ABSTRACT
Plasmids of incompatibility group A/C (IncA/C) are becoming increasingly prevalent within pathogenic Enterobacteriaceae. They are associated with the dissemination of multiple clinically relevant resistance genes, including blaCMY and blaNDM. Current typing methods for IncA/C plasmids offer limited resolution. In this study, we present the complete sequence of a blaNDM-1-positive IncA/C plasmid, pMS6198A, isolated from a multidrug-resistant uropathogenic Escherichia coli strain. Hypersaturated transposon mutagenesis, coupled with transposon-directed insertion site sequencing (TraDIS), was employed to identify conserved genetic elements required for replication and maintenance of pMS6198A. Our analysis of TraDIS data identified roles for the replicon, including repA, a toxin-antitoxin system; two putative partitioning genes, parAB; and a putative gene, 053. Construction of mini-IncA/C plasmids and examination of their stability within E. coli confirmed that the region encompassing 053 contributes to the stable maintenance of IncA/C plasmids. Subsequently, the four major maintenance genes (repA, parAB, and 053) were used to construct a new plasmid multilocus sequence typing (PMLST) scheme for IncA/C plasmids. Application of this scheme to a database of 82 IncA/C plasmids identified 11 unique sequence types (STs), with two dominant STs. The majority of blaNDM-positive plasmids examined (15/17; 88%) fall into ST1, suggesting acquisition and subsequent expansion of this blaNDM-containing plasmid lineage. The IncA/C PMLST scheme represents a standardized tool to identify, track, and analyze the dissemination of important IncA/C plasmid lineages, particularly in the context of epidemiological studies.
KEYWORDS: uropathogenic E. coli, IncA/C plasmid, functional genomics, New Delhi metallo-beta-lactamase, plasmid multilocus sequence typing
INTRODUCTION
IncA/C plasmids are large, low-copy-number, broad-host-range plasmids with varying capacities for conjugation (1). These plasmids represent an increasing threat to public health due to their association with the dissemination of the blaCMY cephalosporinase genes (2) and more recently the blaNDM metallo-beta-lactamase genes (3–5). The first IncA/C plasmids were isolated from aquatic host species, including the fish pathogen Aeromonas salmonicida (6), and pandemic strains of Vibrio cholerae (7, 8). However, recently, there has been a significant increase in the isolation of IncA/C plasmids from Enterobacteriaceae, including Salmonella (9), Klebsiella pneumoniae (10), and Escherichia coli (11).
Plasmids from the IncA/C group were discovered more than 40 years ago and initially assigned to two separate groups, namely, IncA (RA1) (6) and IncC (12). Subsequent investigations into compatibility, exclusion, and phage sensitivity provided strong evidence for combining these two groups into a single group named IncA/C (12–14). Molecular analysis of the IncA/C replicon has again split this group into two distinct types, A/C1 (RA1) and A/C2 (15). The A/C2 type comprises the vast majority of IncA/C plasmids sequenced to date (1). Plasmid backbone comparisons have informed the subtyping of IncA/C2 into type 1 and type 2 plasmids (16). The two types differ in several ways, including two replacement regions (R1 and R2) that lie within rhs and a large coding sequence (CDS) in transfer region 1, respectively, as well as the presence or absence of two small segments (i1 and i2) (16). Common features are also found in relation to resistance gene content, for example, the vast majority of type 1 plasmids possess the antimicrobial resistance island A (ARI-A) located within rhs and an ISEcp1-blaCMY insertion within the large CDS in transfer region 1 (16, 17). An additional resistance island (ARI-B), located upstream of the par locus, is found in both type 1 and 2 plasmids but is not always present.
The IncA/C replicon was first defined and characterized using the archetype plasmid RA1. Thirteen direct repeats (iterons) are located downstream of the repA replication gene, similar to IncP plasmids (18). Both repA and the iterons are required for IncA/C replication (19). Similar to other iteron-controlled replicons, there is an imperfect inverted repeat upstream of repA (18), suggesting autoregulation (20). IncA/C plasmids also possess a putative toxin-antitoxin (TA) system. The TA genes are strongly transcribed, suggesting the system is functionally active in the postsegregation killing of plasmid-free progeny (21). Moreover, attempts to construct deletion mutants of the antitoxin component proved to be lethal to the cell (22).
Partitioning systems are one of the most important factors that contribute to the stable inheritance of large, low-copy-number plasmids (23). Partitioning typically involves three components: a cis-acting DNA binding site (centromere; parS), a centromere binding protein (ParB), and an NTPase (ParA) (24). Together, they facilitate the correct positioning of plasmid molecules during cell division to increase plasmid retention (25). Partitioning systems are classified into different groups based on the characteristics of the NTPase (24). The IncA/C ParA protein contains a Walker-type ATPase, indicating IncA/C plasmids possess a type I partitioning system (1, 25). This ParA protein has similarity to ParA of IncP plasmids, while IncA/C ParB contains both ParB and KorB domains (1). These genes are transcribed at low levels, similar to repA (21). Another putative partitioning gene, stbA, is found in a separate genetic location. StbA has similarity to the ParM partitioning protein from the IncFII plasmid NR1 (1). In addition to these elements, IncA/C plasmids carry a number of other genes putatively involved in replication, including kfrA and ter (1). However, the functions of these genes have not been experimentally determined.
Transposon-directed insertion site sequencing (TraDIS), along with other, similar techniques, including Tn-seq (26), INseq (27), and HITS (28), is a high-throughput whole-genome screening method used to perform bacterial functional genomic analyses (29, 30). A typical TraDIS experiment examines a highly saturated transposon mutant library under a condition of interest, with pre- and postselection libraries subjected to deep sequencing to simultaneously identify all of the transposon insertion sites. After selection, the lack of insertions within a gene is used to determine the importance of that gene for survival under the condition tested. The technique has been applied to identify genes that enable the maintenance and transmission of the IncI1 plasmid pESBL (31) and the essential genes of the IncF plasmid pEC958 (32). Here, we employed TraDIS to identify genetic elements involved in the replication and maintenance of the IncA/C plasmid group. These experimentally validated elements provided a framework for development of a novel plasmid multilocus sequence typing (PMLST) scheme for tracking this important plasmid group.
RESULTS
Genomic analysis of the carbapenem-resistant E. coli strain MS6198.
MS6198 is a carbapenem-resistant uropathogenic Escherichia coli (UPEC) strain. MS6198 is also nonsusceptible to multiple other antibiotics, including beta-lactams, nalidixic acid, ciprofloxacin, gentamicin, kanamycin, sulfamethoxazole, trimethoprim, tetracycline, and tobramycin (see Data Set S1 in the supplemental material). The complete genome of MS6198 was determined and shown to consist of a circular chromosome comprising 5,176,750 base pairs (51.59% G-C content). In silico typing assigned MS6198 to sequence type 648 (ST648), which has been associated with a number of disease outbreaks in Asia and Europe (33–35). MS6198 contained a number of UPEC virulence factors, including genes encoding type 1 fimbriae, Ag43, capsule, and several iron acquisition systems. A list of known UPEC virulence genes found in MS6198 is shown in Data Set S2 in the supplemental material. In addition, MS6198 contained 4 circular plasmids, pMS6198A (IncA/C type; 137,565 bp), pMS6198B (IncFII type; 128,428 bp), pMS6198C (untypeable; 98,242 bp), and pMS6198D (IncI1 type; 50,899 bp). Methylome analysis of MS6198 identified three distinct DNA recognition motifs, indicating the presence of at least three active adenine methyltransferase enzymes (see Data Set S3 in the supplemental material).
Characterization of an IncA/C multidrug resistance plasmid harboring the blaNDM-1 carbapenemase gene.
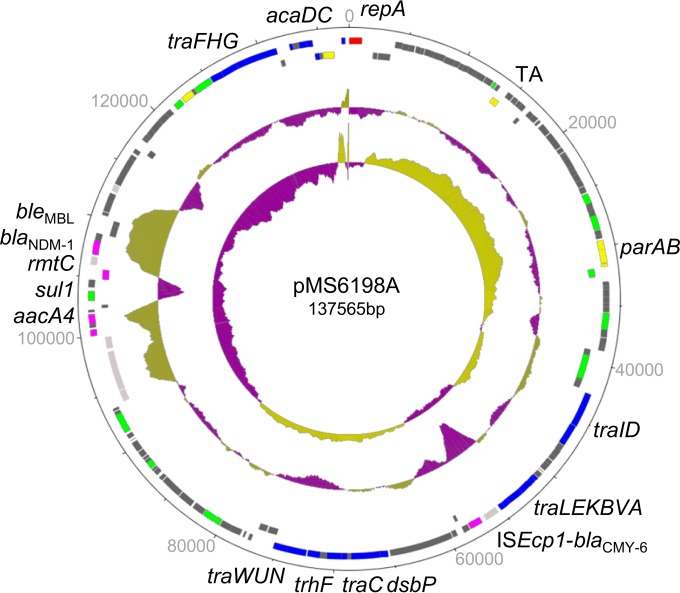
We focused our study on characterization of the blaNDM-1-positive IncA/C plasmid pMS6198A. Plasmid pMS6198A belongs to type 1 of the IncA/C2 group and contains 172 CDSs classified into seven functional groups (Fig. 1). The plasmid contains the typical IncA/C replicon and additional putative maintenance genes, including parAB, parM, kfrA, and a putative TA system. Similarity searches of public sequence databases indicated that pMS6198A contains 19 conjugation genes, including the master regulators acaDC (22). In addition to a typical ISEcp1-blaCMY-6 insertion within transfer region 1, five resistance genes are located within ARI-A—aacA4, rmtC, blaNDM-1, bleMBL, and sul1—along with a truncated qacEΔ1 gene. Plasmid pMS6198A contains 92 CDSs that have no assigned annotation.
FIG 1.
Genetic map of pMS6198A. The outermost rings are CDSs on forward and reverse strands. The CDSs are color coded as follows: replication, red; maintenance, yellow; DNA modification, green; conjugative, blue; mobile element, light pink; antimicrobial resistance, dark pink. The inner rings are GC plot (outer) and GC skew (inner).
Sequence comparison with other IncA/C plasmids showed that pMS6198A displayed ∼99% conservation over its entire sequence with two other blaNDM-1-positive IncA/C plasmids: pKP1-NDM1 (KF992018) from Australia and pNDM10469 (JN861072) from Canada. The genetic structure of blaNDM-1 in pMS6198A is highly similar to those of eight other IncA/C plasmids (see Fig. S1 in the supplemental material). A full complement of transfer genes, as defined previously (1), were present in pMS6198A. Accordingly, pMS6198A was transferable by conjugation in static liquid culture at 37°C to the recipient strain E. coli J53 at a frequency of 10−3 transconjugants per donor. Transfer of pMS6198A to J53 was confirmed by PCR amplification of the IncA/C replicon. Antibiotic resistance profiling of the transconjugant strain (MS6614 [see Data Set S1 in the supplemental material]) showed that pMS6198A was capable of conferring resistance to multiple antibiotics, including cefotaxime-clavulanic acid, ceftazidime-clavulanic acid, piperacillin-tazobactam, amoxicillin-clavulanic acid, cefoxitin, cefpodoxime, ceftriaxone, cephalothin, ampicillin, gentamicin, kanamycin, imipenem, meropenem, ertapenem, and tobramycin. Purification of pMS6198A from MS6614 and electrotransformation into E. coli TOP10 were performed, with subsequent analysis of the transformed TOP10 strain by PCR, conjugation, and antibiotic resistance profiling indicating the integrity of the plasmid was maintained (see Data Set S1 in the supplemental material). Thus, all subsequent analysis of pMS6198A was performed using plasmids purified from MS6614.
Identification of genes required for pMS6198A maintenance and replication.
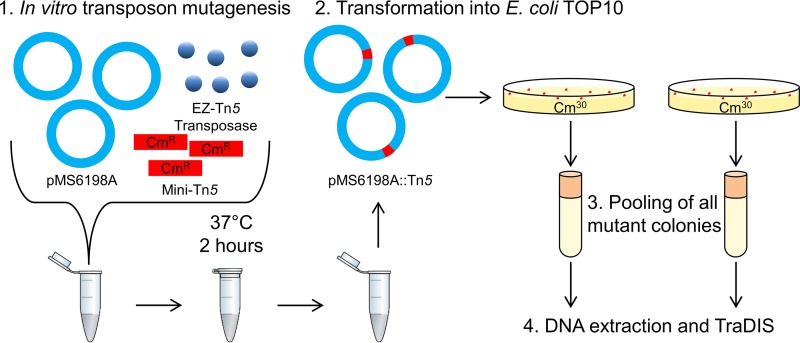
To identify the genes required for the maintenance and replication of pMS6198A in E. coli, we employed in vitro mutagenesis in combination with TraDIS as shown in Fig. 2. First, in vitro mini-Tn5 mutagenesis of pMS6198A was carried out to create a highly saturated mutant plasmid DNA library. This library was transformed into E. coli TOP10 by electroporation and subsequently grown in the presence of chloramphenicol to select for pMS6198A::mini-Tn5-containing transformants. This process was performed in duplicate, resulting in two saturated libraries, each of which contained approximately 10,000 transformants. Purified plasmid DNA was extracted from both libraries and analyzed by TraDIS. Examination by inverse PCR of a subset of individual Tn5 mutant colonies was performed to investigate the randomness of insertion and the number of insertions per plasmid molecule. The majority of mutants examined (17/18; 94%) contained a single insertion, each at a different location on the plasmid (see Fig. S2 in the supplemental material).
FIG 2.
Overview of pMS6198A in vitro mutagenesis and TraDIS analysis. (1) Tn5::Cm transposons were inserted into pMS6198A. (2) The mutagenized plasmid mixture was transformed into E. coli TOP10 by electroporation, and mutant plasmids were selected by growth in the presence of chloramphenicol. (3) All mutant chloramphenicol-resistant colonies were counted, pooled, and stored at −80°C. Two replicate libraries were generated by this method. (4) Genomic DNA was extracted from each library for TraDIS analysis.
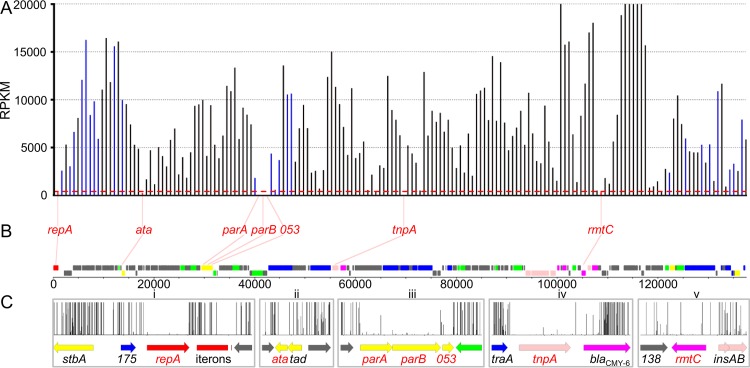
TraDIS identified a total of 10,178 unique insertion sites in pMS6198A from the two libraries, which was equivalent to an average of one insertion site every 13.52 bp. Correlation tests of insertion sites per gene for each library showed high reproducibility for the TraDIS sequencing method (R2 > 0.99) (see Fig. S3 in the supplemental material). The relative abundance of reads mapped to mini-Tn5 insertion sites within each gene (expressed in reads per kilobase per million [RPKM]) was calculated (Fig. 3A; see Data Set S4 in the supplemental material). This enabled us to identify genes required for the maintenance and replication of pMS6198A, as plasmids containing mutations in such genes would be lost and thus underrepresented in the libraries, reflected as a low RPKM value.
FIG 3.
Essential gene TraDIS read data. (A) RPKM for each gene listed on the x axis. The red lines indicate genes below the 418-RPKM threshold; the blue lines indicate genes present in all 82 IncA/C plasmids and above the 418-RPKM threshold; the black lines indicate all other genes above the 418-RPKM threshold. (B) Genes with ≤418 RPKM. The red lines indicate positions on the graph and pMS6198A sequence. (C) Enlarged view of the genetic environment for each gene in panel B. The graphs show the number of transposon reads mapped to each position.
The repA gene is known to be required for IncA/C plasmid replication (19), and we observed a relative abundance of mini-Tn5 insertions in repA (418 RPKM) (Fig. 3Ci) significantly lower than the average insertion abundance for all pMS6198A genes (7,222 RPKM). Therefore, we used 418 RPKM as a biological threshold for genes required for replication and maintenance of pMS6198A. Six additional genes were identified with insertion abundances lower than this threshold: 022, parA, parB, 053, tnpA, and rmtC (Fig. 3B).
The putative TA system of IncA/C plasmids belongs to the tad-ata-like family found in a range of different genetic elements, including ICE SXT, Enterobacteria phage N15, a genomic island of E. coli EDL933, and the plasmid pAMI2 (36). The 022 gene (here referred to as ata, for antitoxin of addiction system) lies immediately adjacent to the 023 gene (referred to as tad, for toxin of addiction system) (Fig. 3Cii). Both tad and ata are 100% conserved in all fully sequenced IncA/C2 plasmids but absent from IncA/C1 plasmids. Tad is a member of the Gp49 family proteins (Pfam identifier PF05973), which is comprised of known toxins. In the phage N15, Gp49 is controlled by the adjacently encoded Gp48 protein (UniProt accession no. O64356), which contains a helix-turn-helix (HTH) DNA binding domain (36). Analysis of the Ata amino acid sequence showed it is 24% identical and 50% similar to that of Gp48 (71% coverage) and possesses an HTH domain (Pfam identifier PF13744), suggestive of a DNA binding function. Our TraDIS data showed that mutation of ata is strongly underrepresented, while mutation of tad is tolerated. Taken together, our data suggest that ata and tad encode an active TA system, in which Ata is the antitoxin.
Three adjacent genes, parA (051), parB (052), and 053, were also identified in our TraDIS analysis (Fig. 3Ciii). The three genes are conserved in all fully sequenced IncA/C plasmids. ParA belongs to an ATPase family (Pfam identifier PF13614), while ParB contains both ParB (Pfam identifier PF02195) and KorB (Pfam identifier PF08535) domains. Based on bioinformatics analysis, parA and parB are homologous to IncP partitioning genes. The third gene in the locus (053) encodes a hypothetical protein of 90 amino acids (aa) containing a winged HTH domain (Pfam identifier PF09904), suggestive of a DNA binding function. Additionally, 053 is present at the same genetic location (after parB) in all IncA/C plasmids.
Two other genes, tnpA and rmtC, were identified in our TraDIS screen (Fig. 3Civ and v). The tnpA gene encodes a transposase from the insertion sequence ISEcp1 (37), while rmtC encodes a 16S ribosomal methyltransferase (UniProt entry Q33DX5) that confers resistance to many aminoglycoside antibiotics. These two genes occupy regions of pMS6198A with the lowest G-C content (see Fig. S4 in the supplemental material) that may be associated with an insertion bias (38). Furthermore, the genes are not conserved among completely sequenced IncA/C plasmids, being present in only 50% and 11%, respectively. Based on these data, they are unlikely to be involved in the maintenance and replication of pMS6198A.
The par locus of IncA/C plasmids, of which 053 is a crucial component, contributes to stability.
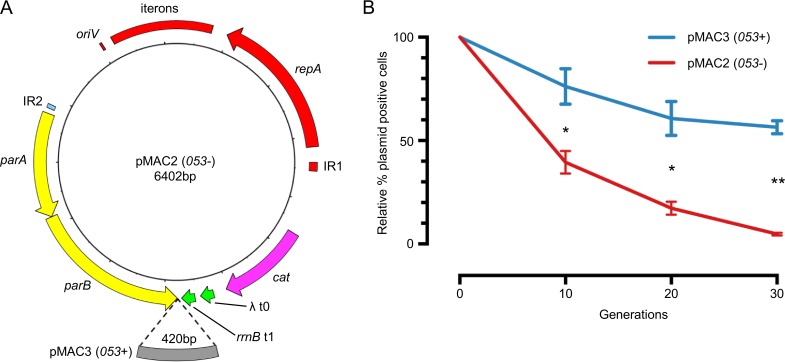
TraDIS analysis identified two putative partitioning genes (parA and parB) and a region containing an open reading frame (ORF) immediately downstream of parB (053) as required for plasmid maintenance. Bioinformatics analysis revealed that the structural organization of parA-parB-053 is completely conserved in all sequenced IncA/C plasmids examined in this study. This led us to hypothesize that the partitioning system in IncA/C is comprised of three components that are all required for plasmid stability in E. coli. To provide evidence to support our hypothesis, we constructed pMS6198A-derived mini-A/C plasmids containing different versions of the partitioning locus and examined their stability in E. coli strain MG1655. The mini-A/C plasmid contained the IncA/C replicon, as previously described (1, 18, 19). A selectable marker (cat cassette, conferring chloramphenicol resistance) was included, along with two transcriptional terminators to prevent transcriptional read-through from the cassette. Two variations of the partitioning locus were incorporated into this mini-A/C plasmid, generating pMAC2 (parAB) and pMAC3 (parAB plus 053) (Fig. 4A).
FIG 4.
Contribution of the partitioning locus to IncA/C stability. (A) Plasmid map of mini-IncA/C constructs pMAC2 and pMAC3. Compared to plasmid pMAC2, plasmid pMAC3 contains an additional 420-bp fragment downstream of parB. (B) Time course plasmid stability assay in broth culture. At each time point, cultures were serially diluted and grown on LB agar with or without Cm to determine the percentage of plasmid-positive cells relative to time zero. Starting cultures consisted of 50% plasmid-positive plus 50% plasmid-free cells. The assays were performed in triplicate. The data points are means ± standard deviations. *, P < 0.005; **, P < 0.0001.
The stability of pMS6198A was initially examined by growth in the absence of selection for three serial passages, each incorporating 10 generations. After 30 generations, no plasmid loss was observable (see Fig. S5 in the supplemental material), highlighting the high stability of the parent plasmid. The same experiment was used to assess the stability of pMAC2 and pMAC3. This showed that pMAC2 was less stable than pMAC3: even at time zero, only 50% of cells retained pMAC2, and by 30 generations, this had dropped to 2% (see Fig. S5 in the supplemental material). In contrast, pMAC3 started at 100% and was reduced to 64% after 30 generations (see Fig. S5 in the supplemental material).
The difference in plasmid stabilities observed between the starting populations of pMAC2 and pMAC3 (time zero) was addressed by mixing cells harboring pMAC3 in a 1:1 ratio with plasmid-free cells, thus mimicking the starting population of cells harboring pMAC2. Analyses using this equivalent starting population resulted in plasmid stability profiles very similar to those observed in the single-strain experiment, with pMAC2-harboring cells reduced to 5% after 30 generations compared to significantly higher (57%) retention of pMAC3 (Fig. 4B).
The TraDIS-identified maintenance genes are conserved in all IncA/C plasmids.
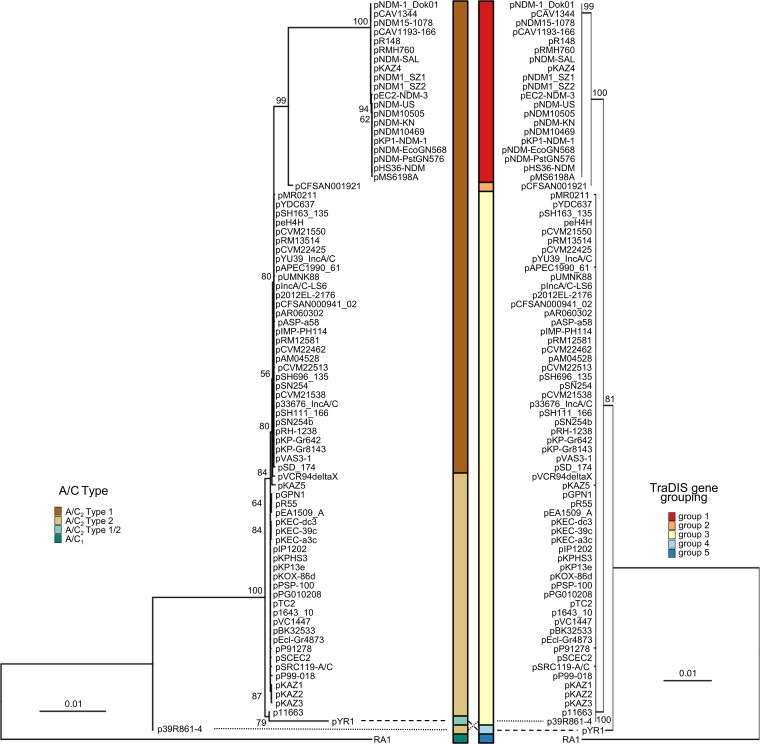
Plasmid multilocus sequence typing was originally developed for the typing of large collections of plasmids using a set of conserved genes (39). In the context of IncA/C plasmid typing, our TraDIS data have provided a defined subset of genes with essential plasmid maintenance functions that could be used in the development of a PMLST scheme. To provide a framework for this analysis, a collection of 82 complete IncA/C plasmid sequences available in the GenBank database were examined for overall sequence conservation (see Data Set S5 in the supplemental material). A total of 28 genes were completely conserved within this collection, which included the four genes identified by TraDIS (repA, parA, parB, and 053) (Table 1). The concatenated sequences of all 28 fully conserved genes from each plasmid were used to build a maximum-likelihood tree (Fig. 5, left). Interestingly, this analysis identified multiple previously defined hybrid plasmid groups, including pYR1 and p39R861-4, and also differentiated between type 1 and 2 A/C2 plasmids (1, 16, 40), with type 1 plasmids also separated into three distinct groups. Using this as a baseline, the discriminatory power of repA, parA, parB, and 053 was examined by using the concatenated sequences of the four genes from each plasmid to build a maximum-likelihood tree (Fig. 5, right). The overall topology of the tree is similar to our analysis of IncA/C plasmids using all 28 conserved genes, including a split between IncA/C1 (group 5) and IncA/C2 (groups 1 to 4). The IncA/C2 lineage was further split into four distinct groups; groups 1 and 2 comprised type 1 plasmids, and group 3 included both type 1 and type 2 plasmids, while group 4 represented the hybrid plasmid pYR1. Although the resolution of the 28 conserved genes separated hybrid plasmid p39R861-4 and both type 1 and type 2 plasmids, their similarity in the sequences of repA-parA-parB-053 clustered the plasmids together in group 3. Furthermore, the preservation of groups 1, 2, 4, and 5 between the two analyses highlights them as distinct plasmid lineages. Taken together, the analysis demonstrates that the sequences of the four essential genes identified by TraDIS are sufficient to capture the phylogenetic relatedness of IncA/C plasmids and could be used for molecular typing.
TABLE 1.
Conserved genes in the IncA/C plasmid database
| Gene | Product | Base rangea | Length (bp)a |
|---|---|---|---|
| repA | Replication protein RepA | 1–1101 | 1,101 |
| 002 | Hypothetical protein | c(2138–2572)b | 1,116 |
| 004 | Hypothetical protein | 3973–4581 | 609 |
| 005 | Hypothetical protein | 4586–5107 | 522 |
| 007 | Hypothetical protein | 5728–6201 | 474 |
| 008 | Hypothetical protein | 6191–6505 | 315 |
| 009 | Hypothetical protein | 6510–7364 | 855 |
| 010 | Signal peptide peptidase | 7364–8323 | 960 |
| 011 | Putative protein-disulfide isomerase | 8339–9199 | 861 |
| 015 | Hypothetical protein | 10521–11039 | 519 |
| 017 | Hypothetical protein | 11256–12113 | 858 |
| 050 | Hypothetical protein | 28936–29247 | 312 |
| parA | Partitioning protein ParA | 29421–30206 | 786 |
| parB | Partitioning protein ParB | 30210–31391 | 1,182 |
| 053 | Putative DNA binding protein | 31440–31712 | 273 |
| 054 | Putative DNA methyltransferase | c(31765-32400) | 636 |
| 055 | Hypothetical protein | c(32492-32635) | 144 |
| 056 | Hypothetical protein | 32962–33339 | 378 |
| 058 | Hypothetical protein | 33588–34262 | 675 |
| 059 | Hypothetical protein | 34255–34761 | 507 |
| xerD | Putative intergrase XerD | 118619–119629 | 1,011 |
| kfrA | Putative DNA binding protein KfrA | 122338–123378 | 1,041 |
| traH | Putative pilus assembly protein TraH | 126329–127762 | 1,434 |
| 167 | Hypothetical protein | c(131427–131789) | 363 |
| 169 | Putative lytic transglycosylase | 132777–133310 | 534 |
| acr2 | H-NS histone family protein | c(134537-134956) | 273 |
| 173 | Hypothetical protein | c(134958-135251) | 294 |
| mobI | Plasmid transfer protein MobI | 136927–137289 | 363 |
As in pMS6198A.
c denotes CDS on reverse strand.
FIG 5.
TraDIS-identified essential genes describe IncA/C phylogeny. Shown is a comparison of IncA/C phylogenies using 28 conserved genes (left) and 4 required genes (right). The brown and green boxes show IncA/C types, and the red and blue boxes show TraDIS gene groupings, as indicated. TraDIS gene groups were assigned based on 99% sequence similarity.
Development of an IncA/C PMLST scheme.
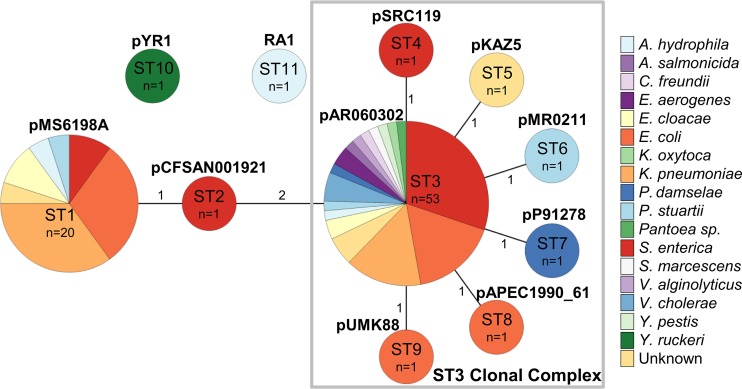
As the repA, parA, parB, and 053 genes could distinguish different groups of IncA/C plasmids, we used these biologically validated genes to develop a PMLST scheme for IncA/C plasmids. Amplification and sequencing primers were designed to target conserved regions within each essential gene (Table 2). PCR and Sanger sequencing using these primers were performed on pMS6198A to validate the methodology. Using the sequences obtained in silico from 82 IncA/C plasmids, different alleles for repA (n = 5), parA (n = 6), parB (n = 7), and 053 (n = 3) were identified, which together form 11 STs, as shown in Fig. 6 (see Data Set S6 in the supplemental material). The minimum spanning tree comprises two singletons, ST10 (pYR1; IncA/C2) and ST11 (RA1; IncA/C1), with the remaining nine STs linked together as single-locus variants (SLVs) or double-locus variants (DLVs). ST3 is the largest group and includes 53 plasmids. ST3 connects to ST4, ST5, ST6, ST7, ST8, and ST9 as SLVs, forming a clonal complex with ST3 as the founder (ST3 clonal complex [ST3CC]). ST3 links with ST2 as a DLV, and ST2 connects to ST1 as an SLV. ST1 is the second largest group and comprises 20 plasmids. ST1 and ST3 together account for 89% of the total IncA/C plasmids investigated.
TABLE 2.
Primers for IncA/C plasmid multilocus sequence typing
| Primer | Sequence (5′–3′) | Primer information | Amplicon size (bp)a |
|---|---|---|---|
| repA-F | AAGAGAACCAAAGACAAAGAC | Amplify repA | 982 |
| repA-R | GCTGCTTACGCTTGTTGGA | ||
| parA-F | AAAAGTAATCAGCTTCGCCA | Amplify parA | 780 |
| parA-R | TAGCCCACCTTCTCTAATAG | ||
| parB-F | TGTCCGAACTTGCTAAAGC | Amplify parB | 1,128 |
| parB-R | CTGACACAGGCACATGAA | ||
| 053-F | AGATCTCACAGGACATGAA | Amplify 053 | 250 |
| 053-R | TTCAAGAACGAAGACCTGT | ||
| repA-Seq1 | TGGAGTTCGTACAGAGTGA | Sequence 5′ region of repA fragment | NA |
| repA-Seq2 | GCTCCAGCTTCTTCCCGAT | Sequence 3′ region of repA fragment | NA |
| parB-Seq1 | CACACAGTCAGGTAGCTT | Sequence 5′ region of parB fragment | NA |
| parB-Seq2 | AAGCTACCTGACTGTGTG | Sequence central region of parB fragment | NA |
| parB-Seq3 | GATGCTCTTCCTCCTCTG | Sequence 3′ region of parB fragment | NA |
NA, not available.
FIG 6.
Minimal spanning tree of IncA/C PMLST. The branch lengths indicate the numbers of allele differences between STs. The pie charts for each ST indicate the species of isolation. The gray rectangle encompasses STs belonging to the ST3 clonal complex. A. hydrophila, Aeromonas hydrophila; C. freundii, Citrobacter freundii; E. aerogenes, Enterobacter aerogenes; E. cloacae, Enterobacter cloacae; K. oxytoca, Klebsiella oxytoca; P. damselae, Photobacterium damselae; P. stuartii, Providencia stuartii; S. marcescens, Serratia marcescens; V. alginolyticus, Vibrio alginolyticus; Y. pestis, Yersinia pestis; Y. ruckeri, Yersinia ruckeri.
To provide higher-resolution typing applicable to next-generation sequencing data, a core gene PMLST (cgPMLST) scheme was also constructed by extending the 4-gene PMLST to 28 conserved genes (see Data Set S7 in the supplemental material). The four loci shared between the two schemes allow backward compatibility from cgPMLST to PMLST. Thus, cgPMLST is capable of subtyping 11 STs into 35 subgroups, including 4 subgroups for ST1 (ST1.1 to ST1.4), 22 subgroups for ST3 (ST3.1 to ST3.22), and 1 subgroup for each of the remaining STs (see Data Set S7 and Fig. S6 in the supplemental material).
IncA/C PMLST highlights a lineage of blaNDM-harboring plasmids.
To determine if the distribution of antibiotic resistance genes among IncA/C plasmids is associated with STs, the resistance gene content of each plasmid was overlaid with the PMLST phylogenetic scheme (Fig. 7). Examination of the resistance gene profiles highlighted distinct patterns between ST1 and ST3CC. ST1 predominantly harbors blaCMY-6, while ST3CC plasmids mainly possess the blaCMY-2 variant (Fig. 7). ST3CC also exhibits a higher prevalence of tetracycline and streptomycin resistance genes. ST1 is strongly associated with the carriage of blaNDM (15/20 plasmids; 75%), while only two plasmids outside ST1 carry blaNDM (one each from ST3 and ST6). Comparison of the blaNDM genetic location has shown multiple genetic organizations (41–46). Within our database, we observed a total of seven distinct blaNDM structures (see Fig. S1 in the supplemental material). Six of them are found in individual plasmids (pNDM-SAL, ST1; pNDM-1_Dok01, ST1; pEC2-NDM-3, ST1; pNDM15-1078, ST1; pRH-1238, ST3; and pMR0211, ST6), and one has been observed within ARI-A of nine plasmids (45, 47). All nine plasmids belong to ST1, subgroups ST1.2 and ST1.4 (see Fig. S6 in the supplemental material). Overall, non-ST1 blaNDM structures are distinct from all others, while the presence of similar structures in 9/15 ST1 plasmids suggests the successful dissemination and diversification of the plasmid lineage.
FIG 7.
Phylogeny and resistance gene profiles of IncA/C plasmids. The tree was built using sequences derived from the PMLST analysis. Plasmids that contain a blaNDM structure similar to that present in pMS6198A (see Fig. S1 in the supplemental material) are indicated by asterisks. The full list of resistance genes is shown in Data Set S8 in the supplemental material in the same order as in the figure.
DISCUSSION
Carbapenem-resistant Enterobacteriaceae have been recognized as an urgent threat to human health (48). Infections caused by these bacteria are often resistant to almost all clinically available antibiotics and are frequently associated with poor health outcomes. New Delhi metallo-β-lactamase is a recently emerged carbapenemase first described in 2009 (49). Since then, several IncA/C plasmids carrying the blaNDM-1 gene (4, 41, 47, 50–52), and recently blaNDM-3 (45), have been reported. Plasmids of incompatibility group A/C have been known for more than 40 years, but they have only recently gained increased interest due in part to their emergence as the major plasmid type carrying the cephalosporinase gene blaCMY (2). However, the broad-host range characteristic of IncA/C plasmids and their roles in the dissemination of multiple antibiotic resistance genes, including blaCMY and blaNDM, are underappreciated. Here, we have used a validated set of essential genes to develop a high-resolution typing scheme to monitor the spread and transmission of IncA/C plasmids.
Previous studies on the replicon of RA1, the prototypical IncA/C plasmid, showed that IncA/C plasmid replication is mediated by the autoregulated repA gene and an iteron upstream of a DnaA box (19). Indeed, our mutagenesis analysis confirmed a requirement for repA and its adjacent iteron region in pMS6198A (Fig. 3Ci). This validated our method to identify required genetic components and also served as a reference point for identification of six other genes, namely, ata, parA, parB, 053, tnpA, and rmtC.
The addiction system of IncA/C plasmids has been the subject of various studies, but its contribution to IncA/C plasmid stability has not been fully established. Recent transcriptome analysis of pAR060302 showed that the system is strongly transcribed (21). Furthermore, multiple attempts to mutate ata were ultimately unsuccessful (22). Our TraDIS data confirmed that ata mutants were highly underrepresented. This is likely a result of uncontrolled toxin expression leading to cell death and demonstrates that the system is active in pMS6198A. The activity of the system may contribute to the variation in stability between the parent plasmid, pMS6198A, and pMAC3 (see Fig. S5 in the supplemental material). Moreover, it is possible that additional stability elements may be present on pMS6198A that exert subtle effects undetected by our stringent TraDIS threshold.
A putative partitioning locus is present in all IncA/C plasmids. It has been noted that the parA and parB genes share similarity to IncP plasmid partitioning and regulatory elements (1). Our TraDIS data strongly support a role for these genes in the maintenance and stability of IncA/C plasmids. Additionally, we identified the adjacent ORF, 053, as a novel element of the locus that has no IncP homolog. We showed that parAB alone were not sufficient for maintenance (Fig. 4, pMAC2) and that plasmid stability improved markedly with the addition of a 420-bp fragment containing ORF 053 (Fig. 4, pMAC3). Our results invoke at least two hypotheses: that a parS cis-acting centromere-like site of the ParAB partitioning system is present within this region or that 053 encodes a novel partitioning protein. Further work is required to elucidate the mechanisms by which this 420-bp region contributes to plasmid stability.
Plasmid pMS6198A carries a number of other genes with putative replication and maintenance functions, including kfrA (161) and stbA (174). However, they were not identified by TraDIS analysis (see Data Set S4 in the supplemental material), suggesting they are not critical to pMS6198A maintenance in E. coli. Conversely, tnpA and rmtC have functions unrelated to plasmid stability yet were identified. Both genes have low GC contents (34% and 41%, respectively, compared to an average of 52%) across the entire pMS6198A sequence. This is consistent with an overall increased mini-Tn5 insertion frequency in high- versus low-GC regions of pMS6198A (see Fig. S4 in the supplemental material). Thus, the identification of these genes may be the result of insertion bias of the Tn5 transposon in the in vitro mutagenesis reaction (38).
Deciphering phylogenetic relationships among plasmids can be challenging due to their mosaic nature. However, within one plasmid incompatibility group, it is expected that replication and maintenance machineries required for its biology should be conserved. This notion has been successfully applied to select genes for multilocus sequence typing schemes of plasmids from many Inc groups, including IncI1 (39), IncHI1 (53), IncHI2 (54), and IncN (55). Here, we propose a new PMLST scheme for IncA/C plasmids based on an experimentally validated set of essential genes. The loci selected for our PMLST scheme were based on the required genes identified by TraDIS analysis and supported by their presence in all IncA/C plasmids investigated. The scheme identified 11 sequence types, demonstrating higher resolution than current typing methods. We suggest that our PMLST scheme could be used to monitor dissemination and diversification patterns of IncA/C plasmids.
Of the 11 IncA/C sequence types, the majority of plasmids belong to two main types: ST1 and ST3. ST3 is the largest group, comprising 53 plasmids that form a clonal complex (ST3CC) with plasmids from ST4, ST5, ST6, ST7, ST8, and ST9 (Fig. 6). The ST3CC plasmids were isolated from 1969 to 2015 in 19 countries and 16 species (Fig. 6; see Data Set S5 and Fig. S7 in the supplemental material), highlighting their wide geographical distribution and broad host range characteristics. Interestingly, the IncA/C2 type 1 plasmids within ST3CC showed strong association with blaCMY (23/31; 74%). As highlighted previously, the high frequency of blaCMY-positive IncA/C plasmids isolated from Salmonella enterica in the United States is an indication of sampling bias within the data set (1).
ST1 is the second-largest group in our data set, with 20 plasmids. The most striking feature of ST1 is the strong association with the carriage of blaNDM, with the majority of ST1 plasmids isolated from E. coli and K. pneumoniae. The clinical relevance of these species and carbapenem resistance has likely contributed to a sampling bias of ST1 plasmids. Nevertheless, it is tempting to hypothesize that ST1 is a newly emerged lineage of IncA/C plasmids, with carbapenem resistance enhancing its selection and dissemination. Further work is needed to test this hypothesis; however, the presence of a common blaNDM structure in specific ST1 plasmid subgroups is supportive of the tenet. The observation of most concern here is the identification of ST1 in nine countries spanning four continents (see Data Set S5 and Fig. S7B in the supplemental material), highlighting the urgent need for surveillance and control of this extensively drug-resistant plasmid lineage.
More in-depth investigations within each ST may provide a better framework for analyzing the evolution of IncA/C plasmids. Inspired by the development of core genome MLST schemes (56), we have also constructed a cgPMLST scheme for IncA/C plasmids using the 28 conserved genes identified in our database (see Fig. S6 and Data Set S7 in the supplemental material). IncA/C cgPMLST is intended as a subtyping scheme complementary to PMLST to increase the discriminatory power suitable for plasmid epidemiology studies. The discriminatory power, measured by Hunter's index (57), of our IncA/C PMLST is 0.53 and is increased to 0.90 for cgPMLST. Other methods, such as that proposed by Harmer and Hall (58), could also be used in conjunction with PMLST to subtype IncA/C plasmids.
While sequence data from next-generation sequencing platforms, especially those from Pacific Biosciences (PacBio), would provide full plasmid backbone sequences for in-depth epidemiology and high-resolution phylogeny analysis, such technologies remain out of reach for many laboratories in developing countries, where surveillance and control measures are most needed. Our PMLST scheme provides a robust method based on PCR and Sanger sequencing for the identification of major lineages of IncA/C plasmids. These major lineages can then be subtyped using cgPMLST when whole-plasmid sequence data are available. Like other MLST-based schemes, both of our schemes are compatible with freely available tools, such as SRST2 (59) and pMLST (60), for in silico determination of plasmid STs, enabling the use of typing data across different settings. Our PMLST and cgPMLST schemes are also available on the public databases for molecular typing and microbial genome diversity (PubMLST) (79).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli MS6198 was isolated from the urine of a patient with a urinary tract infection in Haryana, India, in 2010 (61). The E. coli strain J53 was provided by G. Jacoby (62). The strains were routinely cultured at 37°C under orbital shaking (250 rpm), in liquid or on solid lysogeny broth (LB) medium, supplemented with appropriate antibiotics. The following concentrations were typically used: ampicillin, 100 μg/ml; sodium azide, 100 μg/ml; meropenem, 1 μg/ml; and chloramphenicol, 30 μg/ml. Electrocompetent cells were prepared, and transformations were performed as described previously (29). All the strains were stored in 15% glycerol at −80°C.
DNA purification and analysis.
The PureLink HiPure Midiprep plasmid DNA purification kit (Invitrogen) was used to purify pMS6198A, while vector plasmids (<10 kb) were purified with the QIAprep Spin Miniprep kit (Qiagen). Genomic DNA was obtained with MoBio's Ultraclean microbial DNA isolation kit. DNA concentrations were quantified using a NanoDrop 2000 (Thermo Scientific) and/or Qubit 2.0 (Life Technologies) fluorometer.
PCR and sequencing.
The presence of plasmids was determined by PCR- based replicon typing (63, 64); blaNDM was identified with primers 5′-GGTTTGGCGATCTGGTTTTC-3′ and 5′-CGGAATGGCTCATCACGATC-3′ as previously described (65), using One Taq polymerase (New England BioLabs). All restriction enzymes, T4 ligase, and Antarctic phosphatase were purchased from New England BioLabs. All capillary sequencing reactions were prepared using BigDye Terminator mix and sequenced by the Australian Equine Genetics Research Centre (AEGRC). A full list of primers used in this study is shown in Data Set S9 in the supplemental material.
PMLST PCR and sequencing protocol.
Amplification of the four loci used in PMLST was performed with Kapa HiFi DNA polymerase (Kapa Biosystems) using primers listed in Table 2 with the following cycling program: 95°C for 3 min; 25 cycles of 98°C for 20 s, 60°C for 15 s, and 72°C for 30 s; and a final extension of 72°C for 3 min. Each amplicon was then purified using the QIAgen PCR purification kit and sequenced using BigDye Terminator v3.1 cycle sequencing (Life Technology) with the appropriate sequencing primers listed in Table 2.
Disc diffusion and mating assays.
The disc diffusion assay was performed and interpreted according to the Clinical and Laboratory Standards Institute guidelines (2014). Antimicrobial discs were obtained from Becton Dickinson. For mating assays, the sodium azide-resistant E. coli strain J53 (62) was used as the recipient in all mating assays. Donor and recipient strains were grown to an optical density at 600 nm (OD600) equal to 2.0. The cells were then mixed at a ratio of 1:2 (donors to recipients) in LB and incubated at 37°C for 2 h under static conditions. Total CFU of donors, recipients, and transconjugants were enumerated on LB agar plates with appropriate antibiotic selection (ampicillin for donors and sodium azide for recipients), and the conjugation frequency was calculated as the number of transconjugants per donor.
Plasmid stability assays.
Time course stability assays were performed essentially as previously described (66). Population counts were achieved by 10-fold serial dilutions and 5-μl drop test on LB agar with or without selection. For solid-medium stability assays, strains were grown overnight on LB agar supplemented with antibiotics, and then single colonies were suspended in 0.9% NaCl. Population counts were achieved by 10-fold serial dilutions and 5-μl drop tests on LB agar with or without selection.
In vitro transposon mutagenesis.
A custom mini-Tn5 transposon containing a chloramphenicol resistance cassette (Tn5-Cm) was generated as previously described (30). An in vitro plasmid mutant library was created by incubating 200 ng of pMS6198A DNA and equimolar Tn5-Cm DNA (1.588 ng) with 1 μl of Tn5 transposase (1 U/μl) from an EZ-Tn5 <R6Kγori/KAN-2> insertion kit (Epicentre) at 37°C for 2 h. The reaction was stopped by adding 1 μl 1% SDS and heating at 70°C for 10 min. The mutant plasmid library (1 μl) was transformed into 50 μl of E. coli TOP10 electrocompetent cells. Cells carrying mutant plasmids were selected by plating on LB agar supplemented with chloramphenicol. Mutants were pooled by scraping colonies off agar plates into LB. After addition of glycerol to a final concentration of 15%, the mutant library was stored at −80°C.
Inverse-PCR method.
Purified DNA from individual mutants was digested with BanII for 2 h at 37°C and heat inactivated at 65°C for 20 min. This mixture was ligated with T4 DNA ligase overnight at 16°C. This ligation mixture was used as the template for PCR using OneTaq and cat-specific primers 3748 and 3950, with thermocycling: 94°C for 1 min; 30 cycles of 94°C for 30 s, 62°C for 30 s, and 68°C for 3.5 min; and a final extension time of 5 min.
Transposon-directed insertion site sequencing.
Illumina library preparation was performed using a Nextera DNA Sample Prep kit (Illumina) following the manufacturer's instructions with modifications for TraDIS. Briefly, genomic DNA was fragmented and tagged with adapter sequence via one enzymatic reaction (tagmentation). Following tagmentation, the DNA was purified using the Zymo DNA Clean and Concentrator kit (Zymo Research). The PCR enrichment step was run using index primer 1 (one index per sample) and a custom transposon-specific primer, 4844 (5′-AATGATACGGCGACCACCGAGATCTACACTAGATCGCaacttcggaataggaactaagg-3′ [transposon-specific sequence is in lowercase]) to enrich for transposon insertion sites and allow multiplexing sequencing; the thermocycler program was 72°C for 3 min and 98°C for 30 s, followed by 22 cycles of 98°C for 10 s, 63°C for 30 s, and 72°C for 1 min. Each library was purified using Agencourt Ampure XP magnetic beads. Verification and quantification of the resulting libraries were calculated using a Qubit 2.0 fluorometer, a 2100 Bioanalyzer (Agilent Technologies), and quantitative PCR (qPCR) (Kapa Biosciences). All libraries were pooled in equimolar amounts to a final concentration of 3.2 nM and submitted for sequencing on the MiSeq platform at the Queensland Centre for Medical Genomics (Institute for Molecular Bioscience, University of Queensland). The MiSeq sequencer was loaded with 12 pM of pooled library with 5% PhiX spike-in and sequenced (single-end; 101 cycles) using a mixture of standard Illumina sequencing primer and Tn5-specific sequencing primer 4845 (5′-actaaggaggatattcatatggaccatggctaattcccatgtcagatgtg-3′).
All sequence data analysis and insertion site mapping were performed as previously described (30, 32). The threshold for plasmid maintenance was set to the value of the repA gene (418 RPKM), in accordance with previous work that has demonstrated the gene is required for IncA/C plasmid replication (19).
Construction of mini-A/C plasmids.
The replicon fragment was amplified with primers 7139/7140 from purified pMS6198A, the transcriptional terminator (TT) fragment gene was amplified with primers 7141/7142 from pQE30, and the cat gene was amplified with primers 7143/7144 from pKD3. The primers were designed so that adjacent fragments had 20-bp complementary overhangs, and a multiple-cloning site (MCS) was included between the replicon and TT fragments. All three fragments were mixed in equimolar ratios in a PCR using Kapa HiFi with thermocycling: 95°C; 30 cycles of 98°C for 20 s, 66°C for 20 s, and 72°C for 6 min; and a final extension of 72°C for 8 min. The product of this reaction was electroporated into E. coli TOP10. The desired product was confirmed by PCR screening and NheI digestion of purified plasmid DNA.
The TT fragment contained two terminators, lambda t0 and rrnB t1, separated by a cat gene. The cat gene was removed by PCR amplification of the plasmid using primers 7145/7146, followed by NheI digestion 37°C for 2 h and overnight ligation with T4 DNA ligase. The product was electroporated into E. coli TOP10, and subsequent purification, PCR screening, and NheI digestion confirmed the desired product, which was referred to as pMAC1.
The partitioning fragments were amplified using Kapa HiFi with primers 7147/7149 for pMAC2 and 7147/7148 for pMAC3. Each fragment was cloned into the MCS of pMAC1 using BamHI and HindIII restriction sites. The resultant plasmids (pMAC2 and pMAC3) were confirmed to be correct by sequencing.
Genome sequencing, assembly, and methylome analysis.
Genomic DNA from MS6198 was sequenced on the PacBio RSII (University of Malaya) using the P4 polymerase and C2 sequencing chemistry. The raw sequencing data were assembled de novo using the hierarchical genome assembly process (HGAP) version 2 from the SMRT Analysis software suite (version 2.3.0; Pacific Biosciences) with default parameters. The assembled contigs were visually screened for overlapping sequences on their 5′ and 3′ ends using contiguity (67). These overlapping ends were manually trimmed based on sequence similarity, and the contigs were circularized. The circularized contigs (chromosome and plasmids) were then polished by mapping raw sequencing reads back onto the assembled circular contigs.
The detection of methylated bases and clustering of modified sites to identify methylation-associated motifs was performed as previously reported (68). In brief, raw reads were aligned to the complete genome of MS6198, and interpulse duration (IPD) ratios were calculated using PacBio's in silico kinetic reference computational model.
Sequence analysis, annotation, and in silico typing.
Visualization and annotation of plasmid sequences were performed using PROKKA v1.11 (69) and the Artemis Genome Browser (70). Sequence comparisons were constructed using WebACT (71) and visualized with Easyfig 2.1 (72) and Artemis Comparison Tool (ACT) (73). In silico DNA manipulations and analysis were conducted and visualized in CLC Main Workbench (version 7.0.2; Qiagen Bioinformatics) and Easyfig 2.1 (72). In silico E. coli multilocus sequence typing was performed using the MLST database hosted at the University of Warwick (74). Plasmid Inc types were determined by PCR-based replicon typing (63, 64) or in silico using PlasmidFinder (60).
Collection of IncA/C complete sequences and analysis.
Complete sequences of IncA/C plasmids from GenBank were selected by BLASTn using the sequence of RA1 repA as a reference (>90% identity and >90% query coverage). The BLASTn hits were manually reviewed, and only published sequences were included in our IncA/C plasmid database. Each plasmid sequence was also verified by BLASTn to contain IncA/C replicon-typing primer binding sites (63). With the addition of pMS6198A, this database comprised 82 IncA/C plasmid sequences (as of 9 May 2016) (see Data Set S5 in the supplemental material). The gene annotations of pMS6198A were used as a reference to identify genes present in all 82 IncA/C plasmids, using default BLASTn v2.2.26 settings with the criteria of an expected value of 10−30 and minimum coverage of 95%. The sequence of each conserved gene was extracted from each plasmid using EMBOSS v6.5.7 (75). PMLST minimal spanning trees were built by Phyloviz using the goeBURST algorithm (76). All alignments were constructed in MEGA 6.06 (77) using ClustalW with default settings. Phylogenetic trees were produced in MEGA 6.06 using maximum likelihood with default settings and supported with 1,000 bootstraps. The presence or absence of resistance genes was determined using the BLASTn algorithm (100% identity at 100% coverage) against the resistance gene database ARG-ANNOT (78). IncA/C sequence analysis and metadata have been incorporated into the microreact database (https://microreact.org/project/IncACPlasmids?tt=rc&tns=4&tts=6) (80).
Accession number(s).
The sequences for the MS6198 chromosome, pMS6198A, pMS6198B, pMS6198C, and pMS6198D have been deposited in the NCBI GenBank database under accession numbers CP015834 to CP015838, respectively. Raw PacBio sequence reads for MS6198 and Illumina MiSeq reads for duplicate TraDIS runs have been deposited in the Sequence Read Archive (SRA) under accession numbers SRX1797306, SRX1992326 (replicate 1), and SRX1992327 (replicate 2), respectively.
Supplementary Material
ACKNOWLEDGMENTS
We thank David Miller, Tim Bruxner, and Angelika Christ from the Queensland Centre for Medical Genomics (Institute for Molecular Bioscience, University of Queensland) for technical support with the TraDIS protocol. The protocol was set up in consultation with Brian Fritz (Illumina) and Sabine Eckert, Daniel Turner, and Matthew Mayho (Wellcome Trust Sanger Institute).
This work was supported by grants from the National Health and Medical Research Council (NHMRC) of Australia (GNT1033799 and GNT1067455) and High Impact Research (HIR) grants from the University of Malaya (UM-MOHE HIR grant UM C/625/1/HIR/MOHE/CHAN/14/1, no. H-50001-A000027; UM-MOHE HIR grant UM C/625/1/HIR/MOHE/CHAN/01, no. A000001-50001). M.A.S. is supported by an NHMRC Senior Research Fellowship (GNT1106930), and S.A.B. is supported by an NHMRC Career Development Fellowship (GNT1090456). Teik Min Chong is supported by the Postgraduate Research Fund (PPP) (grant no. PG080-2015B).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01740-16.
REFERENCES
- 1.Harmer CJ, Hall RM. 2015. The A to Z of A/C plasmids. Plasmid 80:63–82. doi: 10.1016/j.plasmid.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 2.Lindsey RL, Fedorka-Cray PJ, Frye JG, Meinersmann RJ. 2009. IncA/C plasmids are prevalent in multidrug-resistant Salmonella enterica isolates. Appl Environ Microbiol 75:1908–1915. doi: 10.1128/AEM.02228-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walsh TR, Weeks J, Livermore DM, Toleman MA. 2011. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis 11:355–362. doi: 10.1016/S1473-3099(11)70059-7. [DOI] [PubMed] [Google Scholar]
- 4.Carattoli A, Villa L, Poirel L, Bonnin RA, Nordmann P. 2012. Evolution of IncA/C blaCMY-2-carrying plasmids by acquisition of the blaNDM-1 carbapenemase gene. Antimicrob Agents Chemother 56:783–786. doi: 10.1128/AAC.05116-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poirel L, Dortet L, Bernabeu S, Nordmann P. 2011. Genetic features of blaNDM-1-positive Enterobacteriaceae. Antimicrob Agents Chemother 55:5403–5407. doi: 10.1128/AAC.00585-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aoki T, Egusa S, Kimura T, Watanabe T. 1971. Detection of R factors in naturally occurring Aeromonas salmonicida strains. Appl Microbiol 22:716–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hedges RW, Jacob AE. 1975. A 98 megadalton R factor of compatibility group C in a Vibrio cholerae El Tor isolate from southern U.S.S.R. J Gen Microbiol 89:383–386. doi: 10.1099/00221287-89-2-383. [DOI] [PubMed] [Google Scholar]
- 8.Rahal K, Gerbaud GR, Chabbert YA. 1973. Properties of a transferable resistance factor in Vibrio cholerae biotype eltor. Ann Microbiol (Paris) 124:283–294. (In French.) [PubMed] [Google Scholar]
- 9.Welch TJ, Fricke WF, McDermott PF, White DG, Rosso ML, Rasko DA, Mammel MK, Eppinger M, Rosovitz MJ, Wagner D, Rahalison L, Leclerc JE, Hinshaw JM, Lindler LE, Cebula TA, Carniel E, Ravel J. 2007. Multiple antimicrobial resistance in plague: an emerging public health risk. PLoS One 2:e309. doi: 10.1371/journal.pone.0000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hazen TH, Zhao L, Boutin MA, Stancil A, Robinson G, Harris AD, Rasko DA, Johnson JK. 2014. Comparative genomics of an IncA/C multidrug resistance plasmid from Escherichia coli and Klebsiella isolates from intensive care unit patients and the utility of whole-genome sequencing in health care settings. Antimicrob Agents Chemother 58:4814–4825. doi: 10.1128/AAC.02573-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernandez-Alarcon C, Singer RS, Johnson TJ. 2011. Comparative genomics of multidrug resistance-encoding IncA/C plasmids from commensal and pathogenic Escherichia coli from multiple animal sources. PLoS One 6:e23415. doi: 10.1371/journal.pone.0023415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Datta N, Hedges RW. 1972. R factors identified in Paris, some conferring gentamicin resistance, constitute a new compatibility group. Ann Inst Pasteur (Paris) 123:849–852. [PubMed] [Google Scholar]
- 13.Hedges RW. 1974. R factors from Providence. J Gen Microbiol 81:171–181. doi: 10.1099/00221287-81-1-171. [DOI] [PubMed] [Google Scholar]
- 14.Sirgel FA, Coetzee JN, Hedges RW, Lecatsas G. 1981. Phage C-1: an IncC group; plasmid-specific phage. J Gen Microbiol 122:155–160. [DOI] [PubMed] [Google Scholar]
- 15.Carattoli A, Miriagou V, Bertini A, Loli A, Colinon C, Villa L, Whichard JM, Rossolini GM. 2006. Replicon typing of plasmids encoding resistance to newer beta-lactams. Emerg Infect Dis 12:1145–1148. doi: 10.3201/eid1207.051555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harmer CJ, Hall RM. 2014. pRMH760, a precursor of A/C2 plasmids carrying blaCMY and blaNDM genes. Microb Drug Resist 20:416–423. doi: 10.1089/mdr.2014.0012. [DOI] [PubMed] [Google Scholar]
- 17.Meinersmann RJ, Lindsey RL, Bono JL, Smith TP, Oakley BB. 2013. Proposed model for the high rate of rearrangement and rapid migration observed in some IncA/C plasmid lineages. Appl Environ Microbiol 79:4806–4814. doi: 10.1128/AEM.01259-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Llanes C, Gabant P, Couturier M, Michel-Briand Y. 1994. Cloning and characterization of the IncA/C plasmid RA1 replicon. J Bacteriol 176:3403–3407. doi: 10.1128/jb.176.11.3403-3407.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Llanes C, Gabant P, Couturier M, Bayer L, Plesiat P. 1996. Molecular analysis of the replication elements of the broad-host-range RepA/C replicon. Plasmid 36:26–35. doi: 10.1006/plas.1996.0028. [DOI] [PubMed] [Google Scholar]
- 20.del Solar G, Giraldo R, Ruiz-Echevarria MJ, Espinosa M, Diaz-Orejas R. 1998. Replication and control of circular bacterial plasmids. Microbiol Mol Biol Rev 62:434–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lang KS, Danzeisen JL, Xu W, Johnson TJ. 2012. Transcriptome mapping of pAR060302, a blaCMY-2-positive broad-host-range IncA/C plasmid. Appl Environ Microbiol 78:3379–3386. doi: 10.1128/AEM.07199-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carraro N, Matteau D, Luo P, Rodrigue S, Burrus V. 2014. The master activator of IncA/C conjugative plasmids stimulates genomic islands and multidrug resistance dissemination. PLoS Genet 10:e1004714. doi: 10.1371/journal.pgen.1004714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Motallebi-Veshareh M, Rouch DA, Thomas CM. 1990. A family of ATPases involved in active partitioning of diverse bacterial plasmids. Mol Microbiol 4:1455–1463. doi: 10.1111/j.1365-2958.1990.tb02056.x. [DOI] [PubMed] [Google Scholar]
- 24.Baxter JC, Funnell BE. 2014. Plasmid partition mechanisms. Microbiol Spectr 2. doi: 10.1128/microbiolspec.PLAS-0023-2014. [DOI] [PubMed] [Google Scholar]
- 25.Gerdes K, Moller-Jensen J, Bugge Jensen R. 2000. Plasmid and chromosome partitioning: surprises from phylogeny. Mol Microbiol 37:455–466. [DOI] [PubMed] [Google Scholar]
- 26.van Opijnen T, Bodi KL, Camilli A. 2009. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods 6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goodman AL, McNulty NP, Zhao Y, Leip D, Mitra RD, Lozupone CA, Knight R, Gordon JI. 2009. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe 6:279–289. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gawronski JD, Wong SM, Giannoukos G, Ward DV, Akerley BJ. 2009. Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc Natl Acad Sci U S A 106:16422–16427. doi: 10.1073/pnas.0906627106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langridge GC, Phan MD, Turner DJ, Perkins TT, Parts L, Haase J, Charles I, Maskell DJ, Peters SE, Dougan G, Wain J, Parkhill J, Turner AK. 2009. Simultaneous assay of every Salmonella typhi gene using one million transposon mutants. Genome Res 19:2308–2316. doi: 10.1101/gr.097097.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phan MD, Peters KM, Sarkar S, Lukowski SW, Allsopp LP, Gomes Moriel D, Achard ME, Totsika M, Marshall VM, Upton M, Beatson SA, Schembri MA. 2013. The serum resistome of a globally disseminated multidrug resistant uropathogenic Escherichia coli clone. PLoS Genet 9:e1003834. doi: 10.1371/journal.pgen.1003834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamaichi Y, Chao MC, Sasabe J, Clark L, Davis BM, Yamamoto N, Mori H, Kurokawa K, Waldor MK. 2015. High-resolution genetic analysis of the requirements for horizontal transmission of the ESBL plasmid from Escherichia coli O104:H4. Nucleic Acids Res 43:348–360. doi: 10.1093/nar/gku1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phan MD, Forde BM, Peters KM, Sarkar S, Hancock S, Stanton-Cook M, Ben Zakour NL, Upton M, Beatson SA, Schembri MA. 2015. Molecular characterization of a multidrug resistance IncF plasmid from the globally disseminated Escherichia coli ST131 clone. PLoS One 10:e0122369. doi: 10.1371/journal.pone.0122369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sherchan JB, Hayakawa K, Miyoshi-Akiyama T, Ohmagari N, Kirikae T, Nagamatsu M, Tojo M, Ohara H, Sherchand JB, Tandukar S. 2015. Clinical epidemiology and molecular analysis of extended-spectrum-beta-lactamase-producing Escherichia coli in Nepal: characteristics of sequence types 131 and 648. Antimicrob Agents Chemother 59:3424–3432. doi: 10.1128/AAC.00270-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ewers C, Bethe A, Stamm I, Grobbel M, Kopp PA, Guerra B, Stubbe M, Doi Y, Zong Z, Kola A, Schaufler K, Semmler T, Fruth A, Wieler LH, Guenther S. 2014. CTX-M-15-D-ST648 Escherichia coli from companion animals and horses: another pandemic clone combining multiresistance and extraintestinal virulence? J Antimicrob Chemother 69:1224–1230. doi: 10.1093/jac/dkt516. [DOI] [PubMed] [Google Scholar]
- 35.Cao X, Zhang Z, Shen H, Ning M, Chen J, Wei H, Zhang K. 2014. Genotypic characteristics of multidrug-resistant Escherichia coli isolates associated with urinary tract infections. APMIS 122:1088–1095. [DOI] [PubMed] [Google Scholar]
- 36.Dziewit L, Jazurek M, Drewniak L, Baj J, Bartosik D. 2007. The SXT conjugative element and linear prophage N15 encode toxin-antitoxin-stabilizing systems homologous to the tad-ata module of the Paracoccus aminophilus plasmid pAMI2. J Bacteriol 189:1983–1997. doi: 10.1128/JB.01610-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karim A, Poirel L, Nagarajan S, Nordmann P. 2001. Plasmid-mediated extended-spectrum beta-lactamase (CTX-M-3 like) from India and gene association with insertion sequence ISEcp1. FEMS Microbiol Lett 201:237–241. [DOI] [PubMed] [Google Scholar]
- 38.Green B, Bouchier C, Fairhead C, Craig NL, Cormack BP. 2012. Insertion site preference of Mu, Tn5, and Tn7 transposons. Mob DNA 3:3. doi: 10.1186/1759-8753-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garcia-Fernandez A, Chiaretto G, Bertini A, Villa L, Fortini D, Ricci A, Carattoli A. 2008. Multilocus sequence typing of IncI1 plasmids carrying extended-spectrum beta-lactamases in Escherichia coli and Salmonella of human and animal origin. J Antimicrob Chemother 61:1229–1233. doi: 10.1093/jac/dkn131. [DOI] [PubMed] [Google Scholar]
- 40.Anantham S, Harmer CJ, Hall RM. 2015. p39R861-4, a type 2 A/C2 plasmid carrying a segment from the A/C1 plasmid RA1. Microb Drug Resist 21:571–576. doi: 10.1089/mdr.2015.0133. [DOI] [PubMed] [Google Scholar]
- 41.McGann P, Hang J, Clifford RJ, Yang Y, Kwak YI, Kuschner RA, Lesho EP, Waterman PE. 2012. Complete sequence of a novel 178-kilobase plasmid carrying blaNDM-1 in a Providencia stuartii strain isolated in Afghanistan. Antimicrob Agents Chemother 56:1673–1679. doi: 10.1128/AAC.05604-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Villa L, Guerra B, Schmoger S, Fischer J, Helmuth R, Zong Z, Garcia-Fernandez A, Carattoli A. 2015. IncA/C plasmid carrying blaNDM-1, blaCMY-16, and fosA3 in a Salmonella enterica serovar Corvallis strain isolated from a migratory wild bird in Germany. Antimicrob Agents Chemother 59:6597–6600. doi: 10.1128/AAC.00944-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarkar A, Pazhani GP, Chowdhury G, Ghosh A, Ramamurthy T. 2015. Attributes of carbapenemase encoding conjugative plasmid pNDM-SAL from an extensively drug-resistant Salmonella enterica serovar Senftenberg. Front Microbiol 6:969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Du H, Chen L, Chavda KD, Pandey R, Zhang H, Xie X, Tang YW, Kreiswirth BN. 2016. Genomic characterization of Enterobacter cloacae isolates from China that coproduce KPC-3 and NDM-1 carbapenemases. Antimicrob Agents Chemother 60:2519–2523. doi: 10.1128/AAC.03053-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wailan AM, Sidjabat HE, Yam WK, Alikhan NF, Petty NK, Sartor AL, Williamson DA, Forde BM, Schembri MA, Beatson SA, Paterson DL, Walsh TR, Partridge SR. 2016. Mechanisms involved in acquisition of blaNDM genes by IncA/C2 and IncFIIY plasmids. Antimicrob Agents Chemother 60:4082–4088. doi: 10.1128/AAC.00368-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mataseje LF, Peirano G, Church DL, Conly J, Mulvey M, Pitout JD. 2016. Colistin-nonsusceptible Pseudomonas aeruginosa sequence type 654 with blaNDM-1 arrives in North America. Antimicrob Agents Chemother 60:1794–1800. doi: 10.1128/AAC.02591-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hudson CM, Bent ZW, Meagher RJ, Williams KP. 2014. Resistance determinants and mobile genetic elements of an NDM-1-encoding Klebsiella pneumoniae strain. PLoS One 9:e99209. doi: 10.1371/journal.pone.0099209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Centers for Disease Control and Prevention. 2013. Antibiotic resistance threats in the United States. http://www.cdc.gov/drugresistance/threat-report-2013/pdf/ar-threats-2013-508.pdf#page=112:53-54.
- 49.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-beta-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sekizuka T, Matsui M, Yamane K, Takeuchi F, Ohnishi M, Hishinuma A, Arakawa Y, Kuroda M. 2011. Complete sequencing of the blaNDM-1-positive IncA/C plasmid from Escherichia coli ST38 isolate suggests a possible origin from plant pathogens. PLoS One 6:e25334. doi: 10.1371/journal.pone.0025334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mulvey MR, Grant JM, Plewes K, Roscoe D, Boyd DA. 2011. New Delhi metallo-beta-lactamase in Klebsiella pneumoniae and Escherichia coli, Canada. Emerg Infect Dis 17:103–106. doi: 10.3201/eid1701.101358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tijet N, Richardson D, MacMullin G, Patel SN, Melano RG. 2015. Characterization of multiple NDM-1-producing Enterobacteriaceae isolates from the same patient. Antimicrob Agents Chemother 59:3648–3651. doi: 10.1128/AAC.04862-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Phan MD, Kidgell C, Nair S, Holt KE, Turner AK, Hinds J, Butcher P, Cooke FJ, Thomson NR, Titball R, Bhutta ZA, Hasan R, Dougan G, Wain J. 2009. Variation in Salmonella enterica serovar Typhi IncHI1 plasmids during the global spread of resistant typhoid fever. Antimicrob Agents Chemother 53:716–727. doi: 10.1128/AAC.00645-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garcia-Fernandez A, Carattoli A. 2010. Plasmid double locus sequence typing for IncHI2 plasmids, a subtyping scheme for the characterization of IncHI2 plasmids carrying extended-spectrum beta-lactamase and quinolone resistance genes. J Antimicrob Chemother 65:1155–1161. doi: 10.1093/jac/dkq101. [DOI] [PubMed] [Google Scholar]
- 55.Garcia-Fernandez A, Villa L, Moodley A, Hasman H, Miriagou V, Guardabassi L, Carattoli A. 2011. Multilocus sequence typing of IncN plasmids. J Antimicrob Chemother 66:1987–1991. doi: 10.1093/jac/dkr225. [DOI] [PubMed] [Google Scholar]
- 56.Maiden MC, Jansen van Rensburg MJ, Bray JE, Earle SG, Ford SA, Jolley KA, McCarthy ND. 2013. MLST revisited: the gene-by-gene approach to bacterial genomics. Nat Rev Microbiol 11:728–736. doi: 10.1038/nrmicro3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hunter PR, Gaston MA. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol 26:2465–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harmer CJ, Hall RM. 2016. PCR-based typing of IncC plasmids. Plasmid 87-88:37–42. doi: 10.1016/j.plasmid.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 59.Inouye M, Dashnow H, Raven LA, Schultz MB, Pope BJ, Tomita T, Zobel J, Holt KE. 2014. SRST2: Rapid genomic surveillance for public health and hospital microbiology labs. Genome Med 6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, Villa L, Moller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, Chaudhary U, Doumith M, Giske CG, Irfan S, Krishnan P, Kumar AV, Maharjan S, Mushtaq S, Noorie T, Paterson DL, Pearson A, Perry C, Pike R, Rao B, Ray U, Sarma JB, Sharma M, Sheridan E, Thirunarayan MA, Turton J, Upadhyay S, Warner M, Welfare W, Livermore DM, Woodford N. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis 10:597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jacoby GA, Han P. 1996. Detection of extended-spectrum beta-lactamases in clinical isolates of Klebsiella pneumoniae and Escherichia coli. J Clin Microbiol 34:908–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 64.Villa L, Garcia-Fernandez A, Fortini D, Carattoli A. 2010. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J Antimicrob Chemother 65:2518–2529. doi: 10.1093/jac/dkq347. [DOI] [PubMed] [Google Scholar]
- 65.Poirel L, Walsh TR, Cuvillier V, Nordmann P. 2011. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis 70:119–123. doi: 10.1016/j.diagmicrobio.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 66.Kramer MG. 2016. Determination of plasmid segregational stability in a growing bacterial population. Methods Mol Biol 1409:125–133. doi: 10.1007/978-1-4939-3515-4_11. [DOI] [PubMed] [Google Scholar]
- 67.Sullivan MJ, Ben Zakour NL, Forde BM, Stanton-Cook M, Beatson SA. 2015. Contiguity: contig adjacency graph construction and visualisation. PeerJ PrePr 3 https://peerj.com/preprints/1037/. [Google Scholar]
- 68.Forde BM, Phan MD, Gawthorne JA, Ashcroft MM, Stanton-Cook M, Sarkar S, Peters KM, Chan KG, Chong TM, Yin WF, Upton M, Schembri MA, Beatson SA. 2015. Lineage-specific methyltransferases define the methylome of the globally disseminated Escherichia coli ST131 Clone. mBio 6:e01602-15. doi: 10.1128/mBio.01602-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 70.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 71.Abbott JC, Aanensen DM, Rutherford K, Butcher S, Spratt BG. 2005. WebACT—an online companion for the Artemis Comparison Tool. Bioinformatics 21:3665–3666. doi: 10.1093/bioinformatics/bti601. [DOI] [PubMed] [Google Scholar]
- 72.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Carver T, Berriman M, Tivey A, Patel C, Bohme U, Barrell BG, Parkhill J, Rajandream MA. 2008. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 24:2672–2676. doi: 10.1093/bioinformatics/btn529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16:276–277. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 76.Francisco AP, Vaz C, Monteiro PT, Melo-Cristino J, Ramirez M, Carrico JA. 2012. PHYLOViZ: phylogenetic inference and data visualization for sequence based typing methods. BMC Bioinformatics 13:87. doi: 10.1186/1471-2105-13-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gupta SK, Padmanabhan BR, Diene SM, Lopez-Rojas R, Kempf M, Landraud L, Rolain JM. 2014. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother 58:212–220. doi: 10.1128/AAC.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jolley KA, Maiden MC. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinforma 11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Argimón S, Abudahab K, Goater RJE, Fedosejev A, Bhai J, Glasner C, Grundmann H, Yeats C, Spratt BG, Aanensen B. 30 November 2016. Microreact: visualising and sharing data for genomic epidemiology and phylogeography. Microb Genom doi: 10.1099/mgen.0.000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.