is not a pattern recognition molecule for these targets. We therefore challenge the view of properdin as a pattern recognition molecule, and argue that properdin typically binds a complement-activating surface subsequent to C3b to stabilize the alternative pathway C3 convertase.
Keywords: complement, properdin, C3, myeloperoxidase, Neisseria meningitidis
Abstract
Two functions have been assigned to properdin; stabilization of the alternative convertase, C3bBb, is well accepted, whereas the role of properdin as pattern recognition molecule is controversial. The presence of nonphysiological aggregates in purified properdin preparations and experimental models that do not allow discrimination between the initial binding of properdin and binding secondary to C3b deposition is a critical factor contributing to this controversy. In previous work, by inhibiting C3, we showed that properdin binding to zymosan and Escherichia coli is not a primary event, but rather is solely dependent on initial C3 deposition. In the present study, we found that properdin in human serum bound dose-dependently to solid-phase myeloperoxidase. This binding was dependent on C3 activation, as demonstrated by the lack of binding in human serum with the C3-inhibitor compstatin Cp40, in C3-depleted human serum, or when purified properdin is applied in buffer. Similarly, binding of properdin to the surface of human umbilical vein endothelial cells or Neisseria meningitidis after incubation with human serum was completely C3-dependent, as detected by flow cytometry. Properdin, which lacks the structural homology shared by other complement pattern recognition molecules and has its major function in stabilizing the C3bBb convertase, was found to bind both exogenous and endogenous molecular patterns in a completely C3-dependent manner. We therefore challenge the view of properdin as a pattern recognition molecule, and argue that the experimental conditions used to test this hypothesis should be carefully considered, with emphasis on controlling initial C3 activation under physiological conditions.
Properdin, also referred to as factor P, was first described in 1954 by Pillemer and colleagues as a component that, in an antibody-independent manner, is able to promote complement activation on zymosan particles and on other carbohydrates (1). These claims were controversial, and properdin-dependent complement activation was dismissed by the scientific community (2–4); however, the “properdin system” was reborn as the alternative pathway (AP) more than 20 y later (3), with properdin described as a stabilizer and positive regulator of the AP C3 convertase (5, 6). Properdin and its possible different roles in complement activation have been a basis for further studies in this area (7–11).
In the current conception, although yet to be proven in vivo, the AP of the complement system is slowly autoactivated via spontaneous or induced formation of fluid-phase AP C3 convertase (12, 13). The C3 moiety within this convertase is C3(H2O) formed on exposure and subsequent hydrolysis of the internal thioester, which is normally protected inside native C3 (14-16). C3(H2O) is “C3b-like”; it still contains C3a, but is conformationally similar to C3b. C3(H2O) can bind factor B, which is cleaved by factor D into Ba and Bb. Bb remains bound to C3(H2O), forming the enzymatic complex that cleaves C3 into C3b and C3a. Surface-bound C3b can form additional AP C3 convertase molecules with Bb, which rapidly cleave more C3 resulting in self-amplification and generation of the C5 convertase C3bBbC3b. The degree of amplification on a surface is determined by the rate of the C3b feedback (i.e., C3 cleavage) and breakdown (i.e., C3b degradation) cycles (17).
We previously reported that amplification via the AP on an unprotected surface contributes to more than 80% of terminal pathway activation after specific initial classical pathway or lectin pathway activation (18, 19). The C3bBb complex is relatively unstable, with a half-life of 90 s under physiological conditions (6, 20); however, properdin can associate with C3bBb and create the more stable C3bBbP complex that is essential for effective AP amplification (5, 21). Recently published electron microcopy images of the C3bBbP complex have shown how properdin is associated with the convertase near the C345C domain of C3b and the von Willebrand factor type A domain of factor B (22).
Discussion of the role of properdin as a pattern recognition molecule and initiator of the AP was renewed with experiments showing that purified unfractionated properdin covalently attached to a biosensor surface could serve as a platform for in situ assembly of the AP C3 convertase (23). This was done in a relatively artificial system by using purified components in buffer milieu. That study was followed by several reports of biological substrates suggested to serve as patterns for the direct recognition by properdin for AP complement activation. Reported patterns include exogenous microorganisms (7, 24), endogenous cells (25–27), and various biological substrates (9, 28, 29); however, many of these experiments were performed in systems permitting either C3 activation with initial C3b deposition or in buffer systems with purified properdin. In the presence of intact C3, it is virtually impossible to demonstrate whether properdin acts in a recognition manner or subsequently binds to C3b. On the other hand, purified properdin is sensitive to aggregation into sticky multimers, and these aggregates, referred to as activated properdin, do not behave as the native form; thus, results with purified properdin must be judged with care (30, 31), especially in unfractionated preparations (32). Early preparations of properdin, such as those used in the initial studies by Pillemer et al. (1), were later shown to contain impurities and high molecular aggregates of properdin, which were lost when adsorption to zymosan was omitted from the purification protocols (30, 33, 34). Furthermore, it is crucially important to distinguish between passive binding of properdin to a surface and subsequent activation of AP as a result of this binding. For this purpose, the methodological conditions should be under strict control.
In previous work, by inhibiting C3 cleavage with the peptide inhibitor compstatin Cp40, we showed that properdin binding to zymosan and Escherichia coli is not a primary event, but is completely dependent on initial C3 deposition (35). In the present study, we aimed to investigate whether the binding of properdin to other tentative targets for pattern recognition, i.e., granulocyte myeloperoxidase (MPO), human umbilical vein endothelial cells (HUVECs), and Neisseria meningitidis, is a primary event or dependent on C3 deposition. For this purpose, we compared normal human serum (NHS) with and without compstatin Cp40, as well as C3-depleted human serum with and without reconstitution of purified C3.
Results
Properdin Binding to MPO-Coated Wells After Incubation with Human Serum.
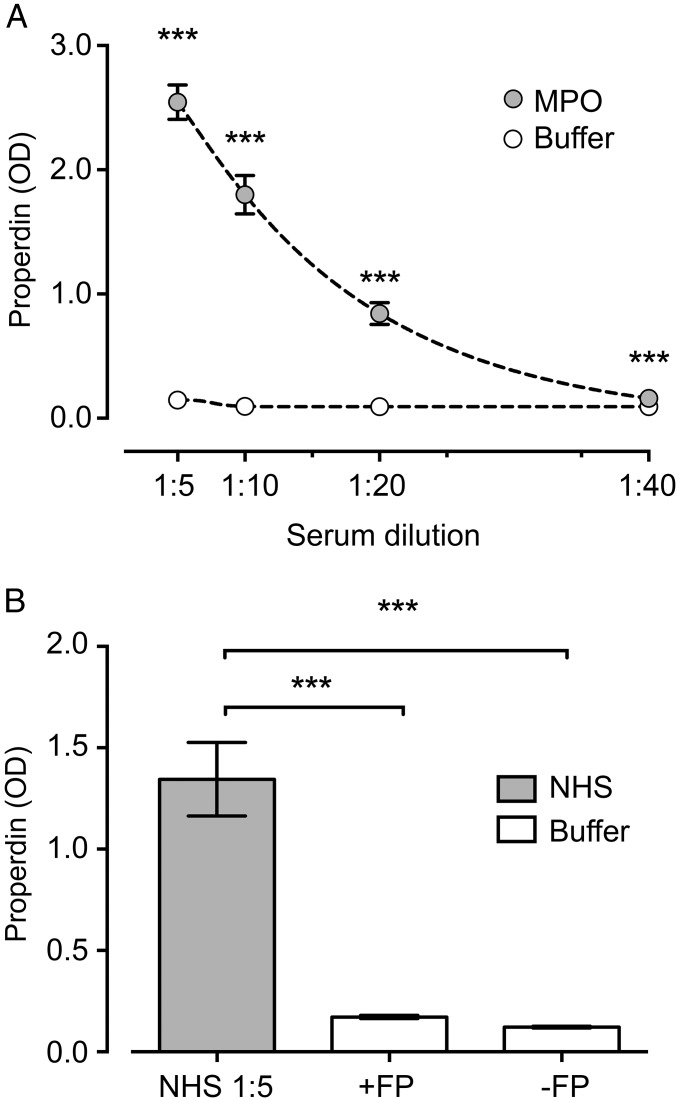
Owing to the retained peroxidase activity of immobilized MPO, the binding of properdin to MPO-coated wells was not possible to evaluate in our regular ELISA with HRP-labeled detection antibody and substrate dependent on peroxidase activity. Thus, a detection system based on alkaline phosphatase-labeled anti-mouse IgG antibody was developed. A pool of NHS incubated in serial dilutions on the MPO coat led to a dose-dependent binding of properdin (Fig. 1A). In contrast to properdin-containing NHS, there was minimal binding of purified properdin using a concentration corresponding to that in NHS (OD = 1.35 vs. 0.17), with a signal similar to that of the buffer control (OD = 0.12) (Fig. 1B).
Fig. 1.
Binding of properdin to immobilized MPO. NHS in serial dilutions (A) or purified properdin diluted in VBS buffer (B) was incubated in microtiter plates coated with human neutrophil MPO dissolved in 0.05 M Na2CO3, pH 9.5, by overnight incubation at room temperature, or incubated solely with 0.05 M Na2CO3 as a control. All wells were subsequently blocked with 1.0% BSA in PBS (pH 7.4) and Tween (0.1%) for 60 min. NHS, serially diluted in 2.5 mM VBS, pH 7.2, with MgEGTA or purified properdin (2 μg/mL in VBS/MgEGTA), was incubated on MPO-coated surfaces or controls for 60 min at 37 °C. Binding of properdin was demonstrated in ELISA with mouse anti-human properdin. Data are presented as mean with 95% CI OD values for six experiments. ***P < 0.001.
MPO-Induced Terminal Pathway Activation and Dependency on C3.
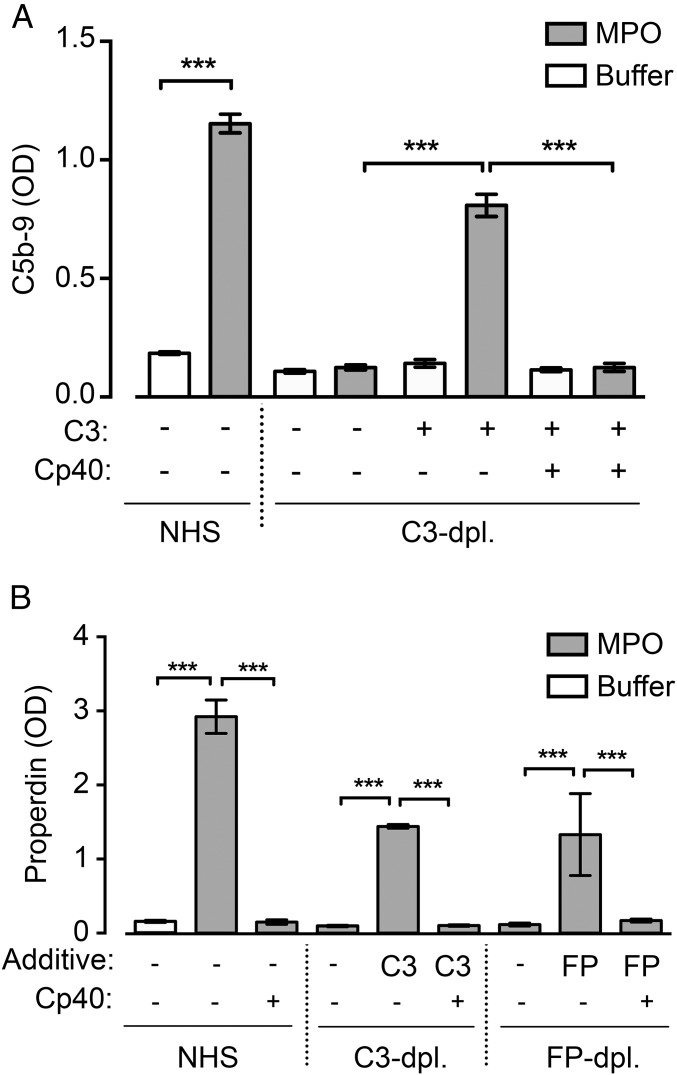
MPO was previously shown to activate complement via the AP (28). MPO coated onto microtiter plates activated complement, as detected by binding of the monoclonal antibody aE11, specific for a C9 neoepitope in the terminal C5b-9 complement complex (36). No generation of C5b-9 on the surface by MPO occurred in human serum depleted of C3 (OD = 0.13); however, on reconstitution with purified C3, a strong deposition of C5b-9 was found (OD = 0.81), which was efficiently blocked by compstatin Cp40 (OD = 0.13) (Fig. 2A).
Fig. 2.
C3 dependence for complement activation and properdin binding on MPO. (A) NHS and C3-depleted human serum diluted 1:5 with and without purified C3 (130 μg/mL) were incubated on MPO-coated wells (gray bars) or uncoated wells (white bars) in the presence or absence of compstatin Cp40. Complement activation triggered by MPO was detected as surface deposition of C5b-9 using the monoclonal antibody aE11, specific for a C9 neoepitope in C5b-9. (B) The same experimental setup as in A, but also including properdin-depleted human serum diluted 1:5 with and without properdin (2 μg/mL), was tested for the binding of properdin to MPO. Properdin was detected as in ELISA with mouse anti-human properdin. Data are presented as mean with 95% CI OD values of six experiments. ***P < 0.001.
C3-Dependent Binding of Properdin to MPO.
We next examined C3 dependence for the binding of properdin to MPO. Binding of properdin in NHS to MPO was completely abolished when C3 cleavage was inhibited with compstatin Cp40 (OD = 2.92 vs. 0.15) (Fig. 2B, Left). Similarly, no binding of properdin occurred from C3-depleted human serum, but binding was restored by reconstitution with purified C3 (OD = 0.10 vs. 1.44) (Fig. 2B, Middle). This binding was abrogated in the presence of compstatin Cp40 (OD = 0.10).
We then assessed binding in properdin-depleted serum before and after reconstitution with purified properdin in the presence and absence of compstatin Cp40 (Fig. 2B, Right). Properdin binding was achieved when properdin-depleted human serum was reconstituted with purified properdin (OD = 1.33), but was again completely dependent on C3 cleavage, as demonstrated by the fact that compstatin Cp40 reduced the binding to background level (OD = 0.17). The purified properdin used was stored at −70 °C and thawed only once for the experiments. Because the composition and quality of the purified properdin preparation used in these experiments are crucial for correct interpretation of the data, we performed a careful characterization of the purified properdin preparation by exclusion chromatography, SDS/PAGE, and Western blot analysis, as described in SI Materials and Methods and shown in Fig. S1.
Fig. S1.
Characterization of purified properdin by gel filtration fractionation, SDS/PAGE, and Western blot analysis. (A) Purified properdin (black and blue curves) and serum (red curve) were separated by size exclusion chromatography. In the purified properdin preparation, total protein was detected in the eluate by absorbance at 280 nm (black curve), and properdin was specifically detected by ELISA (blue curve). In serum, properdin was specifically detected by ELISA (red curve). The majority of properdin in the purified preparation eluted as physiological oligomers (P2–P4), compared with elution of serum properdin, even with the possible presence of some high-order oligomers (P>4). (B) SDS/PAGE (lanes 1 and 2) and Western blot analysis (lane 3) of purified properdin under nonreduced conditions. Purified properdin was loaded at 1 μg (lane 1), 5 μg (lane 2), or 0.4 μg (lane 3) per well. The SDS/PAGE indicates no impurities in the purified preparation, and the Western blot analysis demonstrate that the band visualized slightly above the 50 kDa-molecular marker (lane M) corresponds to monomeric properdin, estimated at 53 kDa.
C3-Dependent Binding of Properdin to HUVECs.
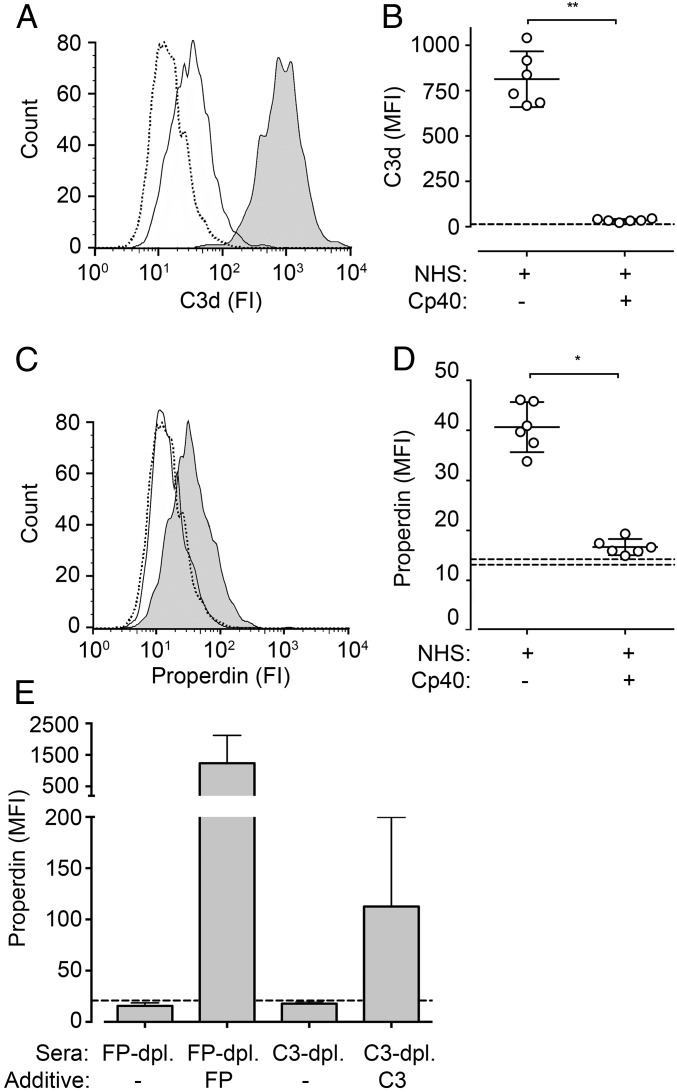
We tested HUVECs incubated with NHS for the binding of C3d and properdin. Incubation for 4 h on HUVECs resulted in substantial deposition of C3d fragments detected by flow cytometry, with a median fluorescence intensity (MFI) of 813 (Fig. 3 A and B). In the presence of compstatin Cp40, C3d deposition was virtually abolished (>97%). Properdin binding on HUVECs followed the same trend and was largely dependent on C3 (Fig. 3 C and D). In the presence of compstatin Cp40, properdin binding from NHS was reduced by 90%, from an MFI of 40.6 to an MFI of 16.7, a value in the same range as the isotype control (MFI = 13.9). Controls with properdin- and C3-depleted human serum showed comparably low values with a distinct increase in properdin binding after reconstitution with purified properdin and C3, respectively (Fig. 3E).
Fig. 3.
C3 deposition and binding of properdin to the surface of HUVECs. Undiluted NHS with and without compstatin Cp40 (A–D) or human serum depleted and reconstituted for C3 or properdin (E) was incubated for 4 h at 37 °C on monolayers of confluent HUVECs. Deposition of C3d fragments (A and B) and binding of properdin (C–E) were analyzed by flow cytometry. A and C show representative curves for binding of C3d and properdin (continuous lines), respectively, to HUVECs in NHS (gray background) or NHS with compstatin Cp40 (white background), or in NHS binding of the isotype control (dotted line). B and D show individual values and mean with 95% CI of six experiments. *P < 0.05; **P < 0.01. Stippled lines represent the range for binding of the isotype control. E shows the mean ± SD value of six experiments.
C3-Dependent Binding of Properdin to N. meningitidis.
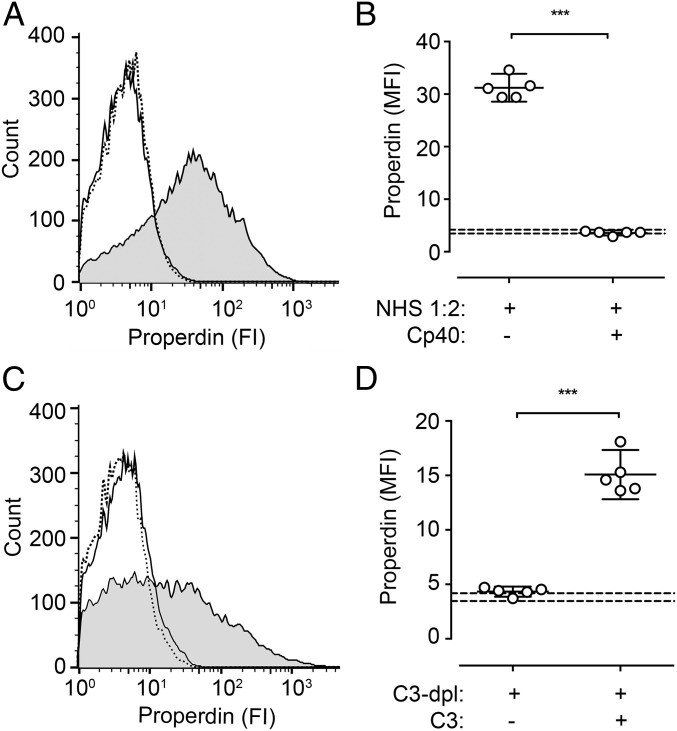
Binding of properdin from NHS to N. meningitidis was evaluated by flow cytometry. Incubation of N. meningitidis in NHS diluted 1:2 yielded a distinct signal for deposition of properdin on the bacteria (MFI = 31.2) (Fig. 4 A and B). The addition of compstatin Cp40 abrogated this deposition completely (MFI = 3.6), showing values similar to the isotype control (MFI = 3.8). Virtually identical findings were seen with C3-depleted human serum (Fig. 4 C and D). After exposure of N. meningitidis to C3-depleted human serum, properdin deposition was in the range of the isotype control (MFI = 4.3). Reconstitution of C3-depleted human serum with purified C3 showed binding approaching the levels in NHS (MFI = 15.1).
Fig. 4.
Binding of properdin to the surface of N. meningitidis. Heat-inactivated N. meningitidis (reference strain 44/76), suspended in VBS with MgCl2 (0.5 mM), CaCl2 (2 mM), and gelatin (0.1%), pH 7.5, were incubated in NHS (diluted 1:2) in the presence or absence of compstatin Cp40 (A and B) or in C3-depleted human serum (diluted 1:2) with and without purified C3 (650 μg/mL) (C and D) at 37 °C for 20 min. Binding of properdin was detected by a monoclonal antibody using flow cytometry. A and C show representative curves for the binding of anti-properdin (continuous line) to bacteria in NHS (gray background) or human serum inhibited/depleted of C3 (white background), or in NHS binding of the isotype control (dotted line). B and D show individual values and the mean with 95% CI value of five experiments. ***P < 0.001. Stippled lines represent the range for binding of the isotype control.
SI Materials and Methods
The purified properdin preparation and serum were subjected to protein size separation by gel filtration chromatography on an ENrich size exclusion column (650, 10 × 300 mm; Bio-Rad) (Fig. S1). The column was equilibrated with Dulbecco’s PBS (DPBS), pH 7.4 (Sigma-Aldrich). Purified properdin (Complement Technology; 100 μg) or NHS (100 μL) was loaded on the column, which was then eluted with DPBS at 1 mL/min. Protein content was continuously detected in the eluate by the absorbance at 280 nm. Fractions were collected at a size of 250 μL. Properdin was specifically detected in collected fractions by ELISA. Fractions were coated on NUNC Maxisorp microtiter plates (Nalgene Nunc) by overnight incubation at room temperature. Wells were then blocked with 1% BSA in PBS (pH 7.4) for 60 min, and properdin was specifically detected with mAb anti-properdin (Quidel; 1:1,000), followed by alkaline phosphatase labeled sheep F(ab′)2 anti-mouse IgG (Sigma-Aldrich; 1:1,000). Purified properdin was run on SDS/PAGE under nonreducing conditions. Proteins were either detected with Coomassie brilliant blue or transferred to PVDF membrane (Bio-Rad). The membrane was incubated with monoclonal anti-properdin (Quidel), followed by HRP-conjugated polyclonal goat anti-mouse (Jackson ImmunoResearch), and then visualized using ECL substrate (Bio-Rad).
Discussion
In the present study, we have demonstrated that properdin is unable to bind directly to granulocyte MPO, to the surface of HUVECs, or to N. meningitidis, but that the interaction is fully dependent on initial C3 activation and thus does not act as a pattern recognition molecule for these targets. We investigated these targets because some interesting observations regarding the interaction between them and properdin have been reported. MPO was recently reported to directly attract serum properdin for initiation of the AP (28). N. meningitidis and endothelial cells were studied based on their central role in properdin pathophysiology. Properdin is significant in the defense against Neisseria species, and Neisseria gonorrhea was claimed to directly bind properdin (7), findings that have been extrapolated to N. meningitidis (37). Endothelial cells are a source of serum properdin (38), and properdin has been shown to interact with various glycosaminoglycans on endogenous cells (25, 39, 40).
In a previous study, we showed that inhibition of C3 cleavage could serve to distinguish between the primary binding of properdin (i.e., pattern recognition function) and properdin binding secondary to initial C3 fragment deposition (i.e., convertase stabilization) (35). Using the C3 inhibitor compstatin Cp40, we consistently found that properdin in human serum and properdin released from activated polymorphonuclear leukocytes did not bind directly to zymosan or E. coli, but that binding of properdin was fully dependent on the initial deposition of C3b. By using the same principle here, properdin binding to MPO, HUVECs, and N. meningitidis was found to be fully dependent on accessible C3. Thus, the addition of a C3 inhibitor, compstatin Cp40, to NHS or the use of C3-depleted serum did not show any direct binding of properdin to these five different tested surfaces and ligands that we tested, of which three were exogenous and two were endogenous, supporting the notion that properdin is not a promiscuous molecule binding to various “danger” ligands.
The background of the present study was the controversy surrounding properdin acting as a pattern recognition molecule in activation of the AP. Properdin lacks the structural homology shared by other pattern recognition molecules of the complement system. C1q and the recognition molecules of the lectin pathway, including mannose-binding lectin, ficolins, and collectins, all have a common structure with a bouquet of globular heads built up of a cysteine-rich N-terminal stretch, a collagen-like domain, and a carbohydrate-recognition domain or fibrinogen-like domain (41). These molecules bind their respective targets with high avidity using multiple sites for interaction (42, 43). This is distinctly different from properdin, which is a linear molecule composed of repeating thrombospondin type I domains (44), a noteworthy difference from the typical fluid-phase pattern recognition molecules of innate immunity. Although this does not completely exclude properdin for recognizing certain structures besides the AP convertase, it is in line with our findings, making it unlikely that properdin can be defined as a recognition molecule.
Indications of the role of properdin as pattern recognition molecule started with experiments showing that purified properdin covalently bound to a biosensor surface could serve as a platform for the in situ assembly of AP C3 convertases (23). Initial data introduced the attractive concept of how properdin could not only stabilize the C3bBb convertase, but also promote surface attraction of preformed fluid-phase C3bBb; however, studies using surface plasmon resonance depend on the use of purified proteins in a buffer system, which introduces a certain degree of artificiality. Properdin in purified preparations is prone to aggregation, and extrapolating findings on covalently bound properdin to native serum properdin interacting on a biological substrate is virtually impossible. Nonetheless, later data indicated that AP is indeed initiated by the noncovalent attachment of properdin on surfaces of different microorganisms, including yeast cell walls, N. gonorrheae, and LPS-negative E. coli (7). However, these experiments were performed in a system permitting autoactivation of C3 (12, 45) and allowing slow, continuous surface C3b deposition, thus making it difficult to determine whether properdin reacts primarily to the activating surface as a true pattern recognition molecule or binds secondarily to C3b.
Agarwal et al. (46) later extensively studied the ability of several strains of N. gonorrheae and N. meningitidis to directly bind properdin. None of the tested strains bound native properdin, but binding did occur with purified preparations of properdin containing high-order oligomers of aggregated properdin, a phenomenon later demonstrated on zymosan and Streptococcus pneumonia as well (46, 47). Aggregates of properdin were shown to appear spontaneously in certain purified preparations on preparation or during long-term storage or freezing and thawing (30, 31). These aggregates are artifacts with different properties than serum properdin; for example, they promote fluid-phase complement activation and C3 depletion, as illustrated by the old term “activated properdin” (30, 31). Ali et al. (47) recently showed that recombinant highly polymerized properdin can be a powerful agent to therapeutically combat infections by N. meningitidis and S. pneumonia. Mice infected with these bacteria were protected when given recombinantly expressed properdin containing high-order oligomers, which, in contrast to low-order oligomers of properdin in plasma, directly interact with the bacterial surface and focus C3b deposition (47).
In contrast to our findings with exogenous ligands, previously shown with zymosan and E. coli (35) and in the present work with N. meningitidis, there is evidence of C3- independent properdin binding to ligands on endogenous cells, such as DNA on late apoptotic or necrotic cells (26), and presumably various glycosaminoglycans on early apoptotic T cells, but not necrotic T cells (25), activated platelets (27), and renal tubular epithelial cells (39, 40). To the best of our knowledge, no data for endothelial cells and properdin binding have been reported previously, even though endothelial cells are a source of serum properdin and can express heparin sulfate proteoglycans, which have been shown to bind properdin (39, 48). In the present study, after incubation of human serum on HUVECs for 4 h, surface complement activation was detected as deposition of C3d fragments on the cells. Properdin was also found on the cells, but, like for N. meningitidis, this binding was dependent on initial C3 activation, because compstatin Cp40 almost entirely inhibited properdin binding.
Recently, MPO was added to the list of suggested endogenous molecules capable of binding properdin directly (28). Thus, MPO could be the molecule that links properdin binding to the surface of activated neutrophils, as has been reported previously (49). By studying properdin binding to immobilized MPO, the authors proposed that MPO promotes complement activation by directly binding properdin (49). We question those findings, however, because they were based on a detection system using HRP-labeled antibodies in which we found it impossible to distinguish substrate turnover caused by peroxidase activity by the antibody-linked HRP and that caused by the immobilized MPO. By changing the enzyme and substrate in the detection system, we showed that properdin was not attracted to MPO in the absence or inhibition of C3; however, immobilized MPO did serve as an excellent complement activator by promoting the formation of C5b-9 on the surface and, in the presence of C3, the binding of properdin.
In conclusion, we challenge the view of properdin as a pattern recognition molecule by providing evidence that it binds to different exogenous and endogenous molecular patterns in only a C3-dependent manner. To support the claim that properdin is a pattern recognition molecule, it should be clearly shown that the initial binding and subsequent activation are C3-independent, and the use of purified properdin preparations should be critically evaluated. Taken together, our data provide further support for the role of properdin as a specific stabilizer of the AP C3 convertase that generally does not act as a recognition molecule.
Materials and Methods
Antibodies and Other Reagents.
Two mouse anti-human properdin IgG1κ antibodies (P1: A233 and P2: A235) were obtained from Quidel, and anti-C3d clone 7C10 was obtained from Abcam. Clone IS7 (IgG1κ, anti-human CD22), from Diatec Monoclonals, served as an isotype control for IgG1κ. Monoclonal Ab aE11, specifically binding a C9 neoepitope in the terminal C5b-9 complement complex (50), was obtained from Diatec Monoclonals. Clone 5A7, from Diatec Monoclonals, served as an IgG2a isotype control. Sheep F(ab′)2 alkaline phosphatase-labeled anti-mouse IgG was obtained from Sigma-Aldrich, F(ab′)2 anti-mouse IgG-FITC was obtained from GE Healthcare UK, goat anti-mouse IgG1-FITC was obtained from Southern Biotech, and goat anti-mouse was obtained from Jackson ImmunoResearch. The compstatin analog Cp40 (dTyr-Ile-[Cys-Val-Trp(Me)-Gln-Asp-Trp-Sar-His-Arg-Cys]-mIle-NH2), used specifically to block C3 cleavage (51), was a kind gift from John D. Lambris, University of Pennsylvania, Philadelphia.
Serum and Purified Proteins.
NHS was collected from nine healthy volunteers, all of whom were normal in classical pathway, lectin pathway, and AP activity, as detected by the Wieslab total complement system screen assay (Euro Diagnostica) (52). Sera were pooled and stored in aliquots at –70 °C. Properdin-depleted human serum and purified properdin were obtained from Complement Technology. The properdin was characterized by size- exclusion chromatography, SDS/PAGE, and Western blot analysis. Properdin was quantified in serum and in C3-depleted human serum as 15.8 and 14.6 μg/mL, respectively, using a human properdin ELISA kit (Hycult Biotech) according to the manufacturer’s instructions. C3-depleted human serum and purified C3 were obtained from Complement Technology and Quidel, respectively. Human neutrophil MPO was obtained from Athens Research & Technology.
Endothelial Cells.
HUVECs were isolated from umbilical cord veins by digestion with 0.1% collagenase A (Roche Diagnostics) and cultured as described previously (53). Confluent HUVECs in passages two through five were used in the experiments.
Bacteria.
The N. meningitidis reference strain 44/76 was used here. It is characterized as B:15:P1.7,16:L3,7,9, belonging to the multilocus sequence type 32 clone (previously electrophoretic type 5). This reference strain was originally isolated from a woman admitted to Ullevål University Hospital, Oslo, Norway, with lethal meningococcal infection in 1976 and is the prototype bacterium of the clone causing the long-lasting Norwegian epidemics. The bacteria were grown overnight on Colombia agar, resuspended in sterile PBS, heat-inactivated at 56 °C for 30 min and frozen at −70 °C until use. Heat-inactivated bacteria were used for safety reasons. Heat inactivation causes only minor alterations to the bacterial membrane, and the biological activity is preserved (54).
Binding of Properdin to MPO.
Microtiter plates (Costar high-binding; Corning) were coated with MPO (20 μg/mL, dissolved in 0.05 M NaCO3, pH 9.5) by overnight incubation at room temperature, or incubated solely with 0.05 M NaCO3 as a control. All wells were subsequently blocked with 1% BSA in PBS (pH 7.4) and Tween (0.1%) for 60 min. NHS, serially diluted in 2.5 mM veronal-buffered saline (VBS), pH 7.2, with MgEGTA (5 mM Mg2+ and 10 mM EGTA) or purified properdin (2 μg/mL in VBS/MgEGTA) were incubated on MPO-coated surfaces or controls for 60 min at 37 °C. In separate sets of experiments, incubation was done on MPO-coated surfaces with NHS (1:5) with or without compstatin Cp40 (8 μM), on C3-depleted serum (1:5) with or without purified C3 (130 μg/mL), and on properdin-depleted serum (1:5) with or without purified properdin (2 μg/mL). Binding of properdin was detected with mAb anti-properdin (1:1,000), followed by alkaline phosphatase-labeled sheep F(ab′)2 anti-mouse IgG (whole molecule) (1:1,000). Deposition of C5b-9 onto MPO surfaces or controls after incubation with NHS with or without compstatin Cp40 or with C3-depleted serum with or without purified C3 was detected with mAb aE11 (1 mg/mL; 1:6,000), followed by alkaline phosphatase-labeled sheep anti-mouse IgG. p-nitrophenyl phosphate (pNPP; Sigma-Aldrich) served as a substrate for alkaline phosphatase.
Binding of Properdin to N. meningitidis.
N. meningitidis were washed by repeated centrifugation at 3,220 × g for 10 min and then resuspended in PBS with 0.1% BSA. Bacteria were then suspended in VBS with MgCl2 (0.5 mM), CaCl2 (2 mM), and gelatin (0.1%), pH 7.5. NHS was preincubated with or without compstatin Cp40 (40 μM) for 5 min and then added to the bacteria (final dilution, 1:2) and incubated at 37 °C for 20 min. Binding of properdin was evaluated with mAb anti-properdin (diluted 1:50) or isotype control clone IS7, followed by F(ab′)2 anti-mouse IgG-FITC (1:50). Samples were washed twice, resuspended in PBS with 1% BSA, and analyzed on a FACSCalibur flow cytometer (BD Biosciences). N. meningitidis was gated on forward-scatter and side-scatter, and 20,000 events were collected. Data were evaluated using FlowJo version 10.
Binding of Properdin to HUVECs.
Confluent HUVECs were washed twice with tempered PBS, pH 7.4. NHS with or without compstatin Cp40 (40 μM) was incubated on a monolayer of HUVECs in cell culture plates for 4 h at 37 °C. HUVECs were also incubated with C3-depleted human serum with and without purified C3 (650 μg/mL) or with properdin-depleted human serum with and without properdin (10 μg/mL) as a control. After incubation, cells were washed twice with ice-cold PBS and then fixed for 2.5 min with 0.5% paraformaldehyde. Binding of C3 fragments and properdin was evaluated using mAbs against C3d and properdin, respectively, or the isotype control clone IS7, all followed by goat anti-mouse IgG1-FITC. Cells were then trypsinated, washed, and run on a FACSCalibur flow cytometer. Data were analyzed with FlowJo version 10.
Statistics.
GraphPad Prism version 5 was used for statistical analyses. Statistical differences were analyzed on normally distributed data using the Student t test or, when more than two conditions were compared, with one-way ANOVA followed by the Bonferroni posttest. A P value <0.05 was considered statistically significant.
Ethics Statement.
The study was designed and performed according to the ethical guidelines from the Declaration of Helsinki. Informed written consent for participation in the study was obtained from all individuals. The study was approved by the regional Ethical Committee of the South-Eastern Norway Regional Health Authority.
Acknowledgments
We thank Grethe Bergseth (Nordlandssykehuset, Bodo, Norway) for technical assistance. This study was financially supported by the Research Council of Norway, the Norwegian Council on Cardiovascular Disease, the Northern Norway Regional Health Authority, the South-Eastern Norway Regional Health Authority, the Odd Fellows Foundation, faculty grants from Linnaeus University, and the European Community's Seventh Framework Program under Grant Agreement 602699 (DIREKT).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1612385114/-/DCSupplemental.
References
- 1.Pillemer L, et al. The properdin system and immunity, I: Demonstration and isolation of a new serum protein, properdin, and its role in immune phenomena. Science. 1954;120(3112):279–285. doi: 10.1126/science.120.3112.279. [DOI] [PubMed] [Google Scholar]
- 2.Ecker EE. Louis Pillemer; 1908-1957. J Immunol. 1958;80(6):415–416. [PubMed] [Google Scholar]
- 3.Lepow IH. Presidential address to American Association of Immunologists in Anaheim, California, April 16, 1980: Louis Pillemer, properdin, and scientific controversy. J Immunol. 1980;125(2):471–475. [PubMed] [Google Scholar]
- 4.Lachmann P. Complement before molecular biology. Mol Immunol. 2006;43(6):496–508. doi: 10.1016/j.molimm.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 5.Fearon DT, Austen KF. Properdin: Binding to C3b and stabilization of the C3b-dependent C3 convertase. J Exp Med. 1975;142(4):856–863. doi: 10.1084/jem.142.4.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Medicus RG, Götze O, Müller-Eberhard HJ. Alternative pathway of complement: Recruitment of precursor properdin by the labile C3/C5 convertase and the potentiation of the pathway. J Exp Med. 1976;144(4):1076–1093. doi: 10.1084/jem.144.4.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spitzer D, Mitchell LM, Atkinson JP, Hourcade DE. Properdin can initiate complement activation by binding specific target surfaces and providing a platform for de novo convertase assembly. J Immunol. 2007;179(4):2600–2608. doi: 10.4049/jimmunol.179.4.2600. [DOI] [PubMed] [Google Scholar]
- 8.Harboe M, Mollnes TE. The alternative complement pathway revisited. J Cell Mol Med. 2008;12(4):1074–1084. doi: 10.1111/j.1582-4934.2008.00350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kimura Y, Miwa T, Zhou L, Song WC. Activator-specific requirement of properdin in the initiation and amplification of the alternative pathway complement. Blood. 2008;111(2):732–740. doi: 10.1182/blood-2007-05-089821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stover CM, et al. Properdin plays a protective role in polymicrobial septic peritonitis. J Immunol. 2008;180(5):3313–3318. doi: 10.4049/jimmunol.180.5.3313. [DOI] [PubMed] [Google Scholar]
- 11.Kemper C, Atkinson JP, Hourcade DE. Properdin: Emerging roles of a pattern-recognition molecule. Annu Rev Immunol. 2010;28:131–155. doi: 10.1146/annurev-immunol-030409-101250. [DOI] [PubMed] [Google Scholar]
- 12.Lachmann PJ, Halbwachs L. The influence of C3b inactivator (KAF) concentration on the ability of serum to support complement activation. Clin Exp Immunol. 1975;21(1):109–114. [PMC free article] [PubMed] [Google Scholar]
- 13.Bexborn F, Andersson PO, Chen H, Nilsson B, Ekdahl KN. The tick-over theory revisited: Formation and regulation of the soluble alternative complement C3 convertase (C3(H2O)Bb) Mol Immunol. 2008;45(8):2370–2379. doi: 10.1016/j.molimm.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tack BF, Harrison RA, Janatova J, Thomas ML, Prahl JW. Evidence for presence of an internal thiolester bond in third component of human complement. Proc Natl Acad Sci USA. 1980;77(10):5764–5768. doi: 10.1073/pnas.77.10.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janssen BJ, et al. Structures of complement component C3 provide insights into the function and evolution of immunity. Nature. 2005;437(7058):505–511. doi: 10.1038/nature04005. [DOI] [PubMed] [Google Scholar]
- 16.Nilsson B, Nilsson Ekdahl K. The tick-over theory revisited: Is C3 a contact-activated protein? Immunobiology. 2012;217(11):1106–1110. doi: 10.1016/j.imbio.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 17.Lachmann PJ. The amplification loop of the complement pathways. Adv Immunol. 2009;104(104):115–149. doi: 10.1016/S0065-2776(08)04004-2. [DOI] [PubMed] [Google Scholar]
- 18.Harboe M, Ulvund G, Vien L, Fung M, Mollnes TE. The quantitative role of alternative pathway amplification in classical pathway induced terminal complement activation. Clin Exp Immunol. 2004;138(3):439–446. doi: 10.1111/j.1365-2249.2004.02627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harboe M, et al. The down-stream effects of mannan-induced lectin complement pathway activation depend quantitatively on alternative pathway amplification. Mol Immunol. 2009;47(2-3):373–380. doi: 10.1016/j.molimm.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 20.Pangburn MK, Müller-Eberhard HJ. The C3 convertase of the alternative pathway of human complement: Enzymic properties of the bimolecular proteinase. Biochem J. 1986;235(3):723–730. doi: 10.1042/bj2350723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fearon DT. Activation of the alternative complement pathway. CRC Crit Rev Immunol. 1979;1(1):1–32. [PubMed] [Google Scholar]
- 22.Alcorlo M, Tortajada A, Rodríguez de Córdoba S, Llorca O. Structural basis for the stabilization of the complement alternative pathway C3 convertase by properdin. Proc Natl Acad Sci USA. 2013;110(33):13504–13509. doi: 10.1073/pnas.1309618110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hourcade DE. The role of properdin in the assembly of the alternative pathway C3 convertases of complement. J Biol Chem. 2006;281(4):2128–2132. doi: 10.1074/jbc.M508928200. [DOI] [PubMed] [Google Scholar]
- 24.Cortes C, Ferreira VP, Pangburn MK. Native properdin binds to Chlamydia pneumoniae and promotes complement activation. Infect Immun. 2011;79(2):724–731. doi: 10.1128/IAI.00980-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kemper C, Mitchell LM, Zhang L, Hourcade DE. The complement protein properdin binds apoptotic T cells and promotes complement activation and phagocytosis. Proc Natl Acad Sci USA. 2008;105(26):9023–9028. doi: 10.1073/pnas.0801015105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu W, et al. Properdin binds to late apoptotic and necrotic cells independently of C3b and regulates alternative pathway complement activation. J Immunol. 2008;180(11):7613–7621. doi: 10.4049/jimmunol.180.11.7613. [DOI] [PubMed] [Google Scholar]
- 27.Saggu G, et al. Identification of a novel mode of complement activation on stimulated platelets mediated by properdin and C3(H2O) J Immunol. 2013;190(12):6457–6467. doi: 10.4049/jimmunol.1300610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Flynn J, Dixon KO, Faber Krol MC, Daha MR, van Kooten C. Myeloperoxidase directs properdin-mediated complement activation. J Innate Immun. 2014;6(4):417–425. doi: 10.1159/000356980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, et al. Properdin contributes to allergic airway inflammation through local C3a generation. J Immunol. 2015;195(3):1171–1181. doi: 10.4049/jimmunol.1401819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farries TC, Finch JT, Lachmann PJ, Harrison RA. Resolution and analysis of “native” and “activated” properdin. Biochem J. 1987;243(2):507–517. doi: 10.1042/bj2430507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pangburn MK. Analysis of the natural polymeric forms of human properdin and their functions in complement activation. J Immunol. 1989;142(1):202–207. [PubMed] [Google Scholar]
- 32.Blatt AZ, Pathan S, Ferreira VP. Properdin: A tightly regulated critical inflammatory modulator. Immunol Rev. 2016;274(1):172–190. doi: 10.1111/imr.12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rothstein F. Some physical and chemical characteristics of properdin. Vox Sang. 1962;8:113–114. [Google Scholar]
- 34.Ensky J, et al. Properties of highly purified human properdin. J Immunol. 1968;100(1):142–158. [PubMed] [Google Scholar]
- 35.Harboe M, et al. The role of properdin in zymosan- and Escherichia coli-induced complement activation. J Immunol. 2012;189(5):2606–2613. doi: 10.4049/jimmunol.1200269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mollnes TE, Lea T, Harboe M, Tschopp J. Monoclonal antibodies recognizing a neoantigen of poly(C9) detect the human terminal complement complex in tissue and plasma. Scand J Immunol. 1985;22(2):183–195. doi: 10.1111/j.1365-3083.1985.tb01870.x. [DOI] [PubMed] [Google Scholar]
- 37.Hourcade DE. Properdin and complement activation: A fresh perspective. Curr Drug Targets. 2008;9(2):158–164. doi: 10.2174/138945008783502458. [DOI] [PubMed] [Google Scholar]
- 38.Bongrazio M, Pries AR, Zakrzewicz A. The endothelium as physiological source of properdin: Role of wall shear stress. Mol Immunol. 2003;39(11):669–675. doi: 10.1016/s0161-5890(02)00215-8. [DOI] [PubMed] [Google Scholar]
- 39.Zaferani A, et al. Identification of tubular heparan sulfate as a docking platform for the alternative complement component properdin in proteinuric renal disease. J Biol Chem. 2011;286(7):5359–5367. doi: 10.1074/jbc.M110.167825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zaferani A, et al. Factor H and properdin recognize different epitopes on renal tubular epithelial heparan sulfate. J Biol Chem. 2012;287(37):31471–31481. doi: 10.1074/jbc.M112.380386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Merle NS, Church SE, Fremeaux-Bacchi V, Roumenina LT. Complement system, part I: Molecular mechanisms of activation and regulation. Front Immunol. 2015;6:262. doi: 10.3389/fimmu.2015.00262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kishore U, et al. Structural and functional anatomy of the globular domain of complement protein C1q. Immunol Lett. 2004;95(2):113–128. doi: 10.1016/j.imlet.2004.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garlatti V, et al. Structural basis for innate immune sensing by M-ficolin and its control by a pH-dependent conformational switch. J Biol Chem. 2007;282(49):35814–35820. doi: 10.1074/jbc.M705741200. [DOI] [PubMed] [Google Scholar]
- 44.Sun Z, Reid KB, Perkins SJ. The dimeric and trimeric solution structures of the multidomain complement protein properdin by X-ray scattering, analytical ultracentrifugation and constrained modelling. J Mol Biol. 2004;343(5):1327–1343. doi: 10.1016/j.jmb.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 45.Pangburn MK, Müller-Eberhard HJ. Initiation of the alternative complement pathway due to spontaneous hydrolysis of the thioester of C3. Ann N Y Acad Sci. 1983;421:291–298. doi: 10.1111/j.1749-6632.1983.tb18116.x. [DOI] [PubMed] [Google Scholar]
- 46.Agarwal S, et al. An evaluation of the role of properdin in alternative pathway activation on Neisseria meningitidis and Neisseria gonorrhoeae. J Immunol. 2010;185(1):507–516. doi: 10.4049/jimmunol.0903598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ali YM, et al. Low-dose recombinant properdin provides substantial protection against Streptococcus pneumoniae and Neisseria meningitidis infection. Proc Natl Acad Sci USA. 2014;111(14):5301–5306. doi: 10.1073/pnas.1401011111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mertens G, Cassiman JJ, Van den Berghe H, Vermylen J, David G. Cell surface heparan sulfate proteoglycans from human vascular endothelial cells: Core protein characterization and antithrombin III binding properties. J Biol Chem. 1992;267(28):20435–20443. [PubMed] [Google Scholar]
- 49.Camous L, et al. Complement alternative pathway acts as a positive feedback amplification of neutrophil activation. Blood. 2011;117(4):1340–1349. doi: 10.1182/blood-2010-05-283564. [DOI] [PubMed] [Google Scholar]
- 50.Mollnes TE, Lea T, Frøland SS, Harboe M. Quantification of the terminal complement complex in human plasma by an enzyme-linked immunosorbent assay based on monoclonal antibodies against a neoantigen of the complex. Scand J Immunol. 1985;22(2):197–202. doi: 10.1111/j.1365-3083.1985.tb01871.x. [DOI] [PubMed] [Google Scholar]
- 51.Risitano AM, et al. Peptide inhibitors of C3 activation as a novel strategy of complement inhibition for the treatment of paroxysmal nocturnal hemoglobinuria. Blood. 2014;123(13):2094–2101. doi: 10.1182/blood-2013-11-536573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seelen MA, et al. Functional analysis of the classical, alternative, and MBL pathways of the complement system: Standardization and validation of a simple ELISA. J Immunol Methods. 2005;296(1-2):187–198. doi: 10.1016/j.jim.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 53.Jaffe EA, Nachman RL, Becker CG, Minick CR. Culture of human endothelial cells derived from umbilical veins: Identification by morphologic and immunologic criteria. J Clin Invest. 1973;52(11):2745–2756. doi: 10.1172/JCI107470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sprong T, et al. Inhibition of C5a-induced inflammation with preserved C5b-9-mediated bactericidal activity in a human whole blood model of meningococcal sepsis. Blood. 2003;102(10):3702–3710. doi: 10.1182/blood-2003-03-0703. [DOI] [PubMed] [Google Scholar]