ABSTRACT
The molecular mechanisms of resistance to fluoroquinolones, tetracyclines, an aminocyclitol, macrolides, a lincosamide, a phenicol, and pleuromutilins were investigated in Mycoplasma bovis. For the identification of mutations responsible for the high MICs of certain antibiotics, whole-genome sequencing of 35 M. bovis field isolates and 36 laboratory-derived antibiotic-resistant mutants was performed. In vitro resistant mutants were selected by serial passages of M. bovis in broth medium containing subinhibitory concentrations of the antibiotics. Mutations associated with high fluoroquinolones MICs were found at positions 244 to 260 and at positions 232 to 250 (according to Escherichia coli numbering) of the quinolone resistance-determining regions of the gyrA and parC genes, respectively. Alterations related to elevated tetracycline MICs were described at positions 962 to 967, 1058, 1195, 1196, and 1199 of genes encoding the 16S rRNA and forming the primary tetracycline binding site. Single transversion at position 1192 of the rrs1 gene resulted in a spectinomycin MIC of 256 μg/ml. Mutations responsible for high macrolide, lincomycin, florfenicol, and pleuromutilin antibiotic MICs were identified in genes encoding 23S rRNA. Understanding antibiotic resistance mechanisms is an important tool for future developments of genetic-based diagnostic assays for the rapid detection of resistant M. bovis strains.
KEYWORDS: antibiotic resistance, cattle, Mycoplasma bovis
INTRODUCTION
Antibiotics are among the most important therapeutic tools in the veterinary and human medicine, but their use is limited since resistance tends to evolve in pathogenic bacteria. The microorganisms are exposed to selective pressure by the use of antimicrobials in medicine and agriculture, favoring the development, survival, and spread of resistant clones (1).
Mycoplasma spp. are members of the class Mollicutes and comprise the simplest life form that can replicate independently from the host (2). Mycoplasma spp. have no cell wall and they have a limited number of metabolic pathways. The greatly reduced genome size and coding capacity of Mycoplasma spp. makes them a good model for genetic studies. Mycoplasma spp. are fast-evolving bacteria with several human and animal pathogens; however, their importance is often underestimated (2). Mycoplasma bovis is a major cause of calf pneumonia, mastitis and arthritis, and it is responsible for significant economic losses (3). Adequate housing and appropriate antibiotic treatment are the main tools in the therapy of mycoplasmosis in cattle since no effective vaccine is available for the control of M. bovis infections (3).
The number of potentially effective antimicrobials is limited since M. bovis is intrinsically resistant to antibiotics acting on cell wall or folate synthesis (4). In general, protein synthesis inhibitor antimicrobial classes are active against M. bovis. Tetracyclines and spectinomycin are primarily binding to the 30S subunit of the ribosome, whereas macrolides, lincosamides, phenicols, and pleuromutilins are mycoplasmastatic antibiotics acting on the 50S ribosomal subunit, preventing the mechanisms of transpeptidation and translation (5). Expanded-spectrum fluoroquinolones (enrofloxacin, danofloxacin, and marbofloxacin) have mycoplasmacidal effect by acting on topoisomerases that inhibit the DNA synthesis of bacteria (6). Among the few antimicrobials licensed for treatment of M. bovis, there is increasing evidence for resistance (5, 7, 8).
In previous studies the mechanisms involved in resistance of human-pathogenic Mycoplasma species were mainly examined, while information about the acquisition and the mechanisms of antibiotic resistance in mycoplasmas with veterinary relevance, including M. bovis, is scarce. Primarily, the mechanisms based on genetic point mutations altering the targets of the antibiotics were described in the case of M. bovis (9–13).
In this study, we investigated the molecular mechanisms involved in the resistance of M. bovis to seven different antimicrobial families (fluoroquinolones, tetracyclines, aminocyclitol, macrolides, lincosamide, phenicol, and pleuromutilins) by using whole-genome sequencing of field isolates and laboratory-derived mutants as well.
RESULTS
Fluoroquinolones.
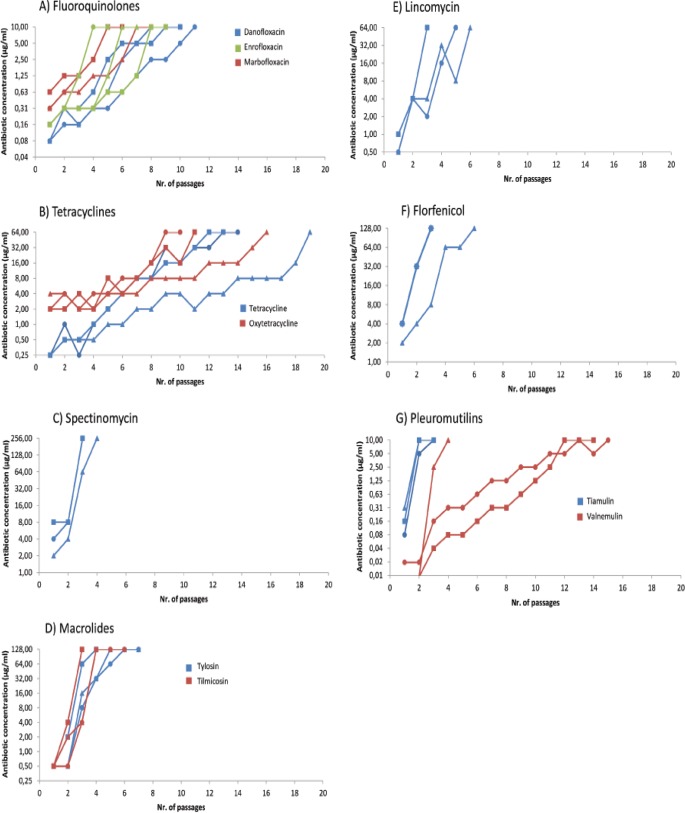
Three strains (MYC44, MYC45, and MYC46) for which the fluoroquinolone MICs were high (>10 μg/ml) were found among the 35 Hungarian field strains (14). In addition, fluoroquinolone-resistant mutants (MIC > 10 μg/ml) were selected in vitro for danofloxacin, enrofloxacin, and marbofloxacin by 7 to 10, 4 to 8, and 5 to 7 passages of the parental strains PG45, MYC52, and MYC53, respectively (Fig. 1A; see also Table S2 in the supplemental material). Mutants remained highly resistant to the selector antibiotic after serial passages in antibiotic-free medium. Moreover, in each case cross-resistance (>10 μg/ml) developed in the resistant mutants to the other fluoroquinolones examined.
FIG 1.
Antibiotic concentrations used during the in vitro selection of resistant M. bovis isolates plotted versus the number of passages. Symbols are used to indicate the parental strains as follows: triangles, M. bovis reference strain (PG45, NCTC 10131); circles, MYC52; and squares, MYC53.
Comparing the whole-genome sequences of strains with extremely high (>10 μg/ml) and low (≤0.625 μg/ml) fluoroquinolone MICs revealed mutations in the quinolone resistance-determining regions (QRDRs) of the DNA gyrase and topoisomerase IV genes (Table 1). All naturally resistant M. bovis strains harbored mutations in gyrA, gyrB, and parC genes. No fluoroquinolone resistance-related mutations were detected in parE.
TABLE 1.
Mutations detected in the gyrA, gyrB, and parC QRDRs of M. bovis field isolates and in vitro-selected mutants with high MICs to fluoroquinolonesa
| Strainb | Mutations associated with high MICs in the current and/or previous studies |
Mutations with unknown effect |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
gyrA |
parC |
gyrA |
gyrB |
|||||||||
| 244, 82 | 248, 83 | 259, 87 | 260, 87 | 232, 78 | 239, 80 | 250, 84 | 1860, 620 | 2038, 680 | 109, 37 | 968, 320 | 1278, 423 | |
| PG45 | G (Asp) | C (Ser)* | G (Glu)* | A (Glu)* | G (Gly)* | G (Ser)* | G (Asp)* | T (Ile) | G (Asp) | C (His) | T (Val) | T (Ile) |
| MYC44-46 | T (Phe) | T (Ile) | C (Ile) | C (Ala) | ||||||||
| MYC52danofloxacin | A (Asn) | A (Asn) | A (Asn) | |||||||||
| MYC53danofloxacin | A (Asn) | A (Lys) | A (Asn) | |||||||||
| PG45danofloxacin | T (Phe) | T (Cys) | ||||||||||
| MYC52marbofloxacin | A (Tyr) | T (Tyr) | A (Asn) | |||||||||
| MYC53marbofloxacin | A (Lys) | C (His) | ||||||||||
| PG45marbofloxacin | A (Lys) | T (Ile) | ||||||||||
| MYC52enrofloxacin | A (Lys) | T (Ile) | ||||||||||
| MYC53enrofloxacin | G (Gly) | A (Asn) | C (His) | |||||||||
| PG45enrofloxacin | A (Lys) | T (Ile) | A (Asn) | |||||||||
Column subheading numbers specify the following: E. coli nucleotide number, E. coli amino acid number. *, Altered positions related to high MICs to fluoroquinolones already described in M. bovis isolates (10, 11, 21, 22).
PG45, M. bovis reference strain NCTC 10131 (MIC = 0.156 μg/ml [danofloxacin and enrofloxacin] and MIC = 0.625 μg/ml [marbofloxacin]); MYC44-46, naturally resistant field isolates (MIC > 10 μg/ml [all tested fluoroquinolones]). Abbreviations of antibiotic-resistant mutants selected in vitro are indicated as follows: straindrug used for selection.
In the gyrA gene in three naturally resistant strains, a silent mutation (T1860C) and a nonsynonymous mutation (C248T), resulting in a Ser83Phe amino acid change, were observed. In the in vitro-selected mutants, position 248 of the gyrA gene was surrounded by additional single-nucleotide polymorphisms (SNPs; G244A, C248T, C248A, G259A, and A260G) that caused the amino acid mutations Asp82Asn, Ser83Tyr, Ser83Phe, Glu87Lys, and Glu87Gly, respectively. In the enrofloxacin-resistant MYC53 mutant strain, a single mutation (G2038C), causing the amino acid alteration Asp680His, was also observed.
In gyrB, a substitution at Escherichia coli nucleotide position 968 was present. This nucleotide, found in naturally resistant M. bovis strains (MYC44, MYC45, and MYC46) but not in selected fluoroquinolone-resistant mutants, results in a Val320Ala alteration. In addition, marbofloxacin- and danofloxacin-resistant MYC52 mutant strains harbored point mutations at C109A and T1278A, causing His37Asn and Ile423Asn amino acid alterations (Table 1).
The SNP G239T of the parC gene of the three naturally fluoroquinolone-resistant strains also caused a Ser80Ile alteration. Resistant mutant strains showed the same mutation (G239T), as well as the following additional mutations of this region of parC: G232T, G250A, G250C, and G250T, resulting in Gly78Cys, Asp84Asn, Asp84His, and Asp84Tyr changes.
Tetracyclines and aminocyclitol.
The tetracycline and oxytetracycline MICs were ≤0.25 and 2 μg/ml for two strains (MYC52 and MYC53), whereas the tetracycline MICs were ≥4 μg/ml and the oxytetracycline MICs were ≥16 for the rest of the strains. Susceptibility to spectinomycin showed a bimodal distribution, with 16/35 strains having an MIC of ≤4 μg/ml and 19/35 strains having an MIC of ≥256 μg/ml, suggesting a high level of resistance to this antibiotic (14).
Mutants with notably increased MICs (>64 μg/ml for tetracyclines and >256 μg/ml for spectinomycin) were obtained with all the selector antibiotics (Fig. 1B and C; see Table S2 in the supplemental material). Tetracycline-, oxytetracycline-, and spectinomycin-resistant strains were selected after 13 to 18, 9 to 15, and 2 to 3 passages, respectively. Resistance to spectinomycin evolved more rapidly in the strains than resistance to tetracyclines. All mutants kept their high-level resistance after serial passages in antibiotic-free medium to the applied antibiotic, with the exception of tetracycline-resistant PG45 mutant (the tetracycline MIC decreased to 8 μg/ml). Cross-resistance between the examined tetracyclines was observed in the mutants. Fourfold increases in MICs to spectinomycin compared to the parental strains were noted in tetracycline-selected mutants (16 μg/ml versus 4 μg/ml). The same results were obtained with spectinomycin-selected mutants, which showed a 4-fold increase in tetracycline MICs (from 0.25 to 1 μg/ml) (see Table S2 in the supplemental material).
Mutations related to tetracycline and spectinomycin resistance were discovered in rrs1 and rrs2 genes encoding the 16S rRNA (Table 2). Mutations A965T and A967T in both rrs1 and rrs2 genes were identified in all field strains with oxytetracycline and tetracycline MICs of ≥16 μg/ml and ≥4 μg/ml, respectively. In tetracycline-resistant mutant strains (with the exception of PG45) mutations were observed at positions 962 to 967, 1058, 1195, and 1196 of the Tet-1 tetracycline binding site located in the 30S ribosomal subunit.
TABLE 2.
Mutations detected in rrs1 and rrs2 genes of M. bovis field isolates and in vitro-selected mutants with high MICs to tetracyclines or spectinomycina
| Strainb | Mutations associated with high MICs in the current and/or previous studies (rrs1-rrs2) |
Mutations with unknown effect (rrs1-rrs2) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 962-964 | 965 | 966 | 967 | 1058 | 1192 | 1195 | 1196 | 1199 | 335 | 859 | 1012 | 1153 | 1268 | |
| PG45 | TGA-TGA | A-A* | G-G* | A-A* | G-G* | C-Cc | C-C | A-A | T-T | C-C | C-C | A-G | G-A | G-A |
| MYC52-53 | ||||||||||||||
| MYC2,42,65-76 | T-T | T-T | T-C | A-A | G-G | G-G | ||||||||
| MYC80 | T-T | T-T | A-A | A-A | G-G | G-G | ||||||||
| MYC30,44-47,77,82-83 | T-T | T-T | A-A | C-T | A-A | G-G | G-G | |||||||
| MYC22,43,48-51,78-79,81,84 | T-T | T-T | A-A | C-C | A-A | G-G | G-G | |||||||
| MYC82 | C-A | A-A | G-G | G-G | ||||||||||
| MYC52tetracycline | T-T | T-T | C-C | |||||||||||
| MYC53tetracycline | T-T | CC-CC | T-T | |||||||||||
| PG45tetracycline | ||||||||||||||
| MYC52oxytetracycline | del-del | |||||||||||||
| MYC53oxytetracycline | T-T | T-T | T-T | |||||||||||
| PG45oxytetracycline | T-T | T-T | ||||||||||||
| MYC52spectinomycin | T-C | |||||||||||||
| MYC53spectinomycin | T-C | |||||||||||||
| PG45spectinomycin | T-C | |||||||||||||
Both SNPs of rrs1 and rrs2 genes are given at certain nucleotide positions, respectively. The E. coli nucleotide number is indicated in the column subheadings. *, Altered positions related to high MICs to tetracyclines already described in M. bovis isolates or in other Mycoplasma species (13, 26). del, deletion.
PG45, M. bovis reference strain NCTC 10131 (MIC = 0.25 μg/ml [tetracycline], MIC = 2 μg/ml [oxytetracycline], and MIC = 4 μg/ml [spectinomycin]); MYC52-53, MIC = 0.25 μg/ml (tetracycline), MIC = 2 μg/ml (oxytetracycline), and MIC = 4 μg/ml (spectinomycin); MYC2,42,65-76, MIC ≥ 4 μg/ml (tetracycline), MIC ≥ 16 μg/ml (oxytetracycline), and MIC ≤ 4 μg/ml (spectinomycin); MYC22,30,43-51,77-84, MIC ≥ 4 μg/ml (tetracycline), MIC ≥ 16 μg/ml (oxytetracycline), and MIC > 256 μg/ml (spectinomycin). Abbreviations of antibiotic-resistant mutants selected in vitro are indicated as follows: straindrug used for selection.
Altered positions related to high MICs to spectinomycin already described in M. bovis (27).
The following SNPs located in the 16S rRNA region not closely associated with Tet-1 or other tetracycline binding sites were also found in all naturally resistant and in vitro-selected mutants: G1012A, A1153G, and A1268G in the rrs2 allele. The same substitutions were also identified in the rrs1 alleles of tylosin-resistant MYC52 and PG45 strains. No tet(M) determinants and derivatives have been identified in the genomes of the examined strains.
All field strains with high spectinomycin MICs (≥256 μg/ml), as well as in vitro-selected mutants, harbored a single mutation at position 1192 of rrs1 gene: a C-to-A mutation in naturally resistant mutants and a C-to-T mutation in resistant mutants, respectively. It should be noted that the substitution C335T was also identified within the rrs1 and rrs2 alleles of field strains with spectinomycin MICs of ≤4 μg/ml but not in the reference strain PG45 (Table 2).
Macrolides, lincosamides, phenicols, and pleuromutilins.
Strains MYC52 and MYC53 were inhibited by low concentrations of macrolides (MIC = 0.5 μg/ml), whereas rest of the strains (33/35) showed tilmicosin MICs of ≥128 μg/ml and tylosin MICs of ≥8 μg/ml. The strains were divided into two groups based on the MICs to lincomycin: strains with MICs of ≥64 μg/ml (19/35) and strains with MICs of ≤2 μg/ml (16/35). Strains with lincomycin MICs of ≥64 μg/ml showed tylosin MICs of ≥128 μg/ml as well (14).
Mutants resistant to tylosin and tilmicosin were obtained after 3 to 6 passages and after 2 to 5 passages, respectively, and remained stable after passages without the selector antibiotics (Fig. 1D; see Table S2 in the supplemental material). Cross-resistance between tylosin and tilmicosin was observed in all macrolide-resistant mutants. At least 8-fold increases in the florfenicol MIC (from 4 to >32 μg/ml) were detected in two cases (the tylosin-resistant mutants MYC53 and PG45), and highly elevated lincomycin MICs (from 0.5 to 1 μg/ml to >64 μg/ml) and tiamulin MICs (from 0.078 to 0.156 μg/ml to 0.625 to 1.25 μg/ml, respectively) were observed in the tilmicosin-resistant MYC52 and MYC53 mutants.
Mutations of rrl1 and rrl2 genes detected in M. bovis strains with elevated MICs to 50S inhibitors are listed in Table 3. In field strains with tilmicosin MICs ≥ 128 μg/ml and tylosin MICs of 8 to 32 μg/ml, substitutions A534T, T1248C, and C1371T in rrl2 and substitution G748A in both rrl1 and rrl2 were detected. In naturally resistant strains with high tylosin MICs (≥128 μg/ml), the additional substitutions G954A in rrl1 and A2059G in both genes were observed. In macrolide-resistant mutants (tylosin and tilmicosin MICs ≥ 128 μg/ml to) the mutations A534T, G748A, G748C, C1371T, A2059G, A2059T, A2063T, C2067T and G2823A were found, as well as the insertion GTG after nucleotide 753 (Table 3).
TABLE 3.
Mutations detected in rrl1, rrl2 genes of M. bovis field isolates and in vitro selected mutants with high MICs to macrolides, lincomycin, florfenicol, or pleuromutilinsa
| Strainb | Mutations associated with high MICs in the current and/or previous studies (rrl1-rrl2) |
Mutations with unknown effect (rrl1-rrl2) |
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 748 | 752 | 2035 | 2059 | 2060 | 2062 | 2063 | 2067 | 2448 | 2500 | 2506 | 2611 | 2612 | 171 | 316 | 534 | 538 | 954 | 1248 | 1371 | 2823 | |
| PG45 | G-G*† | - - | C-C‡ | A-A*† | A-A | G-G† | A-A† | C-C | G-G‡ | C-C‡ | G-G‡ | C-C† | C-C | C-C | C-C | A-A | A-A | G-G | T-T | T-C | G-G |
| MYC52-53 | T-A | ||||||||||||||||||||
| MYC2,42,66-76 | A-A | T-T | T-C | T-T | |||||||||||||||||
| MYC65 | A-A | T-C | T-T | T-C | T-T | ||||||||||||||||
| MYC22,30,43-51,77-79,81-84 | A-A | G-G | T-T | A-G | T-C | T-T | |||||||||||||||
| MYC80 | A-A | G-G | T-T | G-A | A-G | T-C | T-T | ||||||||||||||
| MYC52tylosin | A-A | GTG-GTG | T-T | T-T | |||||||||||||||||
| MYC53tylosin | T-A | T-A | |||||||||||||||||||
| PG45tylosin | T-T | T-T | T-T | T-T | |||||||||||||||||
| MYC52tilmicosin | C-G | G-A | T-A | ||||||||||||||||||
| MYC53tilmicosin | T-A | T-A | |||||||||||||||||||
| PG45tilmicosin | A-G | T-A | A-G | ||||||||||||||||||
| MYC52lincomycin | G-A | T-C | T-A | ||||||||||||||||||
| MYC53lincomycin | T-A | T-A | |||||||||||||||||||
| PG45lincomycin | G-A | T-A | |||||||||||||||||||
| MYC52florfenicol | T-G | T-A | |||||||||||||||||||
| MYC53florfenicol | A-G | T-A | |||||||||||||||||||
| PG45florfenicol | T-A | T-A | |||||||||||||||||||
| MYC52tiamulin | T-G | T-A | |||||||||||||||||||
| MYC53tiamulin | A-G | T-A | |||||||||||||||||||
| PG45tiamulin | T-G | T-A | |||||||||||||||||||
| MYC52valnemulin | T-T | ||||||||||||||||||||
| MYC53valnemulin | A-C | G-A | A-C | T-C | C-T | T-A | |||||||||||||||
| PG45valnemulin | T-G | C-T | T-A | ||||||||||||||||||
Both SNPs of rrl1 and rrl2 genes are given at the certain nucleotide positions, respectively. The E. coli nucleotide number is indicated in the column subheadings. *, Altered positions related to high MICs to macrolides in M. bovis (9); †, positions related to high MICs to macrolides and lincomycin in other Mycoplasma species (33, 35–38); ‡, neighboring mutated positions already described in Mycoplasma gallisepticum (39) and in other bacterial species related to phenicol and pleuromutilin resistance (44, 46).
PG45, M. bovis reference strain NCTC 10131 (MIC = 0.5 μg/ml [tylosin and tilmicosin] and MIC = 1 μg/ml [lincomycin]); MYC52-53, MIC = 0.5 μg/ml (tylosin and tilmicosin) and MIC ≤ 1 μg/ml (lincomycin); MYC2,42,65-76, MIC ≥ 128 μg/ml (tilmicosin), MIC ≤ 32 μg/ml (tylosin), and MIC ≤ 2 μg/ml (lincomycin); MYC22,30,43-51,77-84, MIC ≥ 128 μg/ml (tylosin and tilmicosin) and MIC ≥ 64 μg/ml (lincomycin). Abbreviations of antibiotic-resistant mutants selected in vitro are indicated as follows: straindrug used for selection.
No mutations associated with high macrolide MICs were found in field strains in genes encoding ribosomal proteins L4 and L22. Only tylosin-resistant strain MYC52 harbored a substitution (C191T; according to M. bovis PG45 numbering) causing alteration Ala64Val of the L4 protein (according to PG45 numbering).
Lincomycin-resistant M. bovis mutant strains were obtained after passages 2 to 5 (Fig. 1E; see Table S2 in the supplemental material). Only lincomycin-resistant PG45 mutant showed a decrease in MIC from >64 to 16 μg/ml after serial passages in antibiotic-free medium. Lincomycin-resistant mutant strains (with the exception of the lincomycin-resistant PG45 strain) were associated with highly increased tylosin (from 0.5 to >128 μg/ml), tilmicosin (from 0.5 to >128 μg/ml), and tiamulin (from 0.078 to 0.0156 μg/ml to 1.25 to 5 μg/ml) MICs. In one case (MYC53), a slightly elevated valnemulin MIC (from <0.039 to 0.039 μg/ml) was also observed.
All field strains with lincomycin MICs ≥ 64 μg/ml harbored mutation A2059G in both the rrl1 and the rrl2 alleles, and only the MYC53 mutant harbored this alteration in one rrl1 gene (Table 3). Both lincomycin-resistant MYC52 and PG45 mutants possessed the substitution A2060G in rrl1 gene, and an additional SNP (C2612T) in MYC52 was also detected. Mutants which showed high level cross-resistance to lincomycin (≥64 μg/ml) also had a substitution at either position 2059 or position 2060 of the rrl1 gene, confirming the role of these nucleotides in lincomycin resistance: A2059G in tilmicosin-resistant MYC52, A2059T in tilmicosin-resistant MYC53, and A2060G in valnemulin-resistant MYC53 (Table 3).
Most Hungarian strains showed the same florfenicol MIC (MIC90 = 8 μg/ml) (14). Stable, florfenicol-resistant mutant strains (>32 μg/ml) were selected after passages 2 to 5 (Fig. 1F; see also Table S2 in the supplemental material). In florfenicol-resistant PG45, notable changes in the tylosin (from ≤0.5 to >128 μg/ml) and tilmicosin (from ≤0.5 to 16 μg/ml) MICs and a 2-fold change in the lincomycin MIC (from 1 to 2 μg/ml) were noted. In contrast, in the florfenicol-resistant MYC52 and MYC53 strains only slight increases in the tylosin (from ≤0.5 μg/ml to 2 and 4 μg/ml) and tilmicosin (from ≤0.5 to 8 μg/ml) MICs were observed, but significant increases in the lincomycin (from ≤0.5 and 1 μg/ml to 8 μg/ml), tiamulin (from 0.078 and 0.156 μg/ml to 0.625 and 10 μg/ml), and valnemulin (from ≤0.039 μg/ml to 0.039 and 10 μg/ml) MICs were identified.
Eleven laboratory-selected mutants were described with high florfenicol MICs (≥16 μg/ml); among them, seven strains harbored mutation G2062T or A2063T in at least one allele of the 23S rRNA gene (Table 3; see Table S2 in the supplemental material). In addition, substitution G2506A, described in the florfenicol-resistant MYC52 mutant, showed cross-resistance to tiamulin.
The tiamulin and valnemulin MICs determined by a broth microdilution test are shown in Fig. 2 and Table S1 in the supplemental material. The tiamulin MIC50 and MIC90 values were 0.156 and 0.312 μg/ml, respectively, and the valnemulin MIC50 and MIC90 values were ≤0.039 μg/ml.
FIG 2.
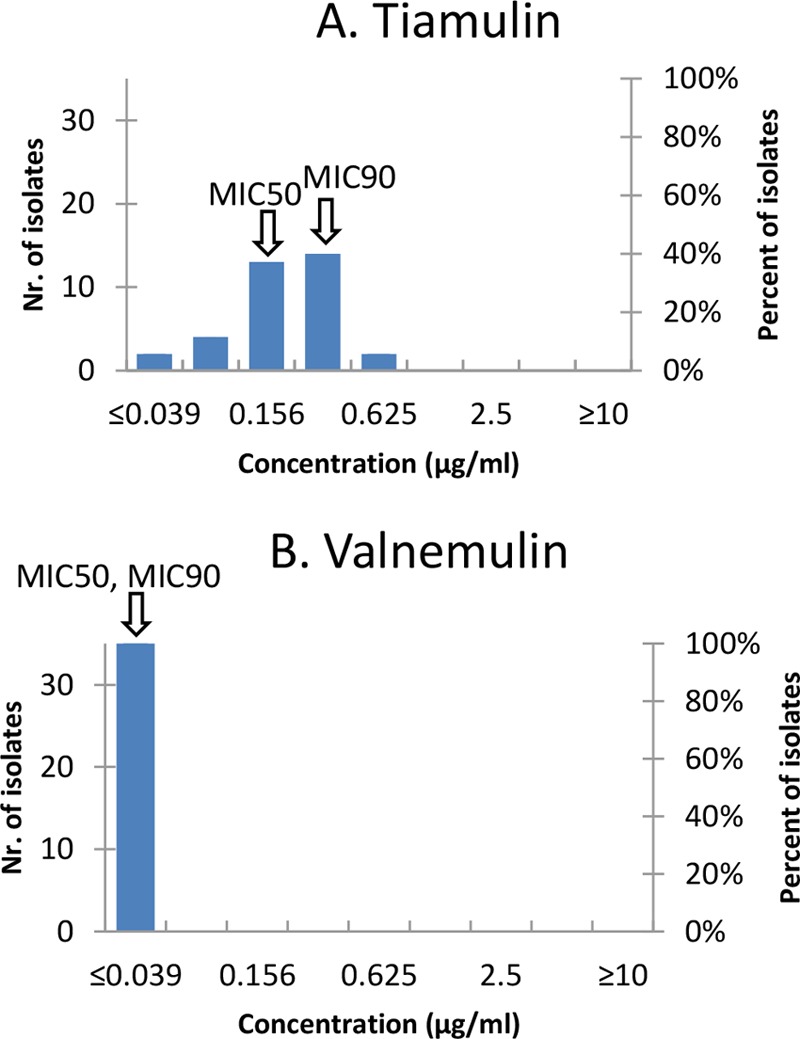
MIC distributions for the 35 Hungarian M. bovis isolates and pleuromutilins tested in this study. (A) Tiamulin; (B) valnemulin. Arrows indicate the MIC50 and MIC90 values.
Tiamulin- and valnemulin-resistant mutant strains were successfully obtained after passages 2 to 3 and passages 3 to 14, respectively (Fig. 1G; see Table S2 in the supplemental material). All tiamulin-resistant mutant strains showed cross-resistance to florfenicol (MIC > 32 μg/ml) and elevated lincomycin MICs (4 to 16 μg/ml). Tiamulin-resistant PG45 became resistant to all of the tested 50S inhibitors except tylosin. The development of valnemulin resistance strongly differed among the strains: PG45 became resistant after only 3 passages, whereas MYC52 and MYC53 needed 14 and 10 passages, respectively. After five passages on antibiotic-free medium, the valnemulin MIC value (0.078 μg/ml) for mutant MYC52 decreased. Highly elevated MICs to all tested 50S inhibitors developed in the valnemulin-resistant MYC53 mutant.
In tiamulin- and valnemulin-resistant mutant strains, the substitutions C2035A, A2060G, G2062T, and C2500A were found; these positions are closely associated with the pleuromutilin binding sites of the 23S rRNA genes. Other mutations were observed in a region not related to the antibiotic binding pocket (C171T and C316A). No chromosomal mutations of the rrl genes were detected in the tiamulin-resistant strain MYC53 and the valnemulin-resistant MYC52 strain. The C/T mutation was observed in the neighboring positions 2611 and 2612 in the valnemulin-resistant MYC53 and the lincomycin-resistant MYC52 mutant strains, respectively (Table 3).
DISCUSSION
Antibiotic resistance is fast evolving in M. bovis strains all over the world (5). Based on our in vitro examinations, pleuromutilins and fluoroquinolones are the most active compounds against M. bovis infection, in agreement with previous observations (5, 7). However, interpretation of the results of the in vitro examinations should be handled with caution since in vivo factors, such as the concentration of the applied antibiotics in the target organs and cells or biofilm formation of the bacteria, can also influence the efficiency of the treatment (15, 16).
Differences were observed between mutations identified in naturally resistant and mutant strains. The spread of M. bovis infection is often reported to be connected to animal transportations or to the introduction of carrier animals to naive herds (3). It has shown that in nature a certain clone which achieved resistance has been dispersed (17), whereas the laboratory-selected mutants achieved their resistance in multiple ways.
The obtained high MICs of mutants to some antibiotics (e.g., tetracycline and valnemulin) tended to decrease rapidly in the absence of the selector agent. The reason for this rapid regaining of susceptibility has not been clarified, but mutations yielding resistance could be reverted or neutralized by additional mutations during passages.
Although fluoroquinolones are considered to be effective agents in the therapy of bovine mycoplasmosis, strains with high MICs (≥8 μg/ml) have been reported from several countries (5, 7, 14, 18). The development of resistance to the fluoroquinolones is usually due to target mutations in QRDRs of the genes encoding topoisomerases, but enhanced efflux of the drug by the overexpression of efflux pumps has also been described (6). The in vitro development of fluoroquinolone resistance in M. bovis in the present study and in other Mycoplasma species in previous studies showed a gradual pattern that suggested multistep mutation events (11, 19, 20). For the increase in fluoroquinolones MICs, substitutions in GyrA are sufficient, and resistance to fluoroquinolones is achieved by an additional change in ParC (10, 11). All M. bovis strains in the present study with high fluoroquinolone MICs (≥10 μg/ml) contained at least one substitution in both gyrA and parC genes. Earlier described mutation hotspots in GyrA and ParC of naturally and artificially selected resistant M. bovis strains have been identified between amino acid positions 81 to 87 and positions 78 to 84, respectively (10, 11, 21, 22). However, the SNP (G244A) causing increased MIC values to fluoroquinolones at position 82 (Asp→Asn) in GyrA protein of M. bovis is described for the first time. An alteration in GyrB at position 320 was observed in all three naturally resistant strains (MYC44, MYC45, and MYC46), which was likely associated with the strains' genetic relatedness (23) instead of fluoroquinolone resistance given the absence of the mutation in the resistant mutant strains. The mutations at the positions, which were not related to the hotspots in the QRDRs (Asp680His of GyrA, as well as His37Asn and Ile423Asn of GyrB) and occurred in only one of the in vitro-selected strains, have unknown influence on fluoroquinolone resistance (Table 1).
Tetracyclines and spectinomycin are protein synthesis inhibitors primarily binding to the 30S ribosomal subunit of the bacterial ribosome (24, 25). In animal pathogenic mycoplasmas, tetracycline and spectinomycin resistance are mainly achieved by point mutations in the 16S rRNA gene altering the tetracycline or spectinomycin binding sites (13, 26, 27), whereas the tetracycline resistance of M. hominis and Ureaplasma spp. are mainly associated with the tet(M) determinant (28–30).
Tetracyclines, and particularly oxytetracycline, are generally used for the treatment of M. bovis-related diseases, but increasing resistance have been reported from Europe and other continents as well (7, 14, 18). The number of mutations in region Tet-1 was shown to correlate with the increase of MICs to tetracycline in M. bovis strains (13). The Tet-1 binding site is located in a pocket formed by segments 1054-1056 and 1196-1200 of helix 34 and segment 964-967 of helix 31 in the 30S ribosomal subunit of Thermus thermophilus. In helix 34, bases 1196 and 1054 form a clamp that holds tetracycline in hydrophobic interactions (24). Similarly to previously examined Mycoplasma species (20, 26), in the present study resistance to tetracyclines developed gradually in M. bovis, suggesting the accumulation of several mutations in the genome. Mutations associated with tetracycline resistance were identified in the Tet-1 site in accordance with previous works (13, 26), e.g., the mutations of helix 34 at positions 1195 to 1199 were described in naturally and artificially resistant strains, whereas the substitution G1058C was observed in only one case (tetracycline-resistant MYC52). Mutations in helix 31 were noted between nucleotides 962 and 967. The effect of SNPs G1012A, A1153G, and A1268G of the rrs2 allele described outside the regions Tet-1 and other tetracycline binding sites identified in all naturally and in vitro selected mutants, and even in tylosin-resistant MYC52 and PG45 strains, has not been clarified (Table 2).
Cross-resistance was observed between tetracycline- and oxytetracycline-resistant mutants, and only slightly elevated MICs to spectinomycin were detected. This finding is in agreement with a previous work suggesting the regional effect of tetracycline, which is caused by its separated binding sites in both secondary and tertiary structures of the 16S rRNA (25).
The use of spectinomycin against M. bovis is widespread, but its decreased effectiveness have been reported all over the world (5, 7, 8, 14). Early studies confirmed the role of position 1192 in 16S rRNA in the antibiotic susceptibility of E. coli: a C-to-A or G-or-T substitution results in spectinomycin resistance (25, 31). The same observations were described for M. bovis strains (27). It is important to note that susceptibility to spectinomycin can be restored in C1192 mutants by an additional substitution at the position T1351C of helix 43 (32). In contrast to the observations of the development of resistance to tetracyclines, resistance to spectinomycin evolved rapidly. This finding is in harmony with the detected spectinomycin susceptibility profile of the 35 Hungarian strains, which either showed low (≤4 μg/ml) or high MICs (≥256 μg/ml) for this antibiotic (14). Whole-genome analysis distinguished two main groups among the strains according to their spectinomycin MICs also. A mutation was found in only one position of the genome: in the rrs1 gene, helix 34, substitution C1192A occurred in naturally resistant strains and substitution C1192T occurred in spectinomycin-resistant mutant strains.
Macrolides, lincosamides, phenicols, and pleuromutilins all belong to the group of protein synthesis inhibitor antibiotics that bind the 50S subunit of the bacterial ribosome. The alteration of the 23S rRNA is the main mechanism of resistance in mycoplasmas to 50S inhibitors (5). Mutations in L4 and L22 ribosomal proteins encoded by the rplD and rplV genes were shown to be responsible for the macrolide and lincosamide resistance of in vitro-selected M. pneumoniae strains (33).
Macrolides have been used traditionally in the therapy of mycoplasmosis in cattle, but now these antibiotics are losing their efficiency against M. bovis in many countries (8, 12, 14, 18). The major components of the macrolide binding site on the 23S rRNA ribosome are the central loop of domain V and the loop of hairpin 35 from domain II (34). One or more point mutations at the macrolide binding site in domain II (positions 748, 752, and 792) and/or domain V (positions 2056 to 2059, positions 2062 to 2064, and positions 2576, 2586, 2597, 2608, and 2611) cause decreased susceptibility to macrolides and/or lincomycin in Mycoplasma species (9, 12, 33, 35–38). In M. bovis the presence of any of the SNPs in positions 748 or 752 (domain II) or positions 2058 or 2059 (domain V) correlated with decreased susceptibility to the 16-membered macrolides tylosin and tilmicosin (9).
In the present study, the rapid evolution of macrolide and lincomycin resistance was observed in the in vitro-selected M. bovis mutants similarly to avian Mycoplasma species (20), presuming the small number of mutation events needed for the change of antibiotic susceptibility in these cases. Mutations (n = 1 to 3/strain) in domain II (position 748 and insertion after nucleotide C752) were necessary to achieve tilmicosin and tylosin MICs of ≥128 and ≤32 μg/ml, respectively, whereas an additional mutation in domain V (positions 2059, 2060, 2063, and 2067) was needed to reach highly elevated tylosin (MIC ≥ 128 μg/ml) and lincomycin (MIC ≥ 64 μg/ml) MICs. The mutation at position 2058 (the most common point mutation in other Mycoplasma species) was not reported here (9, 12, 35, 38, 39). The acquisition of resistance in M. bovis is generally due to the emergence and spread of a single clone decreasing the genetic heterogeneity of antimicrobial-resistant M. bovis isolates (10, 17). The resistant isolates occurring in Hungary harbored the alteration of a neighboring position (position 2059) in rrl genes, an important position in macrolide resistance also (9, 35, 36, 39). The distribution of the strain MIC values for tylosin and tilmicosin were consistent with the distribution of the MICs for new generation macrolides (gamithromycin and tulathromycin [K. M. Sulyok et al., unpublished data]), suggesting similar resistance mechanisms. The alterations found in all lincomycin-resistant mutants emphasize the importance of positions 2059 and 2060 of domain V in the development of lincomycin resistance. Mutation of the L4 ribosomal protein (Ala64Val) was detected in one mutant strain, but its importance has not yet been clarified.
Pleuromutilins are used mainly in veterinary practice, especially in swine and poultry, but valnemulin also proved to be effective in calves infected with M. bovis (40). This antimicrobial group binds to the peptidyl transferase center and also inhibits the peptide bond formation and protein synthesis. Pleuromutilin resistance commonly derives from chromosomal mutations in the 23S rRNA and rplC genes and emerges relatively slowly in a stepwise fashion (41). In the present study, slow evolution of valnemulin resistance was observed in the mutant MYC52 and MYC53 strains, whereas PG45 obtained resistance within three passages. Florfenicol- and tiamulin-resistant mutant strains evolved resistance rapidly also (in two to five steps). The number of mutations correlated with the number of passages needed for the evolution of resistance (e.g., five mutations in MYC53 and one mutation in PG45 in the case of valnemulin resistance).
Some nucleotides of 23S rRNA genes between positions 2044 to 2572 have been described as taking part in the formation of the active pocket of pleuromutilin derivatives in different bacterial species (42–45). The M. bovis pleuromutilin- or florfenicol-resistant mutants of this study harbored mutations at nucleotide positions 2035, 2060, 2062, 2063, 2448, 2500, 2506, and 2611 of the 23 rRNA genes in close association with previously described resistance-related positions, hypothesizing that these mutations have an effect on antibiotic susceptibility (Table 3) (42–46). A mutation at the position 2032 (closely associated with position 2035 of valnemulin-resistant MYC53) was previously reported in B. hyodysenteriae isolates, also with high tiamulin and lincosamides MICs (46). Strains with mutation G2062T of rrl1 gene had highly elevated florfenicol, tiamulin, and valnemulin MICs and increased lincomycin and tilmicosin MICs, with the exception of florfenicol-resistant PG45. Mutants harboring A2063T substitution had eight times higher MIC values to florfenicol than their parental strains, suggesting the impact of this position in florfenicol resistance. The mutations at positions 2500 and 2506 found in the present study are closely related to the ribosomal binding sites of phenicols and pleuromutilins (39, 41). In M. gallisepticum mutation A2503U leads to decreased susceptibility to phenicols and lincomycin beside pleuromutilins (39) and alteration of nucleotides 2498, 2499 2503 and 2504 causes resistance to pleuromutilins or florfenicol in different bacterial species (41, 47). Alteration of position 2448 in tiamulin resistant MYC53 lead to decreased susceptibility to florfenicol and pleuromutilins, and the importance of position 2447 in pleuromutilin resistance was previously described in M. gallisepticum. Previous studies have described the substitution C2611G in domain V of M. pneumoniae and the G2597U and C2611U mutations of M. hyorhinis, confirming their effect on 50S inhibitor resistance (36, 38). The effects of polymorphisms at positions 171 and 316 have not been clarified yet.
Cross-resistance between macrolides and lincomycin has long been described (35, 48), and cross-resistance was also reported between macrolides, pleuromutilins, lincomycin, and florfenicol (39, 41). In the present study, the valnemulin-resistant MYC53 harboring mutations at positions 2035, 2060, 2500, and 2611 had elevated MICs to all of the tested 50S inhibitors. Mutation C2611T was detected in valnemulin-resistant MYC53, which is closely related to mutation C2612T of lincomycin-resistant MYC52, both with elevated MICs to several 50S inhibitors. In two cases (valnemulin-resistant PG45 and MYC53) the rapid development of high level cross-resistance to the tested 50S inhibitors was detected. These results emphasize the importance of responsible antibiotic usage.
Our study described the emergence of antibiotic-resistant M. bovis strains to seven antimicrobial families and showed that mutations of clinical isolates and laboratory-derived mutants in GyrA and ParC proteins, and in 16S rRNA and 23S rRNA genes were responsible for resistance to fluoroquinolones and to 30S and 50S inhibitory antibiotics, respectively. Some of these mutations led to cross-resistance to other classes of antibiotics with similar mechanisms of action. A knowledge of the genetic background of antibiotic resistance is essential for the development of genetic-based diagnostic assays and also for the prevention of the spread of antibiotic-resistant clones.
MATERIALS AND METHODS
M. bovis strains, antimicrobial agents, and susceptibility testing.
A total of 35 M. bovis strains originating from different parts of Hungary were examined in the study (see Table S1 in the supplemental material). The isolation and identification methods of the strains have been previously described (14, 23).
Susceptibility of the M. bovis isolates and the reference strain (M. bovis PG45, NCTC 10131) to certain fluoroquinolones (danofloxacin, enrofloxacin, and marbofloxacin), an aminocyclitol (spectinomycin), tetracyclines (tetracycline and oxytetracycline), macrolides (tylosin and tilmicosin), a phenicol (florfenicol), and a lincosamide (lincomycin) were previously determined by the broth microdilution method (14) according to the recommendation of Hannan (49). Two pleuromutilins (tiamulin and valnemulin) were also included in the present study. Stock solutions (1 mg/ml) of tiamulin (Vetranal; Sigma-Aldrich, Germany) and valnemulin (Vetranal; Sigma-Aldrich) were prepared in sterile-distilled water, and aliquots were stored at −70°C. Twofold dilutions of pleuromutilins in the range 0.039 to 10 μg/ml were freshly prepared for each microtest.
The final MIC value for each strain was defined as the lowest concentration of the antibiotic that completely inhibits the growth in the broth (no pH and color change). The reference strain (M. bovis PG45, NCTC 10131) was used as a quality control of the MIC determination.
Selection of antibiotic-resistant mutants of M. bovis.
Selection of antibiotic-resistant mutants was performed on the M. bovis reference strain (PG45, NCTC 10131) and two field isolates (MYC52 and MYC53) with low MIC values for most of the tested antibiotics (14) (see Table S2 in the supplemental material). The selection was carried out by serial passages of the strains in mycoplasma broth medium (Thermo Fisher Scientific, Inc./Oxoid, Inc., Waltham, MA) containing subinhibitory concentrations (increasing in 2-fold dilutions) of each of the examined antibiotics (19, 33). The culture containing the highest antibiotic concentration with detectable growth (red to orange shift) was used to inoculate another antibiotic dilution panel for the following passage series. Passages were performed until MIC values reached ≥10 μg/ml for fluoroquinolones and pleuromutilins, ≥64 μg/ml for tetracyclines and lincomycin, ≥128 μg/ml for florfenicol, and ≥256 μg/ml for spectinomycin.
Resistant mutants were passed through antibiotic-free medium at least five times, and the MICs of all drugs were determined again by the end of the passages to check whether the phenotype was stable without selection pressure. Cross-resistance was also examined with broth microdilution tests among all examined drugs.
Whole-genome sequencing and sequence analysis.
M. bovis genomic DNAs of the 35 Hungarian strains, the reference strain (PG45, NCTC 10131), and the 36 in vitro-selected mutants were extracted from 2 ml of logarithmic-phase broth cultures by using a QIAamp DNA minikit (Qiagen, Inc., Hilden, Germany) according to the manufacturer's instructions. Two PCR systems were used for the specific amplification of the 23S rRNA genes rrl1 and rrl2 and the 16S rRNA genes rrs1 and rrs2. PCR amplification of the rrl1 and rrs1 genes was performed with MB-282-F and MB-rrl-3R primers according to the method of Lerner et al. (9). To amplify the rrl2 and rrs2 genes, a PCR system was designed in this study. Reactions were carried out in 50-μl final volumes containing 0.4 μl of TaKaRa Taq polymerase (5 U/μl; TaKaRa Bio, Inc., Otsu, Japan), 5 μl of 10× PCR buffer containing 15 mM MgCl2 (TaKaRa Bio), 4 μl of deoxynucleoside triphosphate mixture (2.5 mM; TaKaRa Bio), 2.5 μl of each primer (10 pM/μl; MB-rrs4-F2 [5′-GCA TGT CGA GCG ATG ATA GC-3′] and MB-287-R [5′-CTA ATT CCA AGT GCC ACT AGC G-3′]), and 1 μl of a DNA template. PCRs were carried out in T100 series thermocycler (Bio-Rad, Hercules, CA) according to the following parameters: 95°C for 5 min, followed by 30 cycles of 95°C for 15 s, 56°C for 30 s, and 72°C for 4 min, followed in turn by 72°C for 10 min. Products for sequencing were purified by using a QIAquick gel extraction kit (Qiagen) according to the manufacturer's instructions.
Next-generation sequencing of the 142 PCR products (containing separated rrl and rrs genes) and 71 whole genomes (35 field strains and 36 mutant strains) was performed on ion torrent platform as previously described (50). A total of 100 ng of DNA was subjected to enzymatic fragmentation using the reagents supplied in the NEBNext Fast DNA Fragmentation & Library Prep Set for Ion Torrent kit (New England BioLabs, Hitchin, United Kingdom) according to the manufacturer's instructions. The adaptor ligation was performed using reagents from the same kit, whereas barcoded adaptors were retrieved from the Ion Xpress Barcode Adapters (Life Technologies, Inc., Waltham, MA). The barcoded library DNA samples were column purified using the Gel/PCR DNA fragments extraction kit (Geneaid Biotech, Ltd., Taipei, Taiwan). The eluted library DNA was then run on a 2% precast gel (Thermo Fisher Scientific). Products between 300 and 350 bp were directly used without further purification in the PCR mixture of the NEBNext Fast DNA Fragmentation & Library Prep Set for Ion Torrent kit (New England BioLabs). Library amplification was performed in 12 cycles, and the products were purified by the Gel/PCR DNA fragments extraction kit (Geneaid). The library DNA was eluted in 50 μl of nuclease-free water and quantified fluorometrically on Qubit 2.0 equipment using a Qubit dsDNA BR assay kit (Invitrogen). Subsequently, the library DNA was diluted to 10 to 14 pM and then clonally amplified by emulsion PCR. This step was carried out according to the manufacturer's protocol using the Ion PGM template kit on a OneTouch v2 instrument. Enrichment of the templated beads (on an Ion One Touch ES machine) and further steps for presequencing setup were performed according to the 200-bp protocol of the manufacturer. The sequencing protocol recommended for the Ion PGM sequencing kit on a 316 chip was strictly followed.
Sequence analysis and alignment were performed using DNASTAR software (v12.1.0.145; Lasergene, Inc., Madison, WI). Sequence data were mapped to M. bovis PG45 complete genome sequences (CP002188 and NC_014760.1) from GenBank. Putative SNPs between reference genome and genomes of 35 field strains and the 36 antibiotic-resistant mutants selected in vitro were identified under the following settings: mer size, 19 nucleotides; minimum match percentage, 93%; minimum alignment length, 25; maximum gap size, 30; match score, 10; mismatch penalty, 20; gap penalty, 50; gap extension penalty, 10; alignment cutoff, 200; SNP calculation method, haploid Bayesian; P not reference percentage, 75%; Phred score (Q call), 30; SNP percentage, 75 to 100%; and coverage depth minimum, 5. The average numbers of reads and read lengths were 184,450 reads and 177 bp for whole genomes and 4,985 reads and 169 bp for PCR products. The overall coverage and the depth of coverage were 31.9 and 30.3 for the whole genome per strain and 174.7 and 113.6 for the amplicons per strain. The validity of SNPs was confirmed by manual examination of the assembled sequences.
Numbering of nucleotide and amino acid positions is based on genes and proteins of Escherichia coli strain K-12 substrain MG1655 (GenBank accession number CP014225) unless indicated otherwise. Nomenclature for rrn genes was used throughout the present study as suggested by Amram et al. (51), that is, rrs1 and rrs2 genes instead of rrs3 and rrs4 genes and rrl1 and rrl2 genes instead of rrl3 and rrl4 genes.
Accession number(s).
The database accession numbers for the genes examined in this study are as follows: KR493099 to KR493133 and KX462228 to KX462236 for gyrA, KR493134 to KR493141 and KT218638 to KT218664 for gyrB, KR493142 to KR493176 and KX462237 to KX462245 for parC, KX462246 to KX462298 for rrl1, KX462299 to KX462351 for rrl2, KX462352 to KX462395 for rrs1, and KX462396 to KX462439 for rrs2.
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by the Lendület (Momentum) program of the Hungarian Academy of Sciences and supported by the New National Excellence Program of the Ministry of Human Capacities. The funder had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01983-16.
REFERENCES
- 1.Perron GG, Inglis RF, Pennings PS, Cobey S. 2015. Fighting microbial drug resistance: a primer on the role of evolutionary biology in public health. Evol Appl 8:211–222. doi: 10.1111/eva.12254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Citti C, Blanchard A. 2013. Mycoplasmas and their host: emerging and re-emerging minimal pathogens. Trends Microbiol 21:196–203. doi: 10.1016/j.tim.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 3.Nicholas RAJ, Ayling RD. 2003. Mycoplasma bovis: disease, diagnosis, and control. Res Vet Sci 74:105–112. doi: 10.1016/S0034-5288(02)00155-8. [DOI] [PubMed] [Google Scholar]
- 4.Taylor-Robinson D, Bebear C. 1997. Antibiotic susceptibilities of mycoplasmas and treatment of mycoplasmal infections. J Antimicrob Chemother 40:622–630. doi: 10.1093/jac/40.5.622. [DOI] [PubMed] [Google Scholar]
- 5.Lysnyansky I, Ayling RD. 2016. Mycoplasma bovis: mechanisms of resistance and trends in antimicrobial susceptibility. Front Microbiol 7:595. doi: 10.3389/fmicb.2016.00595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piddock LJ. 1999. Mechanisms of fluoroquinolone resistance: an update 1994-1998. Drugs 58(Suppl 2):S11–S18. [DOI] [PubMed] [Google Scholar]
- 7.Gautier-Bouchardon AV, Ferré S, Le Grand D, Paoli A, Gay E, Poumarat F. 2014. Overall decrease in the susceptibility of Mycoplasma bovis to antimicrobials over the past 30 years in France. PLoS One 9:e87672. doi: 10.1371/journal.pone.0087672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heuvelink A, Reugebrink C, Mars J. 2016. Antimicrobial susceptibility of Mycoplasma bovis isolates from veal calves and dairy cattle in the Netherlands. Vet Microbiol 189:1–7. doi: 10.1016/j.vetmic.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 9.Lerner U, Amram E, Ayling RD, Mikula I, Gerchman I, Harrus S, Teff D, Yogev D, Lysnyansky I. 2014. Acquired resistance to the 16-membered macrolides tylosin and tilmicosin by Mycoplasma bovis. Vet Microbiol 168:365–371. doi: 10.1016/j.vetmic.2013.11.033. [DOI] [PubMed] [Google Scholar]
- 10.Lysnyansky I, Mikula I, Gerchman I, Levisohn S. 2009. Rapid detection of a point mutation in the parC gene associated with decreased susceptibility to fluoroquinolones in Mycoplasma bovis. Antimicrob Agents Chemother 53:4911–4914. doi: 10.1128/AAC.00703-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sato T, Okubo T, Usui M, Higuchi H, Tamura Y. 2013. Amino acid substitutions in GyrA and ParC are associated with fluoroquinolone resistance in Mycoplasma bovis isolates from Japanese dairy calves. J Vet Med Sci 75:1063–1065. doi: 10.1292/jvms.12-0508. [DOI] [PubMed] [Google Scholar]
- 12.Kong LC, Gao D, Jia BY, Wang Z, Gao YH, Pei ZH, Liu SM, Xin JQ, Ma HX. 2016. Antimicrobial susceptibility and molecular characterization of macrolide resistance of Mycoplasma bovis isolates from multiple provinces in China. J Vet Med Sci 78:293–296. doi: 10.1292/jvms.15-0304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amram E, Mikula I, Schnee C, Ayling RD, Nicholas RAJ, Rosales RS, Harrus S, Lysnyanskya I. 2015. 16S rRNA gene mutations associated with decreased susceptibility to tetracycline in Mycoplasma bovis. Antimicrob Agents Chemother 59:796–802. doi: 10.1128/AAC.03876-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sulyok KM, Kreizinger Z, Fekete L, Hrivnák V, Magyar T, Jánosi S, Schweitzer N, Turcsányi I, Makrai L, Erdélyi K, Gyuranecz M. 2014. Antibiotic susceptibility profiles of Mycoplasma bovis strains isolated from cattle in Hungary, Central Europe. BMC Vet Res 10:256. doi: 10.1186/s12917-014-0256-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McAuliffe L, Ellis RJ, Miles K, Ayling RD, Nicholas RAJ. 2006. Biofilm formation by mycoplasma species and its role in environmental persistence and survival. Microbiology 152:913–922. doi: 10.1099/mic.0.28604-0. [DOI] [PubMed] [Google Scholar]
- 16.Reeve-Johnson L. 1999. The impact of mycoplasma infections in respiratory disease in cattle in Europe, p 18–31. In Stipkovits L, Rosengarten R, Frey J (ed), Mycoplasmas of ruminants: pathogenicity, diagnostics, epidemiology, and molecular genetics European Commission Press, Brussels, Belgium. [Google Scholar]
- 17.Becker CA, Thibault FM, Arcangioli MA, Tardy F. 2015. Loss of diversity within Mycoplasma bovis isolates collected in France from bovines with respiratory diseases over the last 35 years. Infect Genet Evol 33:1–9. doi: 10.1016/j.meegid.2015.04.012. [DOI] [PubMed] [Google Scholar]
- 18.Ayling RD, Rosales RS, Barden G, Gosney FL. 2014. Changes in antimicrobial susceptibility of Mycoplasma bovis isolates from Great Britain. Vet Rec 175:486. doi: 10.1136/vr.102713. [DOI] [PubMed] [Google Scholar]
- 19.Gruson D, Pereyre S, Renaudin H, Charron A, Bébéar CM, Bébéar CM, Be C. 2005. In vitro development of resistance to six and four fluoroquinolones in Mycoplasma pneumoniae and Mycoplasma hominis, respectively. Antimicrob Agents Chemother 49:1190–1193. doi: 10.1128/AAC.49.3.1190-1193.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gautier-Bouchardon AV, Reinhardt AK, Kobisch M, Kempf I. 2002. In vitro development of resistance to enrofloxacin, erythromycin, tylosin, tiamulin and oxytetracycline in Mycoplasma gallisepticum, Mycoplasma iowae, and Mycoplasma synoviae. Vet Microbiol 88:47–58. doi: 10.1016/S0378-1135(02)00087-1. [DOI] [PubMed] [Google Scholar]
- 21.Mustafa R, Qi J, Ba X, Chen Y, Hu C, Liu X, Tu L, Peng Q, Chen H, Guo A. 2013. In vitro quinolones susceptibility analysis of Chinese Mycoplasma bovis isolates and their phylogenetic scenarios based upon QRDRs of DNA topoisomerases revealing a unique transition in ParC. Pak Vet J 33:85–92. [Google Scholar]
- 22.Khalil D, Becker CAM, Tardy F. 2015. Alterations in the quinolone resistance determining regions and fluoroquinolone resistance in clinical isolates and laboratory-derived mutants of Mycoplasma bovis: not all genotypes may be equal. Appl Environ Microbiol 82:1060–1068. doi: 10.1128/AEM.03280-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sulyok KM, Kreizinger Z, Fekete L, Jánosi S, Schweitzer N, Turcsányi I, Makrai L, Erdélyi K, Gyuranecz M. 2014. Phylogeny of Mycoplasma bovis isolates from Hungary based on multi locus sequence typing and multiple-locus variable-number tandem repeat analysis. BMC Vet Res 10:108. doi: 10.1186/1746-6148-10-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pioletti M, Schlünzen F, Harms J, Zarivach R, Glühmann M, Avila H, Bashan A, Bartels H, Auerbach T, Jacobi C, Hartsch T, Yonath A, Franceschi F. 2001. Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine, and IF3. EMBO J 20:1829–1839. doi: 10.1093/emboj/20.8.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noah JW, Dolan MA, Wollenzien P, Babin P. 1999. Nucleic acids, protein synthesis, and molecular genetics: effects of tetracycline and spectinomycin on the tertiary structure of ribosomal RNA in the Escherichia coli 30 S ribosomal subunit. J Biol Chem 274:16576–16581. doi: 10.1074/jbc.274.23.16576. [DOI] [PubMed] [Google Scholar]
- 26.Dégrange S, Renaudin H, Charron A, Pereyre S, Bébéar CM, Bébéar CM. 2008. Reduced susceptibility to tetracyclines is associated in vitro with the presence of 16S rRNA mutations in Mycoplasma hominis and Mycoplasma pneumoniae. J Antimicrob Chemother 61:1390–1392. doi: 10.1093/jac/dkn118. [DOI] [PubMed] [Google Scholar]
- 27.Schnee C, Heller M, Lysnyansky I, Gobb F, Roger D, Ayling R, Nicholas AJ, Sachse K. 2014. Elucidation of acquired resistance mechanisms to aminoglycosides in Mycoplasma bovis. 20th Congress of the International Organization for Mycoplasmology, Blumenau, BRazil. [Google Scholar]
- 28.Roberts MC, Koutsky LA, Holmes KK, LeBlanc DJ, Kenny GE. 1985. Tetracycline-resistant Mycoplasma hominis strains contain streptococcal tetM sequences. Antimicrob Agents Chemother 28:141–143. doi: 10.1128/AAC.28.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dégrange S, Renaudin H, Charron A, Bébéar C, Bébéar CM. 2008. Tetracycline resistance in Ureaplasma spp. and Mycoplasma hominis: Prevalence in Bordeaux, France, from 1999 to 2002 and description of two tet(M)-positive isolates of M. hominis susceptible to tetracyclines. Antimicrob Agents Chemother 52:742–744. doi: 10.1128/AAC.00960-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mardassi BBA, Aissani N, Moalla I, Dhahri D, Dridi A, Mlik B. 2012. Evidence for the predominance of a single tet(M) gene sequence type in tetracycline-resistant Ureaplasma parvum and Mycoplasma hominis isolates from Tunisian patients. J Med Microbiol 61:1254–1261. doi: 10.1099/jmm.0.044016-0. [DOI] [PubMed] [Google Scholar]
- 31.Makosky PC, Dahlberg AE. 1987. Spectinomycin resistance at site 1192 in 16S ribosomal RNA of E. coli: an analysis of three mutants. Biochimie 69:885–889. doi: 10.1016/0300-9084(87)90216-1. [DOI] [PubMed] [Google Scholar]
- 32.Dragon F, Spickler C, Pinard R, Carrière J, Brakier-Gringas L. 1996. Mutations of noncanonical base-pairs in the 3′ major domain of Escherichia coli 16 S ribosomal RNA affect the initiation and elongation of protein synthesis. J Mol Biol 259:207–215. doi: 10.1006/jmbi.1996.0313. [DOI] [PubMed] [Google Scholar]
- 33.Pereyre S, Guyot C, Renaudin H, Charron A, Bébéar C, Bébéar CM. 2004. In vitro selection and characterization of resistance to macrolides and related antibiotics in Mycoplasma pneumoniae. Antimicrob Agents Chemother 48:460–465. doi: 10.1128/AAC.48.2.460-465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garza-Ramos G, Xiong L, Zhong P, Mankin A. 2001. Binding site of macrolide antibiotics on the ribosome: New resistance mutation identifies a specific interaction of ketolides with rRNA. J Bacteriol 183:6898–6907. doi: 10.1128/JB.183.23.6898-6907.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lysnyansky I, Gerchman I, Flaminio B, Catania S. 2015. Decreased susceptibility to macrolide-lincosamide in Mycoplasma synoviae is associated with mutations in 23S ribosomal RNA. Microb Drug Resist 21:581–589. doi: 10.1089/mdr.2014.0290. [DOI] [PubMed] [Google Scholar]
- 36.Kobayashi H, Nakajima H, Shimizu Y, Eguchi M, Hata E, Yamamoto K. 2005. Macrolides and lincomycin susceptibility of Mycoplasma hyorhinis and variable mutation of domain II and V in 23S ribosomal RNA. J Vet Med Sci 67:795–800. doi: 10.1292/jvms.67.795. [DOI] [PubMed] [Google Scholar]
- 37.Hong KB, Choi EH, Lee HJ, Lee SY, Cho EY, Choi JH, Kang HM, Lee J, Ahn YM, Kang YH, Lee JH. 2013. Macrolide resistance of Mycoplasma pneumoniae, South Korea, 2000-2011. Emerg Infect Dis 19:1281–1284. doi: 10.3201/eid1908.121455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsuoka M, Narita M, Okazaki N, Yamazaki T, Ouchi K, Suzuki I, Andoh T, Kenri T, Sasaki Y, Shintani M, Arakawa Y, Ohya H, Horino A, Sasaki T. 2004. Characterization and molecular analysis of macrolide-resistant Mycoplasma pneumoniae clinical isolates obtained in Japan. Antimicrob Agents Chemother 48:4624–4630. doi: 10.1128/AAC.48.12.4624-4630.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li BB, Shen JZ, Cao XY, Wang Y, Dai L, Huang SY, Wu CM. 2010. Mutations in 23S rRNA gene associated with decreased susceptibility to tiamulin and valnemulin in Mycoplasma gallisepticum. FEMS Microbiol Lett 308:144–149. doi: 10.1111/j.1574-6968.2010.02003.x. [DOI] [PubMed] [Google Scholar]
- 40.Stipkovits L, Ripley PH, Tenk M, Glávits R, Molnár T, Fodor L. 2005. The efficacy of valnemulin (Econor) in the control of disease caused by experimental infection of calves with Mycoplasma bovis. Res Vet Sci 78:207–215. doi: 10.1016/j.rvsc.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 41.Van Duijkeren E, Greko C, Pringle M, Baptiste KE, Catry B, Jukes H, Moreno MA, Pomba MCMF, Pyörälä S, Rantala M, Ružauskas M, Sanders P, Teale C, Threlfall EJ, Torren-Edo J, Törneke K. 2014. Pleuromutilins: use in food-producing animals in the European Union, development of resistance, and impact on human and animal health. J Antimicrob Chemother 69:2022–2031. doi: 10.1093/jac/dku123. [DOI] [PubMed] [Google Scholar]
- 42.Liu H, Xiao S, Zhang D, Mu S, Zhang L, Wang X, Xue F. 2015. Synthesis and antibacterial activity of novel pleuromutilin derivatives. Biol Pharm Bull 38:1041–1048. doi: 10.1248/bpb.b15-00123. [DOI] [PubMed] [Google Scholar]
- 43.Hillen S, Willems H, Herbst W, Rohde J, Reiner G. 2014. Mutations in the 50S ribosomal subunit of Brachyspira hyodysenteriae associated with altered minimum inhibitory concentrations of pleuromutilins. Vet Microbiol 172:223–229. doi: 10.1016/j.vetmic.2014.04.021. [DOI] [PubMed] [Google Scholar]
- 44.Long KS, Poehlsgaard J, Hansen LH, Hobbie SN, Böttger EC, Vester B. 2009. Single 23S rRNA mutations at the ribosomal peptidyl transferase centre confer resistance to valnemulin and other antibiotics in Mycobacterium smegmatis by perturbation of the drug binding pocket. Mol Microbiol 71:1218–1227. doi: 10.1111/j.1365-2958.2009.06596.x. [DOI] [PubMed] [Google Scholar]
- 45.Davidovich C, Bashan A, Auerbach-Nevo T, Yaggie RD, Gontarek RR, Yonath A. 2007. Induced-fit tightens pleuromutilins binding to ribosomes and remote interactions enable their selectivity. Proc Natl Acad Sci U S A 104:4291–4296. doi: 10.1073/pnas.0700041104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hidalgo A, Carvajal A, Vester B, Pringle M, Naharro G, Rubio P. 2011. Trends towards lower antimicrobial susceptibility and characterization of acquired resistance among clinical isolates of Brachyspira hyodysenteriae in Spain. Antimicrob Agents Chemother 55:3330–3337. doi: 10.1128/AAC.01749-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kehrenberg C, Schwarz S, Jacobsen L, Hansen LH, Vester B. 2005. A new mechanism for chloramphenicol, florfenicol and clindamycin resistance: methylation of 23S ribosomal RNA at A2503. Mol Microbiol 57:1064–1073. doi: 10.1111/j.1365-2958.2005.04754.x. [DOI] [PubMed] [Google Scholar]
- 48.Nitu Y, Hasegawa S, Kubota H. 1974. In vitro development of resistance to erythromycin, other macrolide antibiotics, and lincomycin in Mycoplasma pneumoniae. Antimicrob Agents Chemother 5:513–519. doi: 10.1128/AAC.5.5.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hannan PC. 2000. Guidelines and recommendations for antimicrobial minimum inhibitory concentration (MIC) testing against veterinary mycoplasma species. Vet Res 31:373–395. doi: 10.1051/vetres:2000100. [DOI] [PubMed] [Google Scholar]
- 50.Ronai Z, Kreizinger Z, Dan A, Drees K, Foster JT, Banyai K, Marton S, Szeredi L, Janosi S, Gyuranecz M. 2015. First isolation and characterization of Brucella microti from wild boar. BMC Vet Res 11:147. doi: 10.1186/s12917-015-0456-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Amram E, Borovok I, Nachum-Biala Y, Ayling R, Lerner U, Harrus S, Lysnyansky I. 2016. High prevalence of diverse insertion sequences within the ribosomal RNA operons of Mycoplasma bovis. Appl Environ Microbiol 82:6386–6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.