Abstract
Anticipating the emergence of infectious diseases would be more effective in reducing their toll, rather than reacting to an outbreak. Machine learning and simulation tools that use environmental, epidemiological and molecular data could help manage and analyse the flow or risks, and inform appropriate actions by public health authorities.

Subject Categories: Microbiology, Virology & Host Pathogen Interaction; S&S: Ecosystems & Environment; S&S: Health & Disease
Emerging infectious diseases are among the most destructive and costly natural forces 1: In terms of human and monetary losses, epidemics and pandemics rank with other major natural disasters, such as earthquakes or tsunamis. And like earthquakes and tsunamis, much of the destructive potential of infectious diseases stems from the fact that they often strike unexpectedly, leaving little time for preparation. The best countermeasure is therefore an early warning to give affected regions or communities more time to prepare for the impact. After the devastating earthquake and tsunami in the Indian Ocean that killed 230,000 people in December 2004, the Indian Ocean Tsunami Warning System was installed in 2005 and became operable in 2006: It demonstrated its value after the Banda Aceh earthquake in 2012 when it alerted the affected islands within minutes of the danger. There are some systems for tracking infectious diseases, such as CDC's PulseNet that monitors disease outbreaks across the USA or the global Influenza Surveillance and Response System, but these are focused on particular geographic areas or on specific diseases. As new diseases emerge and old diseases re‐emerge, as pathogens and their vectors are transported worldwide through trade and travel, it is now time to improve global warning systems for emerging infectious diseases in general.
To mitigate the threat of infectious diseases, our main strategy so far has been a strong defense after emergence. Once an outbreak is under control, we improve infrastructure, develop vaccines, and refine our vigilance in order to better respond to the next outbreak so that only a handful people fall ill instead of hundreds, thousands, or tens of thousands. Our ability to put out these proverbial fires has indeed become formidable over time, but it is still reactive. The outbreak of Ebola in West Africa did not become a worldwide pandemic, but it nonetheless wreaked havoc in West Africa. A total of 26,000 humans were infected, 11,300 died, and the outbreak caused losses of about US$2B in the short term 2 with up to US$15B in estimated losses to investment, trade, and tourism over the next couple of years. There are also concerns about the long‐term knock‐on effects on the political and economic stability in Guinea, Liberia, and Sierra Leone 1.
A more efficient approach to emerging infectious disease threats would be anticipatory: responding to disease risk rather than occurrence by managing and reacting to the ebb and flow of risks in real time. Such a strategy would maintain vigilance while simultaneously assessing vulnerabilities: identifying where disease risk is high, and providing decision support analysis (see Sidebar A) to identify which actions could prevent outbreaks or contain epidemics at the outset.
Sidebar A: The use of modeling for disease risk analysis and prediction.
Big Data and Machine Learning have generated a plethora of methodologies that are useful for infectious disease intelligence. For instance, statistical learning algorithms identify certain patterns in datasets and detect anomalies. Decision support analysis incorporates the models identified by such algorithms into an organizational or policy‐making decision process that can align empirical outcomes, such as deaths averted, and possible actions. Mathematical models of social, epidemiologic, and evolutionary dynamics are useful for explaining how individual decisions (such as hospitalization) and events (such as transmission) “scale up” to generate emergent phenomena at the population level. Such models are often too simplistic for tactical use, but may serve as the core for more complex simulations. Simulation models may then be used prospectively for short‐term forecasting (prediction of number of cases in the next one to four weeks) or long‐term scenario analysis. Simulation models can also be used inversely to test hypotheses about the underlying biological mechanisms or to evaluate the plausibility of alternative theories.
Anticipating and responding to disease risk requires interpreting disease events—outbreaks and epidemics—as emergent properties of a complex system from which to gather infectious disease intelligence. The production of intelligence involves identifying actionable and biologically meaningful data patterns, developing predictions about future risk and epidemic trajectories, and characterizing possible losses under a range of intervention scenarios. Infectious disease intelligence therefore relies fundamentally on data from multiple sources to provide a stream of information that can be inspected by modeling and real‐time analytics to make decisions about prevention, surveillance, or emergency responses to outbreaks.
… like earthquakes and tsunamis, much of the destructive potential of infectious diseases stems from the fact that they often strike unexpectedly, leaving little time for preparation.
What data and analytics are most urgently needed to prepare for spillover from animal reservoirs and subsequent spread of infectious diseases? For what populations and regions should these be collected? The answers to these questions vary according to where a particular infectious disease falls along a continuum of risks (Fig 1). To guide the collection of intelligence, we envision the riskscape—the distribution of risk in space—that consists of three threat levels. At risk level I (yellow), there are no detectable human cases, although there may be sources of infection in proximity to human populations. At risk level II (orange), human cases of an infectious disease have been verified. At risk level III (red), the number of human cases is growing so large that it pushes the limits of disease control.
Figure 1. A model for a global warning system for infectious diseases.
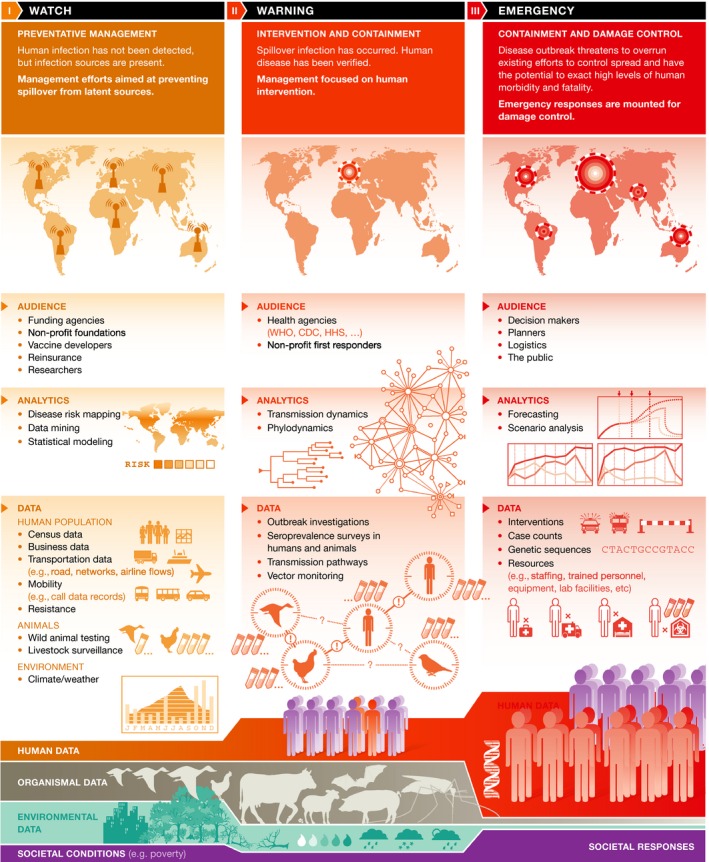
The data and modeling required to assess this riskscape are analogous to that needed for predicting extreme weather events or wildland fires. The risk of a wildfire is quantified at various scales, updated and tracked through time, and coordinated actions are executed in response to analysis of data from multiple sources 3. A fire watch is assigned based on the risk that, if a spark occurs, a fire will catch. If a fire has already started, a warning is issued with continued vigilance and control actions based on the fire's speed and spread. If the fire gets out of control, emergency measures are executed and a coordinated response concentrates on containment and damage control. To apply this analogy to infectious diseases, a watch may be assigned based on empirical quantification of the potential of zoonotic events in an area where conditions would favor a spillover to humans. If a disease is already present in human populations and is increasing in incidence or spreading to new areas, a warning may be issued. At the international level, emergency status is currently assigned when an outbreak threatens to overwhelm existing efforts at controlling the disease that has potential to cause high morbidity and mortality—particularly, the WHO designation of a “Public Health Emergency of International Concern” recognizes that a threat is serious and warrants coordinated international response.
Anticipating and responding to disease risk requires interpreting disease events […] as emergent properties of a complex system from which to gather infectious disease intelligence.
We think that such a system for classifying infectious disease risk would help to guide the development of infectious disease intelligence and to identify best courses of action. In the following sections, we consider data needs and modeling technologies that would serve such activities at each threat level.
During the watch phase, we are primarily concerned with assessing the baseline risk of spillovers from wild animal sources (disease reservoirs) into humans. This would include knowing, for example, which reservoir and vector species occur in an area, and what zoonotic infections they are known to carry 4. While many of these pathogens may not pose an immediate risk, particularly if there is minimal contact between humans and wildlife in this area, quantifying the underlying zoonotic potential is analogous to empirical horizon scanning to assess conditions that would favor the development of a wildfire. Quantifying zoonotic potential would inform management practices and developments that could disturb regions with high but unrealized risk of spillover infections.
To identify conditions that would favor the emergence of an infectious disease, statistical (machine) learning algorithms (see Sidebar A) that are trained on a wide variety of data can identify relationships among interacting variables to characterize how they are associated with background conditions such as specific weather events. Such models can be improved continuously in real time. Quantifying disease risk is analogous to quantifying the amount of flammable tinder in an area and monitoring its accumulation in real time. Current analytical approaches to identifying conditions that predict spillover infection are in their infancy, and our understanding of outbreak prevention is informed primarily by post hoc detective work, carried out on a case‐by‐case basis in the aftermath of costly containment.
The predictive capacity of infectious disease intelligence is not limited by technology. Machine learning methods have already been shown to be effective at harnessing data from multiple sources to characterize the zoonotic potential of particular wildlife species 5. Instead, our capacity to predict spillover events depends on environmental and ecological data, such as the distribution of zoonoses and their vectors and reservoir species, knowledge about pathogens that are not yet known to infect humans, and the assimilation of data from multiple sources to quantify risk and identify trigger conditions early enough for timely intervention. Creating a data infrastructure that would enable real‐time risk quantification would empower the health community to better evaluate the most reasonable preventative investments—such as disrupting plausible transmission chains (prevention), developing vaccines, or improving facilities to better respond to spillover events in high‐risk areas. Such capacity‐building would also add value to ongoing pathogen reconnaissance projects and many investigator‐initiated research programs across a productive and globally distributed scientific community.
At the warning region of the riskscape (risk level II), we are primarily concerned with generating predictions that inform ministries of health and other responders such as the WHO or medical NGOs to react and respond to a disease that has already emerged in a human population. Interventions during the warning phase focus on reducing transmission and mitigating human mortality, economic costs of treatment, and lost productivity. Modeling in the warning phase therefore needs to address various objectives. For example, mathematically modeling transmission dynamics (see Sidebar A) can yield estimates about how quickly the disease may spread, the spatial extent, and epidemiologic outcomes for human populations—the number infected, infection‐induced mortality, recovery rates, and so on—along with the uncertainty in these estimates. The operational time frame of the warning phase scales with the disease based on knowledge of disease transmission. For example, actions following a warning issued for Ebola virus would be carried out within days and focus on the household or the village level 6; this contrasts to a warning issued for helminth parasites for which public health responses may be carried out over the course of months or years to treat chronic infection and prevent transmission 7. Importantly, the downstream consequences of an outbreak could exacerbate the effects of another disease. These complex interactions can be nonlinear and occur at dueling timescales whose dynamical consequences can again be explored using computer models 8.
The hard limits to forecasting are set by the volume and quality of basic scientific information.
In contrast, the goal of phylodynamic modeling is to provide a better understanding of viral evolution. During the Ebola outbreak, molecular analyses helped to answer key epidemiological questions such as confirming the virus and subtype, whether it was introduced multiple times, and the nature of spillover events: This helped to determine that the Kenema cluster in Sierra Leone was the result of a novel spillover event. Integrating epidemiologic and transmission models also enables conclusions about the contact network, the basic reproduction number (R0), and the spread rate. From these data, one can construct transmission trees to determine whether there is overdispersion in contact, depletion of susceptible subjects in the population, or other population‐level phenomena needed for understanding spread. These data streams constitute molecular surveillance. Other pieces of critical information are case reporting, animal surveillance including sentinel species, and environmental monitoring of ecological data such as biodiversity and climatic factors that determine biological interactions.
Finally, emergency measures respond to outbreaks that threaten to overwhelm existing control efforts and that have the potential to exact high levels of human morbidity and fatality (risk level III): The WHO declared both the 2014 Ebola outbreak in West Africa and the ongoing Zika epidemic Public Health Emergencies of International Concern. During disease emergencies, the major goals are quick containment and damage control in the human population. To achieve these objectives, forecasting and scenario analysis must focus on estimating the amount of control or containment efforts—such as the number of treatment centers, the spatial and temporal extent of quarantine, or the mobilization of existing vaccines—needed to achieve a desired outcome, which might be measured in deaths averted, reduction in disability adjusted life years (DALY), or some other societal value.
The network of responders is evolving and improving with each new outbreak to become more efficient and expedient …
The goal of forecasting is to predict the short‐term trajectory of a given situation (see Sidebar A). Data streams that are essential to improving forecasting are real‐time figures about case counts including location data, and results of outbreak investigations, genetic sequences of viral or bacterial isolates, which can then be used to estimate both the evolutionary potential of the pathogen and the actual case burden, and records of actions taken, such as school closures, quarantines, or deployments. Combined, these data can be used to triangulate the current status and trajectory of evolving epidemics.
Scenario analysis, in contrast, does not aim to make quantitative predictions, but explores the possible medium‐ or long‐term outcomes of the available courses of action (see Sidebar A). For instance, to provide useful guidance, modelers need information about infrastructure and equipment such as transportation networks, hospitals, laboratory locations, and capabilities; about available technologies including diagnostic tests and instruments or vaccines; and about supply chains. To predict the potential effectiveness of interventions, it is important to know how effective they are supposed to be. Additionally, effectiveness is modulated by individual behaviors, for instance education about protective measures or government policies, which may have unintended side effects. Most approaches to modeling epidemics are either highly abstract, in which case they may elegantly illuminate the underlying principles governing disease dynamics, but lack the flexibility to represent idiosyncratic conditions on the ground; or they are detailed “tactical models” that characterize the most likely outcomes, but may give a false sense of precision, particularly when data are scarce. During the Ebola epidemic, we developed a new approach, the method of plausible parameter sets, which differs from past approaches in that it aims to characterize the range of plausible outcomes and adapts to the quantity and quality of information available (see Sidebar A) 9.
From the preceding sections, it should be clear that, in our opinion, modeling and analytics are key to generating infectious disease intelligence. But models are not a panacea. One should bear in mind what we might call the First Law of Information: There is no information without data. This version of you‐can't‐get‐something‐from‐nothing states that models cannot make up for ignorance. Modeling is not a magical bridge to cross an information gap. The hard limits to forecasting are set by the volume and quality of basic scientific information.
… modeling can help to inform courses of action in response to the changing and complex appearance of risk as events unfold.
If models cannot make up for lack of information, what can they do? We suggest that models are quantitative tools for structured reasoning. Sometimes, this reasoning is about data: Models can help draw conclusions from data (statistical inference) and extend knowledge of past observations to future conditions (extrapolation). Sometimes, this reasoning is about ideas. For instance, models may serve as a tool for counterfactual investigation: How might transmission be affected by ring vaccination rather than mass vaccination? What is the value of identifying and isolating so‐called super spreaders compared with treating everyone equally, or focusing on protection of the most vulnerable individuals? It is sometimes said that the model is “only as good as the data it's based on”. But, this is too strong. In fact, even data‐free models can serve a useful purpose—as a sanity check, a kind of sophisticated thought experiment to follow an idea through to its logical consequences. Consider an idea like “process P is giving rise to observations O”. For instance, “a depletion of susceptible persons [process] gives rise to a decline in Ebola transmission [observation]”. But our conjecture (P) is complicated: Susceptible depletion is geographically and socially local; there is statistical variation in the number of social contacts each infected person has; and the more connected individuals become infected first, so these patients are differentially removed from the transmission process early in the epidemic. Even though we cannot intuit our way to the implications of P, we can nonetheless determine what their logical entailments are if we encode our key ideas about P into a model. It may remove P from the set of plausible hypotheses, for instance if the required dispersion in social contacts is impossibly large or if the only way for such susceptible depletion to work is an implausible preexisting immunity in the population.
Finally, another useful role for models is to synthesize or stitch together various pieces of information from different places. Continuing with the Ebola example, we could regard information on infectious period, mortality rate, intensity of infection control activities, activities of Red Cross burial teams as parts of a tapestry, and the model as a scaffold on which to hang these pieces and try to make sense of the bigger picture.
The network of responders is evolving and improving with each new outbreak to become more efficient and expedient: The international response to the 2015 outbreak of Zika virus in South America has been faster and more coordinated than the response to Ebola in 2013. However, while this network can rapidly respond once an emerging infectious disease appears on the landscape, a formal and integrated international infectious disease intelligence system has yet to be developed. Such a system should be structured to operate on the entire riskscape and to both support and be informed by modeling and analytics that cross sectors and disciplines. In such a system, data collected during the watch phase would inform decision making during an emergency; postmortem analyses conducted after an emergency will inform how we quantify and respond to future risk 6.
In closing, we consider how infectious disease intelligence relates to three goals of any global response to an emerging disease, as referenced recently in a report by a UN‐appointed panel 2. First, alleviate loss and suffering caused by infectious disease; second, increase global health security and stability; and third, improve global health equity. Achieving these goals requires infectious disease intelligence—knowing what, when, why, and how to respond. Producing this intelligence depends on collecting data and modeling the 356 recognized human infectious diseases, an ambitious task that is already underway for some diseases in some regions 10. Such models can be updated with data collected during emergencies to assess efficacy, costs, and time lags to health improvements.
Similarly, modeling can help to inform courses of action in response to the changing and complex appearance of risk as events unfold. Decision making may be particularly challenging in the transitions from watch to warning and from warning to emergency, with dire consequences: The majority of Ebola deaths in Sierra Leone have been attributed to the lag time in response 6. Forecasting and scenario analyses may be more useful for decision making during rapidly evolving crises than well‐tuned models that quickly go out of date. A pluralistic approach to intelligence is therefore needed. The answers generated by modeling and analytics for surveillance (watch), response (warning), and intervention (emergency) may be imprecise, but they can be improved. Efforts underway within US Government to refine infectious disease analytics, such as infectious disease forecasting (http://bit.do/future-dengue-gov), are a necessary step forward, but greater resources and coordination are needed to improve them, maintain progress, and refine operational utility of analytical results for decision making. Such improvements will require deliberate investment in a quantitative workforce, scaling up systems for data acquisition, an ethic of information sharing, and a culture where decision making and academic modeling are mutually supportive and engaged.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgements
The authors thank Drs. Dylan George and Pejman Rohani for comments on earlier drafts of this article. JMD was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number U01GM110744. The content is solely the responsibility of the authors and does not necessarily reflect the official views of the National Institutes of Health.
References
- 1. World Economic Forum . Global Risk Report 2016. http://wef.ch/1QfL09i
- 2. High‐level Panel on the Global Response to Health Crises (2016) Protecting humanity from future health crises. United Nations, New York, NY, USA. http://www.un.org/News/dh/infocus/HLP/2016-02-05_Final_Report_Global_Response_to_Health_Crises.pdf
- 3. Noonan‐Wright EK, Opperman TS, Finney MA, Zimmerman GT, Seli RC, Elenz LM, Calkin DE, Fiedler JR (2011) Developing the US Wildland Fire Decision Support System. J Combust 2011: e168473 [Google Scholar]
- 4. Han BA, Kramer AM, Drake JM (2016) Global patterns of zoonotic disease in mammals. Trends Parasitol doi: 10.1016/j.pt.2016.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Han BA, Schmidt JP, Bowden SE, Drake JM (2015) Rodent reservoirs of future zoonotic diseases. Proc Natl Acad Sci USA 112: 7039–7044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fang LQ, Yang Y, Jiang JF, Yao HW, Kargbo D, Li XL, Jiang BG, Kargbo B, Tong YG, Wang YW (2016) Transmission dynamics of Ebola virus disease and intervention effectiveness in Sierra Leone. Proc Natl Acad Sci USA 113: 4488–4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Waite RC, Velleman Y, Woods G, Chitty A, Freeman MC (2016) Integration of water, sanitation and hygiene for the control of neglected tropical diseases: a review of progress and the way forward. Int Health 8(Suppl 1): i22–i27 [DOI] [PubMed] [Google Scholar]
- 8. Peters DP, Pielke RA, Bestelmeyer BT, Allen CD, Munson‐McGee S, Havstad KM (2004) Cross‐scale interactions, nonlinearities, and forecasting catastrophic events. Proc Natl Acad Sci USA 101: 15130–15135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Drake JM, Kaul RB, Alexander LW, O'Regan SM, Kramer AM, Pulliam JT, Ferrari MJ, Park AW (2015) Ebola cases and health system demand in Liberia. PLoS Biol 13: e1002056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mundel T (2016) Honing the priorities and making the investment case for global health. PLoS Biol 14: e1002376 [DOI] [PMC free article] [PubMed] [Google Scholar]


