ABSTRACT
Seventy-two (54.5%) out of 132 fecal samples from a group of yellow-legged gulls in Barcelona, Spain, were positive for Escherichia coli producing either extended-spectrum β-lactamases (ESBL) (51.5%), carbapenemase (1.5%), or cephamycinase (1.5%). The isolation of two carbapenemase-producing E. coli strains is a matter of concern.
KEYWORDS: carbapenemases, E. coli, ESBL
TEXT
In the last decade, the number of bacterial pathogens presenting multidrug resistance to antibacterial agents has increased dramatically, becoming an emergent global concern and a major public health problem (1). The main cause behind the increasing rates of resistance can ultimately be found in the abuse and misuse of antibacterial agents, whether used in patients and livestock or released into the environment. Once antimicrobial-resistant bacteria emerge, they can spread locally or globally. The main factors contributing to their spread at a global level comprise migrant birds, globalization of commercial food, and international travel.
There have been several studies about the presence of resistant bacteria in gulls (2, 3), to the extent of being considered an indicator of environmental antibiotic resistance occurrence, as they are distributed almost all around the world (4). Meerburg et al. (5) showed that gull feces contain a greater average concentration of E. coli than other wild animals, and according to Stedt et al. (4), Spain is the country in Europe with the highest levels of gull E. coli isolates resistant to ≥1 antibiotic.
The objective of this study was to investigate the prevalence of extended-spectrum β-lactamase (ESBL)- and/or carbapenemase-producing Enterobacteriaceae from fecal swabs obtained from a group of yellow-legged gulls (Larus michahellis) in Barcelona, Spain.
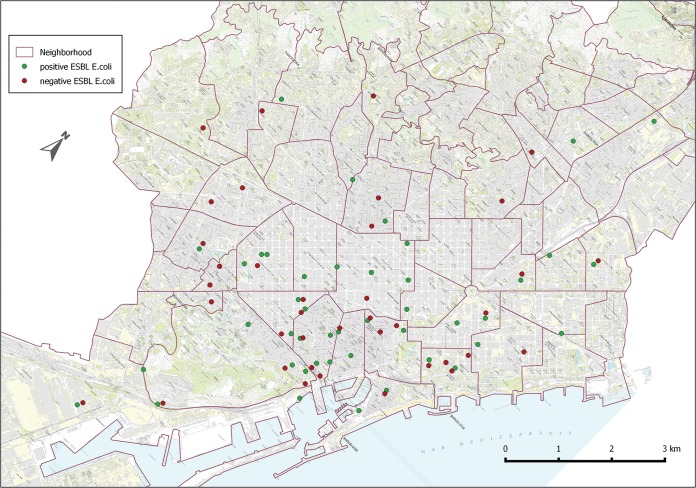
The study was conducted from the beginning of May to late July 2014 in the city of Barcelona, including the breeding period of the yellow-legged gull in the city. The sampling program was part of the sanitary and epidemiological surveillance that is carried out by the Public Health Agency, Barcelona, the institution responsible for the supervision and surveillance of the species. The sampling sites were chosen according to citizens' reports regarding the species nesting on their terraces or high roofs of the city. Every gull chick from each nest found (Fig. 1) was sampled, which amounts to 132 samples in total. All samples were obtained from young specimens born in that same year, and all nests were independent from each other, since the urban structure of cities promotes isolated instead of colonial nesting. Fecal material was obtained by sampling the cloacae of gull chicks with sterile swabs. Each swab was individually preserved in Cary-Blair medium at 2 to 8°C and analyzed within 24 h in the laboratory of the Public Health Agency, Barcelona.
FIG 1.
Location of sites where positive (green) and negative (red) samples were collected (Barcelona, Spain).
The samples were plated on ESBL chromogenic agar (bioMérieux, France), and burgundy red colonies were selected, according to the manufacturer's instructions. Colonies were further identified by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (Bruker Daltonics, Inc., Bremen, Germany). Susceptibility to ampicillin, amoxicillin-clavulanic acid, cefuroxime, ceftriaxone, cefotaxime, meropenem, gentamicin, amikacin, nalidixic acid, and ciprofloxacin was determined by the disk diffusion method, according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines and breakpoints (version 5.0, 2015) (6). The double-disk diffusion technique was performed for phenotypic ESBL detection. The presence of carbapenemases was assessed with the modified Hodge test, according to phenotypic susceptibility results. Characterization of ESBL and carbapenemases genes was performed by PCR, followed by DNA sequencing (7) (for ESBL, the blaTEM, blaSHV, and blaCTX-M genes; for carbapenemases, the blaKPC, blaOXA-48, blaVIM, blaIMP, and blaNDM genes; and for cephamycinases, the blaCMY, blaDHA, blaFOX, blaACC, blaEBC, and blaMOX genes). Seventy-two (54.5%) out of 132 fecal samples were positive for Escherichia coli producing either ESBL (68/132 [51.5%]), carbapenemase (2/132 [1.5%]), or cephamycinase (2/132 [1.5%]) (Table 1), with SHV being the most prevalent group (38/132 [28.8%]). Forty-five strains (62.5%) were resistant to quinolones, 22 strains (30.6%) were resistant to gentamicin, and 9 strains (12.5%) were resistant to amikacin. Repetitive extragenic palindromic-PCR (rep-PCR) showed a high genetic heterogeneity among the strains with up to 57 different clones, with 15 of them containing two different isolates each (data not shown). Agglutination with antiserum O:25 was used to identify CTX-M-15-producing isolates belonging to the high-risk clone O:25b-ST131, but all isolates were negative.
TABLE 1.
Distribution of β-lactamases
| β-Lactamase(s) | No. of strains | % |
|---|---|---|
| SHV group | 38 | 52.8 |
| SHV-12 | 24 | 33.3 |
| SHV-12 + TEM-1 | 13 | 18 |
| SHV-2 | 1 | 1.4 |
| CTX-M group | 30 | 41.6 |
| CTX-M-15 | 11 | 15.3 |
| CTX-M-15 + TEM-1 | 2 | 2.8 |
| CTX-M-1 | 1 | 1.4 |
| CTX-M-1 + TEM-1 | 5 | 6.9 |
| CTX-M-1 + TEM-84 | 1 | 1.4 |
| CTX-M-14 | 4 | 5.5 |
| CTX-M-14 + TEM-1 | 6 | 8.3 |
| VIM-1 + KPC-2 | 2 | 2.8 |
| CMY-2 | 2 | 2.8 |
| Total | 72 | 100 |
Of note, two E. coli isolates carried both the blaKPC-2 and blaVIM genes for carbapenem resistance. Multilocus sequence typing (MLST) (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli) and PCR-based phylogroup analysis (8) identified them as belonging to the sequence type 1011 (ST1011) phylogroup E (strain 40) and ST354 phylogroup F (strain 71), respectively, which have been previously reported in human strains (9, 10). The genetic transference of carbapenemase genes was tested by biparental mating experiments using E. coli J53 AziR as the recipient strain. Transconjugants were selected in Mueller-Hinton agar plates containing 100 μg/ml sodium azide and 1 μg/ml meropenem (Sigma Chemical Co., St. Louis, MO). Successful conjugation was confirmed by specific PCR amplification. Table 2 shows the MICs determined by Etest of different antibiotics for the original and transconjugant strains with the corresponding carbapenemases harbored.
TABLE 2.
In vitro susceptibilities of original strains of E. coli and E. coli transconjugants expressing VIM-1 and/or KPC-2 (Etest, EUCAST)
| Antibiotic | MIC (μg/ml) |
||||||
|---|---|---|---|---|---|---|---|
| Original strains |
Transconjugants |
||||||
| 40 (VIM-1/KPC-2)a | 71 (VIM-1/KPC-2)b | J53c | J53 40T3 (KPC-2)d | J53 40T5 (VIM-1/KPC-2)e | J53 71T1 (VIM-1)f | J53 71T3 (VIM-1/KPC-2)g | |
| Cefoxitin | >256 | >256 | 2 | 8 | 256 | 256 | 64 |
| Cefotaxime | 32 | 24 | 0.094 | 0.75 | 16 | 16 | 64 |
| Ceftazidime | 256 | 96 | 0.125 | 1 | 64 | 64 | 96 |
| Imipenem | 4 | 24 | 0.19 | 1.5 | 3 | 1 | 12 |
| Meropenem | 4 | 32 | 0.023 | 0.75 | 0.5 | 0.25 | 8 |
| Ertapenem | 6 | 12 | 0.008 | 0.38 | 0.38 | 0.64 | 4 |
| Aztreonam | 16 | 128 | 0.064 | 12 | 16 | 0.25 | 256 |
| Ciprofloxacin | >32 | >32 | 0.064 | 0.047 | 0.064 | 0.064 | 0.064 |
| Gentamicin | >32 | >32 | 0.25 | 0.25 | 1 | 2 | 2 |
| Amikacin | 3 | 3 | 1 | 1 | 1.5 | 1.5 | 1.5 |
| Tobramycin | 16 | 12 | 0.125 | 0.125 | 3 | 3 | 3 |
| Colistin | 0.25 | 0.125 | 0.125 | 0.25 | 0.125 | 0.19 | 0.25 |
E. coli strain 40 isolated from a yellow-legged gull.
E. coli strain 71 isolated from a yellow-legged gull.
Sodium azide-resistant E. coli J53 strain used as a recipient in the conjugation experiment.
E. coli transconjugant obtained from strains 40 and J53 that received only blaKPC-2.
E. coli transconjugant obtained from strains 40 and J53 that received both blaKPC-2 and blaVIM-1.
E. coli transconjugant obtained from strains 71 and J53 that received only blaVIM-1.
E. coli transconjugant obtained from strains 71 and J53 that received both blaKPC-2 and blaVIM-1.
Plasmid analysis by S1 nuclease–pulsed-field gel electrophoresis (PFGE) (7) and replicon typing (7) were then performed on both the original strains and transconjugants to determine the size of these plasmids and classify them within the incompatibility groups. Digoxigenin-labeled probes for the blaVIM and blaKPC genes were hybridized against blotted nylon membranes from the S1-PFGE gels. The blaKPC-2 gene was located in a plasmid ca. 60 kb in size in strain 40 and in a plasmid of <50 kb in strain 71, both being nontypeable plasmids. The genetic environment of the blaKPC-2 genes was determined by inverse PCR, leading to the identification of an ISKpn27-ΔTEM-blaKPC-2-ISKpn6-korC genetic arrangement, which was similar to those previously described among different isolates of human origin from China and Taiwan (11, 12). Further analysis based on next-generation sequencing is needed to describe additional elements of these plasmids for a more robust analysis.
The blaVIM-1 gene was located in both strains in an In3103 class I integron, carried in a ca. 100-kb plasmid belonging to the incompatibility group I1-Iγ. This integron also contained an aminoglycoside 6′-N-acetyltransferase (aacA4) gene and a 3′-(10)-O-adenylyltransferase (aadA1) gene. The presence of the blaVIM-1 gene within an In3103 class I integron was also described by at least one report in Spain, albeit in that case, it was located in a nontypeable plasmid of ca. 60 kb and recovered from a human patient (13).
Our data showed a higher percentage of resistant E. coli in gull fecal samples than in previous studies (14–16), but it also represents the first study, to our knowledge, reporting the coexistence of two carbapenemase genes in E. coli recovered from yellow-legged gulls. However, some OXA-48-producing E. coli strains could have been lost due to the methodology that was specifically designed to search for ESBL. The fact that carbapenem-resistant isolates recovered from the fecal samples of gulls share the same sequence types and resistance modules as those recovered from human samples in different parts of the world highlights the potential role of migratory birds in the dissemination and spread of antibiotic resistance genes.
ACKNOWLEDGMENTS
We thank Colomba Control S.L. for their excellent technical assistance in gull control and sampling and Andrea Valsecchi for his collaboration.
REFERENCES
- 1.Roca I, Akova M, Baquero F, Carlet J, Cavaleri M, Coenen S, Cohen J, Findlay D, Gyssens I, Heuer OE, Kahlmeter G, Kruse H, Laxminarayan R, Liébana E, López-Cerero L, MacGowan A, Martins M, Rodríguez-Baño J, Rolain JM, Segovia C, Sigauque B, Tacconelli E, Wellington E, Vila J. 2015. The global threat of antimicrobial resistance: science for intervention. New Microbes New Infect 6:22–29. doi: 10.1016/j.nmni.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hernandez J, Johansson A, Stedt J, Bengtsson S, Porczak A, Granholm S, González-Acuña D, Olsen B, Bonnedahl J, Drobni M. 2013. Characterization and comparison of extended-spectrum β-lactamase (ESBL) resistance genotypes and population structure of Escherichia coli isolates from Franklin's gulls (Leucophaeus pipixcan) and humans in Chile. PLoS One 8:e76150. doi: 10.1371/journal.pone.0076150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aberkane S, Compain F, Barraud O, Ouédraogo AS, Bouzinbi N, Vittecoq M, Jean-Pierre H, Decré D, Godreuil S. 2015. Non-O1/non-O139 Vibrio cholerae avian isolate from France cocarrying the blaVIM-1 and blaVIM-4 genes. Antimicrob Agents Chemother 59:6594–6596. doi: 10.1128/AAC.00400-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stedt J, Bonnedahl J, Hernandez J, McMahon BJ, Hasan B, Olsen B, Drobni M, Waldenstrom J. 2014. Antibiotic resistance patterns in Escherichia coli from gulls in nine European countries. Infect Ecol Epidemiol 1:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meerburg BG, Koene MGJ, Kleijn D. 2011. Escherichia coli concentrations in feces of geese, coots, and gulls residing on recreational water in The Netherlands. Vector Borne Zoonotic Dis 11:601–603. doi: 10.1089/vbz.2010.0218. [DOI] [PubMed] [Google Scholar]
- 6.EUCAST. 2015. Breakpoint tables for interpretation of MICs and zone diameters. European Committee on Antimicrobial Susceptibility Testing (EUCAST), Växjö, Sweden: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_5.0_Breakpoint_Table_01.pdf. [Google Scholar]
- 7.Solé M, Pitart C, Roca I, Fàbrega A, Salvador P, Muñoz L, Oliveira I, Gascón J, Marco F, Vila J. 2011. First description of an Escherichia coli strain producing NDM-1 carbapenemase in Spain. Antimicrob Agents Chemother 55:4402–4404. doi: 10.1128/AAC.00642-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clermont O, Christenson JK, Denamur E, Gordon DM. 2013. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep 5:58–65. doi: 10.1111/1758-2229.12019. [DOI] [PubMed] [Google Scholar]
- 9.Mora A, Blanco M, López C, Mamani R, Blanco JE, Alonso MP, García-Garrote F, Dahbi G, Herrera A, Fernández A, Fernández B, Agulla A, Bou G, Blanco J. 2011. Emergence of clonal groups O1:HNM-D-ST59, O15:H1-D-ST393, O20:H34/HNM-D-ST354, O25b:H4-B2-ST131 and ONT:H21,42-B1-ST101 among CTX-M-14-producing Escherichia coli clinical isolates in Galicia, northwest Spain. Int J Antimicrob Agents 37:16–21. doi: 10.1016/j.ijantimicag.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 10.Guo S, Wakeham D, Brouwers HJM, Cobbold RN, Abraham S, Mollinger JL, Johnson JR, Chapman TA, Gordon DM, Barrs VR, Trott DJ. 2014. Human-associated fluoroquinolone-resistant Escherichia coli clonal lineages, including ST354, isolated from canine feces and extraintestinal infections in Australia. Microbes Infect 17:266–274. [DOI] [PubMed] [Google Scholar]
- 11.Chen YT, Lin JC, Fung CP, Lu PL, Chuang YC, Wu TL, Siu LK. 2014. KPC-2-encoding plasmids from Escherichia coli and Klebsiella pneumoniae in Taiwan. J Antimicrob Chemother 69:628–631. doi: 10.1093/jac/dkt409. [DOI] [PubMed] [Google Scholar]
- 12.Wu W, Feng Y, Carattoli A, Zong Z. 2015. Characterization of an Enterobacter cloacae strain producing both KPC and NDM carbapenemases by whole-genome sequencing. Antimicrob Agents Chemother 59:6625–6628. doi: 10.1128/AAC.01275-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Papagiannitsis CC, Izdebski R, Baraniak A, Fiett J, Herda M, Hrabák J, Derde LP, Bonten MJ, Carmeli Y, Goossens H, Hryniewicz W, Brun-Buisson C, Gniadkowski M, MOSAR WP2, WP3 and WP5 Study Groups. 2015. Survey of metallo-β-lactamase-producing Enterobacteriaceae colonizing patients in European ICUs and rehabilitation units, 2008–11. J Antimicrob Chemother 70:1981–1988. [DOI] [PubMed] [Google Scholar]
- 14.Bonnedahl J, Drobni M, Gauthier-Clerc M, Hernandez J, Granholm S, Kayser Y, Melhus A, Kahlmeter G, Waldenström J, Johansson A, Olsen B. 2009. Dissemination of Escherichia coli with CTX-M type ESBL between humans and yellow-legged gulls in the south of France. PLoS One 4:e5958. doi: 10.1371/journal.pone.0005958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simões RR, Poirel L, Da Costa PM, Nordmann P. 2010. Seagulls and beaches as reservoirs for multidrug-resistant Escherichia coli. Emerg Infect Dis 16:110–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poirel L, Potron A, De La Cuesta C, Cleary T, Nordmann P, Munoz-Price LS. 2012. Wild coastline birds as reservoirs of broad-spectrum-β-lactamase-producing Enterobacteriaceae in Miami Beach, Florida. Antimicrob Agents Chemother 56:2756–2758. doi: 10.1128/AAC.05982-11. [DOI] [PMC free article] [PubMed] [Google Scholar]