ABSTRACT
Toxoplasma gondii is an apicomplexan parasite that causes fatal and debilitating brain and eye disease. Endochinlike quinolones (ELQs) are preclinical compounds that are efficacious against apicomplexan-caused diseases, including toxoplasmosis, malaria, and babesiosis. Of the ELQs, ELQ-316 has demonstrated the greatest efficacy against acute and chronic experimental toxoplasmosis. Although genetic analyses in other organisms have highlighted the importance of the cytochrome bc1 complex Qi site for ELQ sensitivity, the mechanism of action of ELQs against T. gondii and the specific mechanism of ELQ-316 remain unknown. Here, we describe the selection and genetic characterization of T. gondii clones resistant to ELQ-316. A T. gondii strain selected under ELQ-316 drug pressure was found to possess a Thr222-Pro amino acid substitution that confers 49-fold resistance to ELQ-316 and 19-fold resistance to antimycin, a well-characterized Qi site inhibitor. These findings provide further evidence for ELQ Qi site inhibition in T. gondii and greater insight into the interactions of Qi site inhibitors with the apicomplexan cytochrome bc1 complex.
KEYWORDS: Toxoplasma gondii, apicomplexan parasites, cytochrome b, cytochrome bc1, drug targets, experimental therapeutics, mechanisms of action, mitochondria, parasitology, preclinical drug studies
INTRODUCTION
Toxoplasma gondii is a prolific apicomplexan parasite that infects birds and mammals, including one-third of humans (1). T. gondii causes fatal or debilitating brain and eye disease in fetuses and the immunocompromised. In healthy individuals, the prevalence and severity of ocular disease varies geographically, with T. gondii eye disease observed in up to 20% of infected people in areas of Brazil (2). Current first-line therapy with pyrimethamine and sulfadiazine has a high rate of toxic side effects and does not eradicate latent infection from the host (3). These shortcomings are significant, considering that prolonged courses of therapy are required for the treatment of toxoplasmosis in AIDS patients, stem cell transplant patients, and infants. More effective and better-tolerated treatment for toxoplasmosis is needed.
The cytochrome bc1 complex has been a successful drug target for combatting diseases caused by apicomplexans, including T. gondii (4, 5). Located in the inner mitochondrial membrane, cytochrome bc1 is part of the electron transport chain, involved in reducing cytochrome c, transferring protons to the intermembrane space, and generating ubiquinone for pyrimidine biosynthesis. The bc1 complex has two active sites, the Qo site that oxidizes ubiquinol and the Qi site that reduces ubiquinone (6). Atovaquone, a cytochrome bc1 Qo site inhibitor, is combined with proguanil for the prophylaxis and treatment of malaria (5). Atovaquone is a first-line treatment in combination with azithromycin for mild-to-moderate babesiosis and is an alternate treatment and prophylactic for toxoplasmosis (7, 8). Inhibitors of the cytochrome bc1 Qi site, such as antimycin, an antifungal compound made by Streptomyces species, have long been known, but Qi site inhibitors have not been used clinically (9). Recently, antimalarial pyridones, which inhibit the Qi site, were advanced for the first time to human studies but were withdrawn due to cardiotoxicity in rats that was related to activity against host cytochrome bc1 (10). The endochinlike quinolone (ELQ) series of 4(1H)-quinolone-3-diarylethers are suspected to target the Qi site and have been optimized to avoid human cytochrome bc1 inhibition (11).
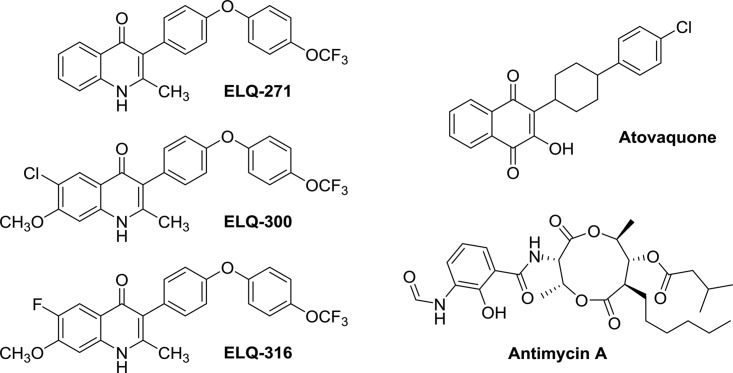
ELQs are efficacious against malaria, babesiosis, and acute and latent experimental toxoplasmosis (11–13). Among the large panel of ELQs that we have synthesized, ELQ-316 was found to have the greatest efficacy against T. gondii and did not inhibit human cytochrome bc1 at concentrations up to 10 μM. Previous genetic mutations in Saccharomyces cerevisiae and Plasmodium falciparum suggest that ELQ-271 and ELQ-300, respectively, inhibit the apicomplexan cytochrome bc1 Qi site (10, 12, 14). However, no direct experimental evidence for the mechanism of action of ELQs in T. gondii or of ELQ-316 has been described. Whereas ELQ-271 inhibited S. cerevisiae growth and cytochrome c reduction, allowing for the investigation of ELQ-271 against a series of S. cerevisiae strains with cytochrome b gene (Cytb) mutations, ELQ-316 did not inhibit S. cerevisiae growth or cytochrome c reduction. This difference in activity is likely related to limitation of the interaction with the Qi pocket of S. cerevisiae due to the bulkier substituents at the sixth and seventh positions of ELQ-316 and ELQ-300 (Fig. 1). An Ile22-Leu mutation in the P. falciparum cytochrome bc1 Qi site was associated with ELQ-300 resistance but not with resistance to ELQ-271 or ELQ-316 (14). In addition, the crystal structures for apicomplexan cytochrome bc1 have not been solved, hindering molecular modeling of compounds directly with the parasite cytochrome bc1. Moreover, targeted mutagenesis of apicomplexan mitochondrial genes has not yet been achieved, preventing definitive evidence of ELQ's mechanism of action through the introduction of single-base-pair mutations. To better understand the mechanism of action and the interaction of ELQ-316 with the cytochrome bc1 Qi site in T. gondii, we selected ELQ-316-resistant T. gondii organisms and characterized the resistant clones. We show that a T. gondii strain possessing a mutation in the Cytb gene that causes a Thr222-Pro amino acid substitution is resistant to ELQ-271 and ELQ-316, as well as the Qi site inhibitor antimycin.
FIG 1.
Chemical structures of cytochrome bc1 inhibitors. ELQ, endochinlike quinolone.
RESULTS
Selection of an ELQ-316-resistant mutant.
ELQ-316-resistant clones of T. gondii were isolated following mutagenesis with N-nitroso-N-ethylurea (ENU) and subsequent exposure to ELQ-316. T. gondii strain RH Δuprt was used for the selection of ELQ-resistant (ER) clones after attempts to isolate T. gondii organisms with sustained ELQ resistance proved unsuccessful, similar to reports of the selection of T. gondii clones resistant to the apicomplexan inhibitor 1-hydroxy-2-dodecyl-4(1H) quinolone (HDQ) (15, 16). T. gondii organisms from the flasks containing 150 nM and 200 nM ELQ-316 were found to have an increased 50% effective concentration (EC50) against ELQ-316 compared to that of the parental strain. Clones ER1 and ER2 were isolated by limiting dilution from each flask for Cytb sequencing and susceptibility testing against inhibitors.
Analysis of Cytb sequence.
Cytochrome b mRNA transcripts of the parental RH Δuprt strain, an ME49 strain, and the ELQ-316-resistant clones (ER1and ER2) were sequenced by reverse transcriptase PCR using primers based on the nucleotide sequence and annotation of the GenBank record with accession number AF015627.1. ER1 Cytb was also sequenced after 45 days (9 passages) of growth without ELQ-316. There are three annotated T. gondii Cytb sequences in GenBank. The T. gondii Cytb sequence with accession number AF015627.1 contains an additional 5′ 117-bp segment compared to the other two sequences, which have accession numbers AF023246.1 and JX473253.1. The 117-bp segment includes an alternative start codon that is 27 bp in frame and upstream from the start codon denoted in the sequence with accession number AF023246.1. The sequence with accession number AF023246.1 was generated by 3′ and 5′ rapid amplification of cDNA ends (RACE) (17). The sequence with accession number JX473253.1 was generated by PCR from DNA using primers based on the sequence with accession number AF023246.1 (18).
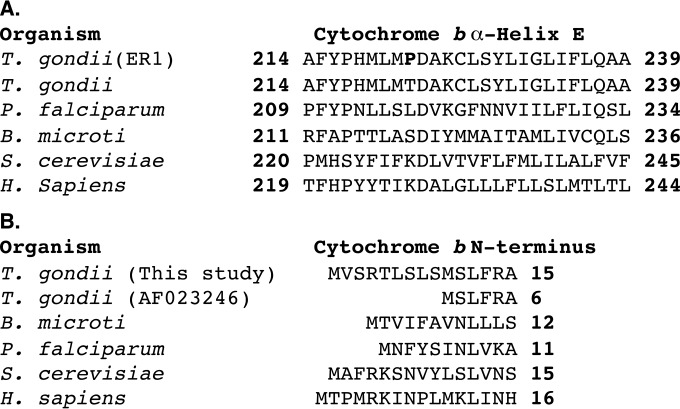
In this study, Cytb amplification from mRNA, using reverse transcriptase PCR, resulted in a single PCR product, indicating that the 27-bp sequence prior to the start codon denoted in the sequence with accession number AF023246.1 is transcribed. The additional 9 N-terminal amino acids are consistent both in length and in the highly conserved arginine within the N-terminal sequence of Cytb proteins of S. cerevisiae, Gallus gallus, and Bos taurus (Fig. 2). Accordingly, we have included the 9 N-terminal amino acids that are excluded from the sequence with accession number AF023246.1 in the putative cytochrome b protein sequence and numbering.
FIG 2.
Cytochrome b protein sequence alignment. (A) Qi site, α-helix E protein sequence alignment between wild-type T. gondii, T. gondii strain ER1, Plasmodium falciparum, Saccharomyces cerevisiae, and human (Homo sapiens) cytochrome b. The Thr222-Pro substitution is in boldface. (B) Putative T. gondii cytochrome b N terminus.
Other than the additional 5′ 27 bp, the Cytb sequences from ER1, ER2, ME49, and the parental RH Δuprt strain were identical to the sequences with accession numbers AF015627.1 and JX473253.1, with the exception of a point mutation of adenine to cytosine at position 664 that results in a Thr222-Pro substitution in ER1 and ER2 (Fig. 2). The Thr222-Pro substitution is located in the Qi site of Cytb adjacent to the highly conserved Asp223 residue and is analogous to the Lys228-Met substitution in S. cerevisiae that causes resistance to the Qi site inhibitors antimycin and HDQ (16). In the context of prior evidence indicating that ELQs act at the Qi site, the association of Thr222-Pro in the Qi site with ELQ and antimycin resistance is highly suggestive that the mechanism of action of ELQ-316 is inhibition of ubiquinone reduction at the Qi site. The presence of Thr222-Pro in the absence of ELQ-316 selection pressure for 45 days of replication indicates that Thr222-Pro is a durable amino acid substitution.
The 222nd amino acid position of cytochrome b is highly conserved across organisms but varies between members of the Apicomplexa (see Fig. S2 in the supplemental material). A broad comparison across organisms, including vertebrates, fungi, plants, insects, and nonapicomplexan Alveolata, reveals that this position is occupied by lysine and adjacent to aspartate at position 223. However, in the suborder Eimeriorina that includes the pathogenic genera Toxoplasma, Sarcocystis, Neospora, Besnoitia, Eimeria, Cyclospora, and Isospora, this position is a threonine. In the order Haemosporida that includes the genus Plasmodium, the analogous amino acid is a leucine. In Piroplasmida, Babesia spp. have a serine or cysteine, while Theileria spp. have a serine or asparagine. The highly divergent cytochrome b of kinetoplastids has an arginine at this position. While the totality of this observation is limited by the breadth and completeness of Cytb sequences available across species, the differing amino acids at this position may contribute to the selectivity of ELQs for apicomplexans over other organisms. In addition, it reveals other apicomplexan pathogens that may be susceptible to ELQs.
Comparison of susceptibilities to ELQ-316, ELQ-271, antimycin, and atovaquone.
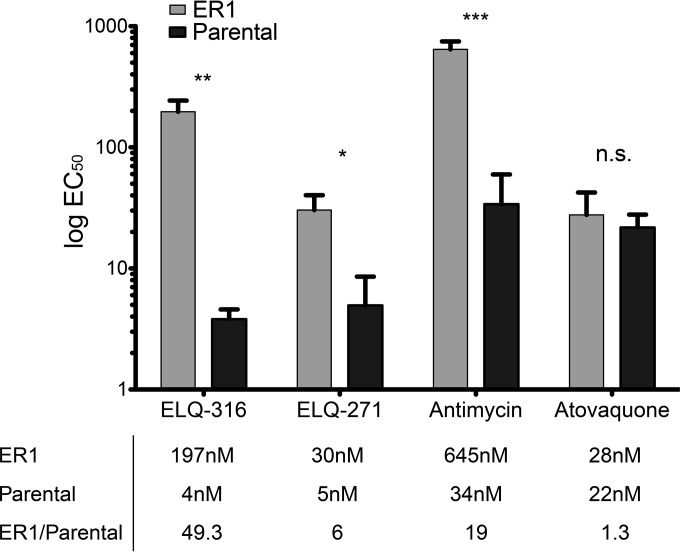
The ELQ-316-selected clone ER1 demonstrated an EC50 of 197 nM against ELQ-316, compared to 4 nM for the parental RH Δuprt strain, a 49-fold difference in susceptibility (Fig. 3). Because ER1 and ER2 were found to have equivalent EC50s against ELQ-316 and both of them had the Thr222-Pro amino acid substitution, ER1 was used for further EC50 testing. ER1 had 6-fold resistance to ELQ-271 (EC50s of 5 nM for the parental strain and 30 nM for ER1) and 19-fold resistance to the Qi site inhibitor antimycin A (EC50s of 39 nM for the parental strain and 645 nM for ER1), but no significant resistance to atovaquone. Cross-resistance between ELQ-271, ELQ-316, and antimycin in the ER1 clone further supports Qi site inhibition as the mechanism of action of ELQs against T. gondii.
FIG 3.
Resistance to cytochrome bc1 inhibitors. Strain ER1, which possesses a Thr222-Pro substitution, is resistant to Qi site inhibitors but not atovaquone compared to the resistances of the parental strain. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; n.s., not significant; error bars show standard deviations.
Comparison of T. gondii cytochrome bc1 susceptibilities to inhibitors.
Isolation of T. gondii mitochondrial membrane fragments provides a means to measure cytochrome c reduction by the T. gondii cytochrome bc1 and chemical inhibition of cytochrome bc1. ELQ-316 inhibited the ER1 cytochrome bc1 with an EC50 of 95 nM, compared to 0.041 nM for wild-type cytochrome bc1 isolated from the parental RH Δuprt strain, resulting in a 2,300-fold difference in inhibition. Antimycin inhibited the ER1 cytochrome bc1 with an EC50 of 98 nM, compared to 2.5 nM for wild-type cytochrome bc1, resulting in a 39-fold difference in inhibition. The steady-state cytochrome c reduction rates were 22 (standard error of the mean [SEM], ±3) μmol cytochrome c mg protein−1 s−1 for the parental cytochrome bc1 and 12 (SEM ±2) μmol cytochrome c mg protein−1 s−1 for ER1, suggesting that the Thr222-Pro mutation decreases the cytochrome bc1 catalytic rate.
DISCUSSION
The association of a Thr222-Pro substitution with resistance to ELQ-316, ELQ-271, and antimycin is highly suggestive that ELQ-316 and ELQ-271 inhibit the cytochrome bc1 Qi site of T. gondii. This mechanism is further supported by previous experiments in S. cerevisiae, Bos taurus, and Babesia microti. A Met221-Gln mutation in the S. cerevisiae cytochrome b Qi site causes resistance to ELQ-271 (12). The Lys228-Met substitution in S. cerevisiae, which is analogous to position 222 in the T. gondii cytochrome b, confers resistance to antimycin, and cocrystallization of antimycin with Bos taurus cytochrome b has shown that the analogous Lys227 forms a key bond with antimycin via a water molecule (16, 19). In addition, a serine-to-tyrosine substitution at the same position in B. microti was associated with the development of ELQ-271 resistance in vivo (13). The degree of resistance associated with Thr222-Pro is consistent with Qi site inhibition being the primary mechanism of action of ELQ-316. The lack of a change in the growth rate of ER1 indicates that this mutation does not alter the fitness of T. gondii under standard in vitro culture conditions. Despite the cumulative evidence for ELQ inhibition of cytochrome b, the inhibition of other enzymes that use ubiquinone as a substrate cannot be fully excluded. Electron transport chain inhibitors with similar structures have been shown to be promiscuous, targeting the Qi site of cytochrome b, dihydroorotate dehydrogenase (DHOD), and NADH:ubiquinone oxidoreductase (14, 15). In addition, the use of chemical mutagenesis raises the possibility of unidentified resistance mutations elsewhere in the genome. However, the resistance of ER1 to antimycin and ELQ-316 observed in the cytochrome c reductase assays makes this possibility less likely. Additionally, the lack of resistance to atovaquone indicates that a generalized mechanism of resistance to cytochrome b inhibitors, such as increased production of cytochrome b, does not account for resistance to ELQs.
The resistance conferred by the Thr222-Pro substitution provides insight into the specificity of ELQs for the apicomplexan Qi site. Threonine is conserved at this position among T. gondii and other medically important coccidian protozoa, such as Neospora, Eimeria, Isospora, Cyclospora, and Sarcocystis. Plasmodium spp. and Babesia spp. possess leucine and serine or cysteine, respectively. In contrast, lysine is broadly conserved at this position across the great majority of organisms. The change from lysine at this position to a smaller residue without a positive charge likely contributes to the selectivity of ELQ-316 for apicomplexans over their host species by allowing bulkier ELQs to occupy the Qi site. This is further supported by the observation that, unlike ELQ-316, ELQ-271 inhibits S. cerevisiae and human cytochrome b, as well as the apicomplexan cytochrome b, and the Thr222-Pro substitution has a much smaller effect on ELQ-271 susceptibility. Prior studies have explored the significance of this amino acid position in the Qi site. Lys228 in S. cerevisiae is proposed to form a water-mediated hydrogen bond with ubiquinone and participate in the Qi site proton uptake pathway (20). If the Lys228 residue does function in this manner, the apicomplexan Qi site has an alternate mechanism of proton uptake, given the tolerance of proline, serine, threonine, asparagine, cysteine, or leucine at this position. Cocrystallization of bovine cytochrome bc1 with 4(1H)-pyridones suggests that the pyridone carbonyl forms a hydrogen bond with the highly conserved adjacent aspartic acid, which has also been proposed to interact with ubiquinone (10, 21, 22). In silico modeling of ELQ-300 docking in this structure suggests similar positioning of ELQ-300 within the Qi pocket (10). Although this in silico model provides insights into possible ELQ positioning, it does not reflect the lack of mammalian cytochrome bc1 inhibition. The change from threonine, a polar residue, to the nonreactive proline suggests either that the reactive hydroxyl group of threonine is involved in ELQ-316 binding in the Qi site or that proline alters the structure of the Qi pocket. The potent activity of ELQ-316 against P. falciparum, which possesses the nonpolar leucine residue at this position, favors the latter possibility.
Qi site inhibitors are a new mechanistic approach for the treatment of apicomplexan-caused diseases. ELQs have excellent efficacy against toxoplasmosis, and the association of Thr222-Pro mutation with resistance to ELQs and antimycin provides evidence that ELQ-316 and ELQ-271 target the T. gondii cytochrome bc1 Qi site. These studies offer further insight into structural interactions of the apicomplexan cytochrome b with Qi site inhibitors and a tool for the evaluation of compounds suspected to act by a similar mechanism.
MATERIALS AND METHODS
Chemicals and T. gondii strains.
ELQs were synthesized as previously described, identified by proton nuclear magnetic resonance (1H-NMR), and determined to be >99% pure by reversed-phase high-performance liquid chromatography (HPLC) (11). Atovaquone and antimycin were obtained from Sigma-Aldrich. The T. gondii RH Δuprt strain, which contains a replacement of the uracil phosphoribosyl transferase (UPRT) gene with the dihydrofolate reductase gene, was kindly provided by David Sibley. The RH and ME49 T. gondii strains were kindly provided by Vern Carruthers.
Selection of ELQ-316-resistant mutants.
N-Nitroso-N-ethylurea (ENU) chemical mutagenesis and ELQ-316 selective pressure were applied to the RH Δuprt strain of T. gondii to select for ELQ-resistant clones as previously described (15, 23). The RH Δuprt strain was used after several attempts to select ELQ-316-resistant clones of RH strain T. gondii resulted in the isolation of clones without sustained resistance. A T25 flask of human foreskin fibroblast (HFF) cells (from American Type Culture Collection) was inoculated with 1 × 107 T. gondii organisms and incubated for 18 h. The medium was replaced with Dulbecco modified Eagle medium (DMEM) containing 0.1% fetal bovine serum (FBS) and 7 mM ENU. After 6 h, T. gondii organisms were collected, washed in phosphate-buffered saline (PBS), and divided into 4 T25 flasks. ELQ-316 at concentrations of 50 nM, 100 nM, 150 nM, and 500 nM was added after 15 h. Parasite replication was observed at 50 nM, 100 nM, and 150 nM. T. gondii organisms in the 100 nM flask were cultured for two passages in 200 nM ELQ-316. Tachyzoites from the 150 nM and 200 nM flasks were tested for ELQ-316 susceptibility. Because T. gondii organisms from both flasks demonstrated ELQ-316 resistance, a T. gondii clone from the 150 nM flask (clone ER1) and one from the 200 nM flask (clone ER2) were isolated by limiting dilution in a 96-well plate containing 150 nM and 200 nM ELQ-316, respectively.
Cytochrome b gene sequence and analysis.
DNA was isolated from the resistant clones ER1 and ER2, the parental strain RH Δuprt, and strain ME49 of T. gondii using the DNeasy blood and tissue purification kit (Qiagen). RNA from these strains was extracted with the Aurum total RNA fatty and fibrous tissue kit (Bio-Rad). RNA was also obtained from the ER1 strain after 9 passages (45 days) without exposure to ELQ-316. cDNA was amplified using the iScript select cDNA synthesis kit (Bio-Rad). The Cytb coding sequences were amplified from genomic DNA and cDNA by PCR with primers 5′ATGGTTTCGAGAACACTCAGT and 3′GTATAAGCATAGAACCAATCCGGT and Phusion DNA polymerase, yielding a single PCR product visualized on an agarose gel. Control PCRs without DNA or with RNA did not yield PCR products. Amplicons were sequenced using sequencing primers 5′CTACCATGGGGACAAATGAGTTTCTGGGGTGCTACAGT and 3′ACCATTCTGGTACGATATGAAGTGGTGTTAC. The nucleotide sequence was submitted to GenBank under accession number KX595194. Protein alignment was performed with MUSCLE (Multiple Sequence Comparison by Log-Expectation) using sequences obtained from UniProtKB.
T. gondii growth inhibition.
The fibroblast lysis assay, like viral plaque assays, measures the death of infected host cells, providing a simple, inexpensive method for comparing drug susceptibility between strains that do not have transgenic markers for growth measurement. Fibroblasts lysed by T. gondii detach from the bottom of the wells, while intact fibroblasts remain attached. The remaining fibroblasts are stained with crystal violet and quantified by image analysis to determine the EC50 for cell lysis.
Compounds were evaluated for comparative growth inhibition of the ELQ-316-exposed clones and the parental RH Δuprt T. gondii strain using a 96-well-plate plaque assay that measures T. gondii host cell lysis. ELQ-316, ELQ-271, and atovaquone were dissolved in dimethyl sulfoxide (DMSO), and antimycin was dissolved in ethanol. Compounds were serially diluted across 96-well plates, leaving the final column with medium only. The final concentrations of DMSO and ethanol in the columns containing the highest inhibitor concentrations were 1.25% (vol/vol). Immediately after the addition of inhibitors, 10,000 T. gondii tachyzoites were added to each well, and plates were incubated for 7 days at 37°C in a humidified 5% CO2 incubator. The supernatant was removed, and wells were washed with PBS. HFF cells were fixed with 5% paraformaldehyde for 5 min at room temperature and then stained with crystal violet solution for 5 min. The crystal violet staining solution consisted of 12.5 g crystal violet dissolved in 125 ml ethanol mixed with 500 ml 1% ammonium oxalate in H2O. After removal of the crystal violet solution, wells were rinsed twice with PBS and air dried. The 96-well plates were inverted, placed on a white light filter transilluminated by UV light, and imaged by using a Bio-Rad Gel Doc imager. The images were analyzed using Image J and the Image J plugin ColonyArea (24). The EC50 based on cell lysis was determined by nonlinear regression analysis of the percentage of cell lysis per well using GraphPad Prism software. The assay was performed in triplicate.
Cytochrome bc1 inhibition.
T. gondii tachyzoites were harvested from lysed HFF cell culture, filtered through a 3-μm polycarbonate filter, and centrifuged at 3,000 × g for 5 min. The T. gondii pellet was resuspended in PBS containing 1 mM phenylmethylsulfonyl fluoride (PMSF) and centrifuged at 3,000 × g for 10 min. The pellet then was resuspended in ice-cold lysis buffer (75 mM sucrose, 225 mM mannitol, 5 mM MgCl2, 5 mM KH2PO4, 1 mM EDTA, 1 mM PMSF, 5 mM HEPES, pH 7.4). This suspension was homogenized in an ice-cold glass Dounce homogenizer. Cell debris was removed by centrifugation at 500 × g for 5 min. The supernatant was then centrifuged at 20,000 × g for 30 min to pellet mitochondrial membrane fragments. The pellet was resuspended in storage buffer (50 mM tricine, 100 mM KCl, 2 mM NaN3, 2.0% N-dodecyl-β-d-maltoside, 30% glycerol, pH 8.0). The total protein concentration was determined by using the bicinchoninic acid assay kit (Thermo Fisher). Aliquots of mitochondrial fragments were frozen in liquid nitrogen and stored at −80°C until needed.
Cytochrome c reduction was monitored as the difference between absorbance at 550 nm and at 542 nm with an Agilent diode array 8453 spectrophotometer. T. gondii mitochondrial membrane fragments (1.5 μg total protein) were added to a cuvette containing 50 μM oxidized cytochrome c (horse heart; Sigma-Aldrich), 50 μM decylubiquinol, 2 mM KCN, 100 mM KCl, and 50 mM tricine at pH 8.0. ELQ-316 was dissolved in DMSO. Decylubiquinone was reduced to decylubiquinol with an excess of sodium borohydride, which was subsequently quenched with hydrochloric acid. Antimycin was dissolved in ethanol. The initial rates of cytochrome c reduction in the presence and absence of inhibitors were measured after the addition of mitochondrial protein and after sufficient time to measure the background reaction between decylubiquinone and cytochrome c. Three replicates were performed at each drug concentration. Plots of log[inhibitor] versus the normalized reduction rate were made in GraphPad Prism software. To determine the EC50 of enzyme inhibition, data were fit to a variable slope model of the Hill equation in which the top and bottom were constrained to 1 and 0, respectively.
Molecular phylogenetic analysis by maximum-likelihood method.
To evaluate amino acid variation at the Thr222 position across cytochrome b sequences, 1,714 reviewed protein sequences from UniProtKB identified by gene name mtcyb, cob, or Cytb were aligned using MUSCLE within the MEGA (Molecular Evolutionary Genetics Analysis) software platform (25). In addition, the NCBI database of nonredundant protein sequences within the taxons Alveolata, Apicomplexa, Kinetoplastida, and Euglenozoa was queried using the T. gondii Cytb protein sequence and Domain Enhanced Lookup Time Accelerated BLAST (26). The NCBI taxonomy browser was used as a taxonomy reference for BLAST queries. The unpublished Cytb sequence from Babesia duncani was obtained from Choukri Ben Mamoun and is included in Fig. S1 in the supplemental material. Partial sequences that did not include the area of interest or that were not annotated as cytochrome b protein sequences were discarded. The resulting sequences from NCBI were aligned using the Constraint-Based Alignment Tool. BLAST was repeated within each query with a representative sequence from each genus. Amino acid variability was identified manually and by using Microsoft Excel software.
The evolutionary history was inferred by using the maximum-likelihood method based on the Whelan and Goldman model (27). The bootstrap consensus tree, inferred from 100 replicates, is taken to represent the evolutionary history of the taxa analyzed (28). Branches corresponding to partitions reproduced in less than 50% of bootstrap replicates are collapsed. The initial tree(s) for the heuristic search were obtained automatically by applying the Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model and then selecting the topology with the superior log-likelihood value. A discrete gamma distribution was used to model evolutionary rate differences among sites (5 categories [+G, parameter = 2.1801]). The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 6.8759% of sites). The analysis involved 39 amino acid sequences. All positions with less than 95% site coverage were eliminated. That is, fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position. There were a total of 308 positions in the final data set. Evolutionary analyses were conducted in MEGA7 (29).
Accession number(s).
Sequence data have been deposited in GenBank under accession number KX595194.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Career Development Award BX002440 to J. Stone Doggett from the U.S. Department of Veterans Affairs Biomedical Laboratory Research and Development. We also acknowledge support for Michael K. Riscoe from NIH R01 AI100569, Peer Reviewed Medical Research Program Project PR130649, and VA Merit Review Funds from the U.S. Department of Veterans Affairs BX003312.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01866-16.
REFERENCES
- 1.Tenter AM, Heckeroth AR, Weiss LM. 2000. Toxoplasma gondii: from animals to humans. Int J Parasitol 30:1217–1258. doi: 10.1016/S0020-7519(00)00124-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grigg ME, Dubey JP, Nussenblatt RB. 2015. Ocular toxoplasmosis: lessons from Brazil. Am J Ophthalmol 159:999–1001. doi: 10.1016/j.ajo.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yan J, Huang B, Liu G, Wu B, Huang S, Zheng H, Shen J, Lun ZR, Wang Y, Lu F. 2013. Meta-analysis of prevention and treatment of toxoplasmic encephalitis in HIV-infected patients. Acta Trop 127:236–244. doi: 10.1016/j.actatropica.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 4.Chirgwin K, Hafner R, Leport C, Remington J, Andersen J, Bosler EM, Roque C, Rajicic N, McAuliffe V, Morlat P, Jayaweera DT, Vilde JL, Luft BJ. 2002. Randomized phase II trial of atovaquone with pyrimethamine or sulfadiazine for treatment of toxoplasmic encephalitis in patients with acquired immunodeficiency syndrome: ACTG 237/ANRS 039 Study. AIDS Clinical Trials Group 237/Agence Nationale de Recherche sur le SIDA, Essai 039. Clin Infect Dis 34:1243–1250. doi: 10.1086/339551. [DOI] [PubMed] [Google Scholar]
- 5.Overbosch D, Schilthuis H, Bienzle U, Behrens RH, Kain KC, Clarke PD, Toovey S, Knobloch J, Nothdurft HD, Shaw D, Roskell NS, Chulay JD, Malarone International Study Team. 2001. Atovaquone-proguanil versus mefloquine for malaria prophylaxis in nonimmune travelers: results from a randomized, double-blind study. Clin Infect Dis 33:1015–1021. doi: 10.1086/322694. [DOI] [PubMed] [Google Scholar]
- 6.Bergdoll L, Ten Brink F, Nitschke W, Picot D, Baymann F. 2016. From low- to high-potential bioenergetic chains: thermodynamic constraints of Q-cycle function. Biochim Biophys Acta 1857:1569–1579. doi: 10.1016/j.bbabio.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 7.Vannier E, Krause PJ. 2012. Human babesiosis. N Engl J Med 366:2397–2407. doi: 10.1056/NEJMra1202018. [DOI] [PubMed] [Google Scholar]
- 8.Panel on Opportunistic Infections in HIV-Infected Adults and Adolescents. 2016. Guidelines for the prevention and treatment of opportunistic infections in HIV-infected adults and adolescents: recommendations from the Centers for Disease Control and Prevention, the National Institutes of Health, and the HIV Medicine Association of the Infectious Diseases Society of America. http://aidsinfo.nih.gov/contentfiles/lvguidelines/adult_oi.pdf Accessed 17 July 2016.
- 9.Potter VR, Reif AE. 1952. Inhibition of an electron transport component by antimycin A. J Biol Chem 194:287–297. [PubMed] [Google Scholar]
- 10.Capper MJ, O'Neill PM, Fisher N, Strange RW, Moss D, Ward SA, Berry NG, Lawrenson AS, Hasnain SS, Biagini GA, Antonyuk SV. 2015. Antimalarial 4(1H)-pyridones bind to the Qi site of cytochrome bc1. Proc Natl Acad Sci U S A 112:755–760. doi: 10.1073/pnas.1416611112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nilsen A, Miley GP, Forquer IP, Mather MW, Katneni K, Li Y, Pou S, Pershing AM, Stickles AM, Ryan E, Kelly JX, Doggett JS, White KL, Hinrichs DJ, Winter RW, Charman SA, Zakharov LN, Bathurst I, Burrows JN, Vaidya AB, Riscoe MK. 2014. Discovery, synthesis, and optimization of antimalarial 4(1H)-quinolone-3-diarylethers. J Med Chem 57:3818–3834. doi: 10.1021/jm500147k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doggett JS, Nilsen A, Forquer I, Wegmann KW, Jones-Brando L, Yolken RH, Bordon C, Charman SA, Katneni K, Schultz T, Burrows JN, Hinrichs DJ, Meunier B, Carruthers VB, Riscoe MK. 2012. Endochin-like quinolones are highly efficacious against acute and latent experimental toxoplasmosis. Proc Natl Acad Sci U S A 109:15936–15941. doi: 10.1073/pnas.1208069109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawres LA, Garg A, Kumar V, Bruzual I, Forquer IP, Renard I, Virji AZ, Boulard P, Rodriguez EX, Allen AJ, Pou S, Wegmann KW, Winter RW, Nilsen A, Mao J, Preston DA, Belperron AA, Bockenstedt LK, Hinrichs DJ, Riscoe MK, Doggett JS, Ben Mamoun C. 2016. Radical cure of experimental babesiosis in immunodeficient mice using a combination of an endochin-like quinolone and atovaquone. J Exp Med 213:1307–1318. doi: 10.1084/jem.20151519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stickles AM, de Almeida MJ, Morrisey JM, Sheridan KA, Forquer IP, Nilsen A, Winter RW, Burrows JN, Fidock DA, Vaidya AB, Riscoe MK. 2015. Subtle changes in endochin-like quinolone structure alter the site of inhibition within the cytochrome bc1 complex of Plasmodium falciparum. Antimicrob Agents Chemother 59:1977–1982. doi: 10.1128/AAC.04149-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hegewald J, Gross U, Bohne W. 2013. Identification of dihydroorotate dehydrogenase as a relevant drug target for 1-hydroxyquinolones in Toxoplasma gondii. Mol Biochem Parasitol 190:6–15. doi: 10.1016/j.molbiopara.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 16.Vallieres C, Fisher N, Antoine T, Al-Helal M, Stocks P, Berry NG, Lawrenson AS, Ward SA, O'Neill PM, Biagini GA, Meunier B. 2012. HDQ, a potent inhibitor of Plasmodium falciparum proliferation, binds to the quinone reduction site of the cytochrome bc1 complex. Antimicrob Agents Chemother 56:3739–3747. doi: 10.1128/AAC.00486-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McFadden DC, Tomavo S, Berry EA, Boothroyd JC. 2000. Characterization of cytochrome b from Toxoplasma gondii and Q(o) domain mutations as a mechanism of atovaquone-resistance. Mol Biochem Parasitol 108:1–12. doi: 10.1016/S0166-6851(00)00184-5. [DOI] [PubMed] [Google Scholar]
- 18.Gjerde B. 2013. Characterisation of full-length mitochondrial copies and partial nuclear copies (numts) of the cytochrome b and cytochrome c oxidase subunit I genes of Toxoplasma gondii, Neospora caninum, Hammondia heydorni and Hammondia triffittae (Apicomplexa: Sarcocystidae). Parasitol Res 112:1493–1511. doi: 10.1007/s00436-013-3296-4. [DOI] [PubMed] [Google Scholar]
- 19.Huang LS, Cobessi D, Tung EY, Berry EA. 2005. Binding of the respiratory chain inhibitor antimycin to the mitochondrial bc1 complex: a new crystal structure reveals an altered intramolecular hydrogen-bonding pattern. J Mol Biol 351:573–597. doi: 10.1016/j.jmb.2005.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hunte C, Palsdottir H, Trumpower BL. 2003. Protonmotive pathways and mechanisms in the cytochrome bc1 complex. FEBS Lett 545:39–46. doi: 10.1016/S0014-5793(03)00391-0. [DOI] [PubMed] [Google Scholar]
- 21.Gao X, Wen X, Esser L, Quinn B, Yu L, Yu CA, Xia D. 2003. Structural basis for the quinone reduction in the bc1 complex: a comparative analysis of crystal structures of mitochondrial cytochrome bc1 with bound substrate and inhibitors at the Qi site. Biochemistry 42:9067–9080. doi: 10.1021/bi0341814. [DOI] [PubMed] [Google Scholar]
- 22.Lange C, Hunte C. 2002. Crystal structure of the yeast cytochrome bc1 complex with its bound substrate cytochrome c. Proc Natl Acad Sci U S A 99:2800–2805. doi: 10.1073/pnas.052704699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coleman BI, Gubbels MJ. 2012. A genetic screen to isolate Toxoplasma gondii host-cell egress mutants. J Vis Exp 2012:e3807. doi: 10.3791/3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guzman C, Bagga M, Kaur A, Westermarck J, Abankwa D. 2014. ColonyArea: an ImageJ plugin to automatically quantify colony formation in clonogenic assays. PLoS One 9:e92444. doi: 10.1371/journal.pone.0092444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boratyn GM, Schaffer AA, Agarwala R, Altschul SF, Lipman DJ, Madden TL. 2012. Domain enhanced lookup time accelerated BLAST. Biol Direct 7:12. doi: 10.1186/1745-6150-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whelan S, Goldman N. 2001. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol 18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 28.Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- 29.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.