ABSTRACT
Chlamydia trachomatis infections present a major heath burden worldwide. The conventional method used to detect C. trachomatis is laborious. In the present study, a novel strategy was utilized to evaluate the impact of antimicrobial agents on the growth of C. trachomatis and its expression of ompA promoter-driven green fluorescence protein (GFP). We demonstrate that this GFP reporter system gives a robust fluorescent display of C. trachomatis growth in human cervical epithelial cells and, further, that GFP production directly correlates to changes in ompA expression following sufficient exposure to antimicrobials. Validation with azithromycin, the first-line macrolide drug used for the treatment of C. trachomatis infection, highlights the advantages of this method over the traditional method because of its simplicity and versatility. The results indicate both that ompA is highly responsive to antimicrobials targeting the transcription and translation of C. trachomatis and that there is a correlation between changing GFP levels and C. trachomatis growth. This proof-of-concept study also reveals that the ompA-GFP system can be easily adapted to rapidly assess antimicrobial effectiveness in a high-throughput format.
KEYWORDS: Chlamydia trachomatis, antimicrobial agents, green fluorescent protein, high throughput, obligate intracellular bacteria, reporter assay
INTRODUCTION
Infectious diseases caused by Chlamydia trachomatis are among the most persistent global health problems (1, 2). These include trachoma, the leading cause of infectious blindness in developing countries. C. trachomatis is also the most frequent cause of urogenital tract infections reported in developed countries. In women in particular, asymptomatic infections are prevalent, and if left untreated, pelvic inflammatory disease and infertility may occur. C. trachomatis, an obligate intracellular bacterium, has a characteristic developmental cycle that involves the interconversion between infectious elementary bodies (EBs) and replicating reticulate bodies (RBs) (3, 4). Both the membrane-bound inclusion in which C. trachomatis replicates and the formidable permeability barrier of the bacterial envelope provide challenges for the targeting and elimination of the bacteria in patients. Currently, there is no effective vaccine. Treatment of C. trachomatis relies solely on cell-penetrating antimicrobial agents, such as azithromycin and doxycycline, of which a relatively long course is required (2, 5). Despite enhanced screening and treatment regimens, the rate of urogenital C. trachomatis infection has not decreased. There is an indication that cure rates might be declining in patients with chronic or recurrent C. trachomatis infections (5, 6). Moreover, the rate of failure of treatment with azithromycin was reported to be 12.8% among heterosexual men with nongonococcal urethritis (7) and as high as 22% among individuals with rectal chlamydia infection (8). It is unclear what causes treatment failure. Although true antimicrobial resistance by genetic mutation has not emerged in human strains, C. trachomatis has the ability to build up a heterotypic resistance, which refers to the replication of a heterogeneous population of resistant and susceptible bacteria from a subculture of a single resistant organism without genetic changes (5, 7–10). The potential role of such nongenetic resistance in the failure of treatment of C. trachomatis infection has been debated and has increasingly drawn a great deal of attention (5, 7–10).
Considering the huge global burden of disease, it is critical to better understand the mechanism underlying C. trachomatis survival against antimicrobials. This highlights the important need for a reliable, robust method for assaying chlamydial susceptibility to antimicrobials. Immunofluorescence assays, which rely on the use of fluorescent dyes linked to primary or secondary antibodies to identify the presence of bacterial antigens, are often used to detect C. trachomatis in fixed cells for laboratory diagnosis and research (11). These assays have proven to be extremely useful in verifying the chlamydial inclusions enumerated using microscopy. However, they are laborious and impractical for providing a dynamic picture of bacterial behavior during infection. Now, emerging tools have made it possible to study important facets of the physiology of Chlamydia spp. in situ. Among these important advances are the use of genetic labeling of C. trachomatis with fluorescent proteins for a variety of purposes, including the tracking of gene insertions made by allelic exchange, deletions, or complementation and quantifying C. trachomatis growth as well as the localization of proteins in live cells (12–15).
Here, we applied a recently developed reporter assay to probe the effects of antimicrobials on the intracellular growth of C. trachomatis in both low- and high-throughput formats. This assay uses the lymphogranuloma venereum (LGV) strain of C. trachomatis L2 that expresses ompA promoter-driven green fluorescence protein (GFP) (16). Approaches that use this reporter and other techniques have been utilized to revisit the transcriptional control of ompA encoding the major outer membrane protein (MOMP). Because it is a predominant surface-exposed protein in both EBs and RBs (17–20), MOMP plays a key role in the maintenance of bacterial integration, mediates initial contact with the host cell (21), and is a promising candidate for vaccine development (22, 23). Three antimicrobials used for treating infections caused by intracellular bacterial pathogens, azithromycin, rifampin, and chloramphenicol, were examined in this study. Due to the relatively well-defined transformation system (13), LGV L2-infected HeLa 229 human cervical epithelial cells were used as a model. Although the LGV L2 strain has a more invasive nature than strains of the common genital serovars D to K, it shares a common developmental cycle with other members of the genus Chlamydia (4). The results indicate that chlamydial ompA is highly responsive to the antimicrobials tested, and this response is linked to GFP dynamics and C. trachomatis growth. Thus, PompA-GFP has considerable potential as a reporter for tracking the changes in bacterial growth induced by antimicrobials. This proof-of-principle study also indicates that this reporter system can be easily adaptable, allowing high-throughput assays for the detection of infection and the rapid evaluation of antimicrobials.
RESULTS AND DISCUSSION
Comparing time courses of GFP-based reporter systems during C. trachomatis infection.
C. trachomatis LGV L2 takes about 48 h to complete a single developmental cycle in human cervical epithelial cells. A successful strategy to overcome issues in recognizing diverse chlamydial forms, including EBs and RBs, involves long-term, robust labeling of the pathogen for easy detection. The L2/PompA-GFP system, which contains ompA promoter-driven GFP, may be best suited for this purpose, as it is constitutively expressed throughout the chlamydial developmental cycle (16). The traceability of the L2/PompA-GFP system was compared with that of the L2/PincD-GFP system (24), which was previously used to track infection (15). Following infection of HeLa 229 cells with C. trachomatis, microscopy and flow cytometry were used to monitor the time course of GFP expression from 8 to 24 h postinfection (hpi) when RBs replicated and/or asynchronously began to transit back to EBs.
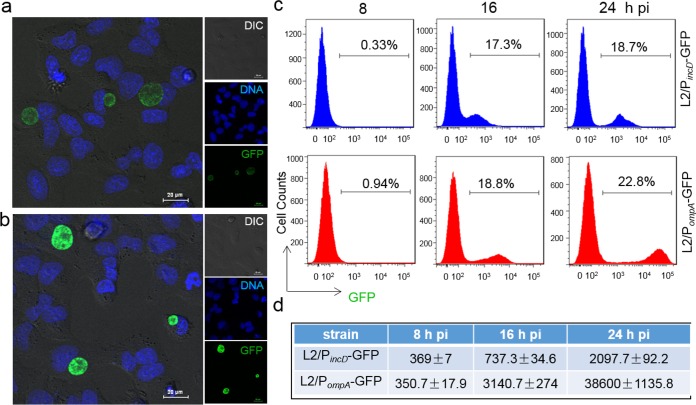
We observed that L2/PompA-GFP formed inclusions displaying fluorescence signals stronger than those of the inclusions formed by L2/PincD-GFP in all cells (Fig. 1a and b). Flow cytometry data indicated that the GFP-positive infected cell population was readily separated from the uninfected cell population at 16 hpi for L2/PompA-GFP-infected cells. At the same time, less separation between infected and uninfected cells was seen for L2/PincD-GFP-infected cells (Fig. 1c and d). The average mean fluorescent intensity (MFI) of the total population of cells analyzed was 4.2- and 18.4-fold higher for L2/PompA-GFP than for L2/PincD-GFP at 16 and 24 hpi, respectively, indicating the higher potency of PompA-GFP in comparison to that of PincD-GFP. These results confirm the effectiveness of PompA-GFP for the labeling of C. trachomatis during infection in HeLa 229 cells. Thus, we chose to investigate L2/PompA-GFP further.
FIG 1.
Performance of C. trachomatis GFP reporter systems. (a and b) Fluorescence microscopy detects GFP-expressing inclusions in HeLa 229 cells infected with L2/PincD-GFP (a) or L2/PompA-GFP (b). Live cells were counterstained for DNA (Hoechst 33342) before visualization. Images were taken at 24 hpi under the same exposure condition for L2/PincD-GFP and L2/PompA-GFP. DIC, differential inference contrast. (c) Quantitative analysis of GFP levels using flow cytometry. Cells (30,000) in each sample were analyzed at the time points indicated. The percentage of GFP-positive cells in each of the samples is shown. (d) MFI obtained from the same samples used in the assay whose results are shown in panel c. Data are representative of those from three independent experiment performed in triplicate for each group. All values were analyzed by FlowJo and GraphPad Prism software.
PompA-GFP is highly responsive to antimicrobials targeting essential processes of C. trachomatis.
Previous studies have reported that Chlamydia spp. are able to adaptively respond to various stressors, including antimicrobials, by rapidly reprogramming their macromolecule synthesis and remodeling their envelope (25–27). We sought to address whether or not PompA-GFP as a reporter can realistically reflect changes in ompA expression. Therefore, the levels of ompA and PompA-GFP expression as well as the morphology of chlamydial inclusions were monitored in the presence of antimicrobials. PompA-GFP contains the ompA P3 promoter (16) and the region of translation initiation from tuf encoding translation elongation factor (EF) Tu (28), so it may be useful in exploring the changes in bacterial growth induced by antimicrobials targeting the processes of bacterial transcription and translation.
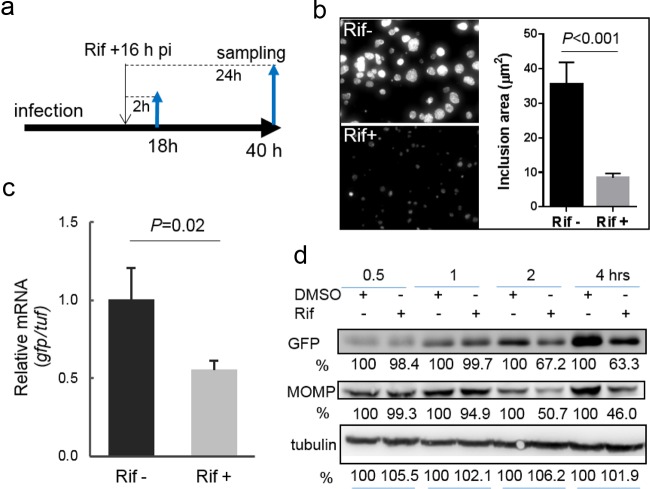
To test this hypothesis, L2/PompA-GFP-infected cells were first exposed to a dose of rifampin (50 ng/ml) administered at 16 hpi, the point at which C. trachomatis is at the exponential phase of its developmental cycle (Fig. 2a). Rifampin exerts its antibacterial action by targeting the β subunit of bacterial RNA polymerase and has been proven to inhibit growth and de novo mRNA synthesis in C. trachomatis (29). GFP signals and chlamydial inclusions were monitored by microscopy. Real-time quantitative reverse transcriptase PCR (qRT-PCR) was used to measure changes in gfp mRNA levels compared with the levels of mRNA for the tuf gene. Exposure to rifampin for 24 h starting at 16 hpi resulted in dramatic decreases in the GFP signal and the inclusion size (Fig. 2b), indicating bacterial growth retardation. In agreement with these findings, gfp mRNA was completely eliminated upon exposure to rifampin for 24 h. Short-course exposure to rifampin for 2 h also resulted in the downregulation of gfp (Fig. 2c). Notably, in contrast to the relatively stable level of tuf mRNA in rifampin-exposed cells, there was an approximately 50% decrease in the gfp mRNA level, suggesting that the ompA P3 transcript is highly sensitive to rifampin. More importantly, the decrease in the amount of the gfp transcript correlated with decreased levels of MOMP and GFP after exposure to rifampin for ≥2 h (Fig. 2d), indicating that the changes in the level of ompA expression and the changes in GFP levels were linked.
FIG 2.
Changes in chlamydial growth and GFP expression induced by rifampin (Rif). (a to c) Experimental schematic for the time of rifampin addition (a) and sample testing by imaging (b) and qRT-PCR analysis (c). (b) Visualization of the inclusion morphology in the absence or presence of rifampin for 24 h starting at 16 hpi. Images were taken at 40 hpi under the same conditions in the absence and presence of rifampin. The graph on the right shows the average size, determined from five different fields using ImageJ software, of inclusions exposed to rifampin relative to that of inclusions that lacked rifampin exposure. The P value was obtained by an unpaired Student t test. (c) Exposure to rifampin for 2 h resulted in a significant decrease in gfp mRNA levels in C. trachomatis, as measured by qRT-PCR (see Materials and Methods). Shown is the ratio of the level of gfp mRNA to the level of tuf mRNA. The P value was obtained by an unpaired Student t test. (d) Quantifying the levels of MOMP and GFP by immunoblotting. Lysates of cells exposed to rifampin or DMSO (control) were paired. GFP or MOMP was probed with specific antibodies, and their levels were quantified by densitometry using Quantity One software. Host cell tubulin was used as a loading control for protein amounts. The values are reported as a percentage by normalization to the corresponding protein intensity from the DMSO control harvested at the same times. Data are representative of those from three independent experiments. In all cases, rifampin was used at a concentration of 50 ng/ml.
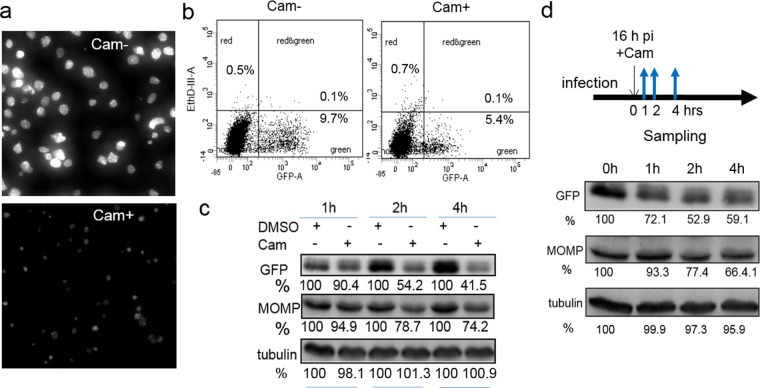
Next, we examined the effects of chloramphenicol, an inhibitor of prokaryotic protein synthesis. Because chloramphenicol can inhibit mitochondrial function and, therefore, cell viability (30), we administered a 60-μg/ml dose of chloramphenicol at 16 hpi. Note that this is less than one-third of the amount previously used for protein synthesis inhibition assays with C. trachomatis (31). The cell morphology and populations were analyzed by microscopy and flow cytometry. To exclude potential chloramphenicol-associated toxic effects, cells were incubated with a cell death probe, ethidium homodimer III (EthD-III), and were then subjected to flow cytometry analysis. EthD-III enters the cells and binds to DNA only when cells die and the plasma membrane becomes disrupted. We observed that the chlamydial growth was dramatically inhibited by exposure to chloramphenicol for 6 and 24 h starting at 16 hpi compared with the growth of the unexposed control (Fig. 3a and b). Consistent with these observations, GFP levels, analyzed by immunoblotting, decreased after chloramphenicol exposure compared to those for the corresponding control at the same time point (Fig. 3c). The levels of MOMP in the same samples also decreased, indicating overall bacterial translational inhibition by chloramphenicol. We verified that this effect was not due to the cytotoxicity of chloramphenicol, because the number of EthD-III-stained dead cells in populations that had been exposed and populations that had not been exposed to chloramphenicol was almost unchanged (Fig. 3b).
FIG 3.
Chloramphenicol (Cam) affects chlamydial growth and GFP expression. (a) Live cell image of L2/PompA-GFP-infected cells that were and were not exposed to chloramphenicol starting at 16 hpi for 24 h. Images were taken at 40 hpi under the same conditions for cells that were and were not exposed to chloramphenicol. (b) Simultaneous quantification of the GFP-positive population and EthD-III-stained dead cells. Cells were harvested after exposure to chloramphenicol (Cam+) for 6 hpi and subjected to flow cytometry. The corresponding cells that lacked exposure to chloramphenicol (Cam−) were harvested at the same time and used as controls. The distribution of cells in the green channel (GFP) and red channel (EthD-III-A) is shown. The percentages of cells displaying no fluorescence (uninfected bystander cells), GFP-positive cells, and/or EthD-III-positive cells relative to the total number of cells initially analyzed are indicated. (c) Measurement of MOMP and GFP levels by immunoblotting. Lysates of L2/PompA-GFP-infected cells exposed to chloramphenicol or DMSO (control) harvested at the same times were compared. GFP, MOMP, or tubulin (loading control) was revealed with specific antibodies. (d) Monitoring of the fate of GFP in C. trachomatis by immunoblotting. Protein bands were quantified by densitometry using Quantity One software. The values are reported as a percentage by normalization of the corresponding protein intensity to that for the DMSO control. Data are representative of those from three independent experiments. In all cases, chloramphenicol was used at a concentration of 60 ng/ml.
The GFP version used in this study has been reported to be quite stable once it is produced (13). To track the fate of GFP, C. trachomatis-infected cells were harvested at 16 hpi prior to chloramphenicol addition and then at 1, 2, or 4 h after the addition of chloramphenicol for immunoblot analysis (Fig. 3d). We found that GFP was ∼50% less intense after chloramphenicol exposure over a period of 2 to 4 h compared to its intensity in the corresponding cells harvested prior to chloramphenicol addition. These data suggest that the half-life of GFP is on the order of hours in C. trachomatis. The gradual decrease in the amount of GFP may reflect the time required for GFP degradation in C. trachomatis, which has a doubling time of 2.5 to 4 h (32, 33). This finding clearly differs from that for many other fast-growing bacteria, such as Escherichia coli, whose replication rate in the laboratory is 15 to 20 min. It is possible that changes in GFP levels induced by short-term antimicrobial exposure may be masked by stable GFP. Nevertheless, in the case of drug exposure over a sufficient period of time, GFP levels decreased, mirroring the growth inhibition of C. trachomatis. These results indicate that the changes in GFP levels are a reflection of the C. trachomatis growth retardation induced by antimicrobials and that the use of the PompA-GFP reporter is well suited for measuring antichlamydial activity.
Impacts of azithromycin on dynamics of C. trachomatis growth and GFP levels.
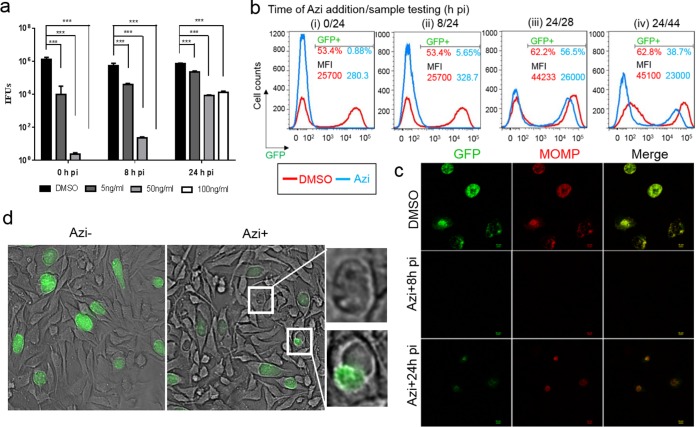
We sought to assess the azithromycin-induced changes in chlamydial growth dynamics at both the single-cell and population levels. Azithromycin, which targets the bacterial 50S ribosome, is often prescribed to treat respiratory infections, ear infections, and sexually transmitted diseases, including C. trachomatis infections (2). Although evidence indicates that plasmid transformation of C. trachomatis does not impair its ability to elicit host cell cytokine production (34), transformed strains display a slight delay of the developmental cycle compared to untransformed strain L2/434/Bu (13, 16). Therefore, we first determined the dose and time response of L2/PompA-GFP to azithromycin by assessing the yield of EBs in HeLa 229 cells using an inclusion-forming unit (IFU) assay (Fig. 4a). The MIC, the lowest azithromycin concentration that inhibited inclusion formation by L2/PompA-GFP for 48 h posttreatment, was identified to be 50 ng/ml. This value is similar to that for L2/434/Bu, suggesting that the introduction of PompA-GFP into C. trachomatis did not significantly influence susceptibility to azithromycin.
FIG 4.
Changes in C. trachomatis growth and the GFP levels induced by azithromycin (Azi). (a) Dose- and time-dependent response of C. trachomatis to azithromycin. HeLa 229 cells infected with C. trachomatis L2/PompA-GFP were exposed to increasing concentrations of azithromycin (0, 5, 50, and 100 ng/ml) starting from 0, 8, or 24 hpi. Cells were harvested at 40 hpi and subjected to the endpoint IFU assay to determine EB yields using immunofluorescence assays with an antibody specific to L2 MOMP. The P value was obtained by one-way ANOVA with a Bonferroni posttest. ***, P < 0.001. (b) Flow cytometry analysis of azithromycin's effect. HeLa 229 cells infected with L2/PompA-GFP were exposed to azithromycin at 0, 8, or 24 hpi and collected at 24, 28, or 44 hpi, as indicated, for flow cytometry. The percentage of the GFP-positive population and the MFI value from DMSO-treated control cells are shown in red, and those values for azithromycin-exposed cells are presented in blue. These data represent averages from three separate experiments, in which triplicates were tested. (c) Results of immunofluorescence assay with fixed cells unexposed or exposed to azithromycin. The L2 MOMP antibody was used to visualize C. trachomatis. Note that there was a correlation between MOMP (red) and GFP (green). (d) Visualization of the effect of azithromycin by live cell imaging. Images were taken at 24 hpi prior to azithromycin addition (left) and then at 42 hpi (middle) after the addition of azithromycin for 18 hpi under the same exposure conditions. Enlarged abnormal inclusions that appeared empty and displayed a lower and uneven GFP signal are shown in the right panels. In all cases with the exception of the assay whose results are presented in panel a, azithromycin was used at a concentration of 50 ng/ml.
Next, C. trachomatis growth was monitored using microscopy and flow cytometry after azithromycin (50 ng/ml) was added at 0, 8, or 24 hpi. In the control experiments, dimethyl sulfoxide (DMSO) was used. We observed that a significant decrease in the GFP-positive cell population was induced by the addition of azithromycin at 0 or 8 hpi (early time points) when the cells were evaluated at 24 hpi; only 0.88% and 5.6% of azithromycin-exposed cells, respectively, were GFP positive, whereas 62.2% of the DMSO-exposed control cells were GFP positive (Fig. 4bi and ii). MFI values were also much lower for the azithromycin-exposed cells than for the control cells, indicating significant inhibition of bacterial growth at early time points. In contrast, exposure to azithromycin starting at 24 hpi appeared to be less effective than exposure at the early points (Fig. 4biii and iv). Upon 4 h of exposure to azithromycin starting at 24 hpi, the proportion of GFP-positive cells decreased from 62.2% to 56.2% when the cells were observed at 28 hpi (Fig. 4biii). In parallel, there was an approximately 2/5 decrease in the MFI value (Fig. 4biii). A 20-h exposure to azithromycin caused a larger fraction of the cells to become GFP negative and a reduction of the MFI value by nearly 1/2 when the cells were observed at 48 hpi (Fig. 4biv). These data were verified by immunofluorescence assays using MOMP antibody staining (Fig. 4c). The profound inhibitory effect of azithromycin on C. trachomatis growth is in line with its ability to impede RB replication, block RB-to-EB differentiation, and prevent subsequent infection by modulating an as-yet-unidentified host cell factor (11, 35). Live cell imaging performed at 42 hpi showed a noticeably considerable variation in the inclusion morphology induced by azithromycin addition starting at 24 hpi (Fig. 4d). Some cells were normal in size and produced a strong GFP signal; others were morphologically aberrant with a weakened GFP signal and fewer GFP-expressing chlamydial organisms. These results suggest that C. trachomatis exists in diverse populations. Such variations were not easily detected in fixed cells by immunofluorescence assay (Fig. 4c). The heterogeneity of the inclusions observed may reflect stage-specific differences in chlamydial physiology and asynchronous growth at the late stage, when RBs, EBs, and intermediate bodies accumulated. Although both EBs and RBs have a metabolic capacity (36, 37), rapidly replicating RBs are more responsive to antimicrobials than EBs (38, 39). Alternatively, a small number of bacteria may have adapted to a viable, nondividing persistent form (9, 26, 39, 40) that was less susceptible to antimicrobials in vitro. Suchland et al. (11) reported that the survival rate of Chlamydia exposed to an effective antimicrobial is approximately one bacterium per 5,000 infected cells. Collectively, these results underscore the growth-stage-dependent susceptibility of C. trachomatis to azithromycin and further verify that dynamic GFP profile correlates with chlamydial growth.
High-throughput assays verified the impacts of antimicrobials on chlamydial growth.
Given its robust performance, we considered the potential use of PompA-GFP in a high-throughput infection assay. As a proof of principle, we assessed whether PompA-GFP could be used to set up experiments to probe antimicrobial effects with the aid of an automatic fluorimeter in a 96-well plate format. HeLa 229 cells were inoculated with a series of L2/PompA-GFP dilutions, permitting infectivity for the host cell to vary over time. The optimized concentrations of azithromycin (50 ng/ml), rifampin (50 ng/ml), and chloramphenicol (60 μg/ml) were added to different wells at 0, 8, or 20 hpi. With these dosages, changes in GFP levels should reflect the antimicrobial effect instead of cytotoxicity. Mock-infected cells were used as autofluorescence baseline controls. The inclusion morphology was monitored by microscopy. The GFP readout was measured by a plate reader.
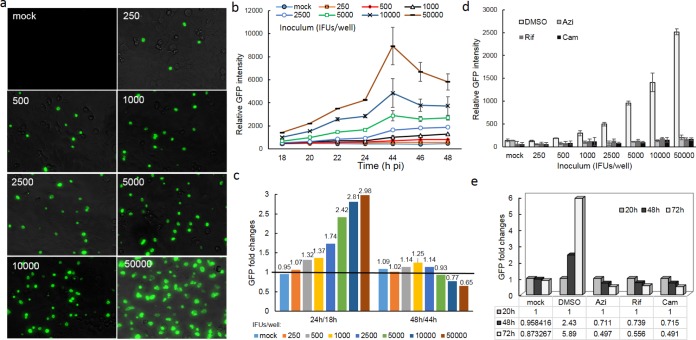
As shown in Fig. 5a, we were able to achieve 5% to 90% C. trachomatis infection in HeLa 229 cells with different-sized inocula. These levels of infection did not cause detectable host cell damage within the 72 h of observation. The GFP signal was low prior to 16 hpi, but in most cases, the fluorescence readout was augmented after 18 hpi as a result of the multiplication and accumulation of C. trachomatis (Fig. 5b). Changes in fluorescence intensity depended on the infectivity and time intervals of detection. The highly infected cells (infected with inocula of 2,500 to 50,000 inclusion-forming units [IFU]/ml) displayed a large increase in GFP intensity (≥1.5-fold) during exponential growth from 18 to 24 hpi (Fig. 5c), a peak GFP intensity at 44 hpi, and a slight or no increase in GFP intensity (0.69- to 1.25-fold) from 44 to 48 hpi. In contrast, the cells infected with a lower inoculum of 250 to 1,000 IFU/well exhibited a constitutive, small increase in fluorescence intensity (<1.5-fold) from 18 to 48 hpi. The disparity in GFP profiling detected between highly and weakly infected cells can be explained by the difference in the amounts of chlamydial organisms that were produced and that accumulated in the inclusions. Indeed, the inclusions tended to be large and contained more chlamydial particles in highly infected cells (Fig. 5a), likely due to the invasion of multiple EBs and homotypic inclusion fusions within a single host cell. Additionally, it was relatively easy to observe the formation of new, small inclusions at 48 hpi and later. In contrast, there were small inclusions in cells infected with a low inoculum. These data support the previous findings that the degree of host infectivity affects the developmental cycle and outcome of C. trachomatis infection. Because the fluorescence signal remained unchanged as a factor of time in mock-infected cells, cellular autofluorescence is unlikely to have a major influence on Chlamydia-derived GFP readouts under our test conditions.
FIG 5.
Fluorimeter-based analysis of C. trachomatis growth and GFP in a 96-well microplate. (a) Microscopic analysis of the progression of infection in HeLa 229 cells with various EB inocula (the inoculum [given as the number of IFU per well] is indicated at the top left of each panel). Live cells were photographed at 20 hpi in the absence of antimicrobials. (b) Tracing of the GFP fluorescence profile with time using a plate reader. Data represent the mean ± standard deviation (SD). (c) Fold changes in fluorescence readouts from 18 to 48 hpi in cells with different inocula. The ratio of GFP intensity at 24 hpi to that at 18 hpi or the ratio of the GFP intensity at 48 hpi to that at 44 hpi is shown. The black line indicates a ratio of 1. (d) The inhibitory effects of azithromycin, chloramphenicol, or rifampin on chlamydial growth correlates with the GFP profile. HeLa 229 cells inoculated with increasing numbers of IFU were exposed to antimicrobials at 8 hpi, and the amount of GFP was measured after antimicrobial exposure for 20 h. Azithromycin (50 ng/ml), rifampin (50 ng/ml), and chloramphenicol (60 μg/ml) were used. (e) Monitoring the fate of GFP. Cells inoculated with 10,000 IFU/ml were exposed to rifampin, chloramphenicol, or azithromycin starting at 20 hpi. Fluorescence was measured at 48 and 72 hpi (or 28 and 52 h after drug exposure) using a plate reader. Mock-infected cells and infected cells exposed to DMSO were used as controls. The ratios of the fluorescence intensity measured at 48 or 72 hpi to that measured at 20 hpi, reflecting the fold change in the amount of GFP, are shown. All graphs indicate the combined average from at least three independent experiments with triplicates in each experiment.
As expected, exposure to azithromycin, rifampin, or chloramphenicol starting from 8 hpi for ≥14 h resulted in dramatic decreases in GFP intensity (Fig. 5d), in agreement with the bacterial growth inhibition shown in Fig. 2 to 4. There was no observed difference in the inhibitory effect when the inoculum sizes of C. trachomatis varied over a range of from 250 to 50,000 IFU/ml. To examine the outcome for GFP, fluorescence intensity was monitored for up to 72 h after antimicrobial exposure starting at 20 hpi. We observed that the GFP signal decreased with time; approximately 30% or 50% of the decrease in the GFP level compared to that at the time that the drug was added was induced by drug exposure for 28 or 52 h. Meanwhile, at 72 hpi GFP levels continued to increase up to ∼6-fold in cells that were not exposed to the drug. Autofluorescence was unchanged in mock-infected cells. Therefore, differences in GFP readouts between drug-exposed and unexposed cells became more obvious with time. More detailed studies of the relationship between fluorescence levels and bacterial growth status are necessary. Nevertheless, these results indicate that genetically encoded GFP in C. trachomatis can produce a strong fluorescence signal that can be measured in <1 min per plate using ordinary fluorescence plate readers or fluorimetry in a 96-well format. All three drugs tested effectively inhibited C. trachomatis growth, on the basis of the decreases in GFP levels, in agreement with the results of non-high-throughput studies (Fig. 2 to 4).
Conclusion.
This study describes a simple, robust GFP reporter assay suitable for the rapid detection of the antichlamydial efficacy of antimicrobials in vitro. This method is based on the well-known dependence of C. trachomatis growth on the expression and proper function of ompA (21, 40). Treatment with three antimicrobials of different classes resulted in considerably decreased levels of GFP in a large fraction of cells, correlating to the killing or growth arrest of chlamydial organisms. We consider this cell-based method to have significant potential to be an efficient tool in screening new antimicrobial agents and measuring neutralizing antibody. The versatility of PompA-GFP also opens numerous possibilities to monitor the dynamic behavior of C. trachomatis, in particular, in scenarios of persistence induced by stressors and reactivation upon removal of the stressors, as well as the host adaptation of C. trachomatis and its related heterotypic resistance (5, 7–10). Such studies will provide new insights into the intracellular survival mechanism of C. trachomatis contributing to C. trachomatis pathogenesis.
Numerous studies have provided examples in which GFP is broadly used to monitor the antimicrobial susceptibility of microbial organisms (41–43). The use of GFP in antimicrobial susceptibility testing is advantageous because it can be easily detected and quantitated using advanced automated techniques, including fluorescence microscopy, flow cytometry, and fluorimetry. We demonstrate the suitability of PompA-GFP as a reporter to assay for antichlamydial efficacy in both low- and high-throughput formats. Due to the inherent stability of the GFP used, GFP alone cannot always provide sensitive readouts of the influence of short-term exposure to drugs. A dynamic GFP profile, as a surrogate for the growth of the bacteria, can be significant and should be accounted for in the studies. Despite the similar trends of the change in fluorescence and the growth pattern of C. trachomatis, the changes in GFP levels do not fully reflect the variety of EBs and RBs, as GFP is present in both. Moreover, our assay provides information only on the in vitro behavior of bacteria; bacteria growing in vivo face much more complex conditions, such as host innate and adaptive immunity, as well as the influence of urogenital tract-associated microbial communities (44).
Despite these considerations and limitations, the GFP reporter assay provides a unique tool with which the dynamic response of C. trachomatis to antimicrobial agents can be rapidly monitored in live cells. When this assay is incorporated with automated detection, it may permit highly parallel, unbiased research to address important biological questions that are otherwise not amenable to study using conventional methods. For example, this assay can facilitate a direct survey of the survival of C. trachomatis exposed to various stressors, including antimicrobials and gamma interferon, the major antichlamydial cytokine. It has been challenging to interpret the antimicrobial susceptibility of C. trachomatis, as determination of the MIC is influenced by many factors, including the strains or serovars tested, the rate of C. trachomatis infection in host cells, the antimicrobial concentration (effective but noncytotoxic), the time of addition, the exposure interval, and the type of cell culture model (11, 39, 45–47). Using these parameters, several studies from different laboratories have shown high degrees of quantitative and qualitative differences in the effects of antimicrobials on growing bacteria as determined by conventional assays (11, 45–47). The use of the GFP reporter in studies of C. trachomatis and the proper interpretation of the data from C. trachomatis studies will require that all these important conditions be satisfied. Careful evaluation of the test parameters and further standardization of test procedures using other antimicrobials, such as the commonly used doxycycline, are warranted. Additionally, this assay can be extended to the non-LGV strains which cause most of the common syndromes associated with C. trachomatis infection.
MATERIALS AND METHODS
Reagents, antimicrobial agents, and antibodies.
Ampicillin, rifampin, azithromycin, chloramphenicol, cycloheximide, and dimethyl sulfoxide (DMSO) were purchased from Sigma-Aldrich (St. Louis, MO, USA). The primary antibodies used in this study included a mouse monoclonal antibody (L2I-45) specific to the LGV L2 MOMP (48), kindly provided by Yun-xun Zhang (Boston University); a rabbit polyclonal GFP antibody (catalog number AG279) from Beyotime (Beijing, China); and a mouse monoclonal antibody to tubulin (catalog number T5168) from Sigma-Aldrich (St. Louis, MO, USA). The secondary antibodies used were Alexa Fluor 568-conjugated goat anti-mouse IgG (catalog number A11004) from Invitrogen (Carlsbad, CA, USA) and horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (catalog number ASS1009) or anti-mouse IgG (catalog number ASS1007) from Abgent (Wuxi, China).
Cell culture and C. trachomatis infection.
HeLa 229 human cervical cancer cells (CCL2; ATCC) were cultured in Dulbecco's modified Eagle's medium (DMEM) containing a high level of glucose (4.5 g/liter) supplemented with 10% fetal bovine serum (Sigma-Aldrich) and l-glutamine (2 mM) (DMEM-10) at 37°C in a humidified incubator with 5% CO2. C. trachomatis strains L2/PompA-GFP and L2/PincD-GFP were generated by transforming plasmid-free C. trachomatis L2/25667R cells with the shuttle plasmid pBOMB-P3 (16) or p2TK2-SW2 IncDProm-RSGFP-IncDTerm (called pPincD-GFP in this study) (24) (kindly provided by Isabelle Derré, University of Virginia), respectively, using the method described previously (16). pBOMB-P3 carried ompA promoter P3-driven GFP (16) and pPincD-GFP carried incD promoter-driven GFP (24). The transformed strain was used to infect HeLa 229 cells, followed by culturing in DMEM-10 containing ampicillin (5 μg/ml) and cycloheximide (1 μg/ml) for 48 h. EBs were then isolated from the other chlamydial forms by density gradient purification as described previously (49), resuspended in sucrose-phosphate-glutamic acid (SPG) buffer (10 mM sodium phosphate [8 mM Na2HPO4, 2 mM NaH2PO4], 220 mM sucrose, 0.5 mM l-glutamic acid), and stored at −80°C until use. The C. trachomatis stocks tested negative for mycoplasma by PCR (Phoenix Research).
Monolayers of HeLa 229 cells in culture plates were infected with C. trachomatis expressing GFP. Infections were synchronized by centrifugation at 1,600 × g for 40 min at 37°C. The medium was changed immediately postinfection to ensure the removal of nonphagocytosed bacteria. C. trachomatis-infected cells were cultured in DMEM-10 containing ampicillin (5 μg/ml) and cycloheximide (0.5 μg/ml) with or without the addition of the test antimicrobial for various time periods, as indicated above for each experimental result. The infectious EB progeny in the culture was evaluated by enumeration of the IFU in 1 ml of cell lysate.
Endpoint inclusion-forming unit assay.
C. trachomatis-infected cells were harvested in sterile water at 44 hpi. Serial dilutions of the harvested cultures were subcultured using HeLa 229 cells. The cells were processed at 44 hpi for indirect immunofluorescence assay as described below. The quantity of infectious EBs was determined by enumeration of the IFU in triplicate wells using fluorescence microscopy. The total numbers are presented as the number of IFU per milliliter.
Indirect immunofluorescence assays.
C. trachomatis-infected HeLa 229 cells were fixed with 2% paraformaldehyde dissolved in phosphate-buffered saline (pH 7.4) for 30 min at room temperature and permeabilized with 0.1% Triton X-100 (Sigma) for an additional 30 min. After the cells were washed and blocked with 3% bovine serum albumin, they were incubated with MOMP antibody (L2I-45) (48) overnight at 4°C, followed by incubation with Alexa Fluor 568-conjugated secondary antibody (Molecular Probes) for 45 min at 37°C. The cells were visualized and photographed using a microscope (see below).
Acquiring and analyzing images using phase-contrast, fluorescence, and confocal microscopy.
A Nikon laser confocal microscope (A1R si) was used to capture images of live or fixed cells. The images were analyzed by use of confocal z-section series and then merged into loss-less montage images using NIS-Elements BR software, version 4.11. In some experiments, the cells were photographed with an inverted fluorescence microscope (Zeiss Axio Observer D1). Images were processed using AxioVision software, version 4.8. ImageJ software, version 1.40g (50), was downloaded from the National Institutes of Health website (the W. S. Rasband website) and used to take measurements of the images.
Flow cytometry.
Cells in six-well plates were detached by trypsinization using a trypsin (0.025%)-EDTA (0.001%) solution (Sigma) at different times, as indicated in the Results and Discussion. Unfixed cells or cells that were fixed with 2% paraformaldehyde at room temperature for 20 min were subjected to flow cytometry analysis using a FACSCanto II flow cytometer (BD Biosciences). Flow cytometry data were recorded for at least 20,000 cells per sample. Mock-infected cells were used as a blank control. Data were analyzed, using FlowJo software, version 7.6 (TreeStar Inc.), for both the percentage of GFP-positive cell populations and the mean fluorescent intensity (MFI) of the total population of cells.
Real-time quantitative reverse transcriptase PCR (qRT-PCR).
Genomic DNA and total RNA were isolated from the same cells using a ZR Duet-DNA/RNA kit (Zymo) according to the manufacturer's instructions. DNase treatment was performed in a column to remove residual DNA. A total of 1 μg of total RNA was reverse transcribed into cDNA, and the genes of interest were amplified using a VeriQuest Fast SYBR green quantitative PCR (qPCR) kit (USB) with the following primer pairs: gfp-rtF (5′-GTATACATCATGGCAGACAAACAAA)/gfp-rtR (5′-TGTTGATAATGGTCTGCTAGTTGAA), tuf-rtF (5′-GTATACATCATGGCAGACAAACAAA)/tuf-rtR (5′-GTAACTCTGCCTGAGGGAATTGA), and 16SF (5′-CGCCAACACTGGGACTGAGA)/16SR (5′-GGCGTCGCTTCGTCAGACTT). The PCR cycle conditions were as follows: 50°C for 2 min, 95°C for 5 min, 95°C for 3 s, and 60°C for 30 s. The last two steps were repeated for 35 to 45 cycles, and fluorescence was detected at the end of each cycle. Transcript levels were normalized to C. trachomatis genomic DNA levels, which were quantified by qPCR with primers specific to the 16S rRNA gene. The 2−ΔΔCT threshold cycle (CT) method was used to obtain relative transcript levels.
Immunoblot analysis.
Cells were lysed in buffer containing 10 mM Tris (pH 8.0) and 8 M urea. The protein content was determined by a bicinchoninic acid (BCA) protein assay kit (Beyotime Biotechnology, Shanghai, China). The optimal amount of protein dissolved in 1× sodium dodecyl sulfate (SDS) loading buffer was separated on a 12% SDS-polyacrylamide gel and transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore) for immunoblotting. The membrane was incubated with primary antibodies against GFP, MOMP, or tubulin, followed by incubation with the secondary antibody horseradish peroxidase-conjugated rabbit anti-mouse or goat anti-rabbit IgG. The protein bands were visualized by use of an enhanced chemiluminescence kit (Bio-Rad).
Measuring GFP intensity using a time-resolved spectrophotometer.
HeLa 229 cells grown overnight in 96-well black microplates (catalog number 655090; Greiner) were infected with C. trachomatis expressing GFP at a dose resulting in infection of 5% to 90% of cells, as indicated above. Antimicrobials were added to triplicate wells at different time points after infection (see the details provided above for each experiment). In a comparable manner, parallel control uninfected cells at the same cellular concentrations received only medium containing a corresponding amount of DMSO. Cultures were incubated in a 37°C incubator, and GFP fluorescence was measured by a plate reader (SynergyMx; BioTek) at appropriate intervals with excitation at 480 ± 20 nm and emission at 530 ± 20 nm. The relative fluorescence intensity was obtained by subtraction of the background reading for the medium from the fluorescence reading for the cells. Measurements were taken at the time points indicated above in the presence or absence of antimicrobials.
Statistical analyses.
Data analyses were performed using GraphPad Prism software (GraphPad Software, Inc.). Statistical significance was determined by two-way analysis of variance (ANOVA) or an unpaired Student t test, as indicated above. P values of <0.05 were considered statistically significant.
ACKNOWLEDGMENTS
This research is supported by grants from the National Natural Science Foundation of China (81370777), NIH/NIAID (AI093565), and the LSU School of Medicine Dean's Research Bridge Funding.
We are grateful to David Martin for critical reading of the manuscript and suggestions and Tan Bing and Constance Porretta for technical assistance.
REFERENCES
- 1.Rekart ML, Gilbert M, Meza R, Kim PH, Chang M, Money DM, Brunham RC. 2013. Chlamydia public health programs and the epidemiology of pelvic inflammatory disease and ectopic pregnancy. J Infect Dis 207:30–38. doi: 10.1093/infdis/jis644. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2015. 2015 sexually transmitted diseases treatment guidelines. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/std/tg2015/chlamydia.htm. [Google Scholar]
- 3.AbdelRahman YM, Belland RJ. 2005. The chlamydial developmental cycle. FEMS Microbiol Rev 29:949–959. doi: 10.1016/j.femsre.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Moulder JW. 1991. Interaction of chlamydiae and host cells in vitro. Microbiol Rev 55:143–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horner PJ. 2012. Azithromycin antimicrobial resistance and genital Chlamydia trachomatis infection: duration of therapy may be the key to improving efficacy. Sex Transm Infect 88:154–156. doi: 10.1136/sextrans-2011-050385. [DOI] [PubMed] [Google Scholar]
- 6.Batteiger BE, Tu W, Ofner S, Van Der Pol B, Stothard DR, Orr DP, Katz BP, Fortenberry JD. 2010. Repeated Chlamydia trachomatis genital infections in adolescent women. J Infect Dis 201:42–51. doi: 10.1086/648734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kissinger PJ, White S, Manhart LE, Schwebke J, Taylor SN, Mena L, Khosropour CM, Wilcox L, Schmidt N, Martin DH. 2016. Azithromycin treatment failure for Chlamydia trachomatis among heterosexual men with nongonococcal urethritis. Sex Transm Dis 43:599–602. doi: 10.1097/OLQ.0000000000000489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khosropour CM, Dombrowski JC, Barbee LA, Manhart LE, Golden MR. 2014. Comparing azithromycin and doxycycline for the treatment of rectal chlamydial infection: a retrospective cohort study. Sex Transm Dis 41:79–85. doi: 10.1097/OLQ.0000000000000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borel N, Leonard C, Slade J, Schoborg RV. 2016. Chlamydial antibiotic resistance and treatment failure in veterinary and human medicine. Curr Clin Microbiol Rep 3:10–18. doi: 10.1007/s40588-016-0028-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sandoz KM, Rockey DD. 2010. Antibiotic resistance in chlamydiae. Future Microbiol 5:1427–1442. doi: 10.2217/fmb.10.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suchland RJ, Geisler WM, Stamm WE. 2003. Methodologies and cell lines used for antimicrobial susceptibility testing of Chlamydia spp. Antimicrob Agents Chemother 47:636–642. doi: 10.1128/AAC.47.2.636-642.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mueller KE, Wolf K, Fields KA. 2016. Gene deletion by fluorescence-reported allelic exchange mutagenesis in Chlamydia trachomatis. mBio 7:e01817-15. doi: 10.1128/mBio.01817-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Kahane S, Cutcliffe LT, Skilton RJ, Lambden PR, Clarke IN. 2011. Development of a transformation system for Chlamydia trachomatis: restoration of glycogen biosynthesis by acquisition of a plasmid shuttle vector. PLoS Pathog 7:e1002258. doi: 10.1371/journal.ppat.1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gong S, Yang Z, Lei L, Shen L, Zhong G. 2013. Characterization of Chlamydia trachomatis plasmid-encoded open reading frames. J Bacteriol 195:3819–3826. doi: 10.1128/JB.00511-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vromman F, Laverrière M, Perrinet S, Dufour A, Subtil A. 2014. Quantitative monitoring of the Chlamydia trachomatis developmental cycle using GFP-expressing bacteria, microscopy and flow cytometry. PLoS One 9:e99197. doi: 10.1371/journal.pone.0099197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cong Y, Gao L, Zhang Y, Xian Y, Hua Z, Elaasar H, Shen L. 2016. Quantifying promoter activity during the developmental cycle of Chlamydia trachomatis. Sci Rep 6:27244. doi: 10.1038/srep27244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caldwell HD, Kromhout J, Schachter J. 1981. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect Immun 31:1161–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skipp PJS, Hughes C, McKenna T, Edwards R, Langridge J, Thomson NR, Clarke IN. 2016. Quantitative proteomics of the infectious and replicative forms of Chlamydia trachomatis. PLoS One 11:e0149011. doi: 10.1371/journal.pone.0149011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frohlich KM, Hua Z, Quayle AJ, Wang J, Lewis ME, Chou C-W, Luo M, Buckner LR, Shen L. 2014. Membrane vesicle production by Chlamydia trachomatis as an adaptive response. Front Cell Infect Microbiol 4:73. doi: 10.3389/fcimb.2014.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Berg EA, Feng X, Shen L, Smith T, Costello CE, Zhang Y-X. 2006. Identification of surface-exposed components of MOMP of Chlamydia trachomatis serovar F. Protein Sci 15:122–134. doi: 10.1110/ps.051616206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su H, Watkins N, Zhang Y-X, Caldwell HD. 1990. Chlamydia trachomatis-host cell interactions: role of the chlamydial major outer membrane protein as an adhesion. Infect Immun 58:1017–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hafner LM, Wilson DP, Timms P. 2014. Development status and future prospects for a vaccine against Chlamydia trachomatis infection. Vaccine 32:1563–1571. doi: 10.1016/j.vaccine.2013.08.020. [DOI] [PubMed] [Google Scholar]
- 23.Gottlieb SL, Deal CD, Giersing B, Rees H, Bolan G, Johnston C, Timms P, Gray-Owen SD, Jerse AE, Cameron CE, Moorthy VS, Kiarie J, Broutet N. 2016. The global roadmap for advancing development of vaccines against sexually transmitted infections: update and next steps. Vaccine 34:2939–2947. doi: 10.1016/j.vaccine.2016.03.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agaisse H, Derré I. 2013. A C. trachomatis cloning vector and the generation of C. trachomatis strains expressing fluorescent proteins under the control of a C. trachomatis promoter. PLoS One 8:e57090. doi: 10.1371/journal.pone.0057090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beatty W, Byrne G, Morrison R. 1993. Morphologic and antigenic characterization of interferon gamma-mediated persistent Chlamydia trachomatis infection in vitro. Proc Natl Acad Sci U S A 90:3998–4002. doi: 10.1073/pnas.90.9.3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peuchant O, Duvert JP, Clerc M, Raherison S, Bébéar C, Bébéar CM, de Barbeyrac B. 2011. Effects of antibiotics on Chlamydia trachomatis viability as determined by real-time quantitative PCR. J Med Microbiol 60:508–514. doi: 10.1099/jmm.0.023887-0. [DOI] [PubMed] [Google Scholar]
- 27.Ouellette SP, Hatch TP, AbdelRahman YM, Rose LA, Belland RJ, Byrne GI. 2006. Global transcriptional upregulation in the absence of increased translation in Chlamydia during IFNγ-mediated host cell tryptophan starvation. Mol Microbiol 62:1387–1401. doi: 10.1111/j.1365-2958.2006.05465.x. [DOI] [PubMed] [Google Scholar]
- 28.Shen L, Shi Y, Douglas AL, Hatch TP, O'Connell CM, Chen JM, Zhang YX. 2000. Identification and characterization of promoters regulating tuf expression in Chlamydia trachomatis serovar F. Arch Biochem Biophys 379:46–56. doi: 10.1006/abbi.2000.1854. [DOI] [PubMed] [Google Scholar]
- 29.Xia M, Suchland RJ, Carswell JA, Van Duzer J, Buxton DK, Brown K, Rothstein DM, Stamm WE. 2005. Activities of rifamycin derivatives against wild-type and rpoB mutants of Chlamydia trachomatis. Antimicrob Agents Chemother 49:3974–3976. doi: 10.1128/AAC.49.9.3974-3976.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li C-H, Cheng Y-W, Liao P-L, Yang Y-T, Kang J-J. 2010. Chloramphenicol causes mitochondrial stress, decreases ATP biosynthesis, induces matrix metalloproteinase-13 expression, and solid-tumor cell invasion. Toxicol Sci 116:140–150. doi: 10.1093/toxsci/kfq085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scidmore MA, Fischer ER, Hackstadt T. 2003. Restricted fusion of Chlamydia trachomatis vesicles with endocytic compartments during the initial stages of infection. Infect Immun 71:973–984. doi: 10.1128/IAI.71.2.973-984.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson DP, Mathews S, Wan C, Pettitt AN, McElwain DLS. 2004. Use of a quantitative gene expression assay based on micro-array techniques and a mathematical model for the investigation of chlamydial generation time. Bull Math Biol 66:523–537. doi: 10.1016/j.bulm.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 33.Lambden PR, Pickett MA, Clarke IN. 2006. The effect of penicillin on Chlamydia trachomatis DNA replication. Microbiology 152:2573–2578. doi: 10.1099/mic.0.29032-0. [DOI] [PubMed] [Google Scholar]
- 34.Hua Z, Frohlich KM, Zhang Y, Feng X, Zhang J, Shen L. 2015. Andrographolide inhibits intracellular Chlamydia trachomatis multiplication and reduces secretion of proinflammatory mediators produced by human epithelial cells. Pathog Dis 73:1–11. doi: 10.1093/femspd/ftu022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Engel JN. 1992. Azithromycin-induced block of elementary body formation in Chlamydia trachomatis. Antimicrob Agents Chemother 36:2304–2309. doi: 10.1128/AAC.36.10.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szaszák M, Steven P, Shima K, Orzekowsky-Schröder R, Hüttmann G, König IR, Solbach W, Rupp J. 2011. Fluorescence lifetime imaging unravels C. trachomatis metabolism and its crosstalk with the host cell. PLoS Pathog 7:e1002108. doi: 10.1371/journal.ppat.1002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Omsland ASJ, Nair V, Sturdevant DE, Hackstadt T. 2012. Developmental stage-specific metabolic and transcriptional activity of Chlamydia trachomatis in an axenic medium. Proc Natl Acad Sci U S A 109:19781–19785. doi: 10.1073/pnas.1212831109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wyrick PB. 2010. Chlamydia trachomatis persistence in vitro: an overview. J Infect Dis 201:S88–S95. doi: 10.1086/652394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reveneau N, Crane DD, Fischer E, Caldwell HD. 2005. Bactericidal activity of first-choice antibiotics against gamma interferon-induced persistent infection of human epithelial cells by Chlamydia trachomatis. Antimicrob Agents Chemother 49:1787–1793. doi: 10.1128/AAC.49.5.1787-1793.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beatty WL, Morrison RP, Byrne GI. 1994. Persistent chlamydiae: from cell culture to a paradigm for chlamydial pathogenesis. Microbiol Rev 58:686–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Webb JS, Barratt SR, Sabev H, Nixon M, Eastwood IM, Greenhalgh M, Handley PS, Robson GD. 2001. Green fluorescent protein as a novel indicator of antimicrobial susceptibility in Aureobasidium pullulans. Appl Environ Microbiol 67:5614–5620. doi: 10.1128/AEM.67.12.5614-5620.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoogenkamp MA, Crielaard W, Krom BP. 2015. Uses and limitations of green fluorescent protein as a viability marker in Enterococcus faecalis: an observational investigation. J Microbiol Methods 115:57–63. doi: 10.1016/j.mimet.2015.05.020. [DOI] [PubMed] [Google Scholar]
- 43.Collins LA, Torrero MN, Franzblau SG. 1998. Green fluorescent protein reporter microplate assay for high-throughput screening of compounds against Mycobacterium tuberculosis. Antimicrob Agents Chemother 42:344–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ziklo N, Huston WM, Hocking JS, Timms P. 2016. Chlamydia trachomatis genital tract infections: when host immune response and the microbiome collide. Trends Microbiol 24:750–765. doi: 10.1016/j.tim.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 45.Siewert K, Rupp J, Klinger M, Solbach W, Gieffers J. 2005. Growth cycle-dependent pharmacodynamics of antichlamydial drugs. Antimicrob Agents Chemother 49:1852–1856. doi: 10.1128/AAC.49.5.1852-1856.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shima K, Szaszák M, Solbach W, Gieffers J, Rupp J. 2011. Impact of a low-oxygen environment on the efficacy of antimicrobials against intracellular Chlamydia trachomatis. Antimicrob Agents Chemother 55:2319–2324. doi: 10.1128/AAC.01655-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mestrović T, Ljubin-Sternak S, Sviben M, Bedenić B, Vranes J, Markotić A, Skerk V. 2016. Antimicrobial sensitivity profile of Chlamydia trachomatis isolates from Croatia in McCoy cell culture system and comparison with the literature. Clin Lab 62:357–364. [DOI] [PubMed] [Google Scholar]
- 48.Baehr W, Zhang YX, Joseph T, Su H, Nano FE, Everett KD, Caldwell HD. 1988. Mapping antigenic domains expressed by Chlamydia trachomatis major outer membrane protein genes. Proc Natl Acad Sci U S A 85:4000–4004. doi: 10.1073/pnas.85.11.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frohlich K, Hua Z, Wang J, Shen L. 2012. Isolation of Chlamydia trachomatis and membrane vesicles derived from host and bacteria. J Microbiol Methods 91:222–230. doi: 10.1016/j.mimet.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]