ABSTRACT
The aim of this study was to characterize the first cases and outbreaks of OXA-48-like-producing Enterobacteriaceae recovered from hospital settings in the Czech Republic. From 2013 to 2015, 22 Klebsiella pneumoniae isolates, 3 Escherichia coli isolates, and 1 Enterobacter cloacae isolate producing OXA-48-like carbapenemases were isolated from 20 patients. Four of the patients were colonized or infected by two or three different OXA-48-like producers. The K. pneumoniae isolates were classified into nine sequence types (STs), with ST101 being predominant (n = 8). The E. coli isolates were of different STs, while the E. cloacae isolate belonged to ST109. Twenty-four isolates carried blaOXA-48, while two isolates carried blaOXA-181 or blaOXA-232. Almost all isolates (n = 22) carried blaOXA-48-positive plasmids of a similar size (∼60 kb), except the two isolates producing OXA-181 or OXA-232. In an ST45 K. pneumoniae isolate and an ST38 E. coli isolate, S1 nuclease profiling plus hybridization indicated a chromosomal location of blaOXA-48. Sequencing showed that the majority of blaOXA-48-carrying plasmids exhibited high degrees of identity with the pOXA-48-like plasmid pE71T. Additionally, two novel pE71T derivatives, pOXA-48_30715 and pOXA-48_30891, were observed. The blaOXA-181-carrying plasmid was identical to the IncX3 plasmid pOXA181_EC14828, while the blaOXA-232-carrying plasmid was a ColE2-type plasmid, being a novel derivative of pOXA-232. Finally, sequencing data showed that the ST45 K. pneumoniae and ST38 E. coli isolates harbored the IS1R-based composite transposon Tn6237 containing blaOXA-48 integrated into their chromosomes. These findings underlined that the horizontal transfer of pOXA-48-like plasmids has played a major role in the dissemination of blaOXA-48 in the Czech Republic. In combination with the difficulties with their detection, OXA-48 producers constitute an important public threat.
KEYWORDS: Klebsiella pneumoniae, Tn1999.2, IncL, OXA-181, OXA-232, ColE2-like, IncX3, Tn1999.5
INTRODUCTION
Since the beginning of the 2000s, carbapenemases of the Ambler class A KPC type or class B type, including IMP- and VIM-like enzymes, were considered to be the most important carbapenemases in Enterobacteriaceae. In 2001, the class D β-lactamase OXA-48, which possesses weak but significant carbapenemase activity, was first detected from a carbapenem-resistant Klebsiella pneumoniae isolate that had been recovered in Istanbul, Turkey (1). Soon, a series of sporadic cases, but also hospital outbreaks, was reported in the main cities of Turkey (2, 3). At about the same time, the blaOXA-48 gene, most often in K. pneumoniae isolates, was also identified in other Middle Eastern and North African countries (4, 5). All those countries can be considered important reservoirs of OXA-48 producers.
Additionally, OXA-48 producers have been identified sporadically in several European countries, including the United Kingdom, Belgium, France, Germany, and the Netherlands (3). The emergence of OXA-48 producers in these countries has been attributed mainly to colonized patients who transferred from North Africa and Turkey (6). These data indicated that the spread of the blaOXA-48 gene was limited to Turkey, the Middle East, and North Africa. However, in countries such as the United Kingdom, France, Belgium, and Germany, recent studies revealed the emergence of OXA-48-producing Enterobacteriaceae in hospital settings, supposing a much more important spread than was previously thought (7–10). Notably, concern was raised by the occurrence of OXA-48 producers in the community in the countries of North Africa and Europe (3, 11). Indeed, the fact that their detection is difficult might have played a significant role in the spread of OXA-like producers, which have somehow been silent. Actually, the expression of the blaOXA-48 gene in the absence of additional resistance mechanisms (e.g., low levels of expression of porins) confers only a low level of resistance to carbapenems. Also, there is no inhibitor-based phenotypic test that can recognize the production of OXA-48-type enzymes. Thus, these two main points do not contribute to the easy recognition of OXA-48-like producers.
In the Czech Republic, the occurrence of carbapenemase-producing Enterobacteriaceae (CPE) was rare, with only a total of two cases being detected in 2009 and 2010 (12). In 2011, the occurrence of CPE increased, and this was mainly due to two hospital outbreaks (13). To contain this increase, in 2012, the Ministry of Health issued national guidelines for the management of patients infected and colonized with CPE (13). In 2012 and 2013, only an outbreak of VIM-producing isolates and four sporadic cases were reported (14). The sporadic cases included two NDM-producing Enterobacteriaceae (15, 16) and the first two OXA-48-producing K. pneumoniae isolates identified in the Czech Republic. These data supposed the success of the national guidelines. However, an increase in the occurrence of CPE was observed during 2014 and 2015, and this was mainly due to the spread of OXA-48-like-producing Enterobacteriaceae in Czech hospitals.
The aim of the present study was to characterize the OXA-48-like producers detected in Czech hospitals in 2014 and 2015.
RESULTS AND DISCUSSION
Carbapenemase-producing Enterobacteriaceae.
A total of 52 Enterobacteriaceae isolates showing carbapenemase activity on a matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) meropenem hydrolysis assay were recovered from Czech hospitals during 2014 (n = 17) and 2015 (n = 35). PCR screening showed that 50 of the isolates were positive for one carbapenemase gene (17 isolates from 2014 [blaKPC, n = 4; blaVIM, n = 4; blaNDM, n = 7; blaOXA-48-like, n = 2] and 33 isolates from 2015 [blaKPC, n = 8; blaVIM, n = 2; blaNDM, n = 2; blaOXA-48-like, n = 21]), while the remaining 2 isolates were positive for the presence of two carbapenemase genes (blaVIM and blaIMP, n = 1; blaOXA-48-like and blaNDM, n = 1).
OXA-48-like-producing isolates.
Altogether, 24 nonrepetitive isolates producing OXA-48-like carbapenemases were isolated from 18 patients in 2014 and 2015. Among them, 20 of the isolates were identified to be K. pneumoniae, 3 were identified to be Escherichia coli, and 1 was identified to be Enterobacter cloacae. Four of the patients were colonized or infected by two or three different OXA-48-like producers (Table 1). Additionally, the two OXA-48-like-producing K. pneumoniae isolates identified in 2013 were studied.
TABLE 1.
Characteristics of OXA-48-like-producing Enterobacteriaceae isolates
| Isolatea | Isolation yr (hospital) | ST | β-Lactamase content | Size of blaOXA-48-like-carrying plasmidb (kb) | Type of plasmid sequence (replicon) | MIC (μg/ml)d |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ctx | Caz | Fep | Imp | Mer | Etp | Gen | Amk | Sxt | Cip | Col | Tgc | ||||||
| K. pneumoniae | |||||||||||||||||
| Kpn-82929 | 2013 (B) | 45 | OXA-48, CTX-M-14 | chrc | 8 | 1 | 8 | 2 | ≤0.12 | 1 | ≤0.12 | ≤0.5 | 1 | ≤0.06 | ≤0.25 | 1 | |
| Kpn-63870 | 2013 (B) | 101 | OXA-48, CTX-M-15, TEM-1 | 63.566 | A0 (IncL) | >8 | >16 | >16 | >16 | 16 | >16 | >16 | 16 | 1 | >8 | 8 | 1 |
| Kpn-74996 | 2014 (B) | 461 | OXA-48 | 63.566 | A0 (IncL) | 0.5 | 1 | 0.5 | 1 | 1 | 2 | ≤0.12 | ≤0.5 | 2 | ≤0.06 | ≤0.25 | 4 |
| Kpn-81700 | 2014 (B) | 461 | OXA-48, TEM-1 | 63.566 | A0 (IncL) | 0.5 | 1 | 0.5 | 4 | 1 | 8 | ≤0.12 | ≤0.5 | 2 | ≤0.06 | 0.5 | 2 |
| Kpn-04976◻ | 2015 (A1) | 1520 | OXA-48 | 63.566 | A0 (IncL) | 0.25 | 0.5 | ≤0.12 | 2 | 0.25 | 4 | ≤0.12 | 1 | 1 | ≤0.06 | ≤0.25 | 1 |
| Kpn-04963◻ | 2015 (A1) | 395 | OXA-48, CTX-M-15, OXA-1, TEM-1 | 63.566 | A0 (IncL) | >8 | >16 | >16 | >16 | 8 | >16 | 0.5 | 8 | >32 | >8 | ≤0.25 | 4 |
| Kpn-05159 | 2015 (A1) | 395 | OXA-48, CTX-M-15, OXA-1, TEM-1 | 63.566 | A0 (IncL) | >8 | >16 | >16 | >16 | 16 | >16 | >16 | 8 | >32 | >8 | ≤0.25 | 4 |
| Kpn-29097 | 2015 (B) | 461 | OXA-48 | 63.566 | A0 (IncL) | 0.25 | 0.5 | 0.25 | 1 | 0.25 | 2 | 0.25 | ≤0.5 | 1 | 1 | ≤0.25 | 4 |
| Kpn-17153● | 2015 (B) | 461 | OXA-48 | 63.566 | A0 (IncL) | 0.5 | 0.25 | 0.25 | 0.5 | 0.25 | 4 | 0.25 | 1 | 0.12 | 0.12 | 0.5 | 4 |
| Kpn-18921● | 2015 (B) | 101 | OXA-48, CTX-M-15, TEM-1 | 63.566 | A0 (IncL) | >8 | >16 | >16 | 0.5 | 0.25 | 4 | >16 | 8 | 1 | >8 | 1 | 0.25 |
| Kpn-20382 | 2015 (B) | 101 | OXA-48, CTX-M-15, TEM-1 | 63.566 | A0 (IncL) | >8 | >16 | >16 | 2 | 0.25 | 4 | >16 | 4 | 0.5 | >8 | ≤0.25 | 0.25 |
| Kpn-23770○ | 2015 (B) | 101 | OXA-48, CTX-M-15, TEM-1 | 63.566 | A0 (IncL) | >8 | >16 | >16 | >16 | 8 | >16 | >16 | 32 | 8 | >8 | ≤0.25 | 0.5 |
| Kpn-23495 | 2015 (B) | 101 | OXA-48, CTX-M-15, TEM-1 | 63.566 | A0 (IncL) | >8 | >16 | >16 | 2 | 0.25 | 2 | >16 | 1 | 2 | >8 | ≤0.25 | 1 |
| Kpn-23482 | 2015 (B) | 101 | OXA-48, CTX-M-15, TEM-1 | 63.566 | A0 (IncL) | >8 | >16 | >16 | 2 | 0.25 | 2 | >16 | 8 | 1 | >8 | ≤0.25 | 0.5 |
| Kpn-24100 | 2015 (B) | 101 | OXA-48, CTX-M-15, TEM-1 | 63.566 | A0 (IncL) | >8 | >16 | >16 | 2 | 0.25 | 2 | >16 | 4 | 0.5 | >8 | ≤0.25 | 0.5 |
| Kpn-29144 | 2015 (C) | 18 | OXA-181, CTX-M-15, OXA-1, TEM-1 | 51.478 | B (IncX3) | >8 | >16 | >16 | >16 | >16 | >16 | >16 | 4 | >32 | >8 | ≤0.25 | 1 |
| Kpn-30715▲ | 2015 (D) | 11 | OXA-48, CTX-M-15 | 65.488 | A1 (IncL) | >8 | >16 | >16 | >16 | >16 | >16 | >16 | >64 | >32 | >8 | ≤0.25 | 1 |
| Kpn-30891▲ | 2015 (D) | 891 | OXA-48, CTX-M-15, TEM-1 | 66.059 | A2 (IncL) | >8 | >16 | 16 | 1 | 0.5 | 4 | >16 | >64 | >32 | >8 | 2 | 2 |
| Kpn-30890 | 2015 (D) | 11 | OXA-48, CTX-M-15 | 65.488 | A1 (IncL) | >8 | >16 | >16 | 2 | 1 | 16 | >16 | >64 | >32 | >8 | 8 | 1 |
| Kpn-31329 | 2015 (D) | 15 | OXA-48, CTX-M-15, OXA-1, TEM-1 | 63.566 | A0 (IncL) | >8 | >16 | >16 | >16 | 1 | 2 | >16 | 2 | 1 | >8 | 0.5 | 0.5 |
| Kpn-31569 | 2015 (D1) | 101 | OXA-48, CTX-M-15, TEM-1 | 63.566 | A0 (IncL) | >8 | >16 | >16 | 2 | 8 | >16 | >16 | 8 | 1 | >8 | ≤0.25 | 1 |
| Kpn-30929 | 2015 (E) | 15 | OXA-232, NDM-1, CTX-M-15, OXA-1 | 12.531 | C (ColE2-like) | >8 | >16 | >16 | >16 | 8 | >16 | >16 | >64 | >32 | >8 | 1 | 0.5 |
| E. coli | |||||||||||||||||
| Eco-32005 | 2015 (A2) | 38 | OXA-48, TEM-1 | chr | 0.25 | ≤0.12 | ≤0.12 | 0.25 | 0.12 | 1 | >16 | 2 | 0.12 | ≤0.06 | ≤0.25 | >32 | |
| Eco-17646● | 2015 (B) | 4956 | OXA-48 | 63.566 | A0 (IncL) | 0.5 | 0.25 | ≤0.12 | 0.5 | 0.12 | 2 | 1 | 2 | 0.06 | 8 | ≤0.25 | 0.25 |
| Eco-26031○ | 2015 (B) | 216 | OXA-48 | 63.566 | A0 (IncL) | 0.12 | ≤0.12 | ≤0.12 | 1 | 0.12 | 0.25 | 0.25 | 1 | 0.03 | ≤0.06 | ≤0.25 | 0.25 |
| E. cloacae Ecl-04292□ | 2015 (A1) | 109 | OXA-48, CTX-M-15, OXA-1, TEM-1 | 63.566 | A0 (IncL) | >8 | 16 | 16 | 4 | 0.5 | >16 | 0.25 | 1 | 4 | ≤0.06 | ≤0.25 | 1 |
White squares, black circles, white circles, and black triangles each indicate the OXA-48-like-producing isolates recovered from the same patient.
Data for plasmids found in transconjugants are shown in bold; data for plasmids observed in transformants are underlined.
chr, chromosomal location of a blaOXA-48 gene.
Ctx, cefotaxime; Caz, ceftazidime; Fep, cefepime; Imp, imipenem; Mer, meropenem; Etp, ertapenem; Gen, gentamicin; Amk, amikacin; Sxt, trimethoprim-sulfamethoxazole; Cip, ciprofloxacin; Col, colistin; Tgc, tigecycline.
OXA-48-like producers were collected from seven Czech hospitals located throughout the Czech Republic. Hospital B was the setting with the highest occurrence of OXA-48 producers. In June 2013, the first OXA-48 producer (Kpn-82929) identified in the Czech Republic was isolated from a newborn. The second OXA-48-producing isolate (Kpn-63870) was recovered from a patient who was directly repatriated from Romania. From April 2014 to March 2015, three further patients colonized or infected with OXA-48-producing K. pneumoniae were identified. Additionally, in hospital B, an outbreak that included six patients diagnosed with OXA-48-producing K. pneumoniae lasted from August to December of 2015. Only two cases of OXA-48-producing K. pneumoniae isolates were reported in hospital A1. The first case, a 1-year-old child, who was directly repatriated from a Russian hospital, was colonized or infected by three OXA-48-producing isolates: K. pneumoniae Kpn-04976 and Kpn-04963 and E. cloacae Ecl-04292. One month later, the transmission of an OXA-48-producing K. pneumoniae isolate (Kpn-05159) to an infant who stayed in the same department was found. An OXA-48 outbreak restricted to three patients occurred in hospital D. A patient that had recently traveled to Ukraine was diagnosed with two OXA-48-producing isolates of K. pneumoniae (Kpn-30715 and Kpn-30891) in August of 2015. Two further patients colonized or infected with OXA-48-producing K. pneumoniae were identified until September. The remaining four cases were detected in four different hospitals. Two of those cases had recently traveled abroad (to India [Kpn-30929] and Tunisia [Kpn-31569]), while no data on whether the other two patients had traveled abroad or had previously been hospitalized were available.
All 26 OXA-48-like producers exhibited resistance to piperacillin and piperacillin-tazobactam (data not shown), while the variations in the MICs of cephalosporins and carbapenems that were observed (Table 1) might reflect the presence of additional resistance mechanisms in some of the isolates. Seventeen of the OXA-48-like producers also exhibited resistance to ciprofloxacin; 16 were resistant to gentamicin, 6 were resistant to tigecycline, and 5 were resistant to amikacin, whereas 2 isolates were resistant to colistin.
The population structure of OXA-48-like-producing isolates studied by multilocus sequence typing (MLST) is shown in Table 1. The K. pneumoniae isolates comprised nine sequence types (STs). ST101 was the most prevalent, accounting for eight isolates. The majority of ST101 isolates (7/8) was recovered from patients hospitalized in hospital B. Ten of the isolates were distributed in STs 461 (n = 4, from hospital B), 11 (n = 2, from hospital D), 15 (n = 2), and 395 (n = 2, from hospital A1). The remaining isolates belonged to distinct STs. STs 11, 15, 45, 101, 395, and 461 have previously been associated with OXA-48-like-producing isolates from several geographical areas (17–19). All three E. coli isolates were of different STs, including the pandemic ST38 (19–21). The E. cloacae isolate was assigned to ST109, previously associated with the production of the CTX-M-15 or SHV-12 enzyme (22).
Sequencing of the PCR products revealed three blaOXA-48-type genes encoding the OXA-48, OXA-181, and OXA-232 enzymes (Table 1) (1, 23, 24). Twenty-four of the isolates were found to produce the OXA-48 β-lactamase, while the ST18 K. pneumoniae isolate produced the OXA-181 enzyme. The remaining K. pneumoniae isolate, which belonged to ST15, coproduced the OXA-232 and NDM-1 carbapenemases. Additionally, most of blaOXA-48-like-positive isolates were confirmed to coproduce the extended-spectrum β-lactamase CTX-M-15 (n = 17) either alone or along with TEM-1 (n = 16) and/or OXA-1 (n = 5), whereas the ST45 OXA-48-producing K. pneumoniae isolate coproduced the CTX-M-14 β-lactamase.
blaOXA-48-like-carrying plasmids.
The blaOXA-48-like genes from 24 out of 26 clinical strains were transferred by conjugation (n = 23) or transformation (n = 1) (Table 1). Neither the ST45 K. pneumoniae isolate nor the ST38 E. coli isolate was capable of transferring the blaOXA-48 gene by either conjugation or transformation. All blaOXA-48-like-positive recombinants exhibited similar resistance phenotypes, showing resistance to piperacillin and piperacillin-tazobactam and decreased susceptibility or resistance to imipenem and ertapenem, while they remained susceptible to cephalosporins and meropenem. Additionally, all blaOXA-48-like-positive recombinants were susceptible to non-β-lactam antibiotics.
Plasmid analysis of OXA-48-producing donor and transconjugant strains revealed the transfer of plasmids, all of which were ∼60 kb (Table 1). The OXA-181-producing transconjugant carried a blaOXA-48-like-positive plasmid with a size of ∼50 kb, while the OXA-232-producing transformant harbored a plasmid of ∼10 kb that hybridized with a blaOXA-48-like probe. Moreover, in the S1 nuclease profiles of the OXA-48-producing ST45 K. pneumoniae and ST38 E. coli isolates, the blaOXA-48-like probe hybridized only with the largest DNA bands, corresponding to the chromosomal material.
Replicon typing showed that all plasmids carrying blaOXA-48 were positive for the IncL allele, whereas the blaOXA-181- and blaOXA-232-carrying plasmids were nontypeable by PCR-based replicon typing (PBRT).
Structure of blaOXA-48-like-carrying plasmids.
The complete sequences of all blaOXA-48-like-carrying plasmids were determined. Illumina sequencing revealed three types of plasmid sequences (types A to C), with type A being the most prevalent and including three subtypes (subtypes A0, A1, and A2).
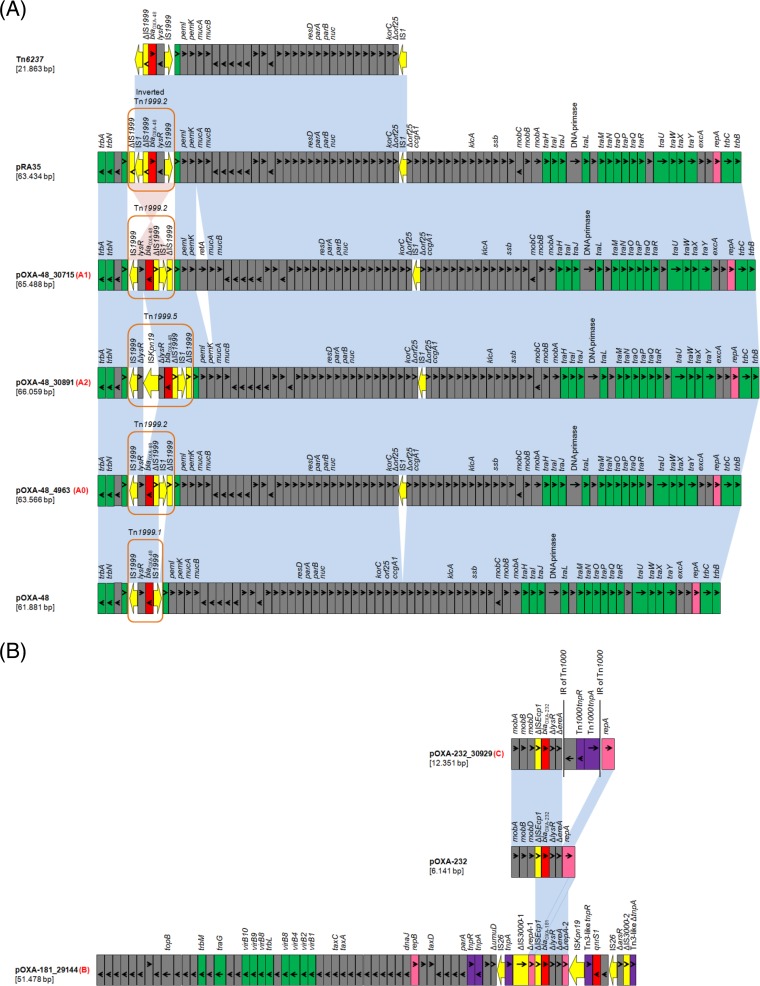
All blaOXA-48-carrying plasmids belonged to type A and were derivatives of the archetypal IncL blaOXA-48-carrying plasmid pOXA-48 (Fig. 1), originally described in the K. pneumoniae 11978 isolate recovered in Turkey in 2001 and then reported worldwide (25). Nineteen out of the 22 sequenced blaOXA-48-carrying plasmids (type A0; Table 1) showed high degrees of similarity to each other and to pE71T (100% coverage, 99% identity), previously characterized from K. pneumoniae E71T isolated in Ireland (26). Plasmid pE71T differed from pOXA-48 by the insertion of two copies of the IS1R element. The carbapenemase gene was part of the Tn1999.2 transposon, which included IS1R integrated in IS1999 located upstream of the blaOXA-48 gene (2). The second IS1R was inserted into orf25. Plasmids pOXA-48_30715 and pOXA-48_30890 (type A1), both of which were isolated from ST11 K. pneumoniae isolates, differed from pE71T by the insertion of a 1,911-bp fragment encoding a reverse transcriptase (RetA) upstream of the mucAB operon. Plasmid pOXA-48_30891 (type A2) was a pE71T derivative carrying a novel variant of the Tn1999.2 transposon (designated Tn1999.5) in which the lysR gene was truncated by the ISKpn19 element. Interestingly, plasmids pOXA-48_30715 and pOXA-48_30891 were characterized from two different K. pneumoniae isolates recovered from the same patient (Table 1). Among all type A blaOXA-48-carrying plasmids, no resistance genes other than blaOXA-48 were identified, as previously described for pOXA-48 and its relatives (25–27).
FIG 1.
Linear maps of the blaOXA-48-like-carrying plasmids. For each plasmid, the type of plasmid sequence is indicated in red next to the plasmid name. (A) Comparison of IncL blaOXA-48-carrying plasmids pOXA-48 (25), pOXA-48_4963, pOXA-48_30715, pOXA-48_30891, and pRA35 and of the composite transposon Tn6237 (29). The boundaries of Tn1999-like transposons are also shown. (B) Comparison of the blaOXA-232-carrying plasmids pOXA-232 (24) and pOXA-232_30929 and of the blaOXA-181-carrying plasmid pOXA-181_29144. Open reading frames (ORFs) are shown as rectangles (arrows within rectangles indicate the direction of transcription). Intact insertion sequences are represented by arrows, while truncated insertion sequence elements appear as rectangles. Replicons of the plasmids are indicated as pink rectangles. Resistance genes, insertion sequence elements, and transposases are shown in red, yellow, and purple, respectively. Green rectangles indicate genes responsible for the conjugative transfer of the plasmids. The remaining genes, including plasmid scaffold regions, are indicated as gray rectangles. Homologous segments (representing ≥99% sequence identity) are indicated by light blue shading, while pink shading shows inverted homologous segments.
pOXA-181_29144 (type B) (Fig. 1), encoding OXA-181, was an IncX3-type plasmid that was identical to pOXA181_EC14828 (100% coverage, 100% identity), which to date has been described only in China from an ST410, E. coli strain (WCHEC14828), isolated in 2014 (28). Similar to pOXA181_EC14828, the qnrS1 gene, conferring low-level resistance to fluoroquinolones, was identified in the sequence of pOXA-181_29144. Finally, plasmid pOXA-232_30929 (type C) appeared to be a derivative of pOXA-232 (Fig. 1), a ColE2-type plasmid originally described from an ST2968 E. coli isolate and two ST14 K. pneumoniae isolates recovered from patients who transferred from India to France in 2011 (24). Only one difference between the two plasmids was observed. A 5,981-bp segment consisting of the Tn1000 transposon was present in pOXA-232_30929 and was found 477 bp upstream of the repA gene.
Finally, de novo assembly obtained a unique contig containing blaOXA-48 for ST45 K. pneumoniae and ST38 E. coli isolates. Sequence analysis showed that these isolates harbored a 21.9-kb plasmid fragment containing blaOXA-48 flanked by IS1R elements integrated into their chromosomes. This plasmidic fragment consisted of the IS1R-based composite transposon (Fig. 1) Tn6237 (29). However, using the Illumina MiSeq platform, we were not able to identify the precise insertion site of Tn6237.
Concluding remarks.
In conclusion, the present study investigated the first cases and outbreaks of OXA-48-like-producing Enterobacteriaceae isolates from the Czech Republic. Five of the patients had recently traveled abroad, with one of them being involved in the initiation of an outbreak (hospital D), while three OXA-48-like isolates (Kpn-82929, Kpn-29114, and Eco-32005) could be described as community acquired since the patients had no history of previous hospitalization or travel abroad. The setting that was most affected was hospital B, in which an outbreak followed a long period with the sporadic occurrence of OXA-48 producers. In hospital B, the outbreak was associated with the spread of K. pneumoniae isolates belonging to ST101. Most of the STs found in isolates of K. pneumoniae (STs 11, 15, 45, 101, 395, and 461) and E. coli (ST38) have previously been associated with OXA-48-like-producing isolates from several geographical areas (17–21).
In four of the patients, two or three different OXA-48 producers were identified during their hospitalization, supposing the in vivo horizontal transfer of the blaOXA-48-carrying plasmid. Sequencing data showed the presence of the same blaOXA-48-carrying plasmid in three of these isolates (Table 1), further confirming this hypothesis. In addition, the same blaOXA-48-carrying plasmid (type A0) was identified in all isolates recovered from patients that were involved in the outbreak which took place in hospital B.
Results from Illumina sequencing showed that pOXA-48-like plasmids played a major role in the dissemination of the blaOXA-48 gene in Czech hospitals. Among our isolates, a highly conserved blaOXA-48-carrying plasmid, which was identical to the previously described pE71T (26), was observed in a polyclonal population of K. pneumoniae isolates (of 5 different STs). Plasmid pE71T was also found in two E. coli isolates of different STs and one E. cloacae isolate. Additionally, two novel pE71T derivatives (plasmids pOXA-48_30715 and pOXA-48_30891) were characterized from K. pneumoniae isolates of STs 11 and 891, respectively. On the other hand, the OXA-181 and OXA-232 carbapenemases were encoded by different types of plasmids belonging to IncX3 and ColE2-like groups, respectively.
The data presented here contribute to the current knowledge of OXA-48-like-producing Enterobacteriaceae. In agreement with the results of previous studies, our findings underline that OXA-48 producers pose an important public threat, mainly due to the difficulties with their detection and the rapid horizontal transfer of pOXA-48-like plasmids.
MATERIALS AND METHODS
Bacterial isolates and confirmation of carbapenemase production.
In 2014 and 2015, Czech hospitals referred a total of 630 Enterobacteriaceae isolates with a meropenem MIC of >0.125 μg/ml (30) to the National Reference Laboratory for Antibiotics. Species identification was confirmed by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) using MALDI Biotyper software (Bruker Daltonics, Bremen, Germany). All isolates were tested for carbapenemase production by the MALDI-TOF MS meropenem hydrolysis assay (31). Isolates that were positive by the MALDI-TOF MS meropenem hydrolysis assay were subjected to metallo-β-lactamase, KPC, and OXA-48 detection using the double-disc synergy test with EDTA, the phenylboronic acid disc test, and the temocillin disc test (9, 32, 33), respectively. Additionally, carbapenemase genes (blaKPC, blaVIM, blaIMP, blaNDM, and blaOXA-48-like) were detected by PCR amplification (1, 34–36). PCR products were sequenced as described below. Isolates positive for blaOXA-48-like genes were further studied. Moreover, the two OXA-48-producing K. pneumoniae isolates, recovered at the University Hospital Pilsen (Pilsen, Czech Republic) during 2013 were included in this study for comparative epidemiological purposes.
Susceptibility testing.
The MICs of piperacillin, piperacillin-tazobactam, cefotaxime, ceftazidime, cefepime, imipenem, meropenem, ertapenem, co-trimoxazole, ciprofloxacin, gentamicin, amikacin, colistin, and tigecycline were determined by the broth dilution method (37). Data were interpreted according to the criteria of the European Committee on Antimicrobial Susceptibility Testing (EUCAST; www.eucast.org).
Typing.
All blaOXA-48-like-positive isolates were typed by multilocus sequence typing (MLST) (38–40). The databases at http://pubmlst.org/ecloacae, http://mlst.warwick.ac.uk/mlst/dbs/Ecoli, and http://bigsdb.web.pasteur.fr/klebsiella were used to assign STs.
Detection of β-lactamases.
The β-lactamase content of all blaOXA-48-like-positive isolates was determined by isoelectric focusing (IEF). Bacterial extracts were obtained by sonication of bacterial cells suspended in 1% glycine buffer and clarified by centrifugation. Sonicated cell extracts were analyzed by IEF in polyacrylamide gels containing ampholytes (pH 3.5 to 9.5; AP Biotech, Piscataway, NJ). The separated β-lactamases were visualized by covering the gel with the chromogenic cephalosporin nitrocefin (0.2 mg/ml; Oxoid Ltd., Basingstoke, United Kingdom) (41).
On the basis of the IEF data, PCR detection of various bla genes was performed by the use of primers specific for blaTEM-1, blaOXA-1, blaSHV, blaCTX-M, and blaCMY, as reported previously (42–45). Both strands of the PCR products were sequenced using an ABI 377 sequencer (Applied Biosystems, Foster City, CA).
Transfer of blaOXA-48-like genes.
Conjugal transfer of blaOXA-48-like genes from the clinical strains was carried out in mixed broth cultures (46), using the rifampin-resistant E. coli A15 laboratory strain as a recipient. Transconjugants were selected on MacConkey agar plates supplemented with rifampin (150 μg/ml) and ampicillin (50 μg/ml). Plasmid DNA from clinical isolates which failed to transfer blaOXA-48-like by conjugation was extracted using a Qiagen maxikit (Qiagen, Hilden, Germany) and used to transform E. coli DH5α cells. The preparation and transformation of competent E. coli cells were done using calcium chloride, as described by Cohen et al. (47). Transformants were selected on Luria-Bertani agar plates with ampicillin (50 μg/ml). Transconjugants or transformants were confirmed to be OXA-48-like producers by PCR (1) and the MALDI-TOF MS meropenem hydrolysis assay (31).
Plasmid analysis.
To define the genetic units of the blaOXA-48-like genes, the plasmid contents of all OXA-48-producing clinical and recombinant strains were analyzed by pulsed-field gel electrophoresis (PFGE) of total DNA digested with S1 nuclease (Promega, Madison, WI, USA) (48). Following PFGE, the DNA was transferred to a BrightStar-Plus positively charged nylon membrane (Applied Biosystems, Foster City, CA) and hybridized with digoxigenin-labeled blaOXA-48-like probes.
Plasmid incompatibility (Inc) groups were determined by the PCR-based replicon typing (PBRT) method (49), using total DNA from transconjugants and transformants. blaOXA-48-like-carrying plasmids were further characterized by a specific IncL PCR assay (27), using the L-FW and L/M-RV primer pair. The forward primer targeted the excA gene of the IncL plasmid type, while the reverse primer targeted the highly conserved repA gene of the IncL and IncM plasmid types (27).
Plasmid and chromosome sequencing.
Plasmid DNAs from transconjugants and transformants were extracted using a Qiagen large-construct kit (Qiagen, Hilden, Germany). Additionally, the genomic DNAs of K. pneumoniae Kpn-82929/13 and E. coli Eco-32005/15 were extracted using a DNA-Sorb-B kit (Sacace Biotechnologies S.r.l., Como, Italy). Plasmids and chromosomes were sequenced using an Illumina MiSeq platform (Illumina Inc., San Diego, CA, USA). Initial paired-end reads were quality trimmed using the Trimmomatic tool (50). For assembly of the plasmids, reads were mapped to the reference E. coli K-12 substrain MG 1655 genome (GenBank accession no. U00096) using the BWA-MEM algorithm (51), in order to filter out the chromosomal DNA. Then, all the unmapped paired-end reads were assembled by use of the de Bruijn graph-based de novo assembler SPAdes (52). The sequence gaps were filled by a PCR-based strategy and Sanger sequencing. For sequence analysis and annotation, the BLAST algorithm (www.ncbi.nlm.nih.gov/BLAST), the ISfinder database (www-is.biotoul.fr/), and the open reading frame (ORF) finder tool (www.bioinformatics.org/sms/) were utilized. Comparative genome alignments were performed using the Mauve (version 2.3.1) program (53).
Accession number(s).
One nucleotide sequence representing each different plasmid type was submitted to GenBank. The nucleotide sequences of the pOXA-48_4963 (type A0), pOXA-48_30715 (type A1), pOXA-48_30891 (type A2), pOXA-181_29144 (type B), and pOXA-232_30929 (type C) plasmids have been deposited in GenBank under accession numbers KX523900, KX523901, KX523902, KX523903, and KX523904, respectively.
ACKNOWLEDGMENTS
We are very thankful to Dana Králová for technical assistance during the present study. Also, we thank all Czech participants of the EARS-Net project for their collaboration and providing the isolates.
This work was supported by the Medical Research Foundation of the Czech Republic (grant number 15-28663A), the National Sustainability Program I (NPU I; grant number LO1503 provided by the Ministry of Education Youth and Sports of the Czech Republic), and the Charles University Research Fund (grant number P36).
We have no conflicts to declare.
REFERENCES
- 1.Poirel L, Heritier C, Tolun V, Nordmann P. 2004. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother 48:15–22. doi: 10.1128/AAC.48.1.15-22.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carrer A, Poirel L, Eraksoy H, Cagatay AA, Badur S, Nordmann P. 2008. Spread of OXA-48-positive carbapenem-resistant Klebsiella pneumoniae isolates in Istanbul, Turkey. Antimicrob Agents Chemother 52:2950–2954. doi: 10.1128/AAC.01672-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poirel L, Potron A, Nordmann P. 2012. OXA-48-like carbapenemases: the phantom menace. J Antimicrob Chemother 67:1597–1606. doi: 10.1093/jac/dks121. [DOI] [PubMed] [Google Scholar]
- 4.Carrër A, Poirel L, Yilmaz M, Akan OA, Feriha C, Cuzon G, Matar G, Honderlick P, Nordmann P. 2010. Spread of OXA-48-encoding plasmid in Turkey and beyond. Antimicrob Agents Chemother 54:1369–1373. doi: 10.1128/AAC.01312-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cuzon G, Naas T, Lesenne A, Benhamou M, Nordmann P. 2010. Plasmid-mediated carbapenem-hydrolysing OXA-48 β-lactamase in Klebsiella pneumoniae from Tunisia. Int J Antimicrob Agents 36:91–93. doi: 10.1016/j.ijantimicag.2010.02.014. [DOI] [PubMed] [Google Scholar]
- 6.Grundmann H, Livermore DM, Giske CG, Canton R, Rossolini GM, Campos J, Vatopoulos A, Gniadkowski M, Toth A, Pfeifer Y, Jarlier V, Carmeli Y, CNSE Working Group. 2010. Carbapenem-non-susceptible Enterobacteriaceae in Europe: conclusions from a meeting of national experts. Euro Surveill 15(46):pii=19711 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19711. [DOI] [PubMed] [Google Scholar]
- 7.Thomas CP, Moore LS, Elamin N, Doumith M, Zhang J, Maharjan S, Warner M, Perry C, Turton JF, Johnstone C, Jepson A, Duncan ND, Holmes AH, Livermore DM, Woodford N. 2013. Early (2008-2010) hospital outbreak of Klebsiella pneumoniae producing OXA-48 carbapenemase in the UK. Int J Antimicrob Agents 42:531–536. doi: 10.1016/j.ijantimicag.2013.08.020. [DOI] [PubMed] [Google Scholar]
- 8.Cuzon G, Ouanich J, Gondret R, Naas T, Nordmann P. 2011. Outbreak of OXA-48-positive carbapenem-resistant Klebsiella pneumoniae isolates in France. Antimicrob Agents Chemother 55:2420–2423. doi: 10.1128/AAC.01452-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glupczynski Y, Huang TD, Bouchahrouf W, Rezende de Castro R, Bauraing C, Gérard M, Verbruggen AM, Deplano A, Denis O, Bogaerts P. 2012. Rapid emergence and spread of OXA-48-producing carbapenem-resistant Enterobacteriaceae isolates in Belgian hospitals. Int J Antimicrob Agents 39:168–172. doi: 10.1016/j.ijantimicag.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 10.Pfeifer Y, Schlatterer K, Engelmann E, Schiller RA, Frangenberg HR, Stiewe D, Holfelder M, Witte W, Nordmann P, Poirel L. 2012. Emergence of OXA-48-type carbapenemase-producing Enterobacteriaceae in German hospitals. Antimicrob Agents Chemother 56:2125–2128. doi: 10.1128/AAC.05315-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barguigua A, El Otmani F, Talmi M, Zerouali K, Timinouni M. 2012. Emergence of carbapenem-resistant Enterobacteriaceae isolates in the Moroccan community. Diagn Microbiol Infect Dis 73:290–291. doi: 10.1016/j.diagmicrobio.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 12.Hrabák J, Niemczyková J, Chudáčková E, Fridrichová M, Studentová V, Cervená D, Urbášková P, Zemličková H. 2011. KPC-2-producing Klebsiella pneumoniae isolated from a Czech patient previously hospitalized in Greece and in vivo selection of colistin resistance. Folia Microbiol (Praha) 56:361–365. doi: 10.1007/s12223-011-0057-6. [DOI] [PubMed] [Google Scholar]
- 13.Hrabak J, Papagiannitsis CC, Studentova V, Jakubu V, Fridrichová M, Zemlickova H, Czech Participants of European Antimicrobial Resistance Surveillance Network. 2013. Carbapenemase-producing Klebsiella pneumoniae in the Czech Republic in 2011. Euro Surveill 18(45):pii=20626. doi: 10.2807/1560-7917.ES2013.18.45.20626. [DOI] [PubMed] [Google Scholar]
- 14.Albiger B, Glasner C, Struelens MJ, Grundmann H, Monnet DL, European Survey of Carbapenemase-Producing Enterobacteriaceae (EuSCAPE) Working Group. 2015. Carbapenemase-producing Enterobacteriaceae in Europe: assessment by national experts from 38 countries, May 2015. Euro Surveill 20(45):pii=30062. doi: 10.2807/1560-7917.ES.2015.20.45.30062. [DOI] [PubMed] [Google Scholar]
- 15.Papagiannitsis CC, Studentova V, Chudackova E, Bergerova T, Hrabak J, Radej J, Novak I. 2013. Identification of a New Delhi metallo-β-lactamase-4 (NDM-4)-producing Enterobacter cloacae from a Czech patient previously hospitalized in Sri Lanka. Folia Microbiol (Praha) 58:547–549. doi: 10.1007/s12223-013-0247-5. [DOI] [PubMed] [Google Scholar]
- 16.Studentova V, Dobiasova H, Hedlova D, Dolejska M, Papagiannitsis CC, Hrabak J. 2015. Complete nucleotide sequences of two NDM-1-encoding plasmids from the same sequence type 11 Klebsiella pneumoniae strain. Antimicrob Agents Chemother 59:1325–1328. doi: 10.1128/AAC.04095-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brañas P, Villa J, Viedma E, Mingorance J, Orellana MA, Chaves F. 2015. Molecular epidemiology of carbapenemase-producing Klebsiella pneumoniae in a hospital in Madrid: successful establishment of an OXA-48 ST11 clone. Int J Antimicrob Agents 46:111–116. doi: 10.1016/j.ijantimicag.2015.02.019. [DOI] [PubMed] [Google Scholar]
- 18.Liapis E, Pantel A, Robert J, Nicolas-Chanoine MH, Cavalié L, van der Mee-Marquet N, de Champs C, Aissa N, Eloy C, Blanc V, Guyeux C, Hocquet D, Lavigne JP, Bertrand X, ONERBA. 2014. Molecular epidemiology of OXA-48-producing Klebsiella pneumoniae in France. Clin Microbiol Infect 20:O1121–O1123. doi: 10.1111/1469-0691.12727. [DOI] [PubMed] [Google Scholar]
- 19.Potron A, Poirel L, Rondinaud E, Nordmann P. 2013. Intercontinental spread of OXA-48 beta-lactamase-producing Enterobacteriaceae over a 11-year period, 2001 to 2011. Euro Surveill 18(31):pii=20549. doi: 10.2807/1560-7917.ES2013.18.31.20549. [DOI] [PubMed] [Google Scholar]
- 20.Zurfluh K, Nüesch-Inderbinen MT, Poirel L, Nordmann P, Hächler H, Stephan R. 2015. Emergence of Escherichia coli producing OXA-48 β-lactamase in the community in Switzerland. Antimicrob Resist Infect Control 4:9. doi: 10.1186/s13756-015-0051-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turton JF, Doumith M, Hopkins KL, Perry C, Meunier D, Woodford N. 2016. Clonal expansion of Escherichia coli ST38 carrying a chromosomally integrated OXA-48 carbapenemase gene. J Med Microbiol 65:538–546. doi: 10.1099/jmm.0.000248. [DOI] [PubMed] [Google Scholar]
- 22.Izdebski R, Baraniak A, Herda M, Fiett J, Bonten MJ, Carmeli Y, Goossens H, Hryniewicz W, Brun-Buisson C, Gniadkowski M, MOSAR WP2, WP3 and WP5 Study Groups. 2015. MLST reveals potentially high-risk international clones of Enterobacter cloacae. J Antimicrob Chemother 70:48–56. doi: 10.1093/jac/dku359. [DOI] [PubMed] [Google Scholar]
- 23.Potron A, Nordmann P, Lafeuille E, Al Maskari Z, Al Rashdi F, Poirel L. 2011. Characterization of OXA-181, a carbapenem-hydrolyzing class D beta-lactamase from Klebsiella pneumoniae. Antimicrob Agents Chemother 55:4896–4899. doi: 10.1128/AAC.00481-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Potron A, Rondinaud E, Poirel L, Belmonte O, Boyer S, Camiade S, Nordmann P. 2013. Genetic and biochemical characterisation of OXA-232, a carbapenem-hydrolysing class D β-lactamase from Enterobacteriaceae. Int J Antimicrob Agents 41:325–329. doi: 10.1016/j.ijantimicag.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 25.Poirel L, Bonnin RA, Nordmann P. 2012. Genetic features of the widespread plasmid coding for the carbapenemase OXA-48. Antimicrob Agents Chemother 56:559–562. doi: 10.1128/AAC.05289-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Power K, Wang J, Karczmarczyk M, Crowley B, Cotter M, Haughton P, Lynch M, Schaffer K, Fanning S. 2014. Molecular analysis of OXA-48-carrying conjugative IncL/M-like plasmids in clinical isolates of Klebsiella pneumoniae in Ireland. Microb Drug Resist 20:270–274. doi: 10.1089/mdr.2013.0022. [DOI] [PubMed] [Google Scholar]
- 27.Carattoli A, Seiffert SN, Schwendener S, Perreten V, Endimiani A. 2015. Differentiation of IncL and IncM plasmids associated with the spread of clinically relevant antimicrobial resistance. PLoS One 10:e0123063. doi: 10.1371/journal.pone.0123063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y, Feng Y, Wu W, Xie Y, Wang X, Zhang X, Chen X, Zong Z. 2015. First report of OXA-181-producing Escherichia coli in China and characterization of the isolate using whole-genome sequencing. Antimicrob Agents Chemother 59:5022–5025. doi: 10.1128/AAC.00442-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beyrouthy R, Robin F, Delmas J, Gibold L, Dalmasso G, Dabboussi F, Hamzé M, Bonnet R. 2014. IS1R-mediated plasticity of IncL/M plasmids leads to the insertion of blaOXA-48 into the Escherichia coli chromosome. Antimicrob Agents Chemother 58:3785–3790. doi: 10.1128/AAC.02669-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.European Committee on Antimicrobial Susceptibility Testing. 2012. EUCAST guidelines for detection of resistance mechanism and specific resistances of clinical and/or epidemiological importance. European Committee on Antimicrobial Susceptibility Testing, Växjö, Sweden: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Consultation/EUCAST_guidelines_detection_of_resistance_mechanisms_121222.pdf. [Google Scholar]
- 31.Papagiannitsis CC, Studentova V, Izdebski R, Oikonomou O, Pfeifer Y, Petinaki E, Hrabák J. 2015. MALDI-TOF MS meropenem hydrolysis assay with NH4HCO3, a reliable tool for the direct detection of carbapenemase activity. J Clin Microbiol 53:1731–1735. doi: 10.1128/JCM.03094-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee K, Lim YS, Yong D, Yum JH, Chong Y. 2003. Evaluation of the Hodge test and the imipenem-EDTA double-disk synergy test for differentiating metallo-β-lactamase-producing isolates of Pseudomonas spp. and Acinetobacter spp. J Clin Microbiol 41:4623–4629. doi: 10.1128/JCM.41.10.4623-4629.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doi Y, Potoski BA, Adams-Haduch JM, Sidjabat HE, Pasculle AW, Paterson DL. 2008. Simple disk-based method for detection of Klebsiella pneumoniae carbapenemase-type beta-lactamase by use of a boronic acid compound. J Clin Microbiol 46:4083–4086. doi: 10.1128/JCM.01408-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Naas T, Cuzon G, Villegas MV, Lartigue MF, Quinn JP, Nordmann P. 2008. Genetic structure at the origin of acquisition of the beta-lactamase blaKPC gene. Antimicrob Agents Chemother 52:1257–1263. doi: 10.1128/AAC.01451-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ellington MJ, Kistler J, Livermore DM, Woodford N. 2007. Multiplex PCR for rapid detection of genes encoding acquired metallo-β-lactamases. J Antimicrob Chemother 59:321–322. [DOI] [PubMed] [Google Scholar]
- 36.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.European Committee on Antimicrobial Susceptibility Testing (EUCAST) of the European Society of Clinical Microbiology and Infectious Diseases (ESCMID). 2003. Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by broth dilution. Clin Microbiol Infect 9:ix–xv. [Google Scholar]
- 38.Miyoshi-Akiyama T, Hayakawa K, Ohmagari N, Shimojima M, Kirikae T. 2013. Multilocus sequence typing (MLST) for characterization of Enterobacter cloacae. PLoS One 8:e66358. doi: 10.1371/journal.pone.0066358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol 43:4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Papagiannitsis CC, Študentová V, Jakubù V, Španělová P, Urbášková P, Žemličková H, Hrabák J. 2015. High prevalence of ST131 among CTX-M-producing Escherichia coli from community-acquired infections, in the Czech Republic. Microb Drug Resist 21:74–84. doi: 10.1089/mdr.2014.0070. [DOI] [PubMed] [Google Scholar]
- 42.Coque TM, Novais A, Carattoli A, Poirel L, Pitout J, Peixe L, Baquero F, Cantón R, Nordmann P. 2008. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum β-lactamase CTX-M-15. Emerg Infect Dis 14:195–200. doi: 10.3201/eid1402.070350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pałucha A, Mikiewicz B, Hryniewicz W, Gniadkowski M. 1999. Concurrent outbreaks of extended-spectrum beta-lactamase-producing organisms of the family Enterobacteriaceae in a Warsaw hospital. J Antimicrob Chemother 44:489–499. doi: 10.1093/jac/44.4.489. [DOI] [PubMed] [Google Scholar]
- 44.Woodford N, Fagan EJ, Ellington MJ. 2006. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum (beta)-lactamases. J Antimicrob Chemother 57:154–155. [DOI] [PubMed] [Google Scholar]
- 45.Pérez-Pérez FJ, Hanson ND. 2002. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol 40:2153–2162. doi: 10.1128/JCM.40.6.2153-2162.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vatopoulos AC, Philippon A, Tzouvelekis LS, Komninou Z, Legakis NJ. 1990. Prevalence of a transferable SHV-5 type beta-lactamase in clinical isolates of Klebsiella pneumoniae and Escherichia coli in Greece. J Antimicrob Chemother 26:635–648. doi: 10.1093/jac/26.5.635. [DOI] [PubMed] [Google Scholar]
- 47.Cohen SN, Chang ACY, Hsu L. 1972. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A 69:2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barton BM, Harding GP, Zuccarelli AJ. 1995. A general method for detecting and sizing large plasmids. Anal Biochem 226:235–240. doi: 10.1006/abio.1995.1220. [DOI] [PubMed] [Google Scholar]
- 49.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 50.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. ArXiv e-Prints 1303:3997. [Google Scholar]
- 52.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comp Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]