Abstract
Objectives:
This study evaluated the prevalence, antibiogram and molecular features of CA-MRSA in Awka, Nigeria.
Methods:
Confirmation of MRSA was done by testing resistance to oxacillin (1µg), cloxacillin (5µg) and cefoxitin (30µg) on sterile Mueller Hinton agar supplemented with 4% sodium chloride. The MRSA strains were subjected to antimicrobial susceptibility testing using Kirby-Bauer disc diffusion method. Minimum inhibitory concentration was determined using agar dilution method. Penicillin binding protein 2a was detected through rapid latex agglutination assay while mecA gene was detected by polymerase chain reaction. A total of 142 S. aureus isolates were obtained from 261 samples sourced from Staff, students and fomites of the Faculty of Pharmaceutical Sciences
Result:
The overall prevalence of MRSA was 22.6%. The carriage rate was higher in females (56.5%) than male (43.5%) and was highest in individuals of 20-30 years of age (57.65%). The MIC of the oxacillin sodium salt ranged from 4-32 μg/ml. The multi-antibiotic resistance indices show that 53.4% had Multiple Antibiotic Resistance Indexing (MARI) higher than 0.2. Penicillin binding protein 2a was detected in 8.4% of MRSA isolates, all from nasal carriage while mecA gene was detected in 5 of isolates.
Conclusion:
This study showed a very high prevalence of MRSA carriage among studied subjects.
Keywords: Antibiogram, Community-associated methicillin resistant Staphylococcus aureus (CA-MRSA), Mec A gene, PBP2a, Prevalence, Nigeria
1. INTRODUCTION
Staphylococcus aureus is a Gram-positive bacterium commonly found on the surfaces of the human skin and mucous membranes and can be carried asymptomatically for weeks or months on mucous membranes. It is a leading cause of community acquired infections particularly in colonized humans [1-3].
Strains of S. aureus have developed resistance to antimicrobial agents and their prevalence outside the hospital is of potential epidemiological threat. Accumulation of resistance factors by S. aureus has rendered the bacterium immune to a variety of commonly used antibiotics, thus increasing the ability of S. aureus to survive in hostile environments [4-6]. S. aureus was initially reported resistant to penicillin when penicillinase-producing S. aureus strains were detected. This led to the development of penicillinase-resistant semi-synthetic penicillins, such as methicillin and oxacillin in 1961 to treat Penicillin resistant S. aureus. Unfortunately, Methicillin-resistant S. aureus (MRSA) was reported 2 years after its introduction in the United Kingdom [2, 7]. MRSA is a strain of S. aureus that is resistant to a large group of antibiotics, called “beta-lactams’. Strains resistant to vancomycin and ciprofloxacin have equally emerged [8, 9]. MRSA is one of the major causes of infections in humans, occurring in both the community and the hospital [10]. It causes skin infections, osteoarthritis and respiratory tract infections. MRSA equally causes abscess in deep organs, and is responsible for toxin mediated diseases. Infections caused by MRSA have been reported to be responsible for increased length of hospital stays, rising health care costs, and a high mortality rate. Carriers of MRSA are also prone to septicemia, wound infections and the occasional toxic shock syndrome [2, 8, 11]. Akerele et al. [8] recently reported that acquisition of MRSA has been associated with two different environments: Community-acquired MRSA (CA-MRSA) and healthcare-acquired MRSA (HA-MRSA). MRSA poses a serious therapeutic problem in Nigeria because information on mechanism of their emergence and spread is limited in the community. Therefore characterization of isolates provides base line information needed to implement strategies for the effective management of MRSA carriage. CA-MRSA is a major global challenge due to the presence of multiple drug resistant genes and colonization has been identified as a major risk factor which influences the rapid spread of this MRSA. This present study was conducted to evaluate the prevalence and molecular features of community acquired-methicillin resistant Staphylococcus aureus from fomites, staff and students of Faculty of Pharmaceutical Sciences, Nnamdi Azikiwe University, Awka, Nigeria.
2. METHODS
2.1. Sampling
Swabs were collected randomly from healthy individuals (Staff and students) and fomites of the Faculty of Pharmaceutical Sciences after obtaining their informed consent. The study protocol was performed according to the Helsinki declaration and approved by management of Nnamdi Azikiwe University Teaching Hospital ethical committee.
2.1.1. Isolation and Identification of Bacteria
Sterile cotton swab was used to collect samples from nasal, hands and fomites. The swab sticks containing various samples were inoculated into Mannitol salt agar (MSA) (Oxoid, England) plates within 1-2 h of collection and incubated at 35oC for 24-48 hours. Streak plate technique was used to obtain culture of each isolate. The isolates were identified using colony morphology, Gram staining and biochemical tests including catalase test, coagulase test according to standard bacteriological procedures [12]. In this study, isolates that were Gram positive cocci in clusters, catalase positive and coagulase positive were considered as S. aureus.
2.2. Detection of MRSA Using Disc Diffusion Method
All the presumptive MRSA isolates were differentiated from other coagulase positive isolates following the methods of Cheesbrough [12]. Briefly, sterile swabs were used to inoculate the test organism onto the sensitivity agar (Mueller Hinton). Sterile forceps were used to carefully distribute the following antibiotic disc, oxacillin (1µg), cloxacillin (5µg), cefoxitin (30µg) and vancomycin (30µg) (Oxoid, Germany) evenly on the inoculated plates at a distance of 30 mm (Fig. 1). The plates were kept on the bench for 30 minutes to allow pre-diffusion of the antibiotics, these were inverted and incubated aerobically at 35oC for 18-24 hours. The zones of inhibition were measured using a meter rule and compared with CLSI guidelines [10].
Fig. (1).
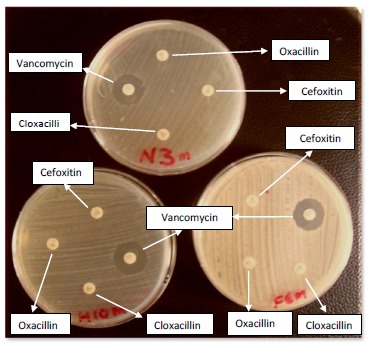
Detection of MRSA using oxacillin, cefoxitin and cloxacillin Oxoid discs.
2.3. Antimicrobial Screening Studies
2.3.1. Antibiotic Sensitivity Test of MRSA
Antibiotic sensitivity of the isolates was determined using Kirby – Bauer disc – diffusion method as described by Okore [13]. The isolates were grown in Nutrient broth at 37oC for 24 hours. Two loopfuls (0.08 mL) of the suspension of each isolate, standardized by matching with 0.5 MCF were inoculated into 20 mL of sterile molten agar in 10 cm diameter Petri dishes and mixed. The plates were allowed to set and the antibiotic sensitivity disc (Oxoid UK) containing ceftriaxone (30µg), ampicillin (10µg), amoxicillin-clavulanic acid (25µg), tetracycline (30µg), ciprofloxacin (5µg), gentamicin (10µg), erythromycin (15µg) and chloramphenicol (30µg) were aseptically placed on their surfaces. The plates were incubated at 37°C for 24 hours and the resultant inhibition zone diameters (IZDs) measured and recorded.
2.3.2. Determination of Minimum Inhibitory Concentration (MIC)
The antibiotics gentamicin, oxacillin sodium salt and vancomycin hydrochloride (Oxoid UK) were used for this assay. Stock solution (128 μg/mL) of each antibiotic was made with distilled water. Adopting the agar dilution method, five serial dilutions of each stock solution were made using 2-fold dilution [13, 14]. Exactly 1 mL from each serial dilution was incorporated into 10 mLof molten nutrient agar and allowed to solidify. A loopful of each suspension of the test organisms was streaked on the plates and incubated at 37°C for 24 hours. The MIC of each antibiotic for each organism was recorded after overnight incubation at 37°C as the lowest concentration yielding no growth or a barely visible haze.
2.4. MRSA Molecular Characterization Studies
2.4.1. Rapid Latex Agglutination
The test tube containing the organism was suspended in microcentrifuge with the extraction reagent and placed in boiling water for 3 min at 95oC to obtain the supernatant. For each supernatant to be tested, 50 μL was placed separately on the circle labelled “Test” and “Control”. One drop of test latex was added, rocked for three minutes and observed for agglutination under normal lighting conditions.
2.4.2. Detection of mecA gene by Polymerase Chain Reaction (PCR): Extraction of DNA and Amplification of mecA gene
The protocol developed by Innis et al. [15], was used for the detection of mecA gene among the isolates. MRSA isolates were cultured on Mueller Hinton agar for 24 hours at 35°C, a single colony from each plate was inoculated in mannitol salt broth and incubated for 24 hours at 35°C. Suspension of 1.5 mL of culture was dispensed into eppendorf tubes, centrifuged at 12000 rpm for 1 minutes and the cell pellet was washed with 200 µL of water. The pellet was vortexed for 5 minutes, heated at 95oC for 10 minutes, washed with 70% ethanol, resuspended in 50 μL of Tris EDTA (TE) elution buffer and kept at –20oC until used. DNA amplification was carried out according to the following protocol: Cycling conditions of the thermo cycler were performed at 94°C for 5 min., followed by 30 cycles of denaturing at 94°C for 30 s and annealing temperature was at 55°C for 30 s. The elongation or extension stage of the mecA gene was at 72°C for 1 min with a final extension temperature at 72°C for 5 min. The PCR products were separated on 1.5% agarose gel electrophoresis and visualised with 0.5 μg/mL ethidium bromide under ultraviolet transilluminator. A 1 kb DNA ladder was used as the molecular weight standard. The PCR amplification was performed in a total volume of 25μL containing the reaction mixture (Table 1).
Table 1.
Composition of reaction mixture for PCR used for amplification of mecA gene.
| Reagents | Vol (μl) |
|---|---|
| 10x buffer | 2.5 |
| dNTP mix (25mM each) | 0.25 |
| MgCl2 (25mM) | 1 |
| Primer mix (130mg/ml each) | 0.25 |
| Taq polymerase (1 U/μl) | 0.25 |
| PCR water | 15.75 |
| Template DNA | 5 |
| Total volume | 25 |
3. RESULTS
3.1. Distribution of Community Isolates of Staphylococcus aureus
In this research, a total of 142 S. aureus were isolated from 261 specimens of different sources (Nasal, hand and fomite). From the nasal and hand isolates, the staphylococcal carriers, 52% were from males and 65% were from females .The percentage of S. aureus isolates was in this order: nasal > hand > fomite having 57%, 25.4% and 17.6% respectively (Table 2).
Table 2.
Distribution of community isolates of S. aureus and MRSA from different (Sources).
| Sources | S. aureus | MRSA (%) | P value |
|---|---|---|---|
| Nose | 81(57%) | 33 (56.0) | P>0.05 |
| Hand | 36(25.4%) | 13 (22.0) | |
| Fomite | 25(17.6%) | 13 (22.0) | |
| Total | 142 | 59 (100) |
3.1.1. Finding of MRSA
A total of 59 methicillin resistant Staphylococcus aureus (MRSA) isolates were detected using these antibiotics: oxacillin, cloxacillin, cefoxitin and vancomycin based on CLSI guide lines (Fig. 1). The 59 MRSA were from Nose (33), Hand (13) and Fomites (13). The overall prevalence of MRSA was 22.6% (59/261). The resistance was 100% in all 59 strains for cefoxitin, cloxacillin and oxacillin disc diffusion test. Out of 59 MRSA isolates, 51 (86.4%) were susceptible to vancomycin while 8 (13.6%) were resistant to vancomycin (Fig. 1).
3.1.2. Distribution of MRSA Isolates in Relation to Gender, Age and Source
MRSA carriage varies according to gender, age and source. Gender based analyses for nasal carriers showed that prevalence of MRSA was higher among females. Out of 46 MRSA isolates, 20 (43.55%) were from males while 26 (56.5%) were from females (Table 3). Twelve (36.4%) out of 33 (40.7%) MRSA nasal carriers were males while 21 (63.6%) were females (Table 3). The difference was not statistically significant, Chi square statistic (with Yates correction: 1.473, d.f.1, P>0.05.) The ages of nasal carriers ranged from ≤ 20 to ≥ 30 years, similarly the order of distribution of MRSA among individuals was 7 (21.2%) in ≤ 20 years, 19 (57.6%) in 20-30 years and 7 (21.2%) in ≥ 30 years (Table 3). On the bases of age, MRSA was found higher among individuals of 20-30 years (57.65%) compared to individuals of ≤ 20 (21.2%) and ≥ 30 years (21.2%). The difference was not statistically significant Chi square statistic (with Yates correction): 0.855, d.f.1, P>0.05.
Table 3.
Gender distribution of MRSA isolates.
| Nasal | Hand | |||
|---|---|---|---|---|
| Sex | S.aureus | MRSA | S.aureus | MRSA |
| Male | 36 | 12 | 16 | 8 |
| female | 45 | 21 | 20 | 5 |
| Total | 81 | 33 | 36 | 13 |
The ages of hand carriers ranged from ≤ 20 to ≥ 30 years, similarly the order of distribution of MRSA among individuals was 2 (15.4%) in ≤ 20, 8 (61.5%) in 20-30 years and 3 (23.1%) in ≥ 30 years .The difference was statistically not significant (Table 4) Chi square statistic (with Yates correction): 1.9990, d.f.1, P>0.05.
Table 4.
MRSA Isolates from different age groups (Nasal and Hand).
| Nasal | Hand | |||
|---|---|---|---|---|
| Age group | S.aureus | MRSA | S.aureus | MRSA |
| ≤20 | 21 | 7 (21.2) | 9 | 2 (15.4) |
| 20-30 | 42 | 19 (57.6) | 22 | 8 (61.5) |
| ≥30 | 18 | 7 (21.2) | 5 | 3 (23.1) |
| Total | 81 | 33 (100) | 36 | 13 (100) |
3.2. Antibiotic Sensitivity Profile of the MRSA Isolates
A total of 8 different antibiotics were used to test for the susceptibility pattern of MRSA isolates on Mueller-Hinton agar (MHA) plates using disc diffusion. The three sources showed variable results. Susceptibility pattern of MRSA nasal isolates was in order: gentamicin (93%)~ erythromycin (93%) > amoxicillin-clavulanic acid (90.9%) > ceftriaxone, (84.8%) > chloramphenicol (81.8%) ~ ciprofloxacin (81.8%) > tetracycline (72.7%) > ampicillin (27.3%) (Fig. 2).
Fig. (2).
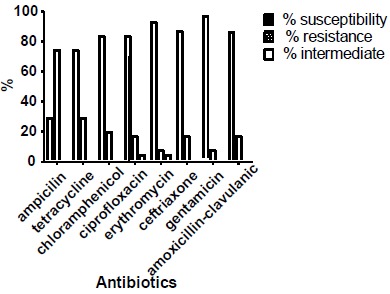
Susceptibility profile of MRSA nasal isolates.
MRSA screened from hand isolates showed that 84.6% were susceptible to gentamicin, 76.9% to chloramphenicol, ciprofloxacin, ceftriaxone and amoxicillin-clauvulanic acid, 69.2% to erythromycin while ampicillin and tetracycline recorded low susceptibility profile of 46.2% and 23.1% respectively (Fig. 3).
Fig. (3).

Susceptibility profile of MRSA hand isolates.
The MRSA isolates from fomites showed 100% sensitivity to ciprofloxacin, gentamicin and amoxicillin-clavulanic acid, 84.6% to ceftriaxone but recorded susceptibility profile of less than 70% to ampicillin, tetracycline, erythromycin and chloramphenicol. Also 69.2% showed an intermediate susceptibility to ampicillin (Fig. 4). Out of 43 isolates from the three sources, 53.4% had MARI higher than 0.2 while 46.5% from the three sources had MARI lower than 0.2 (Table 5).
Fig. (4).
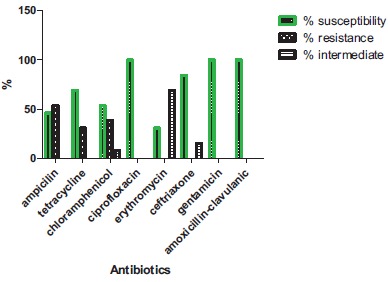
Susceptibility profile of MRSA fomite isolates.
Table 5.
Multiple Antibiotic Resistance (MAR) indices of MRSA.
| Frequency of MAR indices of the three sources | |||
|---|---|---|---|
| MAR Indices | Nose (n=24) | Hand (n=9) | Fomite (n=10) |
| 0 | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| 0.1 | 12 (50.0%) | 4 (44.4%) | 4 (40.0%) |
| 0.2 | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| 0.3 | 6 (25.0%) | 2 (22.2%) | 6 (60.0%) |
| 0.4 | 6 (25.0%) | 2 (22.2%) | 0 (0.0%) |
| 0.5 | 0 (0.0%) | 1(11.1%) | 0 (0.0%) |
| 0.6 | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| 0.7 | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| 0.8 | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| 0.9 | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| 1.0 | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Total | 24 | 9 | 10 |
3.2.1. Minimum Inhibitory Concentration (MIC) of MRSA
In our study, Oxacillin MIC test showed that nasal, hand and fomite isolates tested were resistant with MICs ranging from 4– 32 μg/mL (Table 6) which confirmed the CLSI (1997) criteria of ≥ 4 μg/mL.
Table 6.
Percentage resistance rate of MIC of MRSA nasal isolates using oxacillin sodium salt by agar dilution test.
| Minimum inhibitory Concentration (μg/ml) | ||||
|---|---|---|---|---|
| Source | 4μg/ml | 8μg/ml | 16μg/ml | 32μg/ml |
| Nasal | 21.9% | 34.4% | 18.6% | 6.3% |
| Hand | 0.0% | 16.7% | 33.3% | 16.7% |
| Formite | 7.7% | 23.1% | 38.5% | - |
| Percentage | ||||
For nasal isolates; 21.9% were inhibited at concentration of 4 ug/ml; 34.4% at 8 μg/ml 18.6% at 16 μg/ml and 6.3% at 32 μg/ml, while for hand isolates; no isolates was inhibited at concentration of 4 μg/ml; 16.7% at 8 μg/ml; 33.3% at 16 μg/ml and 16.7% at 32 μg/ml. In the case of fomite isolates; 7.7% were inhibited at concentration of 4 μg/ml; 23.1% at 8 μg/ml; 38.5% at 16 μg/ml and 0% at 32 μg/ml. The organisms expressed high level of resistance to oxacillin.
3.3. Molecular Studies
3.3.1. Result of PBP2a Latex MRSA Screening Test and Detection of mecA gene by Polymerase Chain Reaction (PCR)
The rapid latex kit detected penicillin binding protein 2a in 5 (8.4%) of the MRSA isolates, all from nasal carriage (Table 7) and mecA gene was detected in 5 (8.4%) of the MRSA isolates: N14, N38, N69, N55, and N77.
Table 7.
Latex Agglutination Test for Penicillin Binding Protein (PBP2’) in nasal isolates of methicillin resistant Staphylococcus aureus.
| S/N | Isolates |
Agg present in 3
minutes |
Agg absent in 3
minutes |
Agg present in
Control latex in 3 minutes |
Interpretation of
results |
|---|---|---|---|---|---|
| 1 | N3 | NIL | PBP2’ Negative | NIL | MSSA |
| 2 | N7 | NIL | PBP2’ Negative | NIL | MSSA |
| 3 | N11 | NIL | PBP2’ Negative | NIL | MSSA |
| 4 | N14 | PBP2’ Positive | NIL | NIL | MRSA |
| 5 | N16 | NIL | PBP2’ Negative | NIL | MSSA |
| 6 | N22 | NIL | PBP2’ Negative | NIL | MSSA |
| 7 | N36 | NIL | PBP2’ Negative | NIL | MSSA |
| 8 | N37 | NIL | PBP2’ Negative | NIL | MSSA |
| 9 | N38 | PBP2’ Positive | NIL | NIL | MRSA |
| 10 | N40 | NIL | PBP2’ Negative | NIL | MSSA |
| 11 | N43 | NIL | PBP2’ Negative | NIL | MSSA |
| 12 | N44 | NIL | PBP2’ Negative | NIL | MSSA |
| 13 | N45S | NIL | PBP2’ Negative | NIL | MSSA |
| 14 | N54 | NIL | PBP2’ Negative | NIL | MSSA |
| 15 | N55 | PBP2’ Positive | NIL | NIL | MRSA |
| 16 | N61 | NIL | PBP2’ Negative | NIL | MSSA |
| 17 | N62S | NIL | PBP2’ Negative | NIL | MSSA |
| 18 | N66 | NIL | PBP2’ Negative | NIL | MSSA |
| 19 | N69 | PBP2’ Positive | NIL | NIL | MRSA |
| 20 | N71 | NIL | PBP2’ Negative | NIL | MSSA |
| 21 | N74S | NIL | PBP2’ Negative | NIL | MSSA |
| 22 | N77 | PBP2’ Positive | NIL | NIL | MRSA |
| 23 | N79 | NIL | PBP2’ Negative | NIL | MSSA |
| 24 | N80 | NIL | PBP2’ Negative | NIL | MSSA |
| 25 | N82 | NIL | PBP2’ Negative | NIL | MSSA |
4. DISCUSSION
Community-acquired infections with Staphylococcus aureus have increased and CA-MRSA is a major global challenge due to the presence of multiple drug resistant genes. Because of the ability of this staphylococci strain to acquire antimicrobial resistance over time, MRSA will continue to be a problem in the future.
The overall prevalence of MRSA was 22.6% (59/261). Recent studies indicate that disc diffusion test using cefoxitin is far superior to most of the currently recommended phenotypic methods like oxacillin disc diffusion. It has been reported as surrogate marker of mecA gene, gives clearer end points, easier to read and is more reproducible than tests with Oxacillin disk diffusion. Thus cefoxitin is now an accepted method for detecting MRSA with high efficiency and has been used as an alternative to PCR in resource constrained areas [16]. Also MRSA strains exhibiting inducible resistance to methicillin grow much more readily in the presence of cefoxitin than oxacillin, due to an enhanced induction of PBP 2a by cefoxitin. Other studies have shown such a higher MRSA prevalence ranging from 40.6% to 59.3% [17]. The observed MRSA prevalence of 22.6% is close to 20.1% recorded in a study carried out in National Orthopedic Hospital Enugu also in south eastern Nigeria but higher than 12.5% prevalence recorded in Maiduguri, Northeastern Nigeria [18, 19].Our reported prevalence is lower than the prevalence rate of 34.7% recorded in Ilorin, 30.4% in Ibadan (both in south west Nigeria) and 43.0% reported in Jos North central Nigeria [20-22]. In contrast to our findings, the prevalence of CA-MRSA was found to be very low in Switzerland (0.09%), the United Kingdom (0.005%), Spain and Portugal [23]. However, a very high prevalence of 36.1% frequency of MRSA was reported in Pakistan by Ullah et al. [24]. This confirms the high regional variations in the findings from different countries and cities. The prevalence of 22.6% MRSA in the studied locality is alarming as strains of CA-MRSA that cause soft-tissue infections in healthy people have been recorded worldwide [25, 26]. Similar no significant difference in carriage rates of S. aureus and CA-MRSA isolates among male and female subjects as well as among age groups have previously been reported [7, 11, 27]. Higher MRSA carriage among individuals of 20-30 is very significant in the studied locality. This is because age of most of the students are within the age bracket of 20-30 years and are living in overcrowded hostels thus having members colonized with MRSA will increase the risk of others becoming colonized. Crowded environments have been reported to significantly increase the risk of MRSA colonization. Colonization has been reported as an important step in the chain of events that lead to S. aureus infections [1, 28, 29].
Similarly insignificant variation in MRSA isolation rate from, nasal, fomite and hand (p>0.05) found in this study is similar to findings of Loren and Binh [30], which have suggested that nasal colonization is not the only route for pathogenesis of CA-MRSA infection and that skin-skin and skin-fomite contact could represent important transmission routes. Nasal carriage of MRSA is an important risk factor for subsequent MRSA infection and transmission as the bacterium is transmitted to the nares by contaminated hands and from surfaces where it can survive for months [31].
Al-Ruaily and Khalil [28] equally maintained that contaminated environmental surfaces also contribute to the MRSA transmission. However, transmission occurs mainly through person-to-person contact as the nose and open skin areas are considered the most important sites for colonization.
In this study, MRSA isolates recorded a very high susceptible rate of > 60% to various antibiotics tested based on the three sources. Nasal and hand isolates were susceptible to gentamicin with 93.9% and 84.6% respectively which is in contrast to previous reports [32]. The susceptibility profile (100%) of the fomite MRSA isolates to gentamicin, ciprofloxacin and erythromycin is consistent with previous report of Kadora [33]. Gentamicin is an aminoglycoside which is injectable and thus its usage is highly restricted unlike other antibiotics which are in tablet forms. It was also observed that isolates recorded a very high resistance to ampicillin and thus is in agreement with previous studies in Nigeria [10, 11, 34]. Ampicillin is a beta-lactam antibiotic. Resistance to the above class of antibiotic is normally mediated when there is disruption in the beta-lactam ring by the enzyme beta-lactamase which deactivates the molecule’s antibacterial property [34, 35]. Other causes of resistance may be attributed to non-compliance to prescription among the students. It is also evident that regional habits equally impact the exponential growth of this resistance because most individuals resort to patronizing patent medicine stores without correct prescription, dosage and shelf life in the community. Very low resistance (15.4%) to ciprofloxacin, a fluoroquinolone recorded in our study has been reported by another study with 16.6% [36]. The isolates showed resistance towards commonly used antibiotics such as ampicillin, and tetracycline. This is in agreement with previous reports that most isolates of S. aureus are resistant to a large number of commonly prescribed antibiotics [11, 25]. Self medication and indiscriminate use of antibiotics have severally been reported as promoters of antibiotic resistance and frequent administration of systemic antibiotics modified nasal S. aureus from MSSA to MRSA [11, 37, 38]. A MAR index greater than 0.2 implies that the strains of such bacteria originate from an environment where several antibiotics are used [39]. The MAR indices obtained in this study is similar to the value obtained in our previously published study [39] and this shows that a very greater proportion of the bacterial isolates have been exposed to several antibiotics. Moreover the differences in sensitivity profile of the bacteria isolates among the sources may be attributed to practices of self medication, drug abuse and indiscriminate use of antibiotics. Similarly, Onanuga et al. [3], also confirmed the occurrence of high multiple antibiotic index in their study. This is in conformity with previous observations that most isolates of S. aureus are resistant to a large number of commonly prescribed antibiotics. Methicillin resistance in S. aureus is primarily mediated by the mecA gene which codes for the modified penicillin-binding protein 2a (PBP 2a or PBP 2') [25, 40]. PBP2a is located in the bacterial cell wall and has a low binding affinity for β-lactams. This low percentage rate of PBP2a is in agreement with previous studies about the suitability of latex screen test for MRSA identification [41]. The low percentage rate of PBP2a observed only from nasal carriage can be that the isolates produced small amount of PBP2a which resulted in weak agglutination reaction and that the agglutination occurred very slowly beyond the specified 3 min. Conventional methods of detecting mecA are often limited by the heterogeneous expression of resistance which are in turn influenced by most of in vitro conditions [42, 43]. PCR is considered to be the gold standard among the approaches for the detection of methicillin resistance [44, 45]. Babiker et al ., [46] in their study found a higher prevalence of mecA gene (76%) among MRSA isolates. However a multicenter study in South-western Nigeria by Adesida et al. [47], reported 1.4% mec A gene among the isolates. Resistance in S. aureus was previously reported to involve two mechanisms the expression of beta lactamase and mecA gene [28, 48].
CONCLUSION
Detection of MRSA is important for appropriate implementation of infection control measures. This study recorded overall MRSA prevalence of 22.6%. The carriage rate was higher in females than males and was highest in individuals of 20-30 years age group. Penicillin binding protein 2a and mec A gene were detected in 20% of the MRSA isolates. There is the need to reassess the policies on appropriate use of antibiotics within community and in the hospital environments. Education /awareness on hand washing and utilization of hand sanitizers is necessary to limit the occurrence and spread of MRSA.
ACKNOWLEDGEMENTS
Declared none.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
REFERENCES
- 1.Ugwu M.C., Mokwe N., Ejikeugwu P.C., Gugu T.H., Enemor E.C., Eze C.O., Ugwu B.C. Antibiogram of Staphylococcus aureus from healthy school pupils in Agulu, Southeastern Nigeria. Int. J. Res. Pharm. Biosc. 2015;2(4):5–9. [Google Scholar]
- 2.Kejela T., Bacha K. Prevalence and antibiotic susceptibility pattern of methicillin-resistant Staphylococcus aureus (MRSA) among primary school children and prisoners in Jimma Town, Southwest Ethiopia. Ann. Clin. Microbiol. Antimicrob. 2013;12:11. doi: 10.1186/1476-0711-12-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Onanuga A., Temedie T.C. Nasal carriage of multi-drug resistant Staphylococcus aureus in healthy inhabitants of Amassoma in Niger delta region of Nigeria. Afr. Health Sci. 2011;11(2):176–181. [PMC free article] [PubMed] [Google Scholar]
- 4.Ugwu M.C., Ikegbunam M.N., Nduka S.O., Attama A.A., Ibezim E.C., Esimone C.O. Molecular characterization and efficacy of antibiotic combinations on multiple antibiotic-resistant Staphylococcus aureus isolated from nostrils of healthy human volunteers. Pharm. Sci. Res. 2013;5(1):26–32. [Google Scholar]
- 5.Beceiro A., Tomás M., Bou G. Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world? Clin. Microbiol. Rev. 2013;26(2):185–230. doi: 10.1128/CMR.00059-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desy C.W., Sarasati W., Siti I., Oktavia S. Antibiotic resistance and methicillin resistant Staphylococcus aureus isolated from Bovine, Crossbred Etawa Goat and Human. Asian J. Animal Vet. Adv. 2015;11:122–129. [Google Scholar]
- 7.Sulaiman A. AL Y, Sabry Y, Mahmoud E, Taha M. Prevalence of methicillin-resistant Staphylococcus aureus in Saudi arabia: systemic review and meta-analysis. Afr. J. Cln. Exp. Microbiol. 2013;14(3):146–154. [Google Scholar]
- 8.Akerele J.O., Obasuyi O., Omede D. Prevalence of methicillin-resistant Staphylococcus aureus among healthy residents of Ekosodin Community in Benin-City, Nigeria. Trop. J. Pharm. Res. 2015;14(8):1495–1499. doi: 10.4314/tjpr.v14i8.24. [DOI] [Google Scholar]
- 9.Farhad S., Zienab M., Mohammad T. Vancomycin-resistant Staphylococcus aureus isolates among hospitalized patients; a tertiary medical care center experience from Southern Iran. Cogent Med. 2016;3:1–8. [Google Scholar]
- 10.Ugwu M.C., Anie C.O., Ibezim E.C., Esimone C.O. Antimicrobial evaluation of methicillin-resistant Staphylococcus aureus nasal carriage amongst healthy students in Agbor, Delta State, Nigeria. Arch. Clin. Microbiol. 2016;7(2):1–4. [Google Scholar]
- 11.Okwu M., Sinat B., Wakeel A. Prevalence of nasal carriage of community-associated methicillinresistant Staphylococcus aureus(CA-MRSA) among healthy primary school children in okada, Nigeria. J. Nat Sci Res. 2012;2(4):61–67. [Google Scholar]
- 12.Cheesbrough M. District laboratory practice in tropical countries. Cambridge 374 University Press UK; 2004. pp. 136–142. [Google Scholar]
- 13.Okore V.C. Principles of pharmaceutical microbiology. 2nd ed. Ephrata Publishers; 2009. Evaluation of chemical Antimicrobial agents. pp. 89–95. [Google Scholar]
- 14.Akagha T.N., Gugu T.H., Enemor E.C., Ejikeugwu P.C., Ugwu B.C., Ugwu M.C. Prevalence and antibiogram of salmonella species and Staphylococcus aureus in retail meats sold in Awka metropolis, southeast Nigeria. Int. J. Biol. Pharmaceut. Res. 2015;6(12):924–929. [Google Scholar]
- 15.Innis M.A., Gelfand D.H., Sninsky J.J., White T.J. PCR Protocols: A Guide to Methods and Applications. San Diego, California: Academic Press; 1990. [Google Scholar]
- 16.Faiqa A., Iffet J., Sohaila M., Saeed A. Detection of MecA mediated methicillin resistance in Staphylococcus aureus by cefoxitin disc diffusion method and latex agglutination test. P J M H S. 2016;10(1):106–108. [Google Scholar]
- 17.Muralidharan S. Special article on methicillin resistant Staphylococcus aureus. J Acad Clin Microbiol. 2009;11:15–16. [Google Scholar]
- 18.Udeani T.K., Onyebuchi C.J., Ikpenwa M.C., Ezenwaka U.R. Prevalence and antibiotic susceptibility pattern of methicillin resistant Staphylococcus aureus in burns and pressure ulcer patients. Afr. J. Clin. Exp. Microbiol. 2016;17(2):130–139. doi: 10.4314/ajcem.v17i2.9. [DOI] [Google Scholar]
- 19.Okon K.O., Shittu A.O., Usman H., Adamu N., Balogun S.T., Adesina O.O. Epidemiology and antibiotic susceptibility pattern of methicillin-resistant Staphylococcus aureus recovered from tertiary hospitals in northeastern. Nig J Med Med Sci. 2013;4:214–220. [Google Scholar]
- 20.Taiwo S.S., Bamidele M., Omonigbehin E.A., Akinsinde K.A., Smith S.I., Onile B.A., Olowe A.O. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in Ilorin, Nigeria. West Afr. J. Med. 2005;24(2):100–106. doi: 10.4314/wajm.v24i2.28176. [DOI] [PubMed] [Google Scholar]
- 21.Adetayo T.O., Deji-Agboola A.M., Popoola M.Y., Atoyebi T.J., Egberongbe K.J. Prevalence of methicillin resistant Staphylococcus areus from clinical specimens in ibadan, Nigeria. Int. J. Eng. Sci. 2014;3:1–11. [Google Scholar]
- 22.Ikeh E.I. Methicillin resistant Staphylococcus aureus at Jos University Teaching Hospital (JUTH). Afr J Clin Exp Microbiol. 2003;4(1):52–55. doi: 10.4314/ajcem.v4i1.7324. [DOI] [Google Scholar]
- 23.Pallab R., Vikas G., Rachna S. Methicillin-resistant Staphylococcus aureus (MRSA) in 405 developing and developed countries: implications and solutions. Regional Health Forum 2011; 15: pp. (1)74-82. [Google Scholar]
- 24.Ullah A., Qasim M., Rahman H., Khan J., Haroon M., Muhammad N., Khan A. Noor MHigh frequency of methicillin-resistant Staphylococcus aureus in Peshawar region of Pakistan. Springerplus. 2016;5:1–6. doi: 10.1186/s40064-016-2277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bektas S., Obradovic A., Aljicevic M., Numanovic F., Hodzic D., Sporisevic L. The frequency of community-acquired Methicillin-resistant Staphylococcus aureus (CA-MRSA) among samples in Institute for public health in Canton Sarajevo. Mater. Sociomed. 2016;28(1):61–65. doi: 10.5455/msm.2016.28.61-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Daly J.M., Levy B.T., Ely J.W., Swanson K., Bergus G.R., Jogerst G.J., Smith T.C., Tara C.S. Management of skin and soft tissue infections in community practice before and after implementing a best practice approach: an Iowa Research Network (IRENE) intervention study. J. Am. Board Fam. Med. 2011;24(5):524–533. doi: 10.3122/jabfm.2011.05.110017. [DOI] [PubMed] [Google Scholar]
- 27.Ankur B., Devjyoti M., Barnali P. Prevalence of nasal carriage methicillin-resistant staphylococci in healthy population of Gangtok, East Sikkim. JIMSA. 2008;21(4):191–193. [Google Scholar]
- 28.Al-Ruaily A, Khalil OM. Detection of (mecA)gene in methicillin resistant Staphylococcus 422 aureus (MRSA) at Prince A / Rhman Sidery Hospital . J. Med. Genet. Genomi. 2011;3(3):41–45. doi: 10.1086/430312. [DOI] [PubMed] [Google Scholar]
- 29.Chih-Jung C, Kuang-Hung H, Tzou-Yien L, Hwang K, Chen P, Huang Y . Factors 425 associated with nasal colonization of methicillin-resistant Staphylococcus aureus 426 among healthy children in Taiwan. J. Clin Microb. 2011;49(1):131–137. doi: 10.1128/JCM.01774-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loren GM, Binh AN. Colonization, fomites and virulence. Rethinking the pathogenesis 428 of Community associated methicillin resistant Staphylococcus aureus infection. Clin. Infect. Dis. Soc Am. 2008;46:752–760. doi: 10.1086/526773. [DOI] [PubMed] [Google Scholar]
- 31.Alaklobi F., Aljobair F., Alrashod A., Alhababi R., Alshamrani M., Alamin W., Lyubov L., Alrouki F., Mertz D. The prevalence of community-associated methicillin-resistant Staphylococcus aureus among outpatient children in a tertiary hospital: A prospective observational study in Riyadh, Saudi Arabia. Int. J. Ped. Adoles Med. 2015;2:136–140. doi: 10.1016/j.ijpam.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muhammad A.Y., Hina N., Naeem A., Saira I., Imran J., Abdul H. Antibiotic susceptibility profile of methicillin-resistant Staphylococcus aureus isolated from nasal samples of hospitalised patients. Afr. J. Microbiol. Res. 2010;4(3):204–209. [Google Scholar]
- 33.Kadora I. Antibiotic sensitivity patterns of hospital-acquired and community-acquired methicillin resistant Staphylococcus aureus. Theses, Dissertations and Capstones. Marshall University Paper 100. 2010.
- 34.Nsofor C.A., Nwokenkwo V.N., Ohale C.U. Prevalence and antibiotic susceptibility pattern of Staphylococcus aureus isolated from various clinical specimens in South East Nigeria. MOJ Cell Sci Rep. 2016;3(2):1–5. [Google Scholar]
- 35.Hugo W.C., Russel A.D. Pharmaceutical microbiology. 7th ed. 137. Vol. 17. United Kingdom: BlackWell Science; 2004. pp. 242–244. [Google Scholar]
- 36.Al-Mohana A.M., Al-Charrakh A.H., Nasir A.H., Al-Kudhairy M.K. Community-acquired methicillin-resistant Staphylococcus aureus carrying mecA and Panton-Valentine leukocidin (PVL) genes isolated from the holy shrine in Najaf, Iraq. J. Bacterio Res. 2012;4(2):15–23. doi: 10.5897/JBR12.005. [DOI] [Google Scholar]
- 37.Ghidey F, Igbinosa O, Igbinosa E. Nasal colonization of methicillin resistant Staphylococcus aureus (MRSA) does not predict subsequent infection in the intensive care unit . B e n i - S u e f . Univ J. Basic Appl Sci. 2014;3:81–86. [Google Scholar]
- 38.Ugwu MC, Odimegwu DC, Ibezim EC, Esimone CO. Antibiotic resistance patterns of Staphylococcus aureus isolated from nostrils of healthy human subjects in a southeastern Nigerian locality. Macedonian J. Med. Sci. 2009;12(4):294–300. [Google Scholar]
- 39.Oli A.N., Iyinagolu R.A., Ichoku U.J., Ugwu M.C., Ezeobi I., Ejiofor O.S., Anagu L.O., Esimone C.O. Antibiotic susceptibility profile of community isolates of Staphylococcus aureus. J. Pharma. Res. Opin. 2013;3(7):42–47. [Google Scholar]
- 40.Suner A. Investigation of Nasal colonization of health-care workers with methicillin-resistant Staphylococcus aureus using new generation real-time polymerase chain reaction assay: Discussing of risks. Afr. J. Biotechnol. 2009;8(20):5542–5546. [Google Scholar]
- 41.van Leeuwen W.B., Kreft D.E., Verbrugh H. Validation of rapid screening tests for the identification of methicillin resistance in staphylococci. Microb. Drug Resist. 2002;8(1):55–59. doi: 10.1089/10766290252913764. [DOI] [PubMed] [Google Scholar]
- 42.Geha D.J., Uhl J.R., Gustaferro C.A., Persing D.H. Multiplex PCR for identification of methicillin-resistant staphylococci in the clinical laboratory. J. Clin. Microbiol. 1994;32(7):1768–1772. doi: 10.1128/jcm.32.7.1768-1772.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cabrera-Contreras R., Morelos-Ramírez R., Meléndez-Herrada E. Evaluation of three phenotypic tests vs. duplex (coa & mecA) PCR for detection of methicillin-resistance Staphylococcus aureus (MRSA) strains from patients with nosocomial infections (NI) in a Mexican Hospital. Br. Microbiol. Res. J. 2016;12(3):1–7. doi: 10.9734/BMRJ/2016/23077. [DOI] [Google Scholar]
- 44.Mohanasoundaram K.M., Lalitha M.K. Comparison of phenotypic versus genotypic methods in the detection of methicillin resistance in Staphylococcus aureus. Indian J. Med. Res. 2008;127(1):78–84. [PubMed] [Google Scholar]
- 45.Iraz M., Tekerekoglu M.S., Otlu B., Ay S. Comparison of an automated system with four phenotypic methods for the detection of methicillin-resistant Staphylococcus aureus. Afr. J. Microbiol. Res. 2012;6(4):764–769. [Google Scholar]
- 46.Babiker MAA, Nazlina I, Ti-Fairuz SI, Hazam NM, Iwani MA. Prevalence of methicillin resistant Staphylococcus aureus (MRSA) among healthy university students of Kabanjaar Malaysia. Global J. Bio-Sci. Biotechnol. 2013;2(1):75–81. [Google Scholar]
- 47.Adesida S., Boelens H., Babajide B., Kehinde A., Snijders S., van Leeuwen W., Coker A., Verbrugh H., van Belkum A. Major epidemic clones of Staphylococcus aureus in Nigeria. Microb. Drug Resist. 2005;11(2):115–121. doi: 10.1089/mdr.2005.11.115. [DOI] [PubMed] [Google Scholar]
- 48.Shore A., Deasy E., Slickers P., Brennan G., Connell B., Monecke S., Ehricht R., Coleman D. Detection of staphylococcal cassette chromosome mec Type XI encoding highly divergent mecA, mecI, mecR1, blaZ and ccr genes in human clinical clonal complex 130 methicillin-resistant Staphylococcus aureus. Am Soc Microbiol. 2011;2:1–38. doi: 10.1128/AAC.00187-11. [DOI] [PMC free article] [PubMed] [Google Scholar]


