Abstract
Eukaryotic translation initiation factor 5A (eIF5A) is a protein that is highly conserved and essential for cell viability. This factor is the only protein known to contain the unique and essential amino acid residue hypusine. This work focused on the structural and functional characterization of Saccharomyces cerevisiae eIF5A. The tertiary structure of yeast eIF5A was modeled based on the structure of its Leishmania mexicana homologue and this model was used to predict the structural localization of new site-directed and randomly generated mutations. Most of the 40 new mutants exhibited phenotypes that resulted from eIF-5A protein-folding defects. Our data provided evidence that the C-terminal α-helix present in yeast eIF5A is an essential structural element, whereas the eIF5A N-terminal 10 amino acid extension not present in archaeal eIF5A homologs, is not. Moreover, the mutants containing substitutions at or in the vicinity of the hypusine modification site displayed nonviable or temperature-sensitive phenotypes and were defective in hypusine modification. Interestingly, two of the temperature-sensitive strains produced stable mutant eIF5A proteins – eIF5AK56A and eIF5AQ22H,L93F – and showed defects in protein synthesis at the restrictive temperature. Our data revealed important structural features of eIF5A that are required for its vital role in cell viability and underscored an essential function of eIF5A in the translation step of gene expression.
Keywords: elF5A, hypusine, mutational analysis, structural modeling, translation
The putative eukaryotic translation initiation factor 5A (eIF5A) is a small acidic protein, highly conserved and essential in all organisms from archaea to mammals [1,2]. There are two eIF5A genes in Saccharomyces cerevisiae (TIF51A and TIF51B), which encode two protein isoforms with 90% identity that are differentially transcribed in response to the presence of oxygen [1]. However, these yeast genes, once transcribed, can replace each other, suggesting a functional similarity between them [3].
eIF5A is unique in that it is the only protein known to contain the unusual amino acid residue hypusine [4], which is formed by post-translational modification of a specific lysine residue (K51 for yeast eIF5A). Hypusine synthesis is a two-step process [5]: the enzyme deoxyhypusine synthase (DHS; Dys1 in yeast) transfers a 4-aminobutyl moiety from the poly amine spermidine to one specific lysine residue in eIF5A to form a deoxyhypusine residue, which in turn is hydroxylated by the enzyme deoxyhypusine hydroxylase (DOHH; Lial in yeast; [5]). Inhibition of either DHS or DOHH affects cell growth in mammalian cells [6], and the conservative mutation K51R in yeast eIF5A, which blocks hypusination, cannot complement yeast eIF5A knockout cells [1], demonstrating that deoxyhypusine/hypusine formation is essential for eIF5A function. Curiously, although both steps of hypusination are essential in Drosophila melanogaster, Caenorhabditis elegans and mammals, only the first step, deoxyhypusine synthesis, is required for growth and viability in the budding yeast S. cerevisiae [5].
The importance of eIF5A is evident in that it is essential for cell viability and is highly conserved throughout evolution. However, even after almost three decades of study, its function is still obscure (reviewed in ref. [7]). Initially, eIF5A was characterized as a translation initiation factor, based on its activity in stimulating methionyl-puromycin synthesis, a model assay of the first peptide bond formation [8–10]. Because depletion of eIF5A in yeast causes an increase in the number of G1-arrested cells [11], and inhibitors of DOHH cause cell cycle arrest in mammalian cells at the G1/S boundary [12], it was hypothesized that eIF5A may be important for the translation of mRNAs encoding specific proteins required for cell-cycle progression, probably those involved in S-phase onset [11,13].
It has been demonstrated recently that eIF5A physically interacts with structural components of the 80S ribosome, as well as with the translation elongation factors eEF1A and eEF2 [14,15]. eIF5A was shown to co-fractionate with monosomes in a translation-dependent manner, and eIF5A mutant strains show altered polysome profiles and are sensitive to translation inhibitors [15]. Thus, these results point to a function for eIF5A in translation, although it is still not known whether eIF5A affects the translation of all mRNAs or a subset of specific mRNAs.
The 3D structures determined for eIF5A homologous proteins from three archaea [16–18] and two Leishmania eIF5As (Protein Data Bank ID codes 1XTD and 1X6O) reveal very similar features. The analyses of these structures show that eIF5A is composed of two predominantly β-sheet domains. The N-terminal domain bears the hypusine residue in an exposed loop, is folded in an SH3-like barrel found in other proteins related to translation and harbors a KOW motif proposed to mediate RNA binding. The C-terminal domain forms the OB-fold also present in other translational components (e.g. eIF1A, eIF2α and several ribosomal proteins) and belongs to the nucleic acid-binding protein superfamily, according to Structural Classification of Proteins (http://scop.mrc-lmb.cam.ac.uk/scop). While the RNA-binding activity of the eIF1A OB-fold is attributed to its role in mRNA scanning by the pre-initiation complex, the OB-fold found in eIF2α is characterized as the site of its phosphorylation and regulation by various eIF2 kinases, and the role of its RNA-binding activity in translation initiation is unclear [19]. eIF5A was reported to bind specific oligonucleotide sequences according to the systematic evolution of ligands by exponential enrichment (SELEX) method [20], and subsequent work identified some potential candidate target mRNAs [21]. However, the biological significance of this mRNA-binding capacity of eIF5A is as yet unknown.
Although eubacteria lack the hypusine modification enzymes, the bacterial elongation factor P (EF-P) [22] is closely related to eIF5A in sequence and structure [23]. EF-P is the product of an essential gene, efp [24], encoding a small acidic protein with three domains – I, II and III (domains II and III being repetitive units presumably derived from duplication) [23]. Not only is the primary sequence of EF-P related to eIF5A and its archaeal homologues, but the crystal structure of EP-F domains I and II is superimposable on the N-terminal and C-terminal domains, respectively, of archaeal eIF5A. EF-P is L-shaped and resembles tRNA in structure and size [23]. It stimulates the peptidyl transferase activity of the 70S ribosome in Escherichia coli and enhances the first peptide bond formation in model assays [25]. EF-P was proposed to facilitate the peptide bond synthesis for some aminoacyl-tRNAs that are poor acceptors for the ribosomal peptidyl transferase. Whether eIF5A functionally mimics EF-P or tRNA on ribosomes remains to be explored.
Genetic approaches have been extensively to investigate the function of eIF5A in S. cerevisiae [26–30]. Although various eIF5A mutants have been derived from some of these studies [26,27,29], no comprehensive mutational analysis of yeast eIF5A has been reported. This work exploited two phenotype-based random mutagenesis strategies and site-directed mutagenesis to identify and characterize new eIF5A mutants. These mutants contribute to an understanding of the structural features of eIF5A that essential for cell viability; these structures include the hypusine loop and the C-terminal α-helix. Interestingly, the causative correlation between protein synthesis and growth defects of the two new eIF5A mutants (eIF5AK56A and eIF5AQ22H, L93F) reinforces the involvement of eIF5A in protein synthesis. These mutants may be useful tools in future studies aimed at elucidating the mode of action of eIF5A in translation.
Results
Isolation of new conditional yeast eIF5A mutants
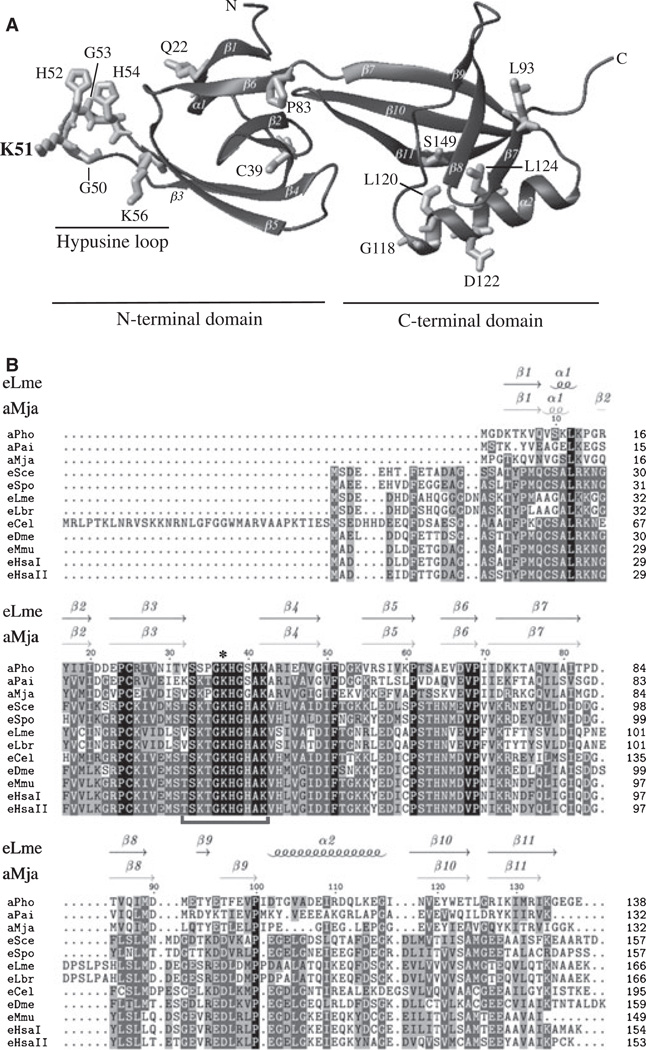
In order to identify structural features that are important for eIF5A function, three different approaches were followed to generate new mutants of yeast eIF5A. First, we used the tif51A-3 (C39Y,G118D) double mutant to identify a third mutation that renders the resulting tif51A allele nonfunctional and thus disables its capacity to complement a tif51A knockout strain. These alleles were obtained from a synthetic lethal screen using the tif51A-3 mutant and the mutations were induced by ethyl methanesulfonate [30]. Second, random PCR-based mutagenesis was conducted to obtain new temperature-sensitive eIF5A mutants. Third, site-directed mutagenesis was performed to derive specific mutants based on features from the 3D model structure of yeast eIF5A generated in this study (Fig. 1A; PMDB PM0075091; see Experimental procedures for details on yeast eIF5A structural modeling). Finally, to gain insight into the contribution of each of the different residues substituted in the alleles obtained by random mutagenesis, most of the triple and double mutations were separated. The model depicted in Fig. 1A highlights some residues modified in the mutated forms of eIF5A discussed herein.
Fig. 1.
Homology modeling of yeast elF5A and multiple alignment of elF5A sequences. (A) The tertiary structure of yeast elF5A was modeled based on the structure of Leishmania mexicana elF5A (PDB ID code 1XTD). The N-terminal and C-terminal domains, as well as the hypusine loop, are indicated by horizontal bars. The residues shown correspond to the locations of the mutations described in this study. The highlighted residue corresponds to the hypusination site. (B) Multiple alignment of the amino acid sequences of elF5A from aPho (Pyrococcus horikoshii), aPai (Pyrobaculum aerophilum), aMja (Methanococcus jannaschii), eSce (Saccharomyces cerevisiae), eSpo (Schizosaccharomyces pombe), eLme (L. mexicana), eLbr (Leishmania braziliensis), eCel (Caenorhabditis elegans), eDme (Drosophila melanogaster), eMmu (Mus musculus), eHsal (Homo sapiens elF5A-1) and eHsall (Homo sapiens elF5A-2) was performed using clustalw (http://www.ebi.ac.uk/clustalw/). The secondary structures of elF5A from eLme and aMja are indicated above the alignment with arrows for β-strands and coils for helices. The amino acid residues completely conserved in all the organisms are highlighted in black and those that were partially conserved are highlighted in grey, the shade of which lightens as the degree of conservation decreases. The horizontal bracket below the sequences indicates the hypusine loop, and an asterisk indicates the lysine residue that undergoes hypusine modification.
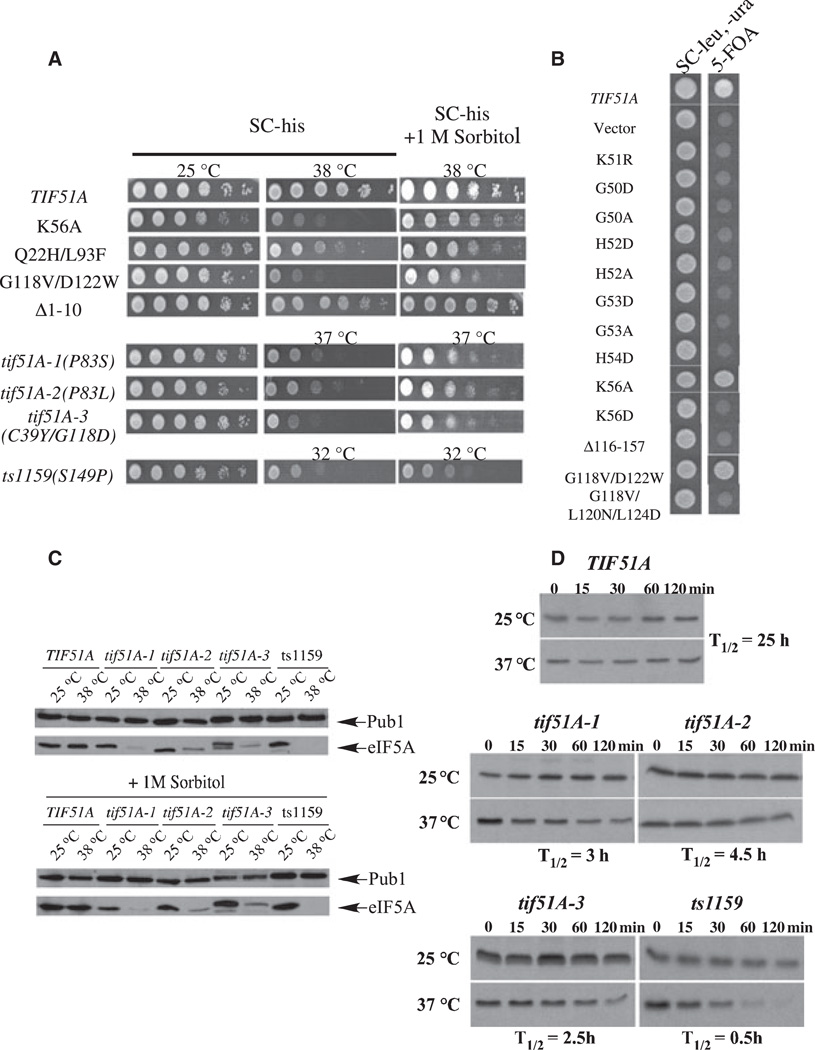
These strategies generated 40 new eIF5A mutants, which were compared for growth-supporting ability at different temperatures. In order to broaden the comparison among eIF5A mutants, the previously identified temperature-sensitive alleles tif51A-1 (P83S), tif51A-2 (P83L), tif51A-3 (C39Y, G118D) and ts1159 (S149P) were included in the analysis. The variety of phenotypes observed ranged from having no discernible growth defect to nonviability. Nineteen out of the 40 new alleles were nonviable, 13 caused temperature sensitivity and eight were temperature resistant (Table 1). Figure 2 shows the growth phenotypes of selected new mutants and previously described temperature-sensitive alleles (Fig. 2A) and some nonviable mutants (Fig. 2B).
Table 1.
Features summary of elF5A mutants. ND, not determined; TS, temperature sensitive; TR, temperature resistant. + indicates levels of elF5A protein.
| Source | Mutation | Growth phenotypea |
Expression of elF5A |
elF5A protein levels at the restrictive temperature |
|---|---|---|---|---|
| TIF51A | ____ | TR | Yes | ++++ |
| tif51A(K51R) [1] | K51R | Nonviable | Yes | ND |
| ts 1159 [26] | S149P | TS 34 °C | Yes | + |
| tif51A-1 [27] | P83S | TS 36 °C | Yes | + |
| tif51A-2 [27] | P83L | TS 37 °C | Yes | +++ |
| tif51A-3 [27] | C39Y/G118D | TS 36 °C | Yes | ++ |
| C39Y | TS 38 °C | Yes | +++ | |
| G118D | TR | Yes | ND | |
| Synthetic lethality | C39Y/P83L/G118D | Nonviable | ND | ND |
| Synthetic lethality | C39Y/T66I/G118D | Nonviable | ND | ND |
| T66I/G118D | TS 37 °C | Yes | +++ | |
| T66I | TS 38 °C | ND | +++ | |
| Synthetic lethality | C39Y/G118D/S140F | Nonviable | ND | ND |
| G118D/S140F | TS 38 °C | Yes | + | |
| S140F | TR | Yes | ND | |
| Synthetic lethality | C39Y/G118D/M142I | Nonviable | ND | ND |
| G118D/M142I | TS 38 °C | Yes | ++ | |
| M142I | TR | Yes | ND | |
| Synthetic lethality | C39Y/P116L/G118D | Nonviable | ND | ND |
| P116L/G118D | TS 38 °C | Yes | ++ | |
| Synthetic lethality | C39Y/G50D/G118D | Nonviable | ND | ND |
| C39Y/G50D | Nonviable | Yes | ND | |
| G50D | Nonviable | Yes | ND | |
| Synthetic lethality | C39Y/G53D/G118D | Nonviable | ND | ND |
| C39Y/G53D | Nonviable | Yes | ND | |
| G53D | Nonviable | Yes | ND | |
| PCR mutagenesis | N29K/M44I/P83S | TS 37 °C | Yes | ++ |
| M44I/P83S | TS 37 °C | Yes | ++ | |
| N29K | TR | Yes | ND | |
| M44I | TR | Yes | ND | |
| PCR mutagenesis | G14D/V57D | TS 37 °C | Yes | +++ |
| G14D | TR | Yes | ND | |
| V57D | TS 37 °C | Yes | +++ | |
| PCR mutagenesis | Q22H/L93F | TS 38 °C | Yes | ++++ |
| L93F | TR | Yes | ND | |
| Q22H | TR | Yes | ND | |
| Site-directed mutagenesis | G50A | Nonviable | Yes | ND |
| Site-directed mutagenesis | H52A | Nonviable | Yes | ND |
| Site-directed mutagenesis | H52D | Nonviable | Yes | ND |
| Site-directed mutagenesis | G53A | Nonviable | Yes | ND |
| Site-directed mutagenesis | H54D | Nonviable | Yes | ND |
| Site-directed mutagenesis | K56D | Nonviable | Yes | ND |
| Site-directed mutagenesis | K56A | TS 37 °C | Yes | ++++ |
| Site-directed mutagenesis | Δ1–10 | TR | Yes | ND |
| Site-directed mutagenesis | Δ116–157 | Nonviable | No | ND |
| Site-directed mutagenesis | S24P | TS 37 °C | Yes | ++ |
| Site-directed mutagenesis | G118V/D122W | TS 37 °C | Yes | ++ |
| Site-directed mutagenesis | G118D/L120N/L124D | Nonviable | No | ND |
The indicated temperatures are the lowest temperature at which the phenotype is observed.
Fig. 2.
Growth analyses of yeast elF5A mutant strains and stability of selected mutant proteins. (A) The tif51A knockout strain containing wild-type TIF51A or the indicated tif51A mutants was grown on synthetic complete minus histidine (SC-his) to D600nm of 0.5. Cells were collected by centrifugation and suspended in one-tenth of the original volume of SC–his. Tenfold serial dilutions of wild-type and mutant strains were spotted onto SC–his or SC-his + 1 M sorbitol plates and incubated at 25 °C or at the minimum specific restrictive temperature for 3 days. pRS315-77F51A was used as a positive growth control. (B) Complementation of a TIF51A knockout strain by mutated elF5A. The wild-type strain and the indicated mutants were grown on SC minus leucine and uracil (–leu, –ura) liquid medium to reach a D value of 0.5 at 600 nm. Cells were spotted onto SC–leu,–ura or onto 5-fluoroorotic acid plates and incubated at 25 °C for 3 days. pRS315-TIF51A was used as a positive growth control, and pRS315 alone and pRS315-tif51AK51R were used as negative growth controls. (C) Levels of selected mutant elF5A proteins in the absence (upper panel) or presence (lower panel) of 1 M sorbitol. The indicated strains were grown on SC–his to reach a D value of 0.5 at 600 nm and collected 3 h after the temperature shift. Cells were lysed, and 5 |ig of total protein was western blotted then probed with anti-elF5A serum. Anti-Pub1 was used as a loading control. (D) Estimation of the half-lives of wild-type and selected mutant elF5A proteins. The indicated strains were subjected to pulse–chase analysis to estimate their elF5A protein half-lives (T1/2) at permissive (25 °C) or nonpermissive (37 °C) temperatures.
As a means of examining the relationship between amino acid residue conservation and mutant phenotype, the conservation of each mutated amino acid residue was examined in all eIF5As and the archaeal homologues (supplementary Fig. S1). In the case of the nonviable alleles with a single mutated residue, there was 100% conservation of this residue in eukaryotes, with the exception of His54. As for the temperature-sensitive alleles with a single mutated residue, this single mutated residue was conserved in 90% of eukaryotes and in 80% of all eIF5As, including the archaeal homologues. Conservation was much lower for the temperature-resistant alleles with a single mutated residue (72% among eukaryotes and 50% among all eIF5As and aIF5As). Thus, there seems to be a strong correlation between the degree of conservation and the severity of mutant phenotypes.
We then determined the protein levels at different temperatures of the temperature-sensitive eIF5A mutants. As summarized in Table 1, most of the new temperature-sensitive alleles (11 out of 13) produced an unstable eIF5A protein at the restrictive temperature. This behavior is similar to that observed for the four previously described conditional eIF5A mutants (Fig. 2C and Table 1) and reveals residues important for eIF5A secondary and tertiary structure. In order to determine if the decreased mutant eIF5A levels are the result of a rapid degradation at the nonpermissive temperature, a pulse–chase assay was carried out to estimate the half-lives of the selected mutant proteins (Fig. 2D). At the permissive temperature, both wild-type and mutant eIF5A proteins showed a long half-life of ~25 h [11]. In contrast, the mutant eIF5A proteins displayed markedly shortened half-lives at the restrictive temperature (Fig. 2D). These results confirm the hypothesis that protein instability is responsible for decreased levels of these mutant eIF5A proteins at restrictive temperature and demonstrate that these tif51A alleles are loss-of-function mutants owing to reduced eIF5A protein.
It was previously reported that sorbitol and/or phenylmethanesulfonyl fluoride suppress the temperature-sensitive phenotypes of certain eIF5A mutants [27,29]. In order to determine whether this is a general phenomenon, we evaluated the effects of sorbitol and phenylmethanesulfonyl fluoride on the temperature sensitivity of all new eIF5A mutants. Addition of phenylmethanesulfonyl fluoride to the culture medium did not improve cell growth of these mutants at the restrictive temperature (data not shown). By contrast, the addition of 1 m sorbitol to the culture medium partially suppressed the conditional mutant phenotype at the minimal restrictive temperature for the selected eIF5A mutants (shown in Fig. 2A) and in all other conditional mutants (data not shown). It is not clear how sorbitol exerts this effect. Because sorbitol did not increase eIF5A protein stability in the mutant eIF5A strains (Fig. 2C), it probably promoted cell viability in cells with a limiting level of eIF5A at the restrictive temperature. However, it could not substitute completely for the function of eIF5A, when tested by a plasmid shuffle assay (data not shown). Moreover, temperature-sensitive eIF5A strains failed to grow indefinitely in the presence of sorbitol at the restrictive temperature (data not shown).
Two new tif51A mutant strains produce stable elF5A proteins at the restrictive temperature and show a severe defect in protein synthesis
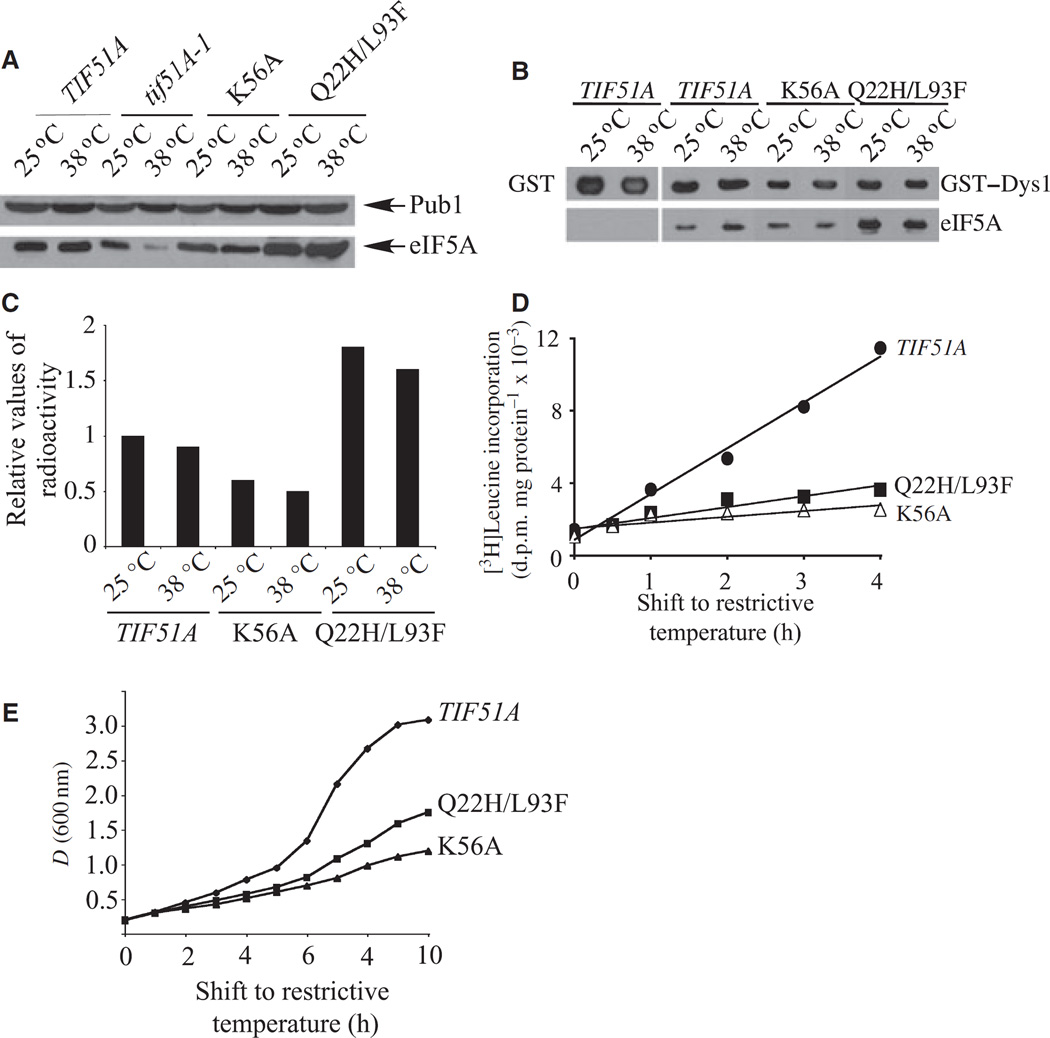
Two new mutant alleles of TIF51A, namely, tif51AK56A and tif51AQ22H, L93F, led to the production of stable eIF5A proteins at the restrictive temperature. Both mutants showed relatively similar temperature sensitivities, with tif51AK56A being a little more temperature-sensitive than tif51AQ22H, L93F (Fig. 2A). As shown in Fig. 3A, unlike the mutant tif51A-1, there is no reduction in eIF5A protein levels in the tif51AK56A and tif51AQ22H, L93F strains at the restrictive temperature. Curiously, the tif51AQ22H, L93F strain showed increased levels of eIF5A protein at both permissive and restrictive temperatures when compared with the wild-type strain (Fig. 3A).
Fig. 3.
Characterization of two new elF5A mutants producing stable elF5A proteins. (A) Determination of mutant elF5A levels. Protein levels were analyzed as in Fig. 2C. (B) GST pulldown of wild-type and mutant elF5A with GST–Dys1 at permissive (25 °C) and nonpermissive (38 °C) temperatures. GST alone was used as a negative control (first pair of lanes). (C) Analysis of elF5A mutant proteins as substrates for hypusine formation. Yeast strains were cultured with [3H]spermidine and analyzed as described in Experimental procedures. The relative values of radioactivity incorporated into protein as deoxyhypusine/hypusine are shown. (D) The effects of wild-type and mutant elF5A on protein synthesis. Protein synthesis was measured as described in Experimental procedures and the degree of [3H]leucine incorporation into protein is shown as disintegrations per minute (d.p.m.)·µg−1 of protein. (E) Growth curves of wild-type and elF5A mutant strains. The strains were grown at 30 °C in rich medium (YPD) to mid-log phase. The cultures were then shifted to 38 °C and growth was monitored for 10 h by spectrophotometry (at 600 nm).
Because eIF5A activity is dependent on its post-translational hypusine modification [5,7], we investigated whether impairment in hypusine formation was responsible for the temperature sensitivity of tif51AK56A and tif51AQ22H,L93F mutants. First, we assayed the ability of eIF5AK56A and eIF5AQ22H,L93F mutant proteins to interact with a functional fusion protein [glutathione S-transferase (GST)–Dys1] of yeast deoxyhypusine synthase, the enzyme that catalyzes the first step in hypusine formation [5]. No discernable decrease in the physical interaction between Dys1 and eIF5A was observed in these mutants at the restrictive temperature (Fig. 3B). Interestingly, considerably more eIF5AQ22H,L93F mutant protein was co-purified with GST–Dys1 (Fig. 3B). This may be a result of the increased amount of eIF5A protein observed in the tif51AQ22H,L93F mutant (Fig. 3A). Next, we tested whether the Dys1–eIF5A physical interaction results in deoxyhypusine/hypusine synthesis in vivo, by measuring the incorporation of a radioactive aminobutyl moiety from spermidine into eIF5A in the tif51AK56A and tif51AQ22H,L93F mutant strains (Fig. 3C). Both of the mutant proteins were effectively modified in cells at both permissive and restrictive temperatures, consistent with the binding results. Whereas the radiolabeling of eIF5AK56A mutant protein was lower than that of the wild-type eIF5A, the eIF5AQ22H,L93F mutant protein showed considerably higher radiolabeling of deoxyhypusine/hypusine than wild-type eIF5A. This finding may be a reflection of the increased amount of the mutant eIF5A protein in the tif51AQ22H, L93F strain (Fig. 3A).
Finally, to understand the ultimate defect caused by Hf51AK56A and Hf51AQ22H,L93F mutations in the cell, we assayed protein synthesis by measuring the amount of [3H]leucine incorporation into proteins. As shown in Fig. 3D, both mutants showed a pronounced reduction in [3H]leucine incorporation into proteins after the shift to the restrictive temperature, as judged by the differences in the slopes of the tif51AK56A and tif51AQ22H,L93F mutant curves compared with that of the wild-type. Interestingly, the protein synthesis defect was evident 1 h after the temperature shift in both mutants (the amounts of [3H]leucine incorporation being ~60% of that of the wild-type), before any differences in the growth curves were apparent (Fig. 3D,E). Moreover, the increase in [3H]leucine incorporation after the first hour was negligibly low for both eIF5A mutants, while the incorporation of radioactive leucine continued to increase in the wild-type strain. The decrease of both protein synthesis and growth was consistently more marked in the tif51AK56A than in the tif51AQ22H,L93F mutant. The finding that protein synthesis was reduced before growth was affected suggests that the depression in protein synthesis in these mutant strains is responsible for their growth arrest at the restrictive temperature.
Substitution of highly conserved residues in the hypusine loop affects hypusine formation and causes growth defects
In the course of our analysis of third-site mutations that render the tif51A-3 (C39Y/G118D) allele nonviable, two new mutations, G50D and G53D, of eIF5A were identified. Interestingly, subsequent characterization of these triple alleles revealed that strains harboring either the G50D or the G53D mutation alone were also nonviable. The residues G50 and G53 are totally conserved and reside in the hypusine loop in eIF5A (Fig. 1 and supplementary Video S1). The hypusine loop is strictly conserved throughout eukaryotic evolution, and contains either positively charged residues or uncharged residues only. For these specific features, we decided to perform a more detailed analysis of residues present in this loop using site-directed mutagenesis.
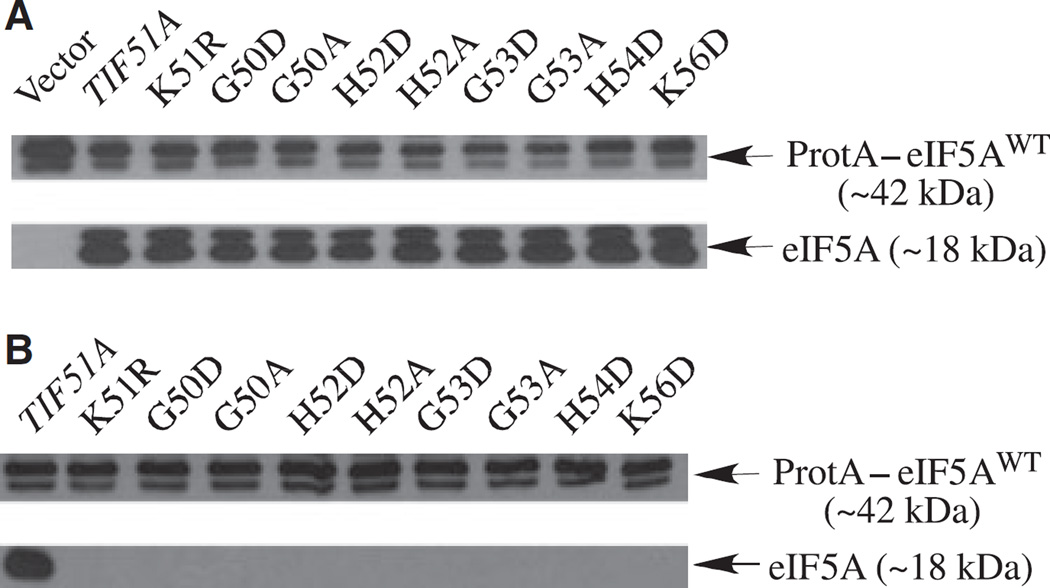
First, in order to test whether the presence of a negatively charged amino acid residue in the hypusine loop was responsible for the nonviability of tif51AG50D and tif51AG53D alleles, we generated mutants with substitution to alanine in residues G50, H52, G53 and K56, and substitution to aspartic acid in residues H52, H54 and K56. These eIF5A mutants were then tested for their ability to support growth of a tif51A knockout strain (Fig. 2B). The results clearly show that only the K56A mutant allele was able to complement the tif51A knockout strain at the permissive temperature. However, it led to a temperature-sensitive phenotype (Fig. 2A).
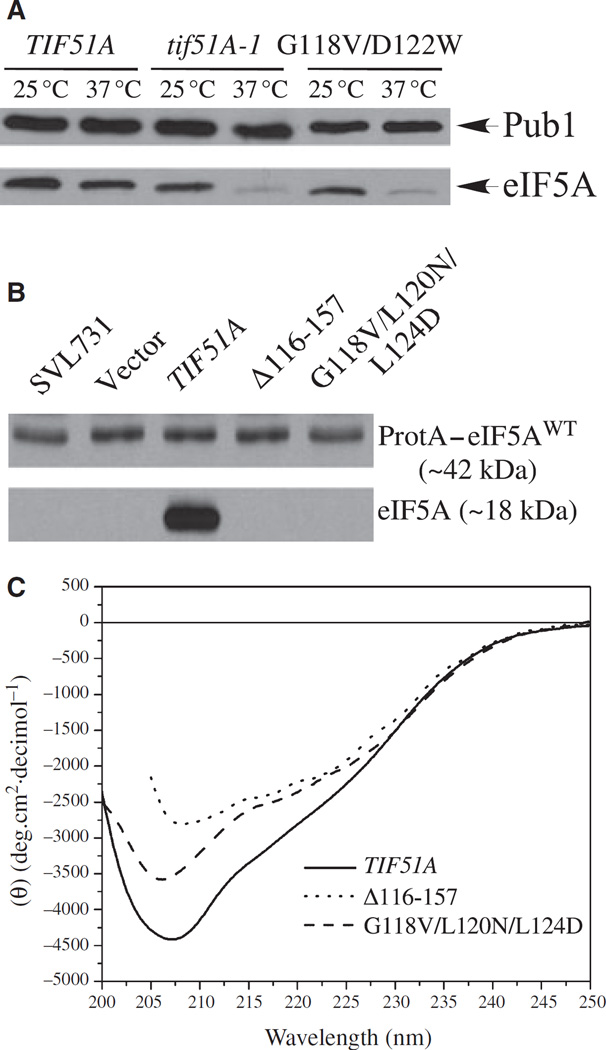
Subsequently, to confirm that the eIF5A mutant protein is produced in these nonviable tif51A alleles, we used a strain expressing the functional fusion protein (protein A–eIF5AWT) as the only source of eIF5A in the cell. As shown in Fig. 4A, all the nonviable hypusine loop mutants produced stable eIF5A, indicating that loss of viability is caused by the lack of eIF5A activity, rather than by the lack of the mutant protein.
Fig. 4.
Effects of substitution of the conserved residues in the hypusine loop on hypusine modification. (A) Expression and stability of the hypusine loop mutant proteins. A strain expressing the functional fusion protein A–wild-type elF5A (SVL731) was transformed with nonviable hypusine loop tif51A alleles and the expression of mutant elF5A proteins was determined by western blot. (B) Analysis of elF5A mutant proteins as substrates for hypusine formation. The same strains as in (A) were grown in 5 ml of SC-leu,-ura liquid medium from to an attenuance (D), at 600 nm, of 0.1–1.2 in the presence of [3H]spermidine (20 µCi). A fluorogram of an SDS gel of labeled cellular protein is shown.
In order to determine whether a defect in hypusine synthesis is responsible for the phenotype of the hypusine loop mutants, we tested these nonfunctional eIF5A mutant proteins (Fig. 4A) as substrates for hypusine modification in cells cultured in the presence of [3H]spermidine. None of the mutant proteins was radiolabeled (Fig. 4B), indicating that they are not substrates for Dys1. No labeling was observed for eIF5AK56D (Fig. 4B), in contrast to K56A, which served as a relatively good substrate for deoxyhypusine/hypusine synthesis (Fig. 3C). Thus, these results demonstrate that amino acid substitutions in the hypusine loop cause a total or a severe impairment in deoxyhypusine/hypusine modification, and thereby in eIF5A function, especially when the substitution involves insertion of a negatively charged residue.
Mutants affecting the C-terminal domain α-helix show defects in eIF5A protein folding
As shown in the yeast eIF5A model (Fig. 1A), the C-terminal domain has an α-helical segment (amino acids 118–130 for the S. cerevisiae eIF5A) and its presence is consistent with the results obtained from CD experiments (Fig. 5C). Curiously, in the solved structures of archaeal eIF5A homologues [16–18], a C-terminal α-helix is not present, not continuous, or much smaller than that observed in the Leishmania eIF5A structures (Protein Data Bank ID codes 1XTD and 1X6O).
Fig. 5.
Defects of mutants in the C-terminal α-helix in the protein folding. (A) Levels of mutant eIF5A protein in the temperature-sensitive tif51AG118V,D122W strain. The indicated strains were grown on SC-his to D600nm of 0.5 and collected 3 h after the temperature shift. Cells were lysed, and 5 µg of total protein was analyzed by western blot with an anti-eIF5A serum. Anti-Pub1 was used as a loading control. (B) Nonviable tif51A alleles lacking the mutant eIF5A protein. A strain expressing the functional fusion protein A– wild-type eIF5A (SVL731) was transformed with the indicated tif51A alleles and the expression of eIF5A mutant proteins was determined by western blot. (C) CD analyses of secondary structures of recombinant eIF5A proteins produced in E. coli. Wild-type and mutant eIF5A were analyzed by CD, as described in Experimental procedures.
In order to verify the existence of this C-terminal α-helix segment and to assess the importance of this secondary structure for eIF5A folding and function, we generated three mutants: G118V/D122W and G118V/L120N/L124D with substitutions in the α-helix segment, preventing formation of the proper protein structure, and Δ116–157 with a deletion of the entire C-terminal extremity, including the α-helix. As shown in Fig. 2A,B, the tif51AG118V,D122W mutant exhibited a temperature-sensitive phenotype, whereas the tif51AG118V,L120N,L124D and tif51AΔ116–157 mutants were nonviable. Western blot analysis of the tif51AG118V,D122W revealed reduced levels of eIF5AG118V,D122W protein at the restrictive temperature (Fig. 5A), suggesting that eIF5A protein folding is impaired in this mutant, consequently causing its temperature-sensitive growth defect. Likewise, no accumulation of eIF5AG118V,L120N,L124D and eIF5AΔ116–157 mutant proteins was observed (Fig. 5B), suggesting that perturbation or deletion of the C-terminal eIF5A α-helix secondary structure disrupts the overall C-terminal structure, causing protein instability.
Finally, in an effort to validate the C-terminal α-helix structure experimentally, we performed CD spectroscopy analyses of recombinant proteins of wild-type and mutant eIF5A expressed in E. coli (Fig. 5C). The CD spectrum of the wild-type yeast eIF5A is characterized by a pronounced minimum centered at approximately 207 ± 1 nm and a broad shoulder at around 216– 223 nm, suggesting the presence of both β-sheet and α-helix. A similar spectrum was also reported for human eIF5A [31]. Both eIF5AG118V,L120N,L124D and eIF5AΔ116–157 showed a decrease in the overall ellipticity (mainly between 216 and 223 nm) when compared with the wild-type eIF5A. These spectral data strongly suggest that the C-terminal domain of eIF5A contains α-helical secondary structure, as predicted in the homology model, and that the perturbation of the sequence of this helix results in significant destabilization of the whole molecule beyond the helix itself.
Another structural difference between eukaryotic eIF5As and their archaeal homologues is the presence of an extended N-terminal extremity in the eukaryotic proteins (Fig. 1B). Curiously, this N-terminal extension in eukaryotes harbors several highly conserved amino acid residues (for example, yeast eIF5A D3, F8 and G14; Fig. 1B). To test whether this extended N terminus of eukaryotic eIF5A is essential, we generated a 10-residue N-terminal truncation of yeast eIF5A (eIF5AΔ1–10) and determined its ability to substitute for wild-type eIF5A. The mutant allele (tif51A Δ1–10) complemented a tif51A knockout strain at all temperatures tested (Fig. 2A). Even though the G14 residue was completely conserved among eukaryotes, the tif51AG14D strain showed no discernible phenotype (Table 1). Thus, the results with Δ1–10 and G14D eIF5A mutants indicate that this N-terminal extension is not required for S. cerevisiae growth and viability and that it is not directly involved in the essential function of eIF5A in yeast.
Discussion
In the present study, we combined structural and mutational analyses of the yeast eIF5A gene to uncover important structural features of the eIF5A protein. We conducted structure/function analyses using a collection of yeast eIF5A mutants, including 40 new yeast eIF5A mutants, and using a yeast eIF5A model (Fig. 1A) constructed on the basis of the crystal structure of the L. mexicana counterpart. Our overall results demonstrated the critical importance of the hypusine loop and the C-terminal α-helix for eIF5A activity and/or eIF5A protein folding, and provided strong evidence for an essential role for eIF5A in translation.
The fact that the gene encoding protein kinase C in yeast (PKC1) is a high-copy suppressor of the tif51A-1 mutant suggests a defect in the cell wall integrity of eIF5A mutant strains. Indeed, the osmotic stabilizer sorbitol [27], which affects cell wall integrity [32], partially suppressed the temperature-sensitive phenotypes of all the eIF5A mutants included in this study. Contrary to the previously reported stabilization of another eIF5A mutant (tsL102A) by sorbitol and also by phenylmethanesulfonyl fluoride [29], our data demonstrated that protein stabilization is not a general mechanism by which sorbitol exerts its phenotypic suppression of eIF5A mutants. Sorbitol failed to stabilize eIF5A produced by any of the conditional mutants reported in this study. Furthermore, sorbitol also suppressed the phenotypes of temperature-sensitive tif51AK56A and tif51AQ22H/L93F mutants, which produce stable eIF5A proteins. Moreover, the addition of phenylmethanesulfonyl fluoride to the culture medium did not suppress the temperature sensitivity of any of our conditional mutants, arguing against phenotypic suppression by eIF5A stabilization. Importantly, these findings reinforce our previous hypothesis of a role for eIF5A in connection with cell wall integrity and actin cytoskeleton organization [27,28] and support the notion that impairment of eIF5A function results in defects of cell wall integrity. Although it is well known that the Pkc1 pathway regulates cell wall integrity in yeast, proteins not directly involved in Pkc1 pathway can also affect cell integrity, like the deacetylase complex Sds3–Rpd3–Sin3, which may act in a parallel pathway to activate genes required for cell wall biosynthesis [33]. Thus, eIF5A may contribute to cell integrity indirectly, by affecting the expression of genes involved in cell wall integrity and/or remodeling during G1/S transition.
Most of the tif51A temperature-sensitive strains showed decreased levels of eIF5A at the restrictive temperature, indicating that the amino acid substitutions affected eIF5A protein folding and stability. The amino acid substitutions in these unstable mutant eIF5As (including S24P, C39Y, V57D, P116L, G118D and D122W) are located in one of the secondary structures (identified in Fig. 1A) and may perturb the proper folding of eIF5A in a temperature-dependent manner. It is also important to note that the accumulation of multiple structural mutations can generate temperature-sensitive and nonviable phenotypes. The best example of this is the nonviable eIF5A mutant C39Y/P83L/G118D that harbors the accumulation of mutations previously described for the tif51A-2 and tif51A-3 alleles [27]. Curiously, the mutations G118D and D122W, located in the C-terminal α-helix of eIF5A, seem to destabilize this secondary structure and thereby the whole C-terminal region. The unstable triple mutant G118D/L120D/L124D showed a significant loss of secondary structure caused by these substitutions in the C-terminal α-helix (Fig. 5B,C). This is the first study to suggest that the C-terminal α-helix, present in eukaryotic eIF5A, is an important structural element – a hypothesis consistent with its conservation among eukaryotic homologues.
Loss of eIF5A function may also result from mutations that do not alter the eIF5A global structure. This is the case for those mutants in the exposed hypusine loop, which has no secondary structure and yet is critical for eIF5A activity. The eIF5A nonviable mutants G50A, G50D, G53A, G53D, H52A, H52D, H54D and K56D produced significant levels of eIF5A protein, and the temperature-sensitive mutant protein K56A is stable at the restrictive temperature. Because hypusine modification is essential for eIF5A activity, the phenotypes of these hypusine loop mutants may be determined by impairment of hypusine formation. Our data clearly show that the lack of hypusine synthesis in all the hypusine loop mutants (except K56A) is the major cause of their phenotypes. Thus, defects of this group of mutants may be influenced by both structural modification of the hypusine loop and the lack of positively charged residues or the presence of negatively charged residues.
Unlike most of the conditional mutants that produced unstable eIF5A, two temperature-sensitive tif51A strains (tif51AK56A and tif51AQ22H/L93F) produced stable mutant proteins. The side chains of both residues Q22 and L93 are exposed to the solvent (supplementary Video S1), which is particularly uncommon for leucine side chains. Considering that the two proteins are stable and that Q22 and L93 are facing the solvent, it is tempting to speculate that these residues may be important for eIF5A–ligand interaction. However, we cannot rule out the possibility that these mutations alter the tertiary structure of eIF5A. Further characterization of K56A and Q22H/L93F eIF5A mutants, and their use in future genetic screenings, may lead to a better understanding of eIF5A function and identification of cellular partners.
A function of eIF5A in protein synthesis has been disputed for a long time. However, substantial evidence that supports its involvement in translation has been reported recently. eIF5A has been shown to co-purify with translation machinery components in a hypusine-dependent manner [14,15], and this physical association of eIF5A seems to occur preferentially with actively translating ribosomes [15]. Furthermore, temperature-sensitive eIF5A mutants (tif51A-1 and tif51A-3) show altered polysome profiles at the restrictive temperature and are sensitive to protein synthesis inhibitors [15]. Measurement of protein synthesis in the two conditional mutants, tif51AK56A and tif51AQ22H/L93F, revealed a rapid depression of protein synthesis at the restrictive temperature, before any growth defects were detectable. A causative relationship between the defects in protein synthesis and cell growth strongly suggests a direct role for eIF5A in translation, rather than an indirect, secondary role.
The structural organization of eIF5A reveals two important features that are associated with nucleic acid binding: the KOW motif at the N-terminal domain and the OB-fold at the C-terminal domain [34,35]. The presence of these elements implicated in nucleic acid binding, together with the physical association of eIF5A with the translational machinery, strongly suggests a role for eIF5A in modulating rRNA, mRNA or tRNA function during protein synthesis. The new mutants described herein will be useful in the search for eIF5A cellular partners and in the elucidation of the precise mechanism of action of this essential factor in protein synthesis.
Experimental procedures
Molecular modeling
The alignment of eIF5A sequences was performed using clustalw (http://www.ebi.ac.uk/clustalw/). Molecular models for eIF5A were built using restraint-based homology modeling, as implemented in the program modeller 9v1 [36]. The alignment of the S. cerevisiae eIF5A protein against the sequence of L. mexicana was initially used as input to the modeller program, together with the atomic coordinates of the latter (Protein Data Bank file 1XTD). Yeast eIF5A presents high sequence identity and similarity when compared with L. mexicana eukaryotic initiation factor 5A (about 50% and 70% respectively) giving us confidence in the use of the comparative protein modeling method. Models were generated by coordinate copying for all atoms that were common between the two sequences, and the models were completed using internal coordinates from the Charm topology library [37]. A 4 Å coordinate randomization was applied to each structure prior to optimization using up to 400 cycles of the variable target function method employing conjugate gradients. Refinement was subsequently performed in Cartesian space using the standard full molecular dynamics algorithm with simulated annealing, as implemented in modeller 9v1. Default limits were applied to the distance restraints. Besides the modeller pseudo-energy-term, the quality of the models generated was also independently evaluated by the programs procheck [38] and verify 3D [39] and by the qualty option [40] of whatif [41], and used to select a representative model for structure analysis. After optimization and refinement, the final model presented excellent stereochemistry with no residue within the disallowed regions of Ramachandran space. Chemical environment analysis( verify 3D) gave an overall score of 52.4, comparable with the expected value of 65.3 for a 144-residue protein and well above the confidence threshold of 29.4. Furthermore, the qualty score of −1.292, which evaluates internal atomic contacts, suggests that the model is reliable. The yeast eIF5A model was used to propose new amino acid substitutions in the eIF5A structure and to localize, in 3D space, the mutations described herein (supplementary Video S1).
Yeast strains manipulation and plasmid cloning
S. cerevisiae strains, plasmids and oligonucleotides used in this work are listed in the supplementary Tables S1–S4. Procedures for cell growth and genetic manipulations were carried out according to standard protocols [42]. Cloning by PCR was performed using Pfx DNA polymerase or HiFi DNA polymerase (Invitrogen, Carlsbad, CA, USA), following standard molecular biology procedures [43].
Isolation and identification of intragenic synthetic lethal mutations of tif51A-3
tif51A mutants that cannot complement the eIF5A knockout strain were generated randomly by ethyl methanesulfonate mutagenesis in a screening of synthetic lethality [30]. Genomic DNA was prepared from the tif51A mutant strains shown in Table S1 (SVL518, SVL519, SVL520, SVL521, SVL522, SVL523 and SVL718), and the tif51A allele of each strain was amplified by PCR using HiFi DNA polymerase (Invitrogen) and primers SVO58 and SVO59, which contain a BamHI site. PCR products were purified and sequenced using the primers SVO49 and SVO50. After identification of mutations, PCR fragments were digested with BamHI, gel purified and ligated into the BamHI site of the vector pUC18. The wild-type TIF51A was cloned into pUC18 using the same strategy. The separation of mutations was achieved by consecutive swapping between parts of mutant and wild-type alleles, using restriction enzyme sites existing in the gene and in the vector. After that, the wild-type TIF51A and the resulting separate mutants were individually subcloned into pRS315. All constructions were confirmed by sequencing.
Isolation of mutants by random-PCR mutagenesis
Mutants of eIF5A were generated by random-PCR mutagenesis, as previously described [44]. In brief, the wild-type TIF51A was used as template in the amplification using Taq DNA polymerase (Invitrogen), and primers SVO58 and SVO59, under mutation-inducing conditions. The plasmid pSV501, harboring the TIF51A gene, was digested with SalI and transformed, together with the mutated tif51A coding sequence, into yeast strain SVL132. After 3 days of growth on synthetic complete minus tryptophan (SC–trp) medium, cells that had lost the wild-type TIF51A-containing plasmids were selected by replica plating onto 5-fluoroorotic acid-containing medium. Colonies were subsequently replica plated onto SC-trp medium and grown for 3 days at 25 or at 38 °C. Only those that failed to grow at 38 °C were selected as new tif51A alleles. Finally, all isolated mutations were identified by sequencing.
Site-directed mutagenesis
The plasmid pRS315, containing TIF51A, was used as a template to generate mutants using the Quick Change site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA), according to the manufacturer’s instructions. The presence of the specific mutations was confirmed by sequencing.
Western blotting
To determine eIF5A levels in tif51A mutant strains, cells were grown to mid-log phase at 25 °C, shifted to a restrictive temperature (37 °C or 38 °C) or left at 25 °C for 3 h, and then lysed in protein extraction buffer (20 mm Tris/HCl, pH 7.5; 2 mm dithiothreitol; 2 mm EDTA and 5 µg·ml−1 of pepstatin, leupeptin, aprotinin and chymostatin) with 2 mm phenylmethanesulfonyl fluoride. Total protein (5 µg) was resolved by SDS-PAGE, transferred to nitrocellulose and eIF5A was detected by immunoblotting with a 1 : 10 000 dilution of rabbit polyclonal antiserum raised against yeast eIF5A [27] and an enhanced chemiluminescence detection system.
Pulse-chase analysis of elF5A
Stationary yeast cultures were diluted in 50 mL of yeast extract peptone dextrose (YPD) to D600nm of 0.5 and grown at 25 °C to reach D600nm of 0.3 D600nm. Cultures were harvested by centrifugation at 3000 g for 4 min, resuspended in 600 µL of fresh SC-methionine medium containing 200 µCi of [35S]methionine and incubated at 25 °C for 15 min. For chasing, cells were harvested, resuspended in SC medium containing nonlabeled methionine (0.2 mg·ml−1) and cycloheximide (0.5 mg·ml−1) and then incubated at 25 or 36 °C. At the given times, 100 µL of culture was mixed with 100 µL of 10% trichloroacetic acid and incubated on ice for 15 min. Cells were washed with 5% trichloroacetic acid and disrupted in 5% trichloroacetic acid using glass beads and a bead beater. Yeast extracts were transferred to a fresh tube and pelleted by spinning at 12 000 g for 5 min. Pellets were washed with acetone, dried and then resuspended in 10 mm Tris/HCl, pH 8.0, containing 1% SDS. Incorporation of [35S]methionine was determined by scintillation counting and the samples were normalized. A portion of yeast extract corresponding to 1 000 000 c.p.m. was diluted in 200 µL of IP buffer (50 mm Hepes, pH 7.5, 150 mm NaCl, 5 mm EDTA, 1% Triton X-100, 0.5 mg·ml−1 BSA, 1 mm dithiothreitol, 1 mm phenylmethanesulfonyl fluoride) containing 5 µL of anti-eIF5A rabbit serum and incubated at 4 °C for 2 h. Suspensions were centrifuged at 12 000 g for 10 min, after which the supernatants were transferred to a fresh tube containing 50 µL of 50% slurry of protein A–Sepharose (in IP Buffer). After incubation at 4 °C for 1 h, the beads were pelleted, washed three times with IP Buffer and suspended in sample buffer. After SDS-PAGE, the gels were stained with Coomassie Blue to visualize the size markers, soaked in autoradiograph enhancer, dried and exposed to autoradiographic film.
Glutathione S-transferase pulldown assay
Plasmids expressing GST and GST–Dys1 were introduced into SVL530, SVL599 or SVL634 yeast strains (Table SI). The resulting strains were used for GST pulldown assays. Briefly, cells were grown at 30 °C to reach a D value, of 0.4, at 600 nm, and then the production of GST fusion proteins was induced with 2% galactose for 3 h. Cells were collected, washed twice in cold NaCl/Pi and suspended in cold buffer A (30 mm HEPES, pH 7.5, 100 mm KAc, 2 mm MgAc, 7 mm l-mercaptoethanol, 5 µg·ml−1 of pepstatin, leupeptin, aprotinin and chymostatin). Cells were lysed by vortex agitation with glass beads for 3 min, incubating the tubes in an ice bath for 1 min after every 1-min agitation. Cell lysates were clarified by centrifugation at 20 000 g for 20 min at 4 °C, and the total protein concentration was determined. Clarified lysates (2 mg total protein) were incubated with 100 µL of glutathione-sepharose beads (50% slurry) for 1 h, at 4 °C. The beads were collected, washed five times with cold buffer A and suspended in 30 µL of SDS-PAGE loading buffer. Proteins were separated by SDS-PAGE (12% gel) and proteins were detected by western blot using polyclonal anti-eIF5A or anti-GST (Sigma, St Louis, MO, USA) and an enhanced chemiluminescence detection system.
Protein synthesis measurement
Yeast strains carrying wild-type or eIF5A mutant alleles were grown to mid-log phase (D600 nm = 0.5) at 30 °C in 10 mL of YPD liquid medium. Then, 20 µCi of [3H]leucine was added and the cells were incubated at 30 °C for 1 h with agitation. Next, 1.5-mL aliquots from cultures were harvested at 4 °C and frozen at −80 °C. The cultures were then shifted to the restrictive temperature (38 °C) for 4 h and 1.5-mL aliquots were collected 1, 2, 3 and 4 h after the temperature shift, harvested at 4 °C and frozen at −80 °C. All frozen cell pellets were resuspended in 15% cold trichloroacetic acid solution and incubated on ice for 15 min. The samples were heated at 100 °C for 15 min and then incubated on ice for 10 min. Trichloroacetic acid precipitates were collected by centrifugation (15 000 g, 4 °C, 10 min) and washed twice with 10% trichloroacetic acid to remove free [3H]leucine. The final washed pellets were resuspended in 0.1 mL of 0.2 m NaOH, and 50-µL aliquots were used to determine the radioactivity incorporated into proteins in a Beckman Scintillation Counter (Beckman Coulter, Pasadena, CA, USA). Total protein concentration was determined by the Bradford protein assay (Bio-Rad, Hercules, CA, USA), using 5-µL aliquots. The amount of protein synthesis was calculated for each sample as d.p.m.·µg−1.
In vivo hypusine synthesis
Yeast knockout strains expressing wild-type or eIF5A mutant proteins (SVL530, SVL634 and SVL599; Table S1) were grown to mid-log phase (D600 nm = 0.5) at 25 °C. [3H]Spermidine (5 µCi·ml−1) was added and each culture was divided into two. One culture was left at 25 °C and the other was shifted to the restrictive temperature (38 °C) for 4 h. Cell pellets were lysed by vortex agitation with glass beads for 2 min, incubating the tubes in an ice bath for 1 min after every 30-s agitation. Cell lysates were clarified by centrifugation (15 000 g, 20 min, 4 °C) and proteins were precipitated using a cold 10% trichloroacetic acid solution containing a mixture of polyamines (1 mm each of putrescine, spermidine and spermine). After extensive washing with 10% trichloroacetic acid containing unlabeled polyamines, the trichloroacetic acid-precipitated proteins were hydrolyzed in 6 N HCl at 110 °C overnight and the radiolabeled hypusine and deoxyhypusine were separated by ion-exchange chromatography [45]. Radioactivity in each fraction was measured in a Beckman Scintillation Counter.
In the case of the hypusination assay for the nonviable mutants (SVL731 harboring one of the plasmids pSV146, pSV39, pSV771, pSV733, pSV785, pSV732, pSV775, pSV731, pSV786 or pSV787), after trichloroacetic acid precipitation, the pellets were resuspended in 0.2 N NaOH. Aliquots were resolved in SDS-PAGE, and the gel was subjected to fluorography, dried and exposed to X-ray film.
CD spectroscopy
Recombinant eIF5A proteins used in CD spectroscopy were produced in E. coli followed by purification. Cell lysate preparation and purification were performed as recommended by the resin manufacturer (GE). Six-histidine-tagged eIF5A wild-type and mutant fusion proteins were purified by affinity chromatography on nickel-nitrilotriacetic acid–Sepharose.
CD spectra for the recombinant six-histidine-tagged eIF5A wild-type and mutant fusion proteins were collected using a Jasco J-715 spectropolarimeter equipped with a temperature control. The protein solution was approximately 25 µm in CD buffer (10 mm Tris/HCl, pH 7.5, 10 mm NaCl). All data were collected using a 1 mm quartz cuvette at 20 °C, using a scan rate of 100 nm·min−1, a spectral bandwidth of 1 nm and a response time of 1 s. The spectra were recorded over the wavelength range from 200 to 250 nm and were determined as an average of 16 scans. Spectra were transformed into molar ellipticity (θ) using the mean weight residue and protein concentration. The buffer contribution was subtracted in all experiments.
Supplementary Material
Acknowledgments
We are grateful to Ana Paula Borges Gregio for technical assistance in obtaining strain SVL731 and to Edith C. Wolff (NIDCR, NIH) for critical reading of the manuscript. This work was supported by grants to S. R. V. from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). We also thank FAPESP, CNPq and CAPES for fellowships awarded to most of the authors.
Abbreviations
- DHS (Dys1 in yeast)
deoxyhypusine synthase
- DOHH (Lia1 in yeast)
deoxyhypusine hydroxylase
- EF-P
elongation factor P
- elF5A
eukaryotic translation initiation factor 5A
- GST
glutathione S-transferase
- SC
synthetic complete
- YPD
yeast extract peptone dextrose
References
- 1.Schnier J, Schwelberger HG, Smit-McBride Z, Kang HA, Hershey JW. Translation initiation factor 5A and its hypusine modification are essential for cell viability in the yeast Saccharomyces cerevisiae . Mol Cell Biol. 1991;11:3105–3114. doi: 10.1128/mcb.11.6.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen KY, Liu AY. Biochemistry and function of hypusine formation on eukaryotic initiation factor 5A. Biol Signals. 1997;6:105–109. doi: 10.1159/000109115. [DOI] [PubMed] [Google Scholar]
- 3.Schwelberger HG, Kang HA, Hershey JW. Translation initiation factor eIF-5A expressed from either of two yeast genes or from human cDNA. Functional identity under aerobic and anaerobic conditions. J Biol Chem. 1993;268:14018–14025. [PubMed] [Google Scholar]
- 4.Cooper HL, Park MH, Folk JE, Safer B, Braverman R. Identification of the hypusine-containing protein hy+ as translation initiation factor eIF-4D. Proc Natl Acad Sci USA. 1983;80:1854–1857. doi: 10.1073/pnas.80.7.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park MH. The post-translational synthesis of a polyamine-derived amino acid, hypusine, in the eukaryotic translation initiation factor 5A (eIF5A) J Biochem (Tokyo) 2006;139:161–169. doi: 10.1093/jb/mvj034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park MH, Wolff EC, Folk JE. Is hypusine essential for eukaryotic cell proliferation? Trends Biochem Sci. 1993;18:475–479. doi: 10.1016/0968-0004(93)90010-k. [DOI] [PubMed] [Google Scholar]
- 7.Zanelli CF, Valentini S. Is there a role for eIF5A in translation? Amino Acids. 2007;33:351–358. doi: 10.1007/s00726-007-0533-0. [DOI] [PubMed] [Google Scholar]
- 8.Kemper WM, Berry KW, Merrick WC. Purification and properties of rabbit reticulocyte protein synthesis initiation factors M2Balpha and M2Bbeta. J Biol Chem. 1976;251:5551–5557. [PubMed] [Google Scholar]
- 9.Benne R, Hershey JW. The mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. J Biol Chem. 1978;253:3078–3087. [PubMed] [Google Scholar]
- 10.Hershey JW, Smit-McBride Z, Schnier J. The role of mammalian initiation factor eIF-4D and its hypusine modification in translation. Biochim Biophys Acta. 1990;1050:160–162. doi: 10.1016/0167-4781(90)90159-y. [DOI] [PubMed] [Google Scholar]
- 11.Kang HA, Hershey JW. Effect of initiation factor eIF-5A depletion on protein synthesis and proliferation of Saccharomyces cerevisiae . J Biol Chem. 1994;269:3934–3940. [PubMed] [Google Scholar]
- 12.Hanauske-Abel HM, Park MH, Hanauske AR, Popowicz AM, Lalande M, Folk JE. Inhibition of the G1-S transition of the cell cycle by inhibitors of deoxyhypusine hydroxylation. Biochim Biophys Acta. 1994;1221:115–124. doi: 10.1016/0167-4889(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 13.Park MH, Wolff EC, Folk JE. Hypusine: its post-translational formation in eukaryotic initiation factor 5A and its potential role in cellular regulation. Biofactors. 1993;4:95–104. [PubMed] [Google Scholar]
- 14.Jao DL, Chen KY. Tandem affinity purification revealed the hypusine-dependent binding of eukaryotic initiation factor 5A to the translating 80S ribosomal complex. J Cell Biochem. 2006;97:583–598. doi: 10.1002/jcb.20658. [DOI] [PubMed] [Google Scholar]
- 15.Zanelli CF, Maragno AL, Gregio AP, Komili S, Pandolfi JR, Mestriner CA, Lustri WR, Valentini SR. eIF5A binds to translational machinery components and affects translation in yeast. Biochem Biophys Res Commun. 2006;348:1358–1366. doi: 10.1016/j.bbrc.2006.07.195. [DOI] [PubMed] [Google Scholar]
- 16.Kim KK, Hung LW, Yokota H, Kim R, Kim SH. Crystal structures of eukaryotic translation initiation factor 5A from Methanococcus jannaschii at 1.8 A resolution. Proc Natl Acad Sci USA. 1998;95:10419–10424. doi: 10.1073/pnas.95.18.10419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peat TS, Newman J, Waldo GS, Berendzen J, Terwilliger TC. Structure of translation initiation factor 5A from Pyrobaculum aerophilum at 1.75 A resolution. Structure. 1998;6:1207–1214. doi: 10.1016/s0969-2126(98)00120-8. [DOI] [PubMed] [Google Scholar]
- 18.Yao M, Ohsawa A, Kikukawa S, Tanaka I, Kimura M. Crystal structure of hyperthermophilic archaeal initiation factor 5A: a homologue of eukaryotic initiation factor 5A (eIF-5A) J Biochem (Tokyo) 2003;133:75–81. doi: 10.1093/jb/mvg011. [DOI] [PubMed] [Google Scholar]
- 19.Hinnebusch AG, Dever TE, Asano K. Mechanism of translation initiation in the yeast Saccharomyces cerevisiae . In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2007. pp. 225–268. [Google Scholar]
- 20.Xu A, Chen KY. Hypusine is required for a sequence-specific interaction of eukaryotic initiation factor 5A with postsystematic evolution of ligands by exponential enrichment RNA. J Biol Chem. 2001;276:2555–2561. doi: 10.1074/jbc.M008982200. [DOI] [PubMed] [Google Scholar]
- 21.Xu A, Jao DL, Chen KY. Identification of mRNA that binds to eukaryotic initiation factor 5A by affinity co-purification and differential display. Biochem J. 2004;384:585–590. doi: 10.1042/BJ20041232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glick BR, Ganoza MC. Identification of a soluble protein that stimulates peptide bond synthesis. Proc Natl Acad Sci USA. 1975;72:4257–4260. doi: 10.1073/pnas.72.11.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanawa-Suetsugu K, Sekine S, Sakai H, Hori-Take-moto C, Terada T, Unzai S, Tame JR, Kuramitsu S, Shirouzu M, Yokoyama S. Crystal structure of elongation factor P from Thermus thermophilus HB8. Proc Natl Acad Sci USA. 2004;101:9595–9600. doi: 10.1073/pnas.0308667101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aoki H, Dekany K, Adams SL, Ganoza MC. The gene encoding the elongation factor P protein is essential for viability and is required for protein synthesis. J Biol Chem. 1997;272:32254–32259. doi: 10.1074/jbc.272.51.32254. [DOI] [PubMed] [Google Scholar]
- 25.Glick BR, Chladek S, Ganoza MC. Peptide bond formation stimulated by protein synthesis factor EF-P depends on the aminoacyl moiety of the acceptor. Eur J Biochem. 1979;97:23–28. doi: 10.1111/j.1432-1033.1979.tb13081.x. [DOI] [PubMed] [Google Scholar]
- 26.Zuk D, Jacobson A. A single amino acid substitution in yeast eIF-5A results in mRNA stabilization. EMBO J. 1998;17:2914–2925. doi: 10.1093/emboj/17.10.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valentini SR, Casolari JM, Oliveira CC, Silver PA, McBride AE. Genetic interactions of yeast eukaryotic translation initiation factor 5A (eIF5A) reveal connections to poly(A)-binding protein and protein kinase C signaling. Genetics. 2002;160:393–405. doi: 10.1093/genetics/160.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zanelli CF, Valentini SR. Pkc1 acts through Zds1 and Gic1 to suppress growth and cell polarity defects of a yeast eIF5A mutant. Genetics. 2005;171:1571–1581. doi: 10.1534/genetics.105.048082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chatterjee I, Gross SR, Kinzy TG, Chen KY. Rapid depletion of mutant eukaryotic initiation factor 5A at restrictive temperature reveals connections to actin cytoskeleton and cell cycle progression. Mol Genet Genomics. 2006;275:264–276. doi: 10.1007/s00438-005-0086-4. [DOI] [PubMed] [Google Scholar]
- 30.Frigieri MC, Thompson GM, Pandolfi JR, Zanelli CF, Valentini SR. Use of a synthetic lethal screen to identify genes related to TIF51A in Saccharomyces cerevisiae . Genet Mol Res. 2007;6:152–165. [PubMed] [Google Scholar]
- 31.Facchiano AM, Stiuso P, Chiusano ML, Caraglia M, Giuberti G, Marra M, Abbruzzese A, Colonna G. Homology modelling of the human eukaryotic initiation factor 5A (eIF-5A) Protein Eng. 2001;14:881–890. doi: 10.1093/protein/14.11.881. [DOI] [PubMed] [Google Scholar]
- 32.Levin DE. Cell wall integrity signaling in Saccharomyces cerevisiae . Microbiol Mol Biol Rev. 2005;69:262–291. doi: 10.1128/MMBR.69.2.262-291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vannier D, Damay P, Shore D. A role for Sds3p, a component of the Rpd3p/Sin3p deacetylase complex, in maintaining cellular integrity in Saccharomyces cerevisiae . Mol Genet Genomics. 2001;265:560–568. doi: 10.1007/s004380100447. [DOI] [PubMed] [Google Scholar]
- 34.Steiner T, Kaiser JT, Marinkovic S, Huber R, Wahl MC. Crystal structures of transcription factor NusG in light of its nucleic acid- and protein-binding activities. EMBO J. 2002;21:4641–4653. doi: 10.1093/emboj/cdf455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murzin AG. OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J. 1993;12:861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 37.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 38.Laskowski RA, MacArthur MW, Thornton JM. Validation of protein models derived from experiment. Curr Opin Struct Biol. 1998;8:631–639. doi: 10.1016/s0959-440x(98)80156-5. [DOI] [PubMed] [Google Scholar]
- 39.Luthy R, Bowie JU, Eisenberg D. Assessment of protein models with three-dimensional profiles. Nature. 1992;356:83–85. doi: 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
- 40.Vriend G, Sander C. Quality control of protein models: directional atomic contact analysis. J Appl Crystallogr. 1993;6:47–60. [Google Scholar]
- 41.Vriend G. WHAT IF: a molecular modeling and drug design program. J Mol Graph. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-v. 29. [DOI] [PubMed] [Google Scholar]
- 42.Guthrie C, Fink GRE. Guide to Yeast Genetics. New York, NY: Academic Press; 1991. [Google Scholar]
- 43.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl KE. Current Protocols in Molecular Biology. New York, NY: Wiley Press; 2005. [Google Scholar]
- 44.Lee MS, Henry M, Silver PA. A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes Dev. 1996;10:1233–1246. doi: 10.1101/gad.10.10.1233. [DOI] [PubMed] [Google Scholar]
- 45.Clement PM, Hanauske-Abel HM, Wolff EC, Kleinman HK, Park MH. The antifungal drug ciclopirox inhibits deoxyhypusine and proline hydroxylation, endothelial cell growth and angiogenesis in vitro . Int J Cancer. 2002;100:491–498. doi: 10.1002/ijc.10515. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.