Abstract
Adipose tissue is distributed in depots throughout the body with specialized roles in energy storage and thermogenesis. PDGFRα is a marker of adipocyte precursors, and increased PDGFRα activity causes adipose tissue fibrosis in adult mice. However, the function of PDGFRα during adipose tissue organogenesis is unknown. Here, by analyzing mice with juxtamembrane or kinase domain point mutations that increase PDGFRα activity (V561D or D842V), we found that PDGFRα activation inhibits embryonic white adipose tissue organogenesis in a tissue-autonomous manner. By lineage tracing analysis, we also found that collagen-expressing precursor fibroblasts differentiate into white adipocytes in the embryo. PDGFRα inhibited the formation of adipocytes from these precursors while favoring the formation of stromal fibroblasts. This imbalance between adipocytes and stromal cells was accompanied by overexpression of the cell fate regulator Zfp521. PDGFRα activation also inhibited the formation of juvenile beige adipocytes in the inguinal fat pad. Our data highlight the importance of balancing stromal versus adipogenic cell expansion during white adipose tissue development, with PDGFRα activity coordinating this crucial process in the embryo.
KEY WORDS: Platelet-derived growth factor, Adipocyte, Lipodystrophy, Myf5, Cell fate, Mouse
Summary: Analysis of PDGFRα gain-of-function mutant mice demonstrates a role in balancing the expansion of fibroblast and adipocyte lineages before birth.
INTRODUCTION
There are two main types of adipose tissue in mammals: white adipose tissue (WAT) and brown adipose tissue (BAT). WAT contains unilocular adipocytes with a large, single lipid droplet for energy storage (Rosen and MacDougald, 2006; Rosen and Spiegelman, 2014). BAT contains multilocular adipocytes that dissipate the electrochemical gradient in mitochondria to generate heat (Harms and Seale, 2013; Sidossis and Kajimura, 2015; Schulz and Tseng, 2013). Recently, a third kind of adipocyte – the beige adipocyte – has been identified within WAT depots (Wu et al., 2012, 2013). Beige adipocytes have low basal levels of thermogenic activity, which can be increased by low temperature or beta-adrenergic stimulation. The adipose tissue is composed of mature adipocytes and a heterogeneous population of stromal-vascular cells that includes preadipocytes, stromal fibroblasts, vascular cells and immune cells. Fibroblasts and preadipocytes are closely related mesenchymal cells, although the former are more specialized for collagen secretion and the latter are specialized for generating new adipocytes. Importantly, the extracellular matrix (ECM) composition of adipose tissue must be carefully controlled to allow hypertrophic expansion of the lipid-storing compartment. Improper ECM remodeling leads to fibrosis, which is associated with metabolic dysfunction as excess ECM impairs adipocyte lipid-handling mechanisms (Sun et al., 2013).
Obesity is the accumulation of excess adipose tissue to the extent that it creates a risk for insulin resistance, type 2 diabetes and other diseases. Lipodystrophy – the absence or degeneration of body fat – is also a cause of severe metabolic dysfunction. Understanding how adipose tissue develops and changes over time is essential to identifying new biomarkers and therapeutic targets to address these diseases. In the adult, adipose tissue mass increases by two mechanisms: adipocyte hypertrophy, which adds lipid into existing adipocytes, and adipocyte hyperplasia, which is the differentiation of new adipocytes (Jo et al., 2009). However, adipose tissue organogenesis is still poorly understood and the exact mechanisms remain to be discovered (Gesta et al., 2007; Han et al., 2011; Hong et al., 2015; Hudak et al., 2014; Jiang et al., 2014). A specific question that has not been explored is how mesenchymal precursor cells adopt an adipocyte fate versus a stromal fibroblast fate to achieve the correct balance between lipid-storing and stromal compartments during organogenesis.
Platelet-derived growth factor (PDGF) is an important extracellular signal for developing mesenchymal cells. PDGF binds the receptor tyrosine kinases PDGFRα and PDGFRβ, which are tightly regulated by an autoinhibitory allosteric conformation. PDGF binding induces receptor dimerization, which relieves this autoinhibition and initiates tyrosine kinase activity to induce downstream signal transduction pathways (Tallquist and Kazlauskas, 2004). Knockout of either receptor has revealed crucial roles for PDGFRα and PDGFRβ in the development of vasculature, craniofacial structures, skeleton and other organs (Andrae et al., 2008; French et al., 2008; Hoch and Soriano, 2003; Soriano, 1994, 1997). PDGFRα is commonly used as a marker for fibroblasts and undifferentiated mesenchymal cells, and it is not expressed by differentiated adipocytes. Recent studies have shown that all adipocytes of juvenile and adult mice are derived from PDGFRα-expressing precursors (Berry and Rodeheffer, 2013; Joe et al., 2010; Lee et al., 2012). However, most of our knowledge about PDGF signaling and adipogenesis comes from cell culture experiments (Artemenko et al., 2005; Fitter et al., 2012; Vaziri and Faller, 1996), and the function of PDGFRα in adipose tissue organogenesis remains to be elucidated in vivo.
We recently found that PDGFRα activation in specific nestin+ fibro-adipogenic progenitor cells causes adult WAT fibrosis by converting progenitor cells into ECM-producing fibroblasts. PDGFRα activation also blocks the differentiation of nestin+ progenitors into adipocytes in vitro (Iwayama et al., 2015), suggesting that PDGFRα regulates the balance between adipogenic and non-adipogenic mesenchymal cell populations. However, nestin+ precursors do not contribute to embryonic adipogenesis that generates newborn fat depots. Homeostatic adipogenesis in the adult and developmental adipogenesis in the embryo are likely to be regulated by different mechanisms. To address the role of PDGFRα in embryonic adipogenesis, we have now analyzed mouse embryos and pups with constitutive activating mutations in PDGFRα during the time period of adipose tissue organogenesis. We found that PDGFRα regulates the balanced formation of stromal and lipid-storing compartments during WAT organogenesis. PDGFRα activation promoted the accumulation of ECM as the result of an enlarged stromal fibroblast population, resulting in lipodystrophy. Furthermore, preadipocyte commitment was disrupted in accordance with overexpression of the anti-adipocyte commitment factor Zfp521 and loss of downstream pro-adipogenic transcription factors. These findings identify PDGFRα as a regulator of cell commitment within the early fibroblast-adipocyte lineage that promotes a stromal fibroblast fate at the expense of generating adipocytes.
RESULTS
Strong PDGFRα activation ablates WAT but not BAT in Myf5-D842V mutants
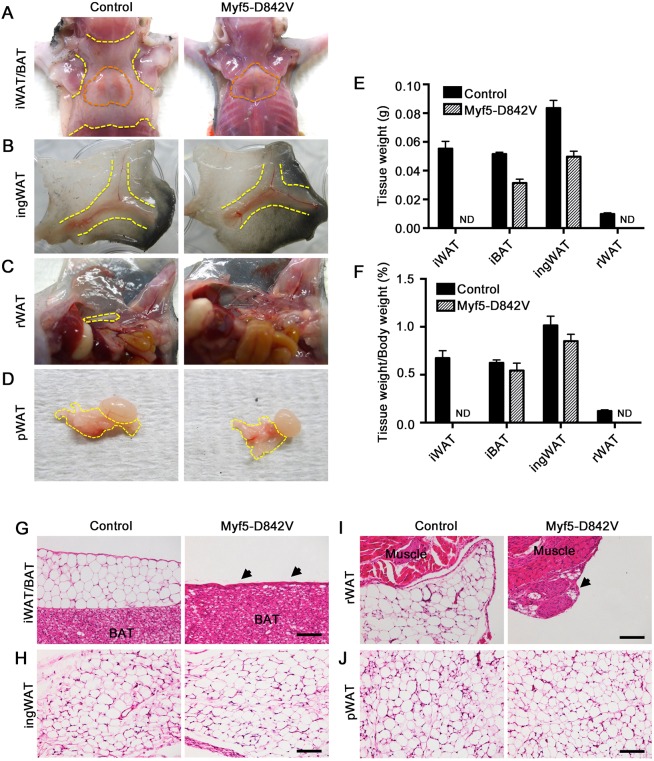
PDGFRαD842V is a strongly hyperactivated isoform associated with human gastrointestinal stromal tumors and inflammatory fibroid polyps (Corless et al., 2004; Schildhaus et al., 2008). Here, we generated mice expressing this mutation in a tissue-specific manner by crossing Myf5-Cre mice with Cre/lox-inducible lox-stop-lox-PDGFRαD842V knockin mice. These knockin mice express PDGFRαD842V from the endogenous Pdgfra gene after Cre/lox recombination removes an intervening stop cassette (Olson and Soriano, 2009). It is well established that Myf5-Cre targets cells that give rise to skeletal muscle and interscapular BAT (iBAT) (Seale et al., 2008). Myf5-Cre also targets interscapular WAT (iWAT) and retroperitoneal WAT (rWAT) but not inguinal WAT (ingWAT) or perigonadal WAT (pWAT) (Sanchez-Gurmaches and Guertin, 2014; Sanchez-Gurmaches et al., 2012). Myf5-Cre;PDGFRα+/D842V mutants (hereafter referred to as Myf5-D842V) were viable, although an overall smaller body size became apparent around postnatal day (P)5. Upon dissection at P18 it was very clear that Myf5+ iWAT and rWAT were missing in Myf5-D842V mutants (Fig. 1A,C), but the Myf5neg ingWAT and pWAT in Myf5-D842V mutants appeared similar in volume to those of control littermates (Fig. 1B,D). The absolute weight of ingWAT was slightly decreased in Myf5-D842V mutants, but the relative weight normalized to total body weight was the same as for control mice (Fig. 1E,F). Morphological analysis of these depots revealed that iWAT and rWAT in Myf5-D842V mutants were replaced by a tiny remnant of stromal tissue (Fig. 1G,I, arrowheads). By contrast, ingWAT and pWAT were fully expanded and indistinguishable between mutants and controls (Fig. 1H,J). There was also no difference in the expression of the adipocyte marker genes Ap2 (also known as Fabp4) and adipsin (Adn; also known as Cfd) in mutant versus control ingWAT (Fig. S1A). Therefore, the effects of specific ablation of iWAT and rWAT of the Myf5+ lineage, without affecting ingWAT or pWAT of the Myf5neg lineage, strongly suggest that PDGFRα activation disrupts WAT organogenesis in a tissue-autonomous manner. We could not perform gene expression analysis on the interscapular and retroperitoneal stromal remnant of Myf5-D842V mice owing to the exceedingly small tissue size.
Fig. 1.
PDGFRα activation inhibits WAT but not BAT development in Myf5-D842V mutants. (A-D) Gross morphology of the indicated adipose tissue depots of a Myf5-D842V mouse and littermate control at P18. Orange outlined areas show the region of iBAT. Yellow outlined areas show the region of WAT. (E) Mass of the indicated tissues at P18. n=3 littermate tissue samples per data point. (F) Mass of the indicated tissues at P18 relative to whole body mass. n=3 littermates per data point. (E,F) Mean±s.e.m. ND, non-detected. (G-J) H&E-stained images of adipose depots derived from Myf5-Cre+ lineage (iWAT, rWAT) or Myf5-Creneg lineage (ingWAT, pWAT) tissues at P21 from a Myf5-D842V mouse and littermate control. Arrows indicate the remnant of stromal tissue that remains in place of WAT. Scale bars: 100 μm.
In contrast to iWAT and rWAT, iBAT was not ablated in Myf5-D842V mutants (Fig. 1A). The absolute weight of Myf5-D842V iBAT was slightly decreased, but the relative weight normalized to total body weight was the same as for control mice (Fig. 1E,F). There was also no difference in the expression of Ap2 and Adn or in the brown adipocyte marker Ucp1 in mutant versus control iBAT (Fig. S1B). We expected PDGFRαD842V to be expressed in iBAT because its expression is under the control of the endogenous Pdgfra gene, which is expressed in iBAT. To confirm this, we compared PDGFRα phosphorylation in cultured iBAT and iWAT stromal vascular cells isolated from Myf5-D842V and control mice. We found that cells from iBAT have a lower basal level of total PDGFRα expression compared with cells isolated from iWAT. However, in Myf5-D842V mutants, cells from both iBAT and iWAT exhibited constitutively phosphorylated PDGFRα, reflecting expression of the activated PDGFRαD842V isoform (Fig. S1C). Therefore, it is possible that the difference in phenotype between iBAT and iWAT is related to the lower expression of PDGFRα in iBAT. Skeletal muscle was not analyzed in our study.
Weak PDGFRα activation causes lipodystrophy and fibrosis
As the WAT phenotype in Myf5-D842V mutants was not amenable to gene expression analysis owing to a lack of tissue, we undertook a detailed examination of a less severe WAT phenotype. PDGFRαV561D is a weakly hyperactivated isoform seen in human tumors and polyps at a lower frequency than PDGFRαD842V. In previous work we showed that PDGFRαV561D generates weaker phenotypes than PDGFRαD842V when activated in adult mice via a tamoxifen-inducible Cre or in development via an epiblast-specific Cre (Olson and Soriano, 2009). Here, we generated mice expressing the V561D mutation in all adipose tissues by crossing Sox2-Cre with lox-stop-lox-PDGFRαV561D knockin mice, where PDGFRαV561D is expressed from the endogenous Pdgfra gene after Cre/lox recombination. Sox2-Cre targets the epiblast and therefore creates a germline Pdgfra mutation in all tissues of the embryo proper (Hayashi et al., 2002). Similar to previous work using Meox2-Cre to target the epiblast (Olson and Soriano, 2009), Sox2-Cre;PDGFRα+/V561D mice (hereafter referred to as Sox2-V561D) were viable for ∼3 weeks.
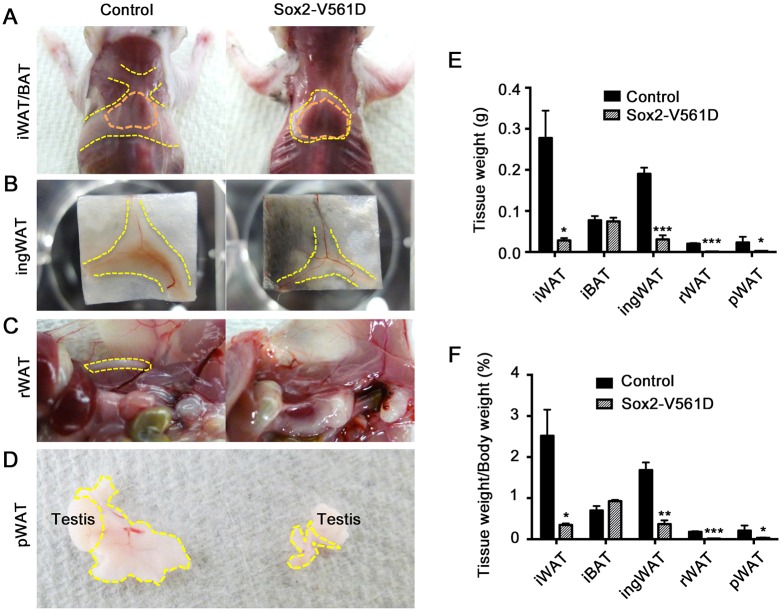
At P18, Sox2-V561D mutants exhibited smaller WAT depots than Sox2-Cre;PDGFRα+/+ control mice. In controls, iWAT covered and partially concealed the underlying iBAT, but Sox2-V561D iBAT was left exposed by smaller and thinner iWAT (Fig. 2A). In Sox2-V561D mutants ingWAT was also reduced and lacked the beige color normally exhibited by control ingWAT at this time point (Fig. 2B). rWAT and pWAT were absent or nearly absent when assessed by anatomical examination in mutant mice (Fig. 2C,D). iWAT and ingWAT in mutant mice weighed ∼90% less than littermate control tissue at P18, both in absolute weight and when normalized to total body weight (Fig. 2E,F). However, consistent with observations in Myf5-D842V mutants, iBAT weight was not diminished by PDGFRα activation.
Fig. 2.
PDGFRα activation causes lipodystrophy in Sox2-V561D mutants. (A-D) Gross morphology of the indicated adipose tissue depots of a Sox2-V561D mouse and littermate control at P18. Orange outlined areas show the region of iBAT. Yellow outlined areas show the region of WAT. (E) Mass of the indicated tissues at P18. n=3 tissue samples per data point. (F) Mass of the indicated tissues at P18 relative to whole body mass. n=3 mice per data point. All data are presented as mean± s.e.m. *P<0.05, **P<0.01, ***P<0.001, Student's t-test.
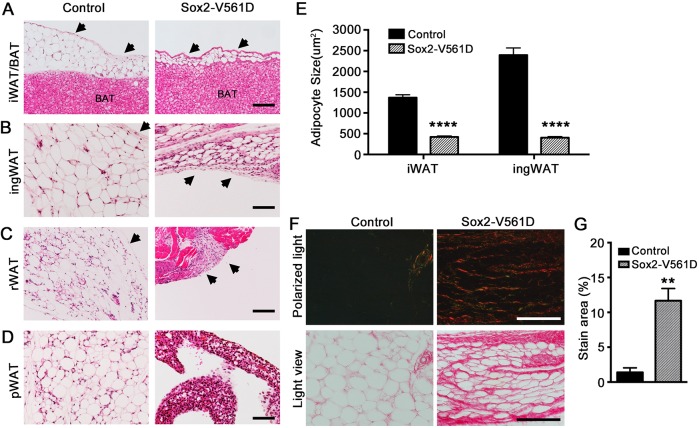
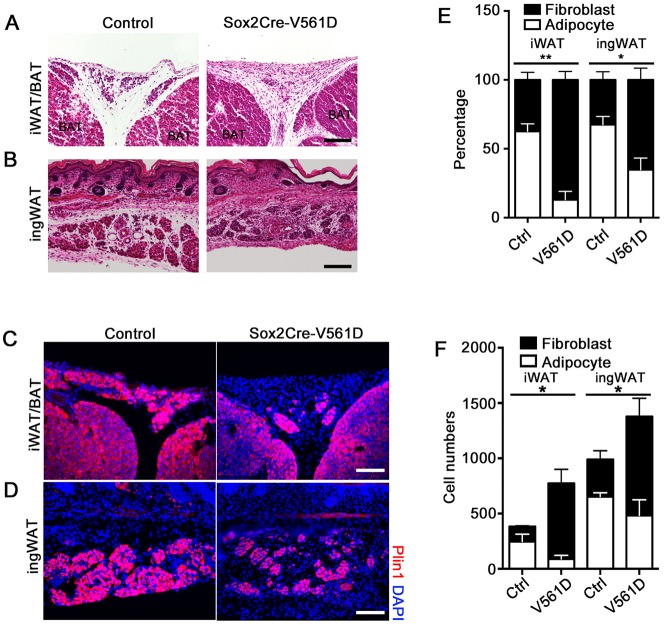
We performed Hematoxylin and Eosin (H&E) staining of paraffin sections to characterize adipose tissue morphology at P18. The smaller iWAT, ingWAT and rWAT in Sox2-V561D mutants contained adipocytes that were smaller than control adipocytes (Fig. 3A,B,E). The stromal margin of the iWAT and ingWAT depots appeared more ECM-rich by H&E staining (Fig. 3A,B, arrows), as did the interstitial space between adipocytes. Mutant rWAT was replaced by a stromal remnant (Fig. 3C). Adipocytes in mutant pWAT were difficult to identify morphologically (Fig. 3D). Immunofluorescence staining for the adipocyte marker perilipin 1 (Plin1) identified Plin1+ cells within the remnant of mutant tissue; however, most cells in the mutant did not express detectable Plin1 (Fig. S2A). Therefore, all WAT depots exhibited an imbalance between the stromal and lipid-storing compartments. Mutant iBAT appeared similar between control and Sox2-V561D mice (Fig. 3A, Fig. S3A), except for more prominent connective tissue septa between the lobules of iBAT, as noted below. Western blotting verified the activation of PDGFRα signaling in Sox2-V561D iBAT stromal vascular cells (Fig. S3B).
Fig. 3.
PDGFRα activation inhibits adipose tissue organogenesis and causes fibrosis in Sox2-V561D mutants. (A-D) H&E-stained images of tissue sections from a Sox2-V561D mouse and littermate control at P18. Arrows indicate the tissue margin enriched for stromal fibroblasts. (E) Quantification of iWAT and ingWAT adipocyte size at P18. n=30 adipocytes per data point. (F) Picrosirius Red-stained images of ingWAT at P18, with polarized light microscopy to visualize collagen fibers. (G) Quantification of collagen fiber area as a percentage of total area. n=3 polarized light images per genotype. All data are presented as mean±s.e.m. **P<0.01, ****P<0.0001, Student's t-test. Scale bars: 100 μm.
The increased stromal area seen by H&E staining of WAT was suggestive of fibrosis. Indeed, Picrosirius Red staining and imaging with polarized light demonstrated an increased density of collagen fibers surrounding adipocytes and in septa-like tracts through the ingWAT (Fig. 3F,G). In mutant iBAT, the septa were more collagen-rich than in controls, but unlike WAT there was no increase in collagen deposition surrounding individual brown adipocytes (Fig. S3C).
We also generated Myf5-V561D mutants and examined fat depots at P18. Once again, the Myf5+ lineage-derived iWAT and rWAT displayed small adipocytes and an increase in stromal area, whereas other tissues appeared normal, including Myf5+ lineage-derived iBAT and ingWAT/pWAT derived from Myf5neg sources (Fig. S2B-D). Taken together, these results demonstrate that WAT develops in Sox2-V561D mice, but tissue expansion is apparently disrupted by hyperplasia of the stromal compartment and dysplasia of the adipocyte compartment. By contrast, iBAT resists the effects of PDGFRαV561D.
Emergence of stromal fibroblasts and adipocytes during organogenesis
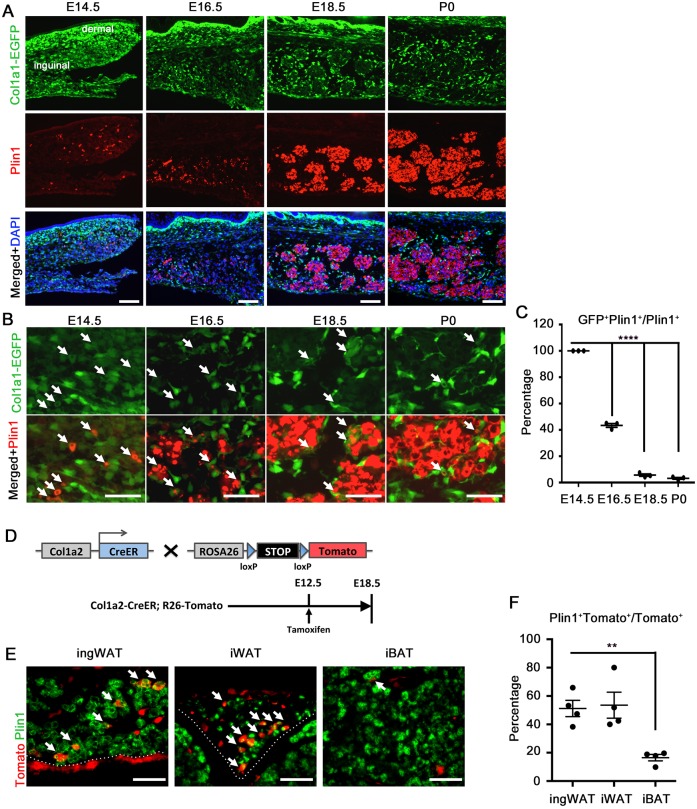
Plin1 expression has been shown to identify early adipocytes in the ingWAT anlage at embryonic day (E)16.5, several days before the cells acquire mature, lipid-filled adipocyte morphology after birth (Hong et al., 2015). To explore the coordinated development of the stromal and lipid-storing compartments in normal ingWAT organogenesis, which has not been described previously, we examined Plin1+ cells in mouse embryos where stromal fibroblasts are marked by EGFP under the control of the collagen type 1, α1 (Col1a1) promoter. At E14.5, the vast majority of the cells in the prospective ingWAT were labeled with EGFP, albeit with variable intensity (Fig. 4A). Among this population were scattered Plin1+ cells colabeled with medium or low EGFP fluorescence (Fig. 4B, arrows indicate double labeling). At E16.5, Plin1+ cells increased in number and formed clusters that expanded in size and cell number through P0 (Fig. 4A), confirming observations from earlier work (Hong et al., 2015). During this morphogenic process between E16.5 and E18.5, most of the remaining cells with high EGFP became restricted to the periphery of the clusters, while cells inside the clusters lost EGFP and developed intense Plin1 staining (Fig. 4B). Plin1/EGFP colabeling dropped from 100% at E14.5 to ∼40% at E16.5 and ∼3% at E18.5 and P0 (Fig. 4C). A few intensely EGFP+ cells remained within the clusters, possibly representing a minor population of interstitial fibroblasts or precursors. Because adipocytes quickly fill with lipid after birth, the distinct morphology of adipocyte clusters surrounded by stromal fibroblasts is not seen in postnatal ingWAT. However, at P4 many Col1a1-EGFP+ cells persisted in a layer along the outer margin of the depot and in the septa dividing tissue lobules (Fig. S4A). We also checked Col1a1-EGFP expression in developing BAT. Unlike the expression in WAT, where ∼30% of the cells were EGFP+, in BAT fewer than 10% were EGFP+ (Fig. S4B,C), suggesting that collagen+ stromal fibroblasts might have a different contribution to WAT and BAT organogenesis. In other E18.5 embryos we examined the expression of a PDGFRα-H2B-GFP knockin reporter in relation to Plin1+ cells. This reporter for PDGFRα+ cells showed a pattern that was highly similar to Col1a1-EGFP (Fig. S4D), with the majority of the labeled cells residing on the periphery of Plin1+ clusters or at the outer tissue margin.
Fig. 4.
Emergence of stromal fibroblasts and adipocytes during embryogenesis. (A) Analysis of ingWAT organogenesis in Col1a1-EGFP mice at the indicated embryonic time points, with immunofluorescence staining for Plin1. (B) Higher magnification of A. Arrows indicate colocalization of Col1a1-EGFP reporter and Plin1. (C) Quantification of the percentage of Plin1+ cells colabeled with EGFP. n=3. (D) Lineage tracing analysis by breeding Col1a2-CreER mice and R26-tdTomato reporter mice, as shown in E,F. (E) Immunofluorescence of Col1a2-CreER;R26-tdTomato lineage tracing in ingWAT, iWAT and iBAT at E18.5, with staining for Plin1. Arrows indicate colocalization of Tomato and Plin1. (F) Quantification of percentage of Tomato+ cells colabeled with Plin1. n=4 embryos. All data are presented as mean±s.e.m. **P<0.01, ****P<0.0001, Student's t-test. Scale bars: 100 μm in A; 50 μm in B,E.
The above observations suggested that stromal fibroblasts and adipocytes separate from common precursors between E14.5 and E16.5 in developing ingWAT. Although Col1a1-EGFP+ cells predominate at E14.5, this population diminishes as the Plin1+ population expands to become the dominant population. In addition, from E16.5-E18.5, the tissue undergoes morphogenesis with clustering of Plin1+ cells and exclusion of most Col1a1-EGFP+ cells to the periphery of the cluster. To directly show the emergence of adipocytes from collagen-expressing precursors, we performed lineage tracing analysis by breeding Col1a2-CreER mice, which express tamoxifen-inducible CreER under control of the Col1a2 promoter (Sonnylal et al., 2007), and R26-tdTomato reporter mice, where Cre/lox deletion of a stop cassette results in indelible labeling of Col1a2-expressing cells and their differentiated progeny (Fig. 4D). We injected tamoxifen into pregnant females at E12.5 and collected embryos at E18.5. Our result show that approximately half of the lineage-traced cells in iWAT and ingWAT became adipocytes with Tomato+ Plin1+ colabeling (Fig. 4E,F). By contrast, only ∼10% of the lineage-traced cells in iBAT were Tomato+ Plin1+, suggesting that collagen-expressing progenitors give rise to brown adipocytes at a low rate during the developmental window between E12.5 and E18.5. In conclusion, many embryonic adipocytes are derived from collagen-expressing precursors, and the separation of stromal fibroblast and adipocyte populations is a normal feature of WAT development.
Disrupted fibroblast-adipocyte balance in newborn Sox2-V561D mice
To understand the origin of the WAT defect that we observed in mutant mice at P18, we examined iWAT and ingWAT in control and Sox2-V561D mice at P0. In H&E-stained tissue sections, control tissue contained cell clusters with eosinophilic cytoplasm (Fig. 5A,B), morphologically corresponding to Plin1+ cells (Fig. 4A, Fig. 5C,D). Plin1+ clusters were also seen in Sox2-V561D tissue, but the clusters appeared smaller. The mutant clusters also appeared less distinct because the inter-cluster space was filled with numerous small cells that did not express Plin1 (Fig. 5C,D). These control and Sox2-V561D mice did not carry the Col1a1-EGFP transgene, but based on their position outside of Plin1+ clusters we presume that most of these interstitial cells correspond to stromal fibroblasts.
Fig. 5.
Disrupted fibroblast-adipocyte balance in newborn Sox2-V561D mice. (A,B) H&E-stained images of iWAT and ingWAT from a Sox2-V561D mouse and littermate control at P0. (C,D) Immunofluorescence analysis of iWAT and ingWAT from a Sox2-V561D mouse and littermate control at P0, stained for Plin1. (E) Quantification of the percentage of Plin1+ adipocytes and Plin1neg fibroblasts from iWAT and ingWAT at P0. n=3-4 sections per data point. (F) Quantification of the total number of Plin1+ adipocytes and Plin1neg fibroblasts from iWAT and ingWAT at P0. n=3-4 sections per data point. All data are presented as mean±s.e.m. *P<0.05, **P<0.01, Student's t-test. Scale bars: 100 μm.
We quantified Plin1+ adipocytes (A) and Plin1neg cells that we call fibroblasts (F) in sections of P0 iWAT and ingWAT. There was a clear shift in the cellularity of the A and F compartments in tissue from mutant versus control mice. When normalized to the total number of cells, the A:F ratio in control tissue was close to 60:40 for both iWAT and ingWAT. The ratio in tissue from mutant mice was 10:90 for iWAT and 25:75 for ingWAT (Fig. 5E). The total number of fibroblasts was always higher in mutants (Fig. 5F). We also examined Plin1+ and Plin1neg cells in Myf5-D842V mutants at P0. In mutants, the A:F ratio was also strongly biased towards fibroblasts in Myf5+ iWAT, but the ratio in Myf5neg ingWAT was similar between tissue from mutant and control (Fig. S5A). Therefore, as an antecedent to lipodystrophy and fibrosis, PDGFRα activation disrupts the balance between stromal fibroblasts and adipocyte lineage cells during adipose tissue organogenesis in both Sox2-V561D and Myf5-D842V mice. In Myf5-D842V mice, the A:F imbalance is restricted to Myf5+ lineage derived (Fig. S5B,C), in keeping with the tissue-autonomous ablation of WAT seen at P18 (Fig. 1).
By P18, the mutant ingWAT was clearly fibrotic (Fig. 3F), but it was not obvious from H&E staining whether ECM deposition was altered at P0. Picrosirius Red staining was also inconclusive at P0 (data not shown). Therefore, we stained P0 ingWAT with antibodies for collagen type 3. In both mutants and controls, collagen 3 immunofluorescence was intense surrounding adipocyte clusters and along the outer margin of the tissue (Fig. S6). There was a clear increase in collagen 3 at the margin of ingWAT from mutant mice, but the amount of collagen 3 within the tissue did not appear to be increased in mutants at P0. Therefore, we conclude that morphological and cellular defects precede fibrosis in Sox2-V561D mutants.
Molecular defects in Sox2-V561D adipose tissue
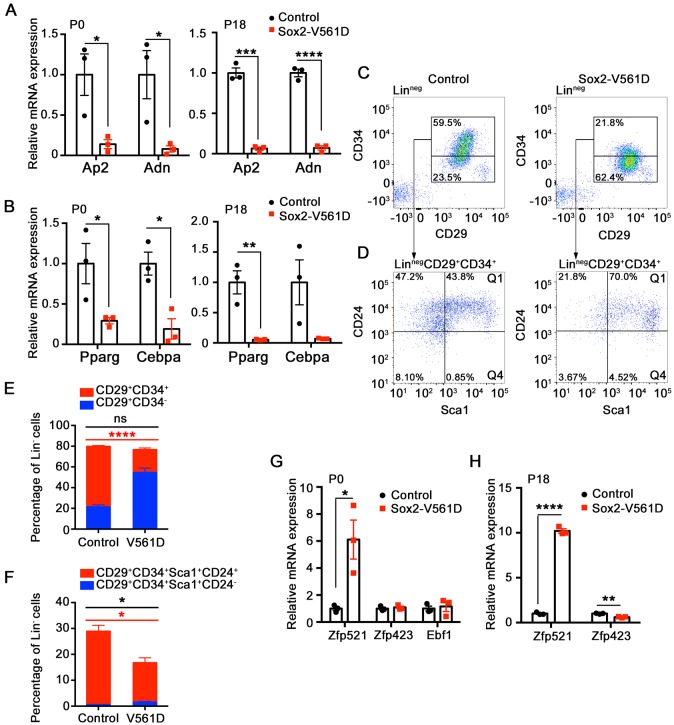
To examine adipocyte differentiation at the mRNA level, we focused on Sox2-V561D and control ingWAT at P0 and P18. At both time points, mutant tissue expressed very low levels of the adipocyte markers Ap2 and Adn compared with control tissue (Fig. 6A). Low expression of the adipocyte markers in mutant ingWAT is consistent with reduced numbers of Plin1+ cells at P0 and lipodystrophy at P18. Preadipocytes express Pparg and Cebpa, two crucial transcription factors that regulate and are required for adipocyte terminal differentiation. These two factors were also downregulated in mutant ingWAT at P0 and P18 (Fig. 6B), which is suggestive of a defect in establishing committed preadipocytes. There were no differences in iBAT expression of Pparg, Cebpa, Prdm16 or Ucp1 between genotypes (Fig. S3D).
Fig. 6.
PDGFRα activation inhibits preadipocyte commitment. (A,B,G,H) Fold change in adipocyte marker and adipogenic transcription factor mRNA levels in ingWAT at the indicated time points, as determined by quantitative RT-PCR. Results are shown as fold change in mutant over control. n=3 tissue samples per genotype. (C) Representative flow cytometry plots of the stromal-vascular fraction from ingWAT in control and Sox2-V561D mice at E18.5. Pseudocolor plots show Linneg (CD45− CD31−) cells. The Linneg (CD29+ CD34+) adipocyte progenitor population and Linneg (CD29+ CD34−) non-adipogenic populations are outlined. (D) Representative flow cytometry plots of Linneg (CD29+ CD34+) cells from C. Pseudocolor plots show the Linneg (CD29+ CD34+ Sca1+ CD24+) Q1 and Linneg (CD29+ CD34+ Sca1+ CD24−) Q4 adipocyte precursor populations. (E,F) Quantification of data in C and D plus additional biological replicates (not shown). n=3 mice per genotype. All data are presented as mean±s.e.m. Red significance identifier refers to subpopulation shown in red; black significance identifier refers to the total cell population. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001, Student's t-test.
We used flow cytometry to monitor preadipocytes based on established cell surface markers (Joe et al., 2010; Macotela et al., 2012; Rodeheffer et al., 2008). After reducing E18.5 control and mutant ingWAT to single cells by enzymatic digestion, we labeled the non-endothelial, non-hematopoietic population (Linneg: CD31− CD45−) with antibodies for preadipocyte antigens CD29 (Itgβ1), CD34, Sca1 (Ly6a) and CD24. We first examined the Linneg fraction in terms of two populations: the CD29+ CD34+ population that is known to be adipogenic in vitro, and the CD29+ CD34− population that is non-adipogenic (Rodeheffer et al., 2008). Flow cytometry analysis showed that 35% of cells in the stromal vascular fraction of control tissue were CD29+ CD34+, whereas only 13% of cells in mutant tissue were CD29+ CD34+ (Fig. 6C,E). Next, we examined the CD29+ CD34+ population in terms of Sca1+ CD24+ and Sca1+ CD24− subpopulations, which are proposed to represent adipocyte precursors at different stages of commitment (Rodeheffer et al., 2008). Owing to the overall reduction in CD29+ CD34+ cells in the mutant, there were severe reductions in the absolute numbers of both subpopulations, as expected (Fig. 6D,F). However, within the overall Linneg fraction, CD24 and Sca1 expression was equivalent between control and mutant, with the CD34 antigen being selectively decreased in mutant tissue (Fig. S7A,B). We also examined the populations of CD29+ CD34+ and CD29+ CD34− cells in the Linneg PDGFRα+ fraction of control and mutant ingWAT, and again found a significant decrease in CD34 expression in the population that otherwise expressed preadipocyte markers (Fig. S7C-E). Therefore, based on low expression of Pparg and Cebpa, plus the loss of CD34, a key cell surface marker of functional preadipocytes, we conclude that PDGFRα activation disrupts preadipocyte commitment.
Recent studies have shown that multipotent mesenchymal cells commit to the adipocyte lineage through the actions of upstream transcription factors. These factors include zinc-finger protein 423 (Zfp423) and early B cell factor 1 (Ebf1), which physically interact with each other and induce Pparg and Cebpa expression (Åkerblad et al., 2002; Festa et al., 2011; Fretz et al., 2010; Gupta et al., 2010; Jimenez et al., 2007). A third factor, Zfp521, exerts an anti-adipogenic role by interacting with Ebf1 to inhibit the Zfp423-Ebf1 functional complex and possibly by also inhibiting Zfp423 expression (Kang et al., 2012). We assessed the expression of these early-acting factors in control and mutant ingWAT. In P0 mutant tissue, Zfp521 was upregulated 6-fold, whereas Zfp423 and Ebf1 were unchanged (Fig. 6G). In P18 mutant tissue, Zfp521 was upregulated 10-fold, whereas Zfp423 was expressed at ∼60% of the normal level (Fig. 6H). These data suggest that Zfp521 expression is regulated by PDGFRα activity. They also point to an early defect in adipocyte commitment, as Zfp521 is the most upstream transcription factor identified in the lineage. High levels of Zfp521 in mutant precursors might disrupt commitment by inhibiting essential functions of Ebf1. The subsequent downregulation of Zfp423 is consistent with fibrotic degeneration of the tissue and progressive loss of adipogenic potential. Zfp521, Zfp423 and Ebf1 expression were unaltered in Sox2-V561D iBAT (Fig. S3E).
PDGFRα inhibits the emergence of beige adipocytes
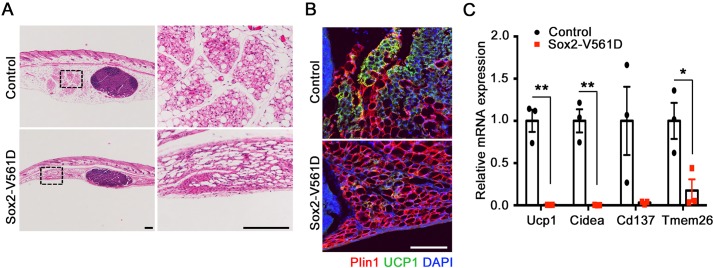
Beige adipocytes have a thermogenic function, like brown adipocytes, but they are found in WAT under conditions of cold exposure or β-adrenergic stimulation (Wu et al., 2012, 2013). In a strain-dependent manner, beige adipocytes emerge transiently in the ingWAT of mice between P15 and P25 (Kozak, 2011). As shown above, the ingWAT depot in control mice is beige at P18, whereas that of Sox2-V561D mice is pale in color (Fig. 2B), suggestive of defective beige adipogenesis in mutant mice. In control ingWAT at P18, H&E staining highlighted some lobules composed of multilocular adipocytes (Fig. 7A). These lobules were consistently located near the central lymph node in the medial-caudal region of the depot. Similar structures could not be identified in mutant ingWAT. Immunostaining for the beige adipocyte marker UCP1 confirmed the identity of the multilocular cells in control ingWAT. However, very few UCP1+ cells were detected in mutant ingWAT (Fig. 7B). We also analyzed the expression of beige adipocyte markers in mRNA isolated from whole ingWAT, and found that Ucp1, Cidea, Cd137 (Tnfrsf9) and Tmem26 were expressed at dramatically reduced levels in mutant tissue (Fig. 7C). Therefore, PDGFRα activation inhibits the endogenous emergence of postnatal beige adipocytes. Other experimental approaches, including the use of a mouse model with inducible PDGFRα activation, will be required to address whether PDGFRα also regulates the formation of beige adipocytes in adult mice in response to cold or β-adrenergic stimulation.
Fig. 7.
PDGFRα inhibits the emergence of beige adipocytes. (A) H&E-stained images of ingWAT from a Sox2-V561D mouse and littermate control at P18. Boxed areas are shown at higher magnification to the right. (B) Immunofluorescence analysis of ingWAT from a Sox2-V561D mouse and littermate control at P18, stained for Plin1 and UCP1. (C) Fold change in beige adipocyte marker mRNA levels in ingWAT at P18, as determined by quantitative RT-PCR. Results are shown as fold change in mutant over control. n=3 tissue samples per genotype. All data are presented as mean±s.e.m. *P<0.05, **P<0.01, Student's t-test. Scale bars: 200 μm in A; 100 μm in B.
DISCUSSION
PDGFRα is expressed on mesenchymal cells of the adipocyte lineage, but whether signaling downstream of the receptor regulates adipose tissue formation during development is unknown. The cells that generate adipose tissue can be separated into Plin1+ cells that differentiate into adipocytes, and Plin1neg stromal fibroblasts that secrete collagen. By analyzing mice with PDGFRα-activating mutations, we provide evidence that PDGFRα regulates the balance between adipocytes and stromal fibroblasts, which is crucial for proper adipose tissue organogenesis. Our results suggest that PDGFRα negatively regulates the emergence of Plin1+ cells from fibroblast-adipocyte progenitors between E14.5 and E16.5. PDGFRα activity inhibits the formation of Plin1+ cells, leading to a decrease in adipocytes and an overabundance of fibroblasts. We show that a strong PDGFRαD842V mutation, driven by tissue-specific Myf5-Cre, ablates iWAT and rWAT, with only a remnant of stromal tissue remaining. A weaker PDGFRαV561D mutation, driven by global Sox2-Cre, allows some, albeit defective, commitment to the adipocyte lineage. However, instead of robust preadipocyte hyperplasia, which is needed to expand the lipid-storing compartment before birth, PDGFRαV561D causes stromal fibroblast hyperplasia. After birth, the enlarged population of stromal fibroblasts ultimately leads to adipose tissue fibrosis.
An important molecular indication of defective adipocyte lineage commitment in Sox2-V561D mice was the downregulation of the transcription factors PPARγ and C/EBPα, which mark preadipocytes and are crucial for adipogenesis (Rosen and MacDougald, 2006). The cell surface marker CD34 was also markedly downregulated in Sox2-V561D ingWAT. CD34 is a highly o-glycosylated single-pass transmembrane protein that serves as a marker for stem or progenitor cells in a variety of tissues (Lee et al., 2000; Trempus et al., 2003; Vanderwinden et al., 2000; Young et al., 1995). In the initial study defining the white adipocyte progenitor population by cell sorting, it was shown that CD34− cells in WAT do not have potential for adipogenic differentiation (Rodeheffer et al., 2008). Taken together, the reduction of PPARγ, C/EBPα and CD34 by E18.5, prior to clear evidence of fibrosis, leads us to conclude that PDGFRα activation antagonizes preadipocyte commitment.
How preadipocyte commitment is regulated is still poorly understood. In a high-throughput screen for commitment-regulating transcription factors, Gupta et al. (2010) identified the zinc-finger protein Zfp423 as a strong inducer of PPARγ. Knockdown of Zfp423 in 3T3-L1 preadipocytes inhibited PPARγ expression and led to failed adipogenesis. Interestingly, Zfp423-deficient mouse embryos exhibited significant reductions in BAT and white adipocyte numbers at E18.5, indicating that Zfp423 is required for adipose tissue organogenesis. Overexpression of Zfp423 in NIH 3T3 fibroblasts was sufficient to confer adipogenic potential in vitro (Gupta et al., 2010). A close paralog to Zfp423 is Zfp521, an established negative regulator of preadipocyte commitment (Kang et al., 2012). Overexpression of Zfp521 in multipotent C3H10T1/2 cells inhibited adipogenesis, whereas knockdown of Zfp521 had the opposite effect (Kang et al., 2012). Furthermore, Zfp521 knockout embryos exhibited enlarged BAT and increased white adipocyte numbers at E18.5. The exact molecular mechanisms by which Zfp521 inhibits adipogenesis are still unclear. Overexpressed Zfp521 can inhibit Zfp423 expression in 3T3-L1 cells, apparently by interacting with Ebf1, which induces Zfp423. However, Zfp521 and Zfp423 can heterodimerize, an effect enhanced by Ebf1, leading to speculation that the presence of Zfp521 might be enough to block the pro-adipogenic function of Zfp423 and Ebf1 (Kang et al., 2012). Our in vivo findings support such a mechanism because we found that a loss of preadipocyte markers at E18.5/P0 was coupled to overexpression of Zfp521, with no reduction in Zfp423 expression until much later. Therefore, the association of Zfp521 and adipocyte fate decisions, together with the observed increase in Zfp521 expression in Sox2-V561D mice, supports a role for a Zfp521-dependent defect in preadipocyte commitment caused by PDGFRα.
Adipocyte differentiation requires major changes in cell size and shape, from the spindle-shaped preadipocyte into the enlarged, round adipocyte. These events are known to be inhibited by collagen and require remodeling of ECM (Sun et al., 2013). For instance, newborn mice deficient for Mmp14, a cell surface matrix metalloproteinase involved in collagen turnover, fail to undergo adipocyte hypertrophy and consequently are lipodystrophic (Chun et al., 2010). The Mmp14−/− preadipocytes were shown to be functionally committed, but lacked the capacity to undergo morphological changes required for adipogenesis in a three-dimensional collagen matrix. Based on previous studies, we knew that PDGFRα activation is sufficient to cause fibrosis in many organs (Olson and Soriano, 2009). We recently found that PDGFRα activation in specific nestin+ fibro-adipogenic progenitor cells causes WAT fibrosis in adult mice by converting adipogenic progenitor cells into ECM-producing fibroblasts (Iwayama et al., 2015). However, whether PDGFRα activity might influence adipose tissue organogenesis was not explored in previous studies. Here, we found lipodystrophy and WAT fibrosis in young mice with two different PDGFRα-activating mutations. Conceivably, these phenotypes could have arisen from an overabundance of ECM, inhibiting adipocyte hypertrophy akin to the Mmp14 mutants. Instead, we found defects in preadipocyte commitment before there was a definitive increase in collagen, strongly suggesting that PDGFRα has a regulatory role in fibroblast-adipocyte cell fate decisions.
As described here, PDGFRα activation has little influence on BAT organogenesis in Sox2-V561D mutants. Even Myf5-D842V mutants with stronger PDGFRα activation lacked Myf5+ WAT but developed essentially normal BAT. Development of BAT occurs earlier than WAT (Schulz and Tseng, 2013) and involves a number of unique signals and transcription factors (Rosen and Spiegelman, 2014; Seale et al., 2009; Sidossis and Kajimura, 2015). Our study indicates that PDGFRα inhibits WAT development by disrupting the population balance between stromal fibroblasts and adipocytes, which are both derived from Col1a1/Col1a2+ progenitors. Meanwhile, our lineage tracing results suggest that the derivation of adipocytes from collagen-expressing precursors predominates in WAT development but not BAT development. Furthermore, our western blotting indicates that PDGFRα is expressed at a lower level in BAT stromal vascular cells compared with WAT. Interestingly, it has been reported that loss of Zfp521 increases white and brown adipogenic potential in development (Kang et al., 2012). However, we found that PDGFRα upregulated Zfp521 expression only in WAT, consistent with our observation of no significant defects in BAT.
Beige adipocytes, which are also known as inducible ‘brown-like’ adipocytes, are a newly discovered type of fat cell that develops in WAT in response to cold exposure and adrenergic stimulation (Rosen and Spiegelman, 2014; Wu et al., 2012, 2013). In some strains of mice, beige adipocytes emerge spontaneously in the rWAT and ingWAT between P15 and P25, or around the time of weaning (Kozak, 2011). Compared with the regulation of white or brown adipogenesis, mechanisms involved in beige adipocyte development are poorly understood. It has been shown that induced beige adipocytes are derived from PDGFRα+ progenitor cells (Lee et al., 2012). Here, we provide evidence that PDGFRα inhibits the development of beige adipocytes in young mice based on analysis of mRNA for beige markers (Ucp1, Cidea, Cd137, Tmem26) and on expression of UCP1 protein. However, whether this effect is direct via blocking beige adipocyte fate commitment/differentiation or is an indirect effect of the fibrotic microenvironment needs to be investigated further with inducible genetic models that avoid WAT fibrosis.
Even though much remains to be learned about the mechanisms underlying beige adipogenesis, this study is the first to suggest a functional link between PDGFRα and beige adipocyte development. This information might lead to a better understanding of the molecular basis of beige adipogenesis in vivo and potentially provide a means to harness beige adipocytes as a new approach to address the obesity epidemic.
MATERIALS AND METHODS
Mice
The mouse strains PDGFRα+/V561D, PDGFRα+/D842V, Sox2-Cre, Myf5-Cre and R26-tdTomato are available at Jackson Laboratories (stock numbers 018432, 018433, 008454, 007893 and 007909). The Col1a1-EGFP and Col1a2-CreER mice were obtained from Dr David Brenner (University of California, San Diego) and Dr James Tomasek (University of Oklahoma Health Sciences Center). Mice were maintained on a mixed C57BL6/129 genetic background, were kept on a daily 12 h light/dark cycle, and fed with a standard chow diet. To study embryonic time points, we performed timed pregnancies with the presence of a vaginal plug regarded as E0.5. All analyses were based on a minimum of three mutants and littermate controls. Males and females were not distinguished in embryos and newborn mice. Only males were used at P18 because of their distinctive pWAT. The Institutional Animal Care and Use Committee of the Oklahoma Medical Research Foundation and/or the University of Oklahoma Health Sciences Center approved all procedures described in this study.
Tamoxifen treatment
Tamoxifen (Cayman, 13258) was dissolved to 20 mg/ml in corn oil (Sigma, C8267) and stored at 4°C until use. Tamoxifen was injected intraperitoneally into pregnant mice at a dose of 80 mg/kg by E12.5. Embryos were harvested by E18.5.
Qualitative histological analysis
For histological analysis, adipose tissues were fixed in Bouin's solution and embedded in paraffin. Sections (8 μm) were de-paraffinized, rehydrated, and stained with Hematoxylin (Electron Microscopy Sciences) and Eosin (Sigma) or Picrosirius Red (Polysciences). For immunofluorescence analysis, adipose tissues were fixed in 4% paraformaldehyde (diluted from 16% stock, Electron Microscopy Sciences), incubated in 25% sucrose in PBS, and embedded in a 1:1 mix of OCT (Tissue-Tek) and Tissue Freezing Medium (TFM, Electron Microscopy Sciences). Immunostaining was performed on 8 μm-thick or 20 μm-thick sections as follows: slides were blocked with 5% donkey serum (Jackson ImmunoResearch) in PBS for 1 h at room temperature, then incubated with primary antibodies in PBS with 5% donkey serum (Jackson ImmunoResearch) overnight at 4°C. On the second day, slides were incubated with secondary antibodies in PBS with 5% donkey serum (Jackson ImmunoResearch) for 1 h at room temperature, followed by nuclear staining with DAPI/PBS (Sigma) for 5 min and mounted with PermaFluor (Thermo Scientific). Antibody against Plin1 was purchased from Novus Biologicals (NB100-60554) or Cell Signaling (9349S) and used at 1:200 or 1:500 dilution, respectively. Antibodies against UCP1 (Ab10983) and collagen III (Ab7778) were purchased from Abcam and used at 1:100 and 1:500, respectively. Purified Alexa Fluor 488-conjugated anti-goat IgG (H+L), Alexa Fluor Cy3-conjugated anti-rabbit IgG (H+L) and Alexa Fluor Cy3-conjugated anti-goat IgG (H+L) were purchased from (Jackson ImmunoResearch) and used at 1:100. All fluorescence images were taken with a Nikon ECLIPSE 80i microscope, except for UCP1 staining of beige adipocytes with an Olympus FV1000 confocal microscope.
Quantitative histological analysis
Adipocyte size quantification was performed on H&E-stained paraffin sections. Representative adipocytes were selected manually and analyzed with NIS-Elements (D3.22) software (Nikon). Adipose tissue fibrotic area was analyzed on Picrosirius Red-stained paraffin sections that were imaged with a polarized light attachment to the ECLIPSE 80i microscope. Signal-positive area was quantified using ImageJ software (NIH) with negative background chosen for the threshold setting. Quantification of Plin1+ and Plin1neg cells at embryonic and perinatal time points was performed on 8 μm sections. The numbers of Plin1+ and Plin1neg cells were counted manually for population percentage analysis with NIS-Elements (D3.22). For lineage tracing with Col1a2-CreER, four Col1a2-CreER;R26-tdTomato embryos were analyzed by manually counting Plin1+ and Tomato+ cells in 20× magnification images using NIS-Elements (D3.22). At least 50 Tomato+ cells were counted for each embryo and each adipose tissue depot analyzed. The percentage of Tomato+ cells expressing Plin1 was then calculated for each tissue sample.
Gene expression analysis
Adipose tissues were removed, flash frozen in liquid nitrogen, and stored at −80°C until use. Total RNA was isolated from adipose tissues using AxyPrep (Axygen) or the RNeasy Kit (Qiagen). Equal amounts of RNA were reverse-transcribed to cDNA. Quantitative RT-PCR was run in 15 μl reactions in a CFX96 real-time PCR system (Bio-Rad) using SYBR Green qPCR ReadyMix (KCQS00-1250RXN, Sigma). Bio-Rad CFX Manager (V2.1) software was used for analyzing cycle threshold (Ct) values and melting curves. Fold differences in mRNA levels were normalized to the expression of the housekeeping gene Timm17b. Forward and reverse primer sequences are listed in Table S1.
Flow cytometry
To isolate the stromal vascular fraction from adipose tissue, ingWAT was isolated and minced with a razor blade. Samples were collected into 2 ml tubes containing 1 ml HBSS containing 0.3 mg/ml collagenase, 3% (w/v) BSA, 1.0 mM MgCl2 and 1.2 mM CaCl2. Tubes were shaken at 37°C for 1 h 30 min for digestion. Digested tissue was filtered through a 40 μm filter into a 50 ml conical tube containing 5 ml FACS buffer followed by centrifugation at 1000 rpm (200 g) at room temperature for 3 min. The cell pellet was collected and resuspended in FACS buffer. The population of preadipocytes was analyzed based on the cell surface markers established previously (Rodeheffer et al., 2008; Berry and Rodeheffer, 2013). Cells were incubated for 15 min on ice with the following antibodies (BioLegend, unless stated otherwise): CD24 (PerCP/Cy5.5, 1:200), CD29 (Alexa Fluor 700, 1:100), CD31 (Pecam1; PE/Cy7, 1:1000), CD34 (Brilliant Violet 421, 1:500), CD45 (Ptprc; APC, 1:200), Sca1 (APC-Cy7, 1:200) and PDGFRα (eBioscience; PE, 1:200). The cells were washed twice, stained with propidium iodide (eBioscience) and then signal obtained with an LSR II flow cytometer (BD Biosciences). All data were analyzed with FlowJo software (Tree Star), with gate setting based on fluorescence minus one controls.
Western blotting
Mouse iBAT was digested using the same collagenase cocktail and conditions as used for flow cytometry. After filtration through a 40 μm filter to remove undigested tissue fragments and gentle centrifugation (200 g) to separate stromal vascular cells from floating adipocytes, the primary stromal vascular cells were cultured in DMEM medium plus 10% FBS and penicillin/streptomycin. The cells were serum starved in DMEM+0.5% FBS for 24 h, and then harvested directly or stimulated with 10 ng/ml PDGF ligand (R&D Systems) for 15 min. Cells were lysed in RIPA buffer and protein concentration was measured by BCA assay (Thermo Scientific). Cell lysates were subjected to western blotting with antibodies to tyrosine phosphorylated PDGFRα (Cell Signaling, 4547; 1:1000), total PDGFRα (Santa Cruz Biotechnology, sc-338; 1:2000) or GAPDH (Santa Cruz Biotechnology, sc-32233; 1:2000). Horseradish peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch) were used at 1:5000.
Statistical analysis
All data are presented as the mean±s.e.m. and calculations were performed using Prism 6 (GraphPad). Unpaired two-tailed Student's t-test was used to determine statistical significance.
Acknowledgements
We thank Cameron Steele, Jang Kim, Eli Rhoades and Andrew Bachman for expert technical assistance; the Oklahoma Medical Research Foundation (OMRF) Centers of Biomedical Research Excellence (COBRE) Flow Cytometry Core for assistance with flow cytometry; and the OMRF COBRE Imaging Core for tissue processing. We also thank Linda Thompson, Courtney Griffin and our laboratory colleagues for critical comments on the manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
C.S. performed all experiments. W.L.B. assisted with lineage tracing experiments. C.S. and L.E.O. designed the experiments, interpreted the data and wrote the manuscript.
Funding
This research was supported by grants from the National Institutes of Health (5P20GM103636), the National Institute of Arthritis and Musculoskeletal and Skin Diseases (1R01AR070235) and the Oklahoma Center for Adult Stem Cell Research (OCASCR) to L.E.O. C.S. was supported by a predoctoral fellowship from the American Heart Association. L.E.O. is a Pew Scholar in Biomedical Research, and this work was supported in part by the Pew Charitable Trusts. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.135962.supplemental
References
- Åkerblad P., Lind U., Liberg D., Bamberg K. and Sigvardsson M. (2002). Early B-cell factor (O/E-1) is a promoter of adipogenesis and involved in control of genes important for terminal adipocyte differentiation. Mol. Cell. Biol. 22, 8015-8025. 10.1128/MCB.22.22.8015-8025.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrae J., Gallini R. and Betsholtz C. (2008). Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 22, 1276-1312. 10.1101/gad.1653708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artemenko Y., Gagnon A., Aubin D. and Sorisky A. (2005). Anti-adipogenic effect of PDGF is reversed by PKC inhibition. J. Cell. Physiol. 204, 646-653. 10.1002/jcp.20314 [DOI] [PubMed] [Google Scholar]
- Berry R. and Rodeheffer M. S. (2013). Characterization of the adipocyte cellular lineage in vivo. Nat. Cell Biol. 15, 302-308. 10.1038/ncb2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun T. H., Inoue M., Morisaki H. and Yamanaka I. (2010). Genetic link between obesity and MMP14-dependent adipogenic collagen turnover. Diabetes 59, 2484-2494. 10.2337/db10-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corless C. L., Fletcher J. A. and Heinrich M. C. (2004). Biology of gastrointestinal stromal tumors. J. Clin. Oncol. 22, 3813-3825. 10.1200/JCO.2004.05.140 [DOI] [PubMed] [Google Scholar]
- Festa E., Fretz J., Berry R., Schmidt B., Rodeheffer M., Horowitz M. and Horsley V. (2011). Adipocyte lineage cells contribute to the skin stem cell niche to drive hair cycling. Cell 146, 761-771. 10.1016/j.cell.2011.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitter S., Vandyke K., Gronthos S. and Zannettino A. C. W. (2012). Suppression of PDGF-induced PI3 kinase activity by imatinib promotes adipogenesis and adiponectin secretion. J. Mol. Endocrinol. 48, 229-240. 10.1530/JME-12-0003 [DOI] [PubMed] [Google Scholar]
- French W. J., Creemers E. E. and Tallquist M. D. (2008). Platelet-derived growth factor receptors direct vascular development independent of vascular smooth muscle cell function. Mol. Cell. Biol. 28, 5646-5657. 10.1128/MCB.00441-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fretz J. A., Nelson T., Xi Y., Adams D. J., Rosen C. J. and Horowitz M. C. (2010). Altered metabolism and lipodystrophy in the early B-cell factor 1-deficient mouse. Endocrinology 151, 1611-1621. 10.1210/en.2009-0987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesta S., Tseng Y.-H. and Kahn C. R. (2007). Developmental origin of fat: tracking obesity to its source. Cell 131, 242-256. 10.1016/j.cell.2007.10.004 [DOI] [PubMed] [Google Scholar]
- Gupta R. K., Arany Z., Seale P., Mepani R. J., Ye L., Conroe H. M., Roby Y. A., Kulaga H., Reed R. R. and Spiegelman B. M. (2010). Transcriptional control of preadipocyte determination by Zfp423. Nature 464, 619-623. 10.1038/nature08816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Lee J.-E., Jin J., Lim J. S., Oh N., Kim K., Chang S.-I., Shibuya M., Kim H. and Koh G. Y. (2011). The spatiotemporal development of adipose tissue. Development 138, 5027-5037. 10.1242/dev.067686 [DOI] [PubMed] [Google Scholar]
- Harms M. and Seale P. (2013). Brown and beige fat: development, function and therapeutic potential. Nat. Med. 19, 1252-1263. 10.1038/nm.3361 [DOI] [PubMed] [Google Scholar]
- Hayashi S., Lewis P., Pevny L. and McMahon A. P. (2002). Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech. Dev. 119, S97-S101. 10.1016/S0925-4773(03)00099-6 [DOI] [PubMed] [Google Scholar]
- Hoch R. V. and Soriano P. (2003). Roles of PDGF in animal development. Development 130, 4769-4784. 10.1242/dev.00721 [DOI] [PubMed] [Google Scholar]
- Hong K. Y., Bae H., Park I., Park D.-Y. and Kim K. H. (2015). Perilipin+ embryonic preadipocytes actively proliferate along growing vasculatures for adipose expansion. Development 142, 2623-2632. 10.1242/dev.125336 [DOI] [PubMed] [Google Scholar]
- Hudak C. S., Gulyaeva O., Wang Y., Park S.-M., Lee L., Kang C. and Sul H. S. (2014). Pref-1 marks very early mesenchymal precursors required for adipose tissue development and expansion. Cell Rep. 8, 678-687. 10.1016/j.celrep.2014.06.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwayama T., Steele C., Yao L., Dozmorov M. G., Karamichos D., Wren J. D. and Olson L. E. (2015). PDGFRα signaling drives adipose tissue fibrosis by targeting progenitor cell plasticity. Genes Dev. 29, 1106-1119. 10.1101/gad.260554.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Berry D. C., Tang W. and Graff J. M. (2014). Independent stem cell lineages regulate adipose organogenesis and adipose homeostasis. Cell Rep. 9, 1007-1022. 10.1016/j.celrep.2014.09.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez M. A., Åkerblad P., Sigvardsson M. and Rosen E. D. (2007). Critical role for Ebf1 and Ebf2 in the adipogenic transcriptional cascade. Mol. Cell. Biol. 27, 743-757. 10.1128/MCB.01557-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo J., Gavrilova O., Pack S., Jou W., Mullen S., Sumner A. E., Cushman S. W. and Periwal V. (2009). Hypertrophy and/or hyperplasia: dynamics of adipose tissue growth. PLoS Comput. Biol. 5, e1000324 10.1371/journal.pcbi.1000324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joe A. W. B., Yi L., Natarajan A., Le Grand F., So L., Wang J., Rudnicki M. A. and Rossi F. M. V. (2010). Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat. Cell Biol. 12, 153-163. 10.1038/ncb2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S., Akerblad P., Kiviranta R., Gupta R. K., Kajimura S., Griffin M. J., Min J., Baron R. and Rosen E. D. (2012). Regulation of early adipose commitment by Zfp521. PLoS Biol. 10, e1001433 10.1371/journal.pbio.1001433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak L. P. (2011). The genetics of brown adipocyte induction in white fat depots. Front. Endocrinol. 2, 64 10.3389/fendo.2011.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. Y., Qu-Petersen Z., Cao B., Kimura S., Jankowski R., Cummins J., Usas A., Gates C., Robbins P., Wernig A. et al. (2000). Clonal isolation of muscle-derived cells capable of enhancing muscle regeneration and bone healing. J. Cell Biol. 150, 1085-1100. 10.1083/jcb.150.5.1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.-H., Petkova A. P., Mottillo E. P. and Granneman J. G. (2012). In vivo identification of bipotential adipocyte progenitors recruited by β3-adrenoceptor activation and high-fat feeding. Cell Metab. 15, 480-491. 10.1016/j.cmet.2012.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macotela Y., Emanuelli B., Mori M. A., Gesta S., Schulz T. J., Tseng Y.-H. and Kahn C. R. (2012). Intrinsic differences in adipocyte precursor cells from different white fat depots. Diabetes 61, 1691-1699. 10.2337/db11-1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson L. E. and Soriano P. (2009). Increased PDGFRα activation disrupts connective tissue development and drives systemic fibrosis. Dev. Cell 16, 303-313. 10.1016/j.devcel.2008.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodeheffer M. S., Birsoy K. and Friedman J. M. (2008). Identification of white adipocyte progenitor cells in vivo. Cell 135, 240-249. 10.1016/j.cell.2008.09.036 [DOI] [PubMed] [Google Scholar]
- Rosen E. D. and MacDougald O. A. (2006). Adipocyte differentiation from the inside out. Nat. Rev. Mol. Cell Biol. 7, 885-896. 10.1038/nrm2066 [DOI] [PubMed] [Google Scholar]
- Rosen E. D. and Spiegelman B. M. (2014). What we talk about when we talk about fat. Cell 156, 20-44. 10.1016/j.cell.2013.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Gurmaches J. and Guertin D. A. (2014). Adipocytes arise from multiple lineages that are heterogeneously and dynamically distributed. Nat. Commun. 5, 4099 10.1038/ncomms5099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Gurmaches J., Hung C.-M., Sparks C. A., Tang Y., Li H. and Guertin D. A. (2012). PTEN loss in the Myf5 lineage redistributes body fat and reveals subsets of white adipocytes that arise from Myf5 precursors. Cell Metab. 16, 348-362. 10.1016/j.cmet.2012.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schildhaus H.-U., Cavlar T., Binot E., Büttner R., Wardelmann E. and Merkelbach-Bruse S. (2008). Inflammatory fibroid polyps harbour mutations in the platelet-derived growth factor receptor alpha (PDGFRA) gene. J. Pathol. 216, 176-182. 10.1002/path.2393 [DOI] [PubMed] [Google Scholar]
- Schulz T. J. and Tseng Y.-H. (2013). Brown adipose tissue: development, metabolism and beyond. Biochem. J. 453, 167-178. 10.1042/BJ20130457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P., Bjork B., Yang W., Kajimura S., Chin S., Kuang S., Scimè A., Devarakonda S., Conroe H. M. and Erdjument-Bromage H. (2008). PRDM16 controls a brown fat/skeletal muscle switch. Nature 454, 961-967. 10.1038/nature07182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P., Kajimura S. and Spiegelman B. M. (2009). Transcriptional control of brown adipocyte development and physiological function - of mice and men. Genes Dev. 23, 788-797. 10.1101/gad.1779209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidossis L. and Kajimura S. (2015). Brown and beige fat in humans: thermogenic adipocytes that control energy and glucose homeostasis. J. Clin. Invest. 125, 478-486. 10.1172/JCI78362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnylal S., Denton C. P., Zheng B., Keene D. R., He R., Adams H. P., VanPelt C. S., Geng Y. J., Deng J. M., Behringer R. R. et al. (2007). Postnatal induction of transforming growth factor β signaling in fibroblasts of mice recapitulates clinical, histologic, and biochemical features of scleroderma. Arthritis Rheum. 56, 334-344. 10.1002/art.22328 [DOI] [PubMed] [Google Scholar]
- Soriano P. (1994). Abnormal kidney development and hematological disorders in PDGF beta-receptor mutant mice. Genes Dev. 8, 1888-1896. 10.1101/gad.8.16.1888 [DOI] [PubMed] [Google Scholar]
- Soriano P. (1997). The PDGF alpha receptor is required for neural crest cell development and for normal patterning of the somites. Development 124, 2691-2700. [DOI] [PubMed] [Google Scholar]
- Sun K., Tordjman J., Clément K. and Scherer P. E. (2013). Fibrosis and adipose tissue dysfunction. Cell Metab. 18, 470-477. 10.1016/j.cmet.2013.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallquist M. and Kazlauskas A. (2004). PDGF signaling in cells and mice. Cytokine Growth Factor Rev. 15, 205-213. 10.1016/j.cytogfr.2004.03.003 [DOI] [PubMed] [Google Scholar]
- Trempus C. S., Morris R. J., Bortner C. D., Cotsarelis G., Faircloth R. S., Reece J. M. and Tennant R. W. (2003). Enrichment for living murine keratinocytes from the hair follicle bulge with the cell surface marker CD34. J. Invest. Dermatol. 120, 501-511. 10.1046/j.1523-1747.2003.12088.x [DOI] [PubMed] [Google Scholar]
- Vanderwinden J.-M., Rumessen J. J., De Laet M.-H., Vanderhaeghen J.-J. and Schiffmann S. N. (2000). CD34 immunoreactivity and interstitial cells of Cajal in the human and mouse gastrointestinal tract. Cell Tissue Res. 302, 145-153. 10.1007/s004410000264 [DOI] [PubMed] [Google Scholar]
- Vaziri C. and Faller D. V. (1996). Down-regulation of platelet-derived growth factor receptor expression during terminal differentiation of 3T3-L1 pre-adipocyte fibroblasts. J. Biol. Chem. 271, 13642-13648. 10.1074/jbc.271.23.13642 [DOI] [PubMed] [Google Scholar]
- Wu J., Boström P., Sparks L. M., Ye L., Choi J. H., Giang A.-H., Khandekar M., Virtanen K. A., Nuutila P., Schaart G. et al. (2012). Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell 150, 366-376. 10.1016/j.cell.2012.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Cohen P. and Spiegelman B. M. (2013). Adaptive thermogenesis in adipocytes: is beige the new brown? Genes Dev. 27, 234-250. 10.1101/gad.211649.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young P. E., Baumhueter S. and Lasky L. A. (1995). The sialomucin CD34 is expressed on hematopoietic cells and blood vessels during murine development. Blood 85, 96-105. [PubMed] [Google Scholar]