ABSTRACT
ColE1 plasmids are small mobilizable replicons that play an important role in the spread of antibiotic resistance in Pasteurellaceae. In this study, we describe how a natural single nucleotide polymorphism (SNP) near the origin of replication of the ColE1-type plasmid pB1000 found in a Pasteurella multocida clinical isolate generates two independent plasmid variants able to coexist in the same cell simultaneously. Using the Haemophilus influenzae Rd KW20 strain as a model system, we combined antibiotic susceptibility tests, quantitative PCRs, competition assays, and experimental evolution to characterize the consequences of the coexistence of the pB1000 plasmid variants. This coexistence produced an increase of the total plasmid copy number (PCN) in the host bacteria, leading to a rise in both the antibiotic resistance level and the metabolic burden produced by pB1000. Using experimental evolution, we showed that in the presence of ampicillin, the bacteria maintained both plasmid variants for 300 generations. In the absence of antibiotics, on the other hand, the bacteria are capable of reverting to the single-plasmid genotype via the loss of one of the plasmid variants. Our results revealed how a single mutation in plasmid pB1000 provides the bacterial host with a mechanism to increase the PCN and, consequently, the ampicillin resistance level. Crucially, this mechanism can be rapidly reversed to avoid the extra cost entailed by the increased PCN in the absence of antibiotics.
KEYWORDS: ColE1, experimental evolution, Haemophilus influenzae, plasmid stability
INTRODUCTION
pB1000 is a small ColE1-like plasmid carrying the blaROB-1 β-lactamase gene and conferring high-level resistance to β-lactams. pB1000 has been described in multiple pathogens from Pasteurellaceae, including Haemophilus influenzae (1–7).
The replication of ColE1 plasmids is under the control of a preprimer RNA (RNAII) which is negatively regulated by a small antisense RNA (RNAI) (8). Mutations affecting these RNAs encoded near the origin of replication (oriV) of the plasmid alter both the plasmid copy number (PCN) and the compatibility of ColE1 plasmids. Single mutations can give rise to new compatible plasmids that are able to coexist in the same cell, increasing the global PCN (7, 8). These mutations have been widely described in vitro (9) and arise naturally by chance due to the high plasmid copy number exhibited by ColE1-like plasmids (10), but to the best of our knowledge, there are no natural examples of cohabiting ColE1 plasmids differentiated by only a single nucleotide polymorphism (SNP).
In this work we analyze a heterozygous pB1000 differentiated only by an SNP near the oriV and isolated from a high-level ampicillin-resistant clinical isolate of Pasteurella multocida (5). We show that this SNP creates a new variant of the plasmid, which coexists with the original pB1000, increasing PCN and the ampicillin resistance level. Using experimental evolution, we demonstrate that plasmid coexistence can easily revert, modulating the resistance level and metabolic burden produced by pB1000.
RESULTS AND DISCUSSION
pB1000 variants act as independent plasmids.
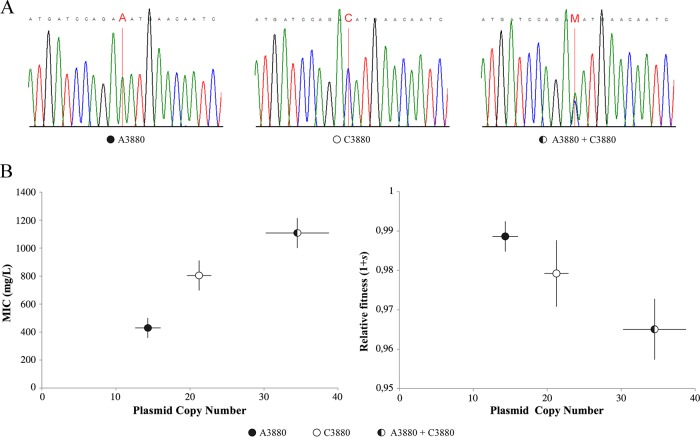
Recently, we recovered a P. multocida strain (BB1038) from a pig in Spain carrying pB1000 and exhibiting very high-level resistance to ampicillin and cefaclor (5). We decided to perform an in-depth analysis of the plasmid sequence from single colonies, and we observed the presence of a heterozygous SNP (C3880A) close to the putative oriV (Fig. 1A). To test if this mutation could be responsible for the generation of independent coexisting plasmids, we transformed H. influenzae Rd KW20 (Rd) with a pB1000 preparation obtained from BB1038, and we selected strains bearing both pB1000 variants individually and together. We measured pB1000 PCNs in the different H. influenzae transformants by quantitative PCR (qPCR) (5, 11). The strain bearing both plasmid alleles carried 34.51 copies (standard error of the mean [SEM], ±4.26) while the PCNs of the variants C3880 and A3880 alone in the bacteria were 14.31 (SEM, ±1.72) and 21.23 (SEM, ±1.63), respectively (Fig. 1B). The PCNs varied significantly among the three strains (analysis of variance [ANOVA], P = 0.029; F = 6.05; df = 1, 12) and between the strains carrying each allele individually (two-tailed t test, P = 0.003; t = 2.04; df = 28). Results suggested that both variants maintained their own PCNs when they were present together since the total PCN in the strain bearing both alleles was the sum of the PCNs of the two alleles when they were carried independently [two-tailed t test, P = 0.830, t = 2.04, and df = 28; Pearson's test r(13) = 0.942 and P = 1.57 × 10−7).
FIG 1.
Phenotypic and genotypic effects of a natural heterozygous SNP in the oriV of pB1000. A heterozygous SNP in the oriV of pB1000 produces an increase in PCN, which leads to a higher ampicillin resistance level and to an increase in the metabolic cost of the plasmid. (A) Chromatograms of the different variants (A3880, C3880, and A3880 plus C3880 in heterozygosis). Notice that the corresponding substitutions are highlighted in bold red letters in the chromatograms. M denotes the heterozygosity of variants A and C. (B) Correlation between the plasmid copy number and resistance level (left) and between the plasmid copy number and relative fitness w (right). Black, white, and black-white dots denote strains bearing variant A3880, variant C3880, and both variants in heterozygosis, respectively. Bars represent the SEMs.
Plasmid coexistence increases ampicillin resistance but decreases bacterial fitness in the absence of antibiotics.
It has been shown in previous works that gene copy number modulates antibiotic resistance (for a review, see reference 12). Specifically, it has been shown that the PCN correlates with the antibiotic resistance level in the case of plasmid-encoded β-lactamases (13). Therefore, we measured the resistance level to ampicillin in the different constructions by determining the MIC of ampicillin (14). The MIC for the C3880 variant was 424 mg/liter (SEM, ±75), and the MIC for the strain carrying the A3880 allele was 836 mg/liter (SEM, ±133). The MIC for the strain carrying the coexisting variants was 1,109 mg/liter (SEM, ±112). Resistance levels varied among the three strains (ANOVA, P = 0.005; F = 10.66; df = 2, 23) and between the strains carrying each variant individually (two-tailed t test, P = 0.017; t = 2.13; df = 15). In addition, there was a significant linear correlation between the PCN and the resistance level [Pearson's test r(25) = 0.853; P = 1.62 × 10−8] (Fig. 1B), suggesting that the resistance phenotype produced by both alleles was the sum of the resistance conferred by each allele individually.
We observed how the increase in pB1000 PCN led to an increase in the ampicillin resistance level due to the increase in the gene dosage. However, in the absence of antibiotics, an increase in the PCN usually produces an increase in the fitness cost produced by a plasmid (10, 15). To analyze this possibility we determined the biological cost of the different plasmid alleles alone and cohabiting using the coefficient of selection (s) as previously described (11). The relative fitness (w) of the strain carrying both plasmids was −0.966 (SEM, ±0.007). The w of the strain carrying the C3880 or the A3880 variant was −0.989 (SEM, ±0.003) or −0.98 (SEM, ±0.008), respectively. There was a significant correlation between the PCN and the biological cost (s) produced by the plasmids [Pearson's test r(7) = 0.879; P = 0.001] (Fig. 1B).
Rapid reversion to single-plasmid genotype in the absence of ampicillin.
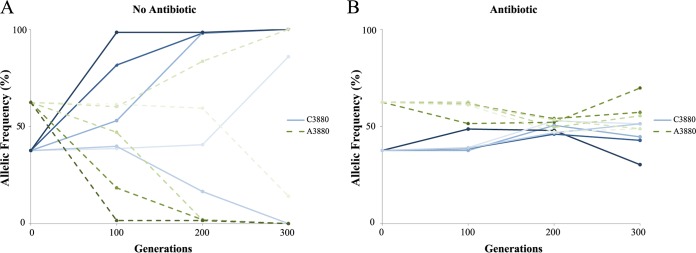
Our data showed that the increase in PCN entailed a fitness cost in the absence of ampicillin, but it also enhanced the MIC of ampicillin, producing a potential advantage in the presence of this antibiotic. Therefore, one may expect both plasmid variants to be maintained in the presence of ampicillin. In the absence of ampicillin, on the other hand, we predicted that the relatively frequent random plasmid loss during cell division affecting small multicopy plasmids (16) would purge populations from one or both variants of the plasmid. To test this hypothesis, we propagated five populations of H. influenzae bacteria bearing both plasmid variants over 300 generations with and without antibiotic pressure of 64 mg/liter of ampicillin. This ampicillin concentration corresponds to the peak concentration achieved in patients under standard ampicillin dosing regimens (11, 17, 18), and it is approximately 1/6 the MIC for the strain carrying the variant conferring the lowest resistance level. At generations 0, 100, 200, and 300, we tested the presence of the different plasmid variants in the different evolving populations (Fig. 2), as well as the MIC of ampicillin and plasmid copy number (Fig. 3). In the populations evolved without ampicillin, four out of five populations completely lost one of the pB1000 variants after 300 generations: three of them lost the more costly variant (A3880) while one population lost the C3880 variant (Fig. 2A). On the other hand, in the populations evolved in the presence of ampicillin, all populations carried both alleles at generations 100, 200, and 300, which supports the hypothesis of the positive fitness effects being associated with the increased PCN in the presence of ampicillin (Fig. 2B).
FIG 2.
Frequency of plasmid variants in the populations during the experimental evolution. The frequencies of pB1000 variants in the different populations estimated during the experimental evolution in the absence (A) or presence (B) of antibiotic pressure are shown. The frequency of each allele is represented; therefore, when one of the alleles progresses to fixation (100% frequency), the other allele disappears from the population (0% frequency). The five populations in each treatment initially carried both plasmid variants and are represented by five different shades of blue lines (variant C3880) and green dotted lines (variant A3880), as indicated. Note that in the presence of ampicillin all the populations maintained both plasmid variants, whereas in the absence of ampicillin, 4 out of 5 populations completely lost one of the variants.
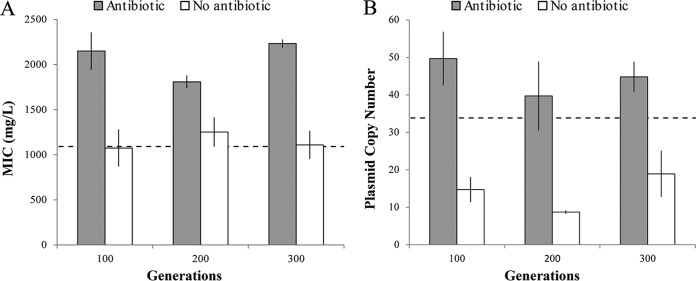
FIG 3.
Ampicillin resistance level and plasmid copy number during experimental evolution. The presence of ampicillin is associated with a higher PCN and ampicillin resistance level in the evolving populations. Graphs represent the resistance level (A) and plasmid copy number (B) during the experimental evolution of H. influenzae Rd carrying both pB1000 variants in the presence and absence of ampicillin. Bars represent the SEMs. The black dashed lines represent the resistance level (A) and the plasmid copy number (B) of the parental H. influenzae Rd KW20 carrying both pB1000 variants at the beginning of the experiment. Note that the populations evolved in the absence of ampicillin showed a decrease in plasmid copy number compared to that of the parental strain (two-tailed t test; P < 0.005; t = 2.04; df = 28), but they showed no decrease in the ampicillin resistance level compared to that of the ancestral strain (two-tailed t test; P = 0.608; t = 2.07; df = 28). This effect is probably due to an increase in the fitness in the evolving populations compared to that of the parental strain (due to general adaptation to the experimental conditions) which, as we reported before, also produces a moderate increase in the ampicillin resistance level (11).
As expected from the previous results, we observed a difference in PCNs and ampicillin resistance levels between the populations propagated in the presence and absence of ampicillin (Fig. 3). Populations evolving under ampicillin pressure presented both higher PCNs and higher ampicillin resistance levels than those propagated without antibiotics at generations 100, 200, and 300 (two-tailed t test for 100 generations, P < 0.005; t = 2.30; df = 8; two-tailed t test for 200 generations, P = 0.013; t = 2.30; df = 8; two-tailed t test for 300 generations, P < 0.005; t = 2.30; df = 8).
The results of this experiment lead to a very interesting conclusion: the increase in resistance mediated by an SNP in pB1000 can be reverted without mutations. This reversion is achieved through plasmid loss, which occurs at a higher frequency than potential mutations that revert the plasmid to the original genotype or compensate the cost associated with the plasmids. Actually, the theoretical rate of plasmid loss per generation for the plasmid alleles is 102- to 104-fold higher than the mutation rate per generation in H. influenzae (16, 19, 20). Therefore, this is a highly plastic strategy to modulate plasmid-mediated antibiotic resistance levels, and we argue that it could be more common in nature than previously thought.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and antibiotic susceptibility tests.
Pasteurella multocida (BB1038) was recovered from an infected pig, and it presented high-level ampicillin and cefaclor resistance (5). By analysis of DNA chromatograms obtained from sequencing of PCR amplicons from single colonies (forward primer, AATTGGTTGGACAATAACGCA; reverse primer, AATCTCGCTTATCAGGTGTGC), two different pB1000 (GenBank accession number GU080069) variants, C3880 and A3880, were discovered coexisting in every colony sequenced from the population.
H. influenzae Rd KW20 (Rd)was transformed by electroporation with both pB1000 variants from P. multocida BB1038 as previously described (5) using a Gene Pulser apparatus (Bio-Rad, USA). By random selection of colonies, PCR amplification (as described above), sequencing, and subsequent analysis of the chromatograms, we isolated Rd strains bearing both pB1000 variants individually and together.
H. influenzae was cultured in Haemophilus test medium (HTM) broth (Siemens Healthcare Diagnostics, Inc., USA) and on chocolate agar PolyViteX plates (bioMérieux, France) at 37° and 5% CO2. Antibiotic susceptibility was determined in HTM using the MIC of ampicillin (Sigma-Aldrich, United Kingdom) by broth microdilution according to the CLSI guidelines (14). We used H. influenzae ATCC 49247 and ATCC 49766 as controls. Results are the means of three independent experiments (with three technical replicates per experiment).
Fitness determination.
The fitness cost of pB1000 was determined by direct competition experiments between H. influenzae Rd and H. influenzae Rd bearing the pB1000 variants individually and together (11). Strains were grown for 16 h at 37°C in 5% CO2 and HTM; then, 106 CFU of each strain was mixed in 2 ml of HTM. The mix was cultured at 37°C in 5% CO2 at 125 rpm for 24 h, and 106 CFU was transferred to 2 ml of HTM (dilution of 1:1,000; approximately 10 generations) every 24 h during 6 days. Samples were taken at time zero (t0) and each 24 h. For each sample, aliquots were plated onto nonselective chocolate agar, and by replica plating 50 to 100 colonies were plated on chocolate agar plates containing 100 mg/liter of ampicillin. The competition index (CI) was calculated as the ratio between the numbers of CFU of the resistant and susceptible strains at each time (each 24 h) divided by the same ratio at t0 (21). The selection coefficient (s) that estimates the difference between the relative fitnesses of the two competitors over the entire competition experiment was calculated as the slope of the linear regression model s = ln(CI)/t, where time (t) was measured in bacterial generations, calculated as the log2 of the dilution factor (22). The relative fitness (w) was calculated with the formula w = 1 + s. Results are the average of three to six independent experiments.
Plasmid copy number quantification.
The average plasmid copy number per cell was determined by quantitative PCR (qPCR) basically as described by San Millan et al. (11). To determine the plasmid copy number, three independent DNA extractions were performed for each strain, and qPCR was then carried out in quintuplicate for each extraction. Each strain was grown in 2 ml of fresh HTM, and the DNA was extracted at an optical density at 600 nm (OD600) of approximately 0.9 using a QIAamp DNA minikit (Qiagen, Inc., Chatworth, CA, USA). The DNA was quantified using a NanoDrop instrument. Following the method of Providenti et al. (23), 1 μg of DNA of each sample was digested with 20 units of PstI (TaKaRa, Japan) for 2 h at 37°C; PstI was inactivated at 60°C for 15 min.
qPCRs were performed using a My iQ single-color real-time PCR detection system (Bio-Rad laboratories) with iQ SYBR green supermix (Bio-Rad Laboratories) at a final DNA concentration of 10 pg/μl. We used a specific qPCR for the plasmid-carried blaROB-1 gene (forward primer, CCAATTCTGTTCATTCGGTAAC; reverse primer, CATAAGCAAAGCGTTCATCTG; amplicon size, 195 bp). In order to determine the average plasmid copy number per chromosome, the monocopy gene rpoB was amplified to compare the ratio of plasmid to chromosomal DNA (forward primer, AAGGCTATAAGAACCTGTTGAC; reverse primer, CGAGACGAAGCGGAAATC; amplicon size, 199 bp). The reaction efficiency was calculated for each qPCR based on the standard curve generated by performing a qPCR with five 8-fold dilutions of the template DNA in triplicate (a working range of DNA concentrations of ∼0.2 ng/μl to 50 fg/μl). Results of reactions with an efficiency lower than 97% and higher than 100% with an R2 lower than 0.998 were discarded. The amplification conditions were as follows: initial denaturation for 10 min at 94°C, followed by 30 cycles of denaturation for 1 min at 94°C, annealing for 1 min at 52°C (rpoB) or 58.7°C (blaROB-1), and extension for 1 min at 72°C. Interrun calibration samples were used to normalize the results from different plates of each qPCR. To calculate plasmid copy number per chromosome, we used the formula described in San Millan et al. (24): cn = (1 + Ec)Ctc/(1 + Ep)Ctp × Sc/Sp, where cn is the plasmid copy number per chromosome, Sc and Sp are the sizes of the chromosomal and plasmid amplicons (in bp), respectively, Ec and Ep are the efficiencies of the chromosomal and plasmid qPCRs (relative to 1), respectively, and Ctc and Ctp are the threshold cycles of the chromosomal and plasmid reactions, respectively.
Experimental evolution.
Five replicate populations of H. influenzae Rd bearing both variants of pB1000 in heterozygosis (in a ratio of approximately 45:55 for the C3880 variant versus the A3880 variant) were propagated for 300 generations (30 days) with a daily transfer using a dilution factor of 1:1,000 (approximately 10 generations per day) in HTM at 37° in 5% CO2 at 125 rpm. In parallel, five replicate populations were propagated as described above with subinhibitory concentrations (64 mg/liter) of ampicillin. At days 10, 20, and 30 (100, 200, and 300 generations, respectively) the proportion of the different pB1000 variants was determined as follows: whole population plasmid extractions of all evolved populations was performed (QIAprep Spin Miniprep kit; Qiagen, USA); then the complete replicon from each preparation was sequenced by Sanger sequencing using the plasmid extraction as a template. Finally, the pB1000 variant frequencies of the pooled plasmid DNA were estimated by analyzing the chromatograms with QSVanalyser software (25). Apart from the polymorphism in position 3880, no changes in the plasmid sequence were identified throughout the experiments.
Statistical analysis.
Comparisons of the results from two and three strains were performed using two-tailed t tests and ANOVA, respectively. To analyze the contribution of both plasmid variants to the total PCN during cohabitation, a two-tailed t test was performed comparing the addition of the PCN of both variants inhabiting the cell alone to the PCN of both variants cohabiting in the same cell as measured by qPCR. Correlations between the variables PCN-fitness and PCN-MIC were determined using the Pearson product-moment correlation; to perform the statistical analysis we compared all the samples of fitness and MICs with the same number of samples of PCNs chosen randomly from all the replicates.
ACKNOWLEDGMENTS
We thank Rafael Martin-Nieto and Carlos Quesada for their mathematical analysis. We acknowledge Natalia Montero for her excellent technical assistance.
A.S.-L., A.S.M., and B.G.-Z. conceived and designed the experiments; A.S.-L., C.B.-B., and M.A.-A. performed the experiments; A.S.-L., A.S.M., and B.G.-Z. analyzed the data; R.O.-H. contributed reagents/materials analysis tools; A.S.-L., A.H., A.S.M., and B.G.-Z. wrote the paper.
This work was funded by REDEEX-2 (MICINN, BFU 2011-14145-E), by Ministerio de Economía y Competitividad (BIO2014-51806-R), by EU projects EvoTAR (282004-FP7) and EFFORT (613754-FP7), by an FPU grant from the Spanish Ministry of Education (FPU13/06215), and by an Indo-Spanish research project (Department of Science and Technology, India; DST/INT/SPAIN/P-28/2011 C) and the Spanish Ministry of Science and Innovation (PRI-PIBIN-2011-0915). A.S.M. is supported by a Miguel Servet fellowship from the Instituto de Salud Carlos III (MS15/00012) cofinanced by the European Social Fund and The European Development Regional Fund “A Way to Achieve Europe” (ERDF).
REFERENCES
- 1.Moleres J, Santos-López A, Lázaro I, Labairu J, Prat C, Ardanuy C, González-Zorn B, Aragon V, Garmendia J. 2015. Novel blaROB-1-bearing plasmid conferring resistance to β-lactams in Haemophilus parasuis isolates from healthy weaning pigs. Appl Environ Microbiol 81:3255–3267. doi: 10.1128/AEM.03865-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.San Millan A, Giufré M, Escudero JA, Hidalgo L, Gutierrez B, Cerquetti M, Gonzalez-Zorn B. 2011. Contribution of ROB-1 and PBP3 mutations to the resistance phenotype of a β-lactamase-positive amoxicillin/clavulanic acid-resistant Haemophilus influenzae carrying plasmid pB1000 in Italy. J Antimicrob Chemother 66:96–99. doi: 10.1093/jac/dkq392. [DOI] [PubMed] [Google Scholar]
- 3.San Millan A, Garcia-Cobos S, Escudero JA, Hidalgo L, Gutierrez B, Carrilero L, Campos J, Gonzalez-Zorn B. 2010. Haemophilus influenzae clinical isolates with plasmid pB1000 bearing blaROB-1: fitness cost and interspecies dissemination. Antimicrob Agents Chemother 54:1506–1511. doi: 10.1128/AAC.01489-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tristram SG, Littlejohn R, Bradbury RS. 2010. blaROB-1 presence on pB1000 in Haemophilus influenzae is widespread, and variable cefaclor resistance is associated with altered penicillin-binding proteins. Antimicrob Agents Chemother 54:4945–4947. doi: 10.1128/AAC.00263-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.San Millan A, Escudero JA, Gutierrez B, Hidalgo L, Garcia N, Llagostera M, Dominguez L, Gonzalez-Zorn B. 2009. Multiresistance in Pasteurella multocida is mediated by coexistence of small plasmids. Antimicrob Agents Chemother 53:3399–3404. doi: 10.1128/AAC.01522-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.San Millan A, Escudero JA, Catalan A, Nieto S, Farelo F, Gibert M, Moreno MA, Dominguez L, Gonzalez-Zorn B. 2007. Beta-lactam resistance in Haemophilus parasuis is mediated by plasmid pB1000 bearing blaROB-1. Antimicrob Agents Chemother 51:2260–2264. doi: 10.1128/AAC.00242-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lacatena RM, Cesareni G. 1981. Base pairing of RNA I with its complementary sequence in the primer precursor inhibits ColE1 replication. Nature 294:623–626. doi: 10.1038/294623a0. [DOI] [PubMed] [Google Scholar]
- 8.Tomizawa JI, Ohmori H, Bird RE. 1977. Origin of replication of colicin E1 plasmid DNA. Proc Natl Acad Sci U S A 74:1865–1869. doi: 10.1073/pnas.74.5.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Camps M. 2010. Modulation of ColE1-like plasmid replication for recombinant gene expression. Recent Pat DNA Gene Seq 4:58–73. doi: 10.2174/187221510790410822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.San Millan A, Escudero JA, Gifford DR, Mazel D, MacLean RC. 7 November 2016. Multicopy plasmids potentiate the evolution of antibiotic resistance in bacteria. Nat Ecol Evol doi: 10.1038/s41559-016-0010. [DOI] [PubMed] [Google Scholar]
- 11.San Millan A, Santos-Lopez A, Ortega-Huedo R, Bernabe-Balas C, Kennedy SP, Gonzalez-Zorn B. 2015. Small plasmid-mediated antibiotic resistance in Haemophilus influenzae is enhanced by increases in plasmid copy number and bacterial fitness. Antimicrob Agents Chemother 59:3335–3341. doi: 10.1128/AAC.00235-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sandegren L, Andersson DI. 2009. Bacterial gene amplification: implications for the evolution of antibiotic resistance. Nat Rev Microbiol 7:578–588. doi: 10.1038/nrmicro2174. [DOI] [PubMed] [Google Scholar]
- 13.Martinez JL, Cercenado E, Rodriguez-Creixems M, Vincente-Perez MF, Delgado-Iribarren A, Baquero F. 1987. Resistance to beta-lactam/clavulanate. Lancet ii:1473. [DOI] [PubMed] [Google Scholar]
- 14.CLSI. 2007. Performance standards for antimicrobial susceptibility testing; 17th informational supplement. Approved standard M100-S17. CLSI, Wayne, PA. [Google Scholar]
- 15.Harrison E, Koufopanou V, Burt A, Maclean RC. 2012. The cost of copy number in a selfish genetic element: the 2-μm plasmid of Saccharomyces cerevisiae. J Evol Biol 25:2348–2356. doi: 10.1111/j.1420-9101.2012.02610.x. [DOI] [PubMed] [Google Scholar]
- 16.Ayala-Sanmartin J, Gomez-Eichelmann MC. 1989. Stability of ColE1-like and pBR322-like plasmids in Escherichia coli. Mol Microbiol 3:1745–1752. doi: 10.1111/j.1365-2958.1989.tb00160.x. [DOI] [PubMed] [Google Scholar]
- 17.Lacy MK, Lu W, Xu X, Tessier PR, Nicolau DP, Quintiliani R, Nightingale CH. 1999. Pharmacodynamic comparisons of levofloxacin, ciprofloxacin, and ampicillin against Streptococcus pneumoniae in an in vitro model of infection. Antimicrob Agents Chemother 43:672–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meyers BR, Wilkinson P, Mendelson MH, Walsh S, Bournazos C, Hirschman SZ. 1991. Pharmacokinetics of ampicillin-sulbactam in healthy elderly and young volunteers. Antimicrob Agents Chemother 35:2098–2101. doi: 10.1128/AAC.35.10.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.San Millan A, Peña-Miller R, Toll-Riera M, Halbert ZV, McLean AR, Cooper BS, MacLean RC. 2014. Positive selection and compensatory adaptation interact to stabilize non-transmissible plasmids. Nat Commun 5:5208. doi: 10.1038/ncomms6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Power PM, Sweetman WA, Gallacher NJ, Woodhall MR, Kumar GA, Moxon ER, Hood DW. 2009. Simple sequence repeats in Haemophilus influenzae. Infect Genet Evol 9:216–228. doi: 10.1016/j.meegid.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bjorkman J, Andersson DI. 2000. The cost of antibiotic resistance from a bacterial perspective. Drug Resist Updat 3:237–245. doi: 10.1054/drup.2000.0147. [DOI] [PubMed] [Google Scholar]
- 22.Lenski RE. 1991. Quantifying fitness and gene stability in microorganisms. Biotechnology 15:173–192. [DOI] [PubMed] [Google Scholar]
- 23.Providenti MA, O'Brien JM, Ewing RJ, Paterson ES, Smith ML. 2006. The copy-number of plasmids and other genetic elements can be determined by SYBR-Green-based quantitative real-time PCR. J Microbiol Methods 65:476–487. doi: 10.1016/j.mimet.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 24.San Millan A, Heilbron K, MacLean RC. 2014. Positive epistasis between co-infecting plasmids promotes plasmid survival in bacterial populations. ISME J 8:601–612. doi: 10.1038/ismej.2013.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carr IM, Robinson JI, Dimitriou R, Markham AF, Morgan AW, Bonthron DT. 2009. Inferring relative proportions of DNA variants from sequencing electropherograms. Bioinformatics 25:3244–3250. doi: 10.1093/bioinformatics/btp583. [DOI] [PubMed] [Google Scholar]