ABSTRACT
A vaginal ring containing dapivirine (DPV) has shown moderate protective efficacy against HIV-1 acquisition, but the activity of DPV against efavirenz (EFV)- and nevirapine (NVP)-resistant viruses that could be transmitted is not well defined. We investigated DPV cross-resistance of subtype C HIV-1 from individuals on failing NVP- or EFV-containing antiretroviral therapy (ART) in South Africa. Plasma samples were obtained from individuals with >10,000 copies of HIV RNA/ml and with HIV-1 containing at least one non-nucleoside reverse transcriptase (NNRTI) mutation. Susceptibility to NVP, EFV, and DPV in TZM-bl cells was determined for recombinant HIV-1LAI containing bulk-amplified, plasma-derived, full-length reverse transcriptase sequences. Fold change (FC) values were calculated compared with a composite 50% inhibitory concentration (IC50) from 12 recombinant subtype C HIV-1LAI plasma-derived viruses from treatment-naive individuals in South Africa. A total of 25/100 (25%) samples showed >500-FCs to DPV compared to treatment-naive samples with IC50s exceeding the maximum DPV concentration tested (132 ng/ml). A total of 66/100 (66%) samples displayed 3- to 306-FCs, with a median IC50 of 17.6 ng/ml. Only 9/100 (9%) samples were susceptible to DPV (FC < 3). Mutations L100I and K103N were significantly more frequent in samples with >500-fold resistance to DPV compared to samples with a ≤500-fold resistance. A total of 91% of samples with NNRTI-resistant HIV-1 from individuals on failing first-line ART in South Africa exhibited ≥3-fold cross-resistance to DPV. This level of resistance exceeds expected plasma concentrations, but very high genital tract DPV concentrations from DPV ring use could block viral replication. It is critically important to assess the frequency of transmitted and selected DPV resistance in individuals using the DPV ring.
KEYWORDS: dapivirine, NNRTI, HIV-1, antiretroviral therapy, cross-resistance, drug resistance, human immunodeficiency virus, non-nucleoside reverse transcriptase inhibitor
INTRODUCTION
Sustained-release formulations of antiretrovirals could substantially decrease the incidence of HIV-1 infection in sub-Saharan Africa by improving product adherence compared to daily preexposure prophylaxis (PrEP) (1). Dapivirine (DPV) is a potent di-aryl-pyrimidine (DAPY) derivative in the non-nucleoside reverse transcriptase inhibitor (NNRTI) class of antiretrovirals. The development of DPV for antiretroviral therapy was halted due to suboptimal oral pharmacokinetics (2). Instead, it was formulated as a 25-mg slow-release vaginal ring for HIV-1 prevention because of its favorable safety profile and physical and chemical attributes. Recent phase III DPV ring studies in over 4,500 HIV-1-negative female participants in ASPIRE (MTN-020) (3) and the Ring Study (IPM-027) showed a minimum of 27% efficacy overall and up to 61% efficacy in compliant women over 25 years of age (4, 5).
A significant concern is whether DPV used as a PrEP agent will be active against circulating NNRTI-resistant HIV-1 variants in sub-Saharan Africa. There is potential for overlapping resistance profiles between DPV and other NNRTIs used for first-line ART and for prevention of mother-to-child transmission (6). In vitro selection experiments and drug susceptibility assays have revealed that the major mutations associated with DPV resistance or cross-resistance are V90I, L100I, K103N, V106I, E138K, Y181C, and Y188L (7, 8). Mutations outside the polymerase domain of reverse transcriptase in the connection and/or RNase H domains of reverse transcriptase, including G335D, N348I, T369I, A371V, and A376S, have been shown to cause cross-resistance to NNRTIs (9–13). Such mutations are missed by most standard population-based genotyping tests and require full-length sequencing of reverse transcriptase.
This study evaluated the activity of DPV against subtype C HIV-1 from individuals on failing first-line NNRTI-containing regimens in South Africa and determined how often high-level cross-resistance to DPV occurs that could result in breakthrough infection in the context of vaginal fluid DPV concentrations during ring use and after ring removal. Mutations in the entire RT coding region and combinations of mutations that are most frequently associated with DPV cross-resistance were analyzed. Overall, this study provides critical information about the potential activity of DPV as a PrEP agent in regions where NNRTI-resistant subtype C HIV-1 may be circulating.
RESULTS
Sample characteristics.
One hundred of the 126 samples collected from individuals on failing NNRTI-based first-line ART were included in the study (Fig. 1). Twenty-six samples were excluded: 25 due to testing failure (6 failed extractions, 14 failed PCR amplifications, 3 were amplified but not cloned into the xxLAI vector, and 2 had no NNRTI resistance mutations) and 1 due to unavailable treatment history. Eighty-eight samples were collected from individuals on EFV-based ART, and 11 samples were collected from individuals on NVP-based ART. One individual on EFV-based ART had also taken single dose NVP to prevent mother-to-child transmission of HIV-1. No samples were from individuals exposed to the DAPY class inhibitors etravirine or rilpivirine.
FIG 1.
Study population characteristics.
Frequency of HIV-1 reverse transcriptase mutations in plasma virus from individuals on failing first-line antiretroviral therapy.
Plasma samples contained a median of 3 (range, 1 to 7) NNRTI-associated drug resistance mutations (DRMs) (Stanford HIVdb v7.0), which included A98G, L100I, K101E/H, K103N/S, V106M, V108I, E138A/K, V179D/E Y181C, Y188L/C, G190A, H221Y, P225H, F227L, and M230L. The most frequent EFV and NVP DRM were K103N in 55 of 100 samples (55%), V106M (44%), and G190A (26%). All 100 samples also carried HIV-1 NRTI mutations, including M184V (82%), K65R (35%), L74I (19%), M41L (17%), and D67N (17%). Resistance mutations (Stanford HIVdb v7.0) in the connection and RNase H domains of RT spanning amino acids 320 to 560 (14) included Y318F (3%) and N348I (14%). Additional subtype C resistance-associated C-terminal mutations/polymorphisms observed were G335D (85%), A371V (17%) and A376S (10%) (see Table S1 in the supplemental material).
Cross-resistance to dapivirine of plasma-derived virus from individuals on failing first-line antiretroviral therapy.
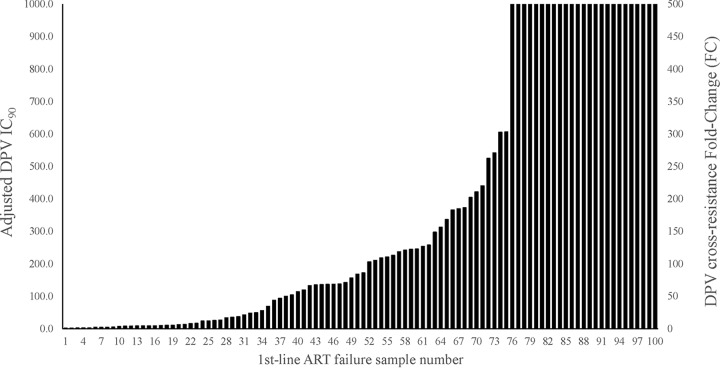
Recombinant viruses derived from plasma samples from 12 treatment-naive individuals yielded a median DPV 50% inhibitory concentration (IC50) of 0.26 ng/ml (range, 0.10 to 0.45 ng/ml) that was used as the wild-type, DPV-susceptible control value. A total of 25 of 100 (25%) recombinant viruses containing full-length RT from first-line ART failures had DPV IC50s exceeding the upper limit of the assay (>IC50 of 132 ng/ml or >505 FCs compared to the wild type). A total of 66 of 100 (66%) had 3.5 to 306 FCs with a median IC50 of 17.6 ng/ml. Only 9 of 100 (9%) samples had DPV IC50s that were within three standard deviations of the maximum IC50 for the wild-type, control viruses (Table 1). The IC50s were converted to protein-adjusted IC90 values to estimate susceptibility in the presence of human serum for comparison to in vivo drug levels of DPV (Table 1 and Fig. 2).
TABLE 1.
Cross-resistance to dapivirine of plasma-derived viruses from individuals on failing first-line NNRTI-based ART
| Resistance category | No. (%) of samples (n = 100) | Median (range) |
||
|---|---|---|---|---|
| Standard IC50 (ng/ml) | Adjusted IC90 (ng/ml)a | Fold changeb | ||
| >500-fold | 25 (25) | ≥132 | ≥1,000 | ≥505 |
| 3- to 500-fold | 66 (66) | 17.6 (0.915–79.8) | 134 (6.95–606) | 67.4 (3.51–306) |
| <3-fold | 9 (9) | 0.383 (0.228–0.636) | 2.91 (1.74–4.83) | 1.47 (0.876–2.44) |
The DPV IC90 was calculated from the IC50 and adjusted for human serum protein binding using a factor of 3.8× as described in Materials and Methods.
Calculated using the IC50 values from 13 wild-type, plasma-derived subtype C HIV-1 viruses from South Africa as the denominator.
FIG 2.
Dapivirine cross-resistance of 100 first-line antiretroviral treatment failures. The adjusted IC90 was calculated by multiplying the IC50 by 2 to approximate IC90 values and applying a DPV human serum binding factor (3.8×) as determined in dapivirine susceptibility experiments comparing HS and FBS (data not shown). Dapivirine FC values were determined by dividing the IC50 generated for each plasma-derived virus by a composite IC50 from 12 treatment-naive plasma-derived viruses collected form the same geographical region. The IC50 values of samples 76 through 100 exceeded the highest concentration of DPV that could be tested in TZMbl cells without cytotoxicity and are reported as >500 FC.
NNRTI resistance mutations responsible for conferring DPV cross-resistance.
DPV susceptibility cutoffs have not been established. The frequency of each Stanford HIVdb-defined RT resistance mutation was compared using the FC categories of >500 (n = 25) and 0 to 500, including the treatment-naive group (n = 87). The choice of FC cutoff for resistance was evaluated in a sensitivity analysis for the association between specific mutations and DPV resistance. The use of a variety of FC cutoffs, ranging from 500 to 10, did not alter the associations with the specific mutations described above (see Tables S2A to S2E in the supplemental material).
L100I and K103N were both overrepresented in the >500-FC group (odds ratio [OR] > 1; false discovery rate (FDR)-adjusted P value [q] < 0.12; Table 2), whereas V106M was underrepresented in the >500-FC group (OR = 0.22; q = 0.1; Table 2). An assessment of all pairwise combinations of NNRTI DRMs showed that 12 of 13 samples with L100I also had K103N and that samples having both L100I and K103N were significantly more likely to have >500-FC DPV resistance compared to those samples having only one of the two mutations (OR = 16.4; q = 0.02; P < 0.001). This analysis also suggested that having mutations in both codons 103 and 179 may be associated with higher DPV resistance (P = 0.009); however, when corrected for multiple comparisons, this mutation pair was not robust (q = 0.5) (Table 3). The number of NNRTI mutations per genotype showed only a trend with level of DPV resistance (P = 0.064).
TABLE 2.
Association of NNRTI mutations with dapivirine cross-resistancec
| Codon | Mutant amino acid | Fold change cutoff (%) |
Odds ratio | Pa | qb | |
|---|---|---|---|---|---|---|
| 0–500 (n = 87) | >500 (n = 25) | |||||
| 98 | G | 10 (11) | 4 (16) | 1.5 | 0.510 | 0.832 |
| 100 | I | 3 (3) | 10 (40) | 18.7 | 1.07E–05 | 4.82E–04 |
| 101 | E | 8 (9) | 2 (8) | 0.9 | 1.000 | 1.000 |
| 101 | H | 3 (3) | 0 (0) | 0.0 | 1.000 | 1.000 |
| 103 | N | 34 (39) | 21 (84) | 8.2 | 7.84E–05 | 0.002 |
| 103 | S | 5 (6) | 1 (4) | 0.7 | 1.000 | 1.000 |
| 106 | M | 40 (46) | 4 (16) | 0.2 | 0.0097 | 0.114 |
| 108 | I | 7 (8) | 6 (24) | 3.6 | 0.069 | 0.345 |
| 138 | K | 2 (2) | 1 (4) | 1.8 | 0.535 | 0.832 |
| 138 | A | 7 (8) | 2 (8) | 1.0 | 1.000 | 1.000 |
| 179 | E | 1 (1) | 2 (8) | 7.5 | 0.125 | 0.510 |
| 179 | D | 10 (11) | 3 (12) | 1.1 | 1.000 | 1.000 |
| 181 | C | 11 (13) | 5 (20) | 1.7 | 0.346 | 0.832 |
| 188 | C | 4 (5) | 0 (0) | 0.0 | 0.573 | 0.832 |
| 188 | L | 4 (5) | 2 (8) | 1.8 | 0.614 | 0.863 |
| 190 | A | 22 (25) | 4 (16) | 0.6 | 0.426 | 0.832 |
| 221 | Y | 6 (7) | 2 (8) | 1.2 | 1.000 | 1.000 |
| 225 | H | 9 (10) | 4 (16) | 1.7 | 0.482 | 0.832 |
| 227 | L | 12 (14) | 0 (0) | 0.0 | 0.065 | 0.345 |
| 230 | L | 6 (7) | 5 (20) | 3.4 | 0.066 | 0.345 |
| 318 | F | 2 (2) | 1 (4) | 1.8 | 0.535 | 0.832 |
| 348 | I | 10 (11) | 3 (12) | 1.1 | 1.000 | 1.000 |
P values were determined by using the Fisher exact test.
q values were computed by using the method of Benjamini and Hochberg.
Significance is indicated with boldfacing.
TABLE 3.
Combinatorial effect of NNRTI mutations on DPV cross-resistance
| Codon combinationa |
Fold change cutoff (%)b |
Odds ratio | Pc | qd | ||||
|---|---|---|---|---|---|---|---|---|
| 0–500 (n = 87) |
>500 (n = 25) |
|||||||
| Codon 1 | Codon 2 | Both codons | One codon | Both codons | One codon | |||
| 100 | 103 | 2 (2) | 36 (41) | 10 (40) | 11 (44) | 16.4 | 0.0002 | 0.025 |
| 103 | 179 | 0 (0) | 48 (55) | 4 (16) | 19 (76) | Inf | 0.009 | 0.478 |
| 181 | 190 | 6 (7) | 22 (25) | 4 (16) | 2 (8) | 7.3 | 0.048 | 1 |
| 98 | 225 | 0 (0) | 19 (22) | 2 (8) | 4 (16) | Inf | 0.050 | 1 |
| 108 | 348 | 0 (0) | 17 (19) | 2 (8) | 5 (20) | Inf | 0.076 | 1 |
| 103 | 188 | 0 (0) | 46 (53) | 2 (8) | 19 (76) | Inf | 0.095 | 1 |
A total of 105 mutation combinations were tested. Only those with P values of <0.1 are shown.
That is, the number of samples in each group in which resistance mutations were observed in two (both codons) or only one (one codon) tested positions. Inf, infinite.
P values were determined by using the Fisher exact test.
q values were computed by using the method of Benjamini and Hochberg.
No novel connection domain mutations were significantly associated with >500-FC DPV resistance. Only the RNase H mutation S468P was overrepresented among the >500-FC group; however, correction for multiple comparisons resulted in loss of statistical significance (q = 0.45). Three of five samples with S468P also had L100I and/or K103N, whereas one of five samples had S468P with G190E.
Risk of breakthrough HIV-1 infection based on plasma and vaginal fluid DPV concentrations during ring use and after ring removal.
Adjusted DPV IC90 of the first-line ART failure viruses were compared to published mean concentrations of DPV found in plasma and vaginal fluid in the cervix, introitus, and area near the ring from two studies (15, 16). The lowest IC90 (1.74 ng/ml) of the 100 viruses tested was 6-fold higher than the mean plasma Cmax (0.29 ng/ml), indicating that plasma DPV concentrations were insufficient to block viral replication. In contrast, reported vaginal fluid concentration on day 28 of DPV ring use (prior to ring removal) of 14,900 to 65,000 ng/ml (15, 16) exceeded the adjusted IC90 of 75% of samples by a minimum of 15-fold. The exact IC90 could not be determined for 25 samples that displayed >500-FC DPV resistance (Fig. 2). About 30 to 49% of the 100 viruses tested had adjusted IC90s higher than the estimated DPV concentrations found in vaginal fluid 3 days after ring removal (186 to 834 ng/ml) (15).
DISCUSSION
The protective efficacy provided by a DPV vaginal ring against transmitted NNRTI-resistant HIV-1 is unknown. In vitro, we observed frequent cross-resistance to DPV among subtype C HIV-1 from individuals experiencing failure of first-line NNRTI-containing ART, with 91% of samples harboring NNRTI-resistant variants and having a ≥3-fold decrease in susceptibility to DPV compared to wild-type HIV-1. In addition, 25% of samples had IC50s exceeding the maximum concentration of dapivirine (132 ng/ml) that could be tested in our susceptibility assay without cytotoxicity. The reported vaginal DPV concentrations with continual monthly ring use (15) exceed the adjusted IC90 of 75% of the DPV cross-resistant viruses by ≥15-fold, which suggests that local concentrations of DPV may be sufficiently high to block infection of NNRTI-resistant HIV-1. The remaining 25% of viruses exceed the capacity of our assays to measure accurate IC90 values, and thus the protective efficacy of the DPV levels provided by the DPV ring against these viruses is uncertain.
The highest risk of NNRTI resistant virus breakthrough could occur during a small window period after ring removal (without replacement with a new ring) when the amount of local DPV falls to low but still detectable levels. Clearance of DPV occurs relatively rapidly, with cervicovaginal fluid concentrations falling to undetectable in 11 of 12 participants within 1 week of ring removal and cervical tissue levels falling below the lower limit of quantification within 3 days of ring removal (16). The concentration of DPV needed in the vaginal fluid and tissue for full protection against wild-type and NNRTI-resistant HIV-1 infection is not known.
The recombinant viruses used in this study were from individuals experiencing viral failure on first-line ART with high viral loads (>10,000 copies/ml) and highly resistant virus containing a median of three NNRTI mutations found by standard population-based genotype. The contribution of minority variants to DPV cross-resistance were not assessed. Only two mutations, L100I and K103N in combination, were significantly associated with high-level (>500-fold) DPV cross-resistance. This mutation combination has also been implicated in high-level rilpivirine resistance (17). Both K103N and L100I result in steric hindrance of the NNRTI binding pocket (18). The in vitro-selected DPV resistance mutation E138K (19) was uncommon in this sample set, being observed in only 3 of 100 samples, none of which had >500-fold resistance to DPV.
A novel and unique system of cloning and phenotyping plasma-derived recombinant virus with full-length RT that included the polymerase, RNase H, and connection domains was used. The C-terminal domain mutation N348I has been associated with decreased susceptibility to NVP, EFV, and etravirine (ETR) in non-subtype B HIV-1 (9, 20), and Y318F has been linked to delavirdine resistance (21). Of the 19 samples with the major C-terminal domain mutations Y318F or N348I (Stanford HIVdb v7.0), none were associated with increased cross-resistance to DPV, indicating that C-terminal mutations selected by first-line ART may not be a major factor for DPV breakthrough.
A limitation of this study is that the IC50 of 25% of the donor-derived clones exceeded the highest DPV concentration that could be tested without cytotoxicity in TZMbl cells (132 ng/ml). It is unknown whether in vivo cell or tissue concentrations of DPV would be sufficient to inhibit these resistant viruses. Assessment of DPV levels in the genital tract has been imperfect, due to limited sampling from vaginal or cervical biopsy specimens (15, 16). Evaluating the susceptibility to DPV in tissue explant models that can achieve higher DPV levels will add critical data on the risk of breakthrough infection with NNRTI-resistant virus.
Overall, these results indicate that cross-resistance to DPV is common for NNRTI-resistant viruses from individuals experiencing viral failure to a first-line regimen. It is not known whether the high levels of DPV in the genital tract achieved with the intravaginal DPV ring will be sufficient to prevent infection by NNRTI-resistant variants. Consistent ring adherence and avoiding ring removal will be critical for protection against HIV-1 and minimizing risk of breakthrough infection with resistant virus. Careful monitoring of DVP ring users is needed to determine the frequency of breakthrough infections with DPV-resistant virus.
MATERIALS AND METHODS
Clinical samples.
Plasma from 129 HIV-1-infected individuals whose samples were sent for routine HIV-1 drug resistance testing to Lancet Laboratories (Johannesburg, South Africa) was selected for this study based on the following criteria: (i) infection with subtype C HIV-1, (ii) experiencing virologic failure defined as having >10,000 HIV-1 RNA copies/ml after 6 months of NNRTI-containing ART, and (iii) viral population-based genotype showing at least one major NNRTI resistance mutation in HIV-1 reverse transcriptase as defined by Stanford HIVdb v7.0. Samples from 12 subtype C HIV-1-infected treatment-naive individuals also collected by Lancet Laboratories from the same geographical location were used as controls. All donor samples were anonymized and testing was approved by the South African Medical Association Research Ethics Committee and the Institutional Review Board of the University of Pittsburgh.
Generation of recombinant HIV-1xxLAI containing plasma-derived full-length RT sequences.
Viral RNA was extracted from plasma samples using an open-mode automated extraction program on the m2000sp. (Abbott Molecular). The full-length HIV-1 RT gene (amino acids 1 to 560) was amplified using a SuperScript III One-Step RT-PCR system with Platinum Taq DNA polymerase (Invitrogen) and the gag- and integrase-specific primers Bcl1_CT(+) (5′-TAAG ACA GTA TGA TCA AAT ACT TAT AGA AAT TTG TGG-3′) and 4232(−) (5′-CC TGA CTT TGG GGA TTG TAG GGA AT −3′). The RNA secondary structure was relaxed by incubating the RNA in the presence of primers for 5 min (min) at 65°C and then chilled rapidly on an ice block. Reverse transcription-PCR was performed for 60 min at 50°C and 2 min at 94°C, followed by 40 cycles of 94°C for 15 s, 50°C for 30 s, and 68°C for 127 s, with a final extension for 5 min at 68°C. Amplified DNA products were treated with ExoSAP-IT (Affymetrix) to remove unused primers and further amplified (PCR2) using the Platinum Taq DNA polymerase high-fidelity (Invitrogen) and hemi-nested primers Bcl1_CT(+) and Xho1_CT(−) (5′-TAA CTT TTC CCT CGA GAT GTG TAC AAT CTA ATT GCC-3′) to generate 15-bp homologous ends for cloning purposes under the following touchdown PCR (22) cycling conditions: an initial incubation at 94°C for 2 min, followed by 26 cycles of 94°C for 15 s, 63°C (descending 0.5°C each subsequent cycle) for 30 s, and 68°C for 2 min. This was followed by 14 cycles of 94°C for 15 s, 50°C for 30 s, and 68°C for 2 min and then a final extension for 5 min at 68°C. PCR2 product was purified after separation on a 1% agarose gel using NucleoSpin and PCR Clean-Up kit (Clontech).
The xxLAI viral vector (23) was modified to insert an XhoI site at nucleotide 4765 using the QuikChange XL kit (Agilent Technologies). Plasma-derived full-length RT was bulk cloned into xxLAIxhoI using the In-Fusion HD cloning system (Clontech) and plasmid DNA was purified using the PureYield plasmid midiprep system (Promega). Lipofectamine 2000 (Life Technologies) transfection of 293T cells with plasmid DNA was performed to generate infectious virus.
HIV-1 phenotyping.
A normalized input of 100 relative light units was used to infect untreated or DPV-treated TZM-bl cells in a luciferase-based single-round infection drug susceptibility assay (Britelite Plus; Perkin-Elmer) as previously described (24). DPV was kindly provided by International Partnership for Microbicides (Silver Spring, MD). Four parameter, nonlinear regression for curve fitting was used to generate IC50s using GraphPad Prism 6 software (GraphPad Software, Inc.). Fold change (FC) values were determined by dividing the IC50 generated for each donor-derived virus by a composite IC50 from the 12 treatment naive plasma-derived subtype C viruses collected from the same geographical region. The IC90 was estimated as double the IC50.
HIV-1 population genotyping.
An in-house population genotyping assay was used to sequence HIV-1 from donor plasma at BARC-Lancet Laboratories, South Africa, as previously described (25). Full-length HIV-1 RT from plasma-derived xxLAI viral stocks were also sequenced using six primers, yielding bidirectional sequencing coverage of the entire length of RT. Mixed bases were called at a threshold of 20% as estimated by peak height ratios using Sequencher (GeneCodes) software. Phylogenetic analysis was done to ensure that plasma-derived cloned virus sequence clustered with virus in the original plasma sample.
Protein binding adjustments.
To better estimate protein binding of DPV in the presence of human serum, drug susceptibility assays were set up as described above in the presence of various amounts (5, 10, and 20%) of human serum (HS) and fetal bovine serum (FBS). Two viral stocks, one showing no resistance and one showing low level resistance (5-fold) in the TZM-bl drug susceptibility assays, were used for these experiments. IC50s were generated for all conditions, and the mean FC differences between FBS and HS (3.8-fold change in IC50s) were used to adjust for human protein binding (data not shown).
Statistical analysis.
A Fisher exact test (FET) was used to assess differences in the prevalence of individual NRTI and NNRTI resistance mutations (as defined by Stanford HIVdb v7.0) between samples with >500-FC DPV resistance and samples with lower resistance, including those from treatment-naive individuals (FC ≤ 500). The additive effect of individual mutations was investigated in all pairwise combinations of NNRTI resistance-associated positions. The frequency of samples containing resistance mutations at two codons and at a single codon was compared across groups using the FET. In an exploratory analysis, the FET was used to test the difference in prevalence of mutations in RT codons 440 and higher between the two groups, corresponding to the RNase H domain. A mutation was included in the analysis if it was present at ≥25% frequency by population sequencing within a sample and if there were at least three occurrences of the mutation in the entire study data set. Correction for multiple comparisons was performed by controlling for the false discovery rate (FDR) using the method of Benjamini and Hochberg (26). Briefly, the FDR estimates the expected proportion of false discoveries among the rejected null hypotheses. An FDR-adjusted P value (or “q value”) of <0.15 was considered statistically significant. Statistical analyses were performed in R (v3.1.2) with the glmnet library.
Supplementary Material
ACKNOWLEDGMENTS
We thank Maritsa Scoulos-Hanson for help in developing the full-length phenotyping assay. We also gratefully acknowledge Jeremy Nuttall and Analene Nel from International Partnership for Microbicides for helpful discussions on dapivirine pharmacokinetics.
This manuscript has received the approval of the LANCET Publications Committee based on a review of its scientific and data interpretation. This study was supported by a grant from the Bill and Melinda Gates Foundation (OPP1019228).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01805-16.
REFERENCES
- 1.van der Straten A, Montgomery ET, Cheng H, Wegner L, Masenga G, von Mollendorf C, Bekker L, Ganesh S, Young K, Romano J, Nel A, Woodsong C. 2012. High acceptability of a vaginal ring intended as a microbicide delivery method for HIV prevention in African women. AIDS Behav 16:1775–1786. doi: 10.1007/s10461-012-0215-0. [DOI] [PubMed] [Google Scholar]
- 2.Devlin B, Nuttall J, Wilder S, Woodsong C, Rosenberg Z. 2013. Development of dapivirine vaginal ring for HIV prevention. Antiviral Res 100(Suppl):S3–S8. doi: 10.1016/j.antiviral.2013.09.025. [DOI] [PubMed] [Google Scholar]
- 3.Palanee-Phillips T, Schwartz K, Brown ER, Govender V, Mgodi N, Kiweewa FM, Nair G, Mhlanga F, Siva S, Bekker LG, Jeenarain N, Gaffoor Z, Martinson F, Makanani B, Naidoo S, Pather A, Phillip J, Husnik MJ, van der Straten A, Soto-Torres L, Baeten J. 2015. Characteristics of women enrolled into a randomized clinical trial of dapivirine vaginal ring for HIV-1 prevention. PLoS One 10:e0128857. doi: 10.1371/journal.pone.0128857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baeten JM, Palanee-Phillips T, Brown ER, Schwartz K, Soto-Torres LE, Govender V, Mgodi NM, Matovu Kiweewa F, Nair G, Mhlanga F, Siva S, Bekker LG, Jeenarain N, Gaffoor Z, Martinson F, Makanani B, Pather A, Naidoo L, et al. . 22 February 2016. Use of a vaginal ring containing dapivirine for HIV-1 prevention in women. N Engl J Med doi: 10.1056/NEJMoa1506110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nel A, Kapiga S, Bekker LG, Devlin B, Borremans M, Rosenberg Z. 2016. Safety and efficacy of dapivirine vaginal ring for HIV-1 prevention in African women, abstr 110LB. Conference on Retroviruses and Opportunistic Infections, Boston, MA http://www.croiconference.org/sessions/safety-and-efficacy-dapivirine-vaginal-ring-hiv-1-prevention-african-women. [Google Scholar]
- 6.Sluis-Cremer N. 2014. The emerging profile of cross-resistance among the nonnucleoside HIV-1 reverse transcriptase inhibitors. Viruses 6:2960–2973. doi: 10.3390/v6082960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schader SM, Oliveira M, Ibanescu RI, Moisi D, Colby-Germinario SP, Wainberg MA. 2012. In vitro resistance profile of the candidate HIV-1 microbicide drug dapivirine. Antimicrob Agents Chemother 56:751–756. doi: 10.1128/AAC.05821-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fletcher P, Harman S, Azijn H, Armanasco N, Manlow P, Perumal D, de Bethune M-P, Nuttall J, Romano J, Shattock R. 2009. Inhibition of human immunodeficiency virus type 1 infection by the candidate microbicide dapivirine, a nonnucleoside reverse transcriptase inhibitor. Antimicrob Agents Chemother 53:487–495. doi: 10.1128/AAC.01156-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brehm JH, Koontz DL, Wallis CL, Shutt KA, Sanne I, Wood R, McIntyre JA, Stevens WS, Sluis-Cremer N, Mellors JW, Team C-SPS. 2012. Frequent emergence of N348I in HIV-1 subtype C reverse transcriptase with failure of initial therapy reduces susceptibility to reverse-transcriptase inhibitors. Clin Infect Dis 55:737–745. doi: 10.1093/cid/cis501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paredes R, Puertas MC, Bannister W, Kisic M, Cozzi-Lepri A, Pou C, Bellido R, Betancor G, Bogner J, Gargalianos P, Banhegyi D, Clotet B, Lundgren J, Menendez-Arias L, Martinez-Picado J, Euro SSG. 2011. A376S in the connection subdomain of HIV-1 reverse transcriptase confers increased risk of virological failure to nevirapine therapy. J Infect Dis 204:741–752. doi: 10.1093/infdis/jir385. [DOI] [PubMed] [Google Scholar]
- 11.Muniz CP, Soares MA, Santos AF. 2014. Early selection of resistance-associated mutations in HIV-1 RT C-terminal domains across different subtypes: role of the genetic barrier to resistance. J Antimicrob Chemother 69:2741–2745. doi: 10.1093/jac/dku214. [DOI] [PubMed] [Google Scholar]
- 12.Xu HT, Colby-Germinario SP, Oliveira M, Han Y, Quan Y, Zanichelli V, Wainberg MA. 2014. The connection domain mutation N348I in HIV-1 reverse transcriptase enhances resistance to etravirine and rilpivirine but restricts the emergence of the E138K resistance mutation by diminishing viral replication capacity. J Virol 88:1536–1547. doi: 10.1128/JVI.02904-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tachedjian G, Sluis-Cremer N. 2013. Role of RNase H activity in NRTI/NNRTI drug resistance, p 281–304. In LeGrice S, Götte M (ed), Human immunodeficiency virus reverse transcriptase: a bench-to-bedside success. Springer, New York, NY. [Google Scholar]
- 14.Tang MW, Liu TF, Shafer RW. 2012. The HIVdb system for HIV-1 genotypic resistance interpretation. Intervirology 55:98–101. doi: 10.1159/000331998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nel A, Haazen W, Nuttall J, Romano J, Rosenberg Z, van Niekerk N. 2014. A safety and pharmacokinetic trial assessing delivery of dapivirine from a vaginal ring in healthy women. AIDS 28:1479–1487. doi: 10.1097/QAD.0000000000000280. [DOI] [PubMed] [Google Scholar]
- 16.Chen BA, Panther L, Marzinke MA, Hendrix CW, Hoesley CJ, van der Straten A, Husnik MJ, Soto-Torres L, Nel A, Johnson S, Richardson-Harman N, Rabe LK, Dezzutti CS. 2015. Phase 1 safety, pharmacokinetics, and pharmacodynamics of dapivirine and maraviroc vaginal rings: a double-blind randomized trial. J Acquir Immune Defic Syndr 70:242–249. doi: 10.1097/QAI.0000000000000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Azijn H, Tirry I, Vingerhoets J, de Bethune MP, Kraus G, Boven K, Jochmans D, Van Craenenbroeck E, Picchio G, Rimsky LT. 2010. TMC278, a next-generation nonnucleoside reverse transcriptase inhibitor (NNRTI), active against wild-type and NNRTI-resistant HIV-1. Antimicrob Agents Chemother 54:718–727. doi: 10.1128/AAC.00986-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iyidogan P, Anderson KS. 2014. Current perspectives on HIV-1 antiretroviral drug resistance. Viruses 6:4095–4139. doi: 10.3390/v6104095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu HT, Colby-Germinario SP, Huang W, Oliveira M, Han Y, Quan Y, Petropoulos CJ, Wainberg MA. 2013. Role of the K101E substitution in HIV-1 reverse transcriptase in resistance to rilpivirine and other nonnucleoside reverse transcriptase inhibitors. Antimicrob Agents Chemother 57:5649–5657. doi: 10.1128/AAC.01536-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Basson AE, Rhee SY, Parry CM, El-Khatib Z, Charalambous S, De Oliveira T, Pillay D, Hoffmann C, Katzenstein D, Shafer RW, Morris L. 2015. Impact of drug resistance-associated amino acid changes in HIV-1 subtype C on susceptibility to newer nonnucleoside reverse transcriptase inhibitors. Antimicrob Agents Chemother 59:960–971. doi: 10.1128/AAC.04215-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harrigan PR, Salim M, Stammers DK, Wynhoven B, Brumme ZL, McKenna P, Larder B, Kemp SD. 2002. A mutation in the 3′ region of the human immunodeficiency virus type 1 reverse transcriptase (Y318F) associated with nonnucleoside reverse transcriptase inhibitor resistance. J Virol 76:6836–6840. doi: 10.1128/JVI.76.13.6836-6840.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Korbie DJ, Mattick JS. 2008. Touchdown PCR for increased specificity and sensitivity in PCR amplification. Nat Protoc 3:1452–1456. doi: 10.1038/nprot.2008.133. [DOI] [PubMed] [Google Scholar]
- 23.Shi C, Mellors JW. 1997. A recombinant retroviral system for rapid in vivo analysis of human immunodeficiency virus type 1 susceptibility to reverse transcriptase inhibitors. Antimicrob Agents Chemother 41:2781–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Melody K, McBeth S, Kline C, Kashuba AD, Mellors JW, Ambrose Z. 2015. Low frequency of drug-resistant variants selected by long-acting rilpivirine in macaques infected with simian immunodeficiency virus containing HIV-1 reverse transcriptase. Antimicrob Agents Chemother 59:7762–7770. doi: 10.1128/AAC.01937-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wallis CL, Papathanasopoulos MA, Lakhi S, Karita E, Kamali A, Kaleebu P, Sanders E, Anzala O, Bekker LG, Stevens G, de Wit TF, Stevens W. 2010. Affordable in-house antiretroviral drug resistance assay with good performance in non-subtype B HIV-1. J Virol Methods 163:505–508. doi: 10.1016/j.jviromet.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benjamini YaH Yosef. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.