Abstract
High-grade glioma is the most aggressive and severe brain tumor that leads to death of almost 50% patients in 1–2 years. Thus, accurate prognosis for glioma patients would provide essential guidelines for their treatment planning. Conventional survival prediction generally utilizes clinical information and limited handcrafted features from magnetic resonance images (MRI), which is often time consuming, laborious and subjective. In this paper, we propose using deep learning frameworks to automatically extract features from multi-modal preoperative brain images (i.e., T1 MRI, fMRI and DTI) of high-grade glioma patients. Specifically, we adopt 3D convolutional neural networks (CNNs) and also propose a new network architecture for using multi-channel data and learning supervised features. Along with the pivotal clinical features, we finally train a support vector machine to predict if the patient has a long or short overall survival (OS) time. Experimental results demonstrate that our methods can achieve an accuracy as high as 89.9% We also find that the learned features from fMRI and DTI play more important roles in accurately predicting the OS time, which provides valuable insights into functional neuro-oncological applications.
1 Introduction
Brain tumors are one of the most lethal and difficult-to-treat cancers. The most deadly brain tumors are known as the World Health Organization (WHO) high-grade (III and IV) gliomas. The prognosis of glioma, often measured by the overall survival (OS) time, varies largely across individuals. Based on histopathology, OS is relatively longer for WHO-III, while shorter for WHO-IV gliomas. For instance, there is a median survival time of approximately 3 years for anaplastic astrocytoma while only 1 year for glioblastoma [2].
Tumor WHO grading, imaging phenotype, and other clinical data have been studied in their relationship to OS [1,6]. Bisdas et al. [1] showed that the relative cerebral blood volume in astrocytomas is predictive for recurrence and 1-year OS rate. However, this conclusion cannot be extended to other higher-grade gliomas, where prognosis prediction is more important. Lacroix et al. [6] identified five independent predictors of OS in glioblastoma patients, including age, Karnofsky Performance Scale score, extent of resection, degree of necrosis and enhancement in preoperative MRI. However, one may question the generalization ability of such models.
Recently, researchers have been using informative imaging phenotypes to study prognosis [8,9,11]. Contrast-enhanced T1 MRI has been widely used for glioblastoma imaging. Previous studies [9] have shown that T1 MRI features can contribute largely to the prognostic studies for survival of glioblastoma patients. For instance, Pope et al. [9] analyzed 15 MRI features and found that several of them, such as non-enhancing tumor and infiltration area, are good predictors of OS. In addition, diffusion tensor imaging (DTI) provides complementary information, e.g., white matter integrity. For instance, authors in [11] concluded that DTI features can help discriminate between short and long survival of glioblastoma patients more efficiently than using only the histopathologic information. Functional MRI (fMRI) has also been used to measure cerebrovascular reactivity, which is impaired around the tumor entity and reflects the angiogenesis, a key sign of malignancy [8].
These approaches mostly used the handcrafted and engineered features according to the previous medical experiences. This often limits the ability to take full advantage of all the information embedded in MR images, since handcrafted features are usually bound to the current limited knowledge of specific field. On the other hand, the features exploited in some previous methods, e.g., [11], were often obtained in unsupervised manners, which could introduce many useless features or even ignore some useful clues. To overcome such problems, we propose a novel method to predict survival time for brain tumor patients by using high-level imaging features captured in a supervised manner.
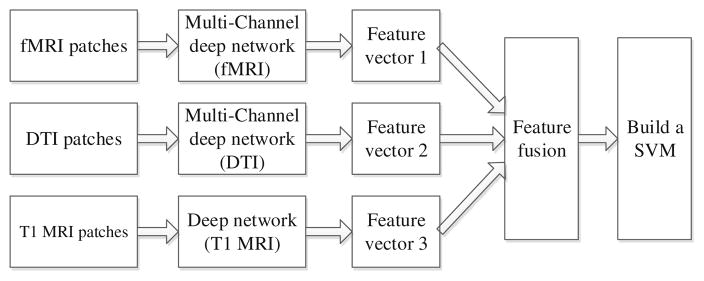
Specifically, we do not use only the handcrafted features; rather, we propose a 3D deep learning approach to discover high-level features that can better characterize the virtue of different brain tumors. Particularly, in the first step, we apply a supervised deep learning method, i.e., convolutional neural network (CNN) to extract high-level tumor appearance features from T1 MRI, fMRI and DTI images, for distinguishing between long and short survival patients. In the second step, we train a SVM [3] with the above extracted features (after feature selection and also some basic clinical information) for predicting the patient’s OS time. The whole pipeline of our system is shown in Fig. 1. Note that the MR images are all 3D images and, therefore, we adopt a 3D CNN structure on the respective 3D patches. Furthermore, since both fMRI and DTI data include multiple channels (as explained later), we further propose a multi-channel CNNs (mCNNs) to properly fuse information from all the channels of fMRI or DTI.
Fig. 1.
The flow chart for our survival prediction system
2 Data Acquisition and Preprocessing
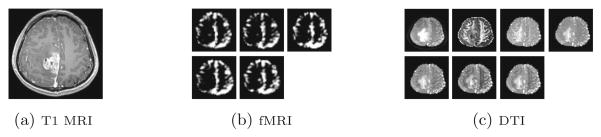
The data acquired from the tumor patients include the contrast-enhanced T1 MRI, resting-state fMRI and DTI images. The images from a sample subject are shown in Fig. 2, in which we can see single-channel data for T1 MRI, and multi-channel images for both fMRI and DTI. These three modalities of images are preprocessed following the conventional pipelines. Briefly, they are first aligned. Then, for T1 MRI, intensity normalization is performed. For DTI, diffusion tensor modeling is done, after which diffusion metrics (i.e., FA, MD, RD, lambda 1/2/3) are calculated in addition to the B0 image. For fMRI data, frequency-specific blood oxygen level-dependent (BOLD) fluctuation powers are calculated in five non-overlapping frequency bands within 0–0.25 Hz.
Fig. 2.
A sample subject with glioblastoma in our dataset.
We collected the data from 69 patients, who have all been diagnosed with WHO-III or IV. Exclusion criteria were (1) any surgery, radiation therapy, or chemotherapy of brain tumor, before inclusion in the study, (2) missing data, and (3) excessive head motion or presence of artifacts. All patients were treated under the same protocol based on clinical guideline. The whole study was approved by a local ethical committee.
Patch Extraction
The tumor often appears in a certain region of the brain. We want to extract features not only from the contrast-enhanced regions, but also from the adjacent regions, where edema and tumor infiltration occur. In this paper, we manually label and annotate the tumor volume in T1 MRI that has the highest resolution. Specifically, we define a cuboid by picking the 8 vertices’ coordinates that confine the tumor and its neighboring areas. For fMRI and DTI data, we locate the tumor region according to the tumor mask defined in the T1 MRI data. With the extracted tumor region, we rescale the extracted tumor region of each subject to a predefined size (i.e., 64 × 64 × 64), from which we can extract many overlapping 32 × 32 × 32 patches to train our CNN/mCNNs.
Definition of Survival Time
OS time is defined as the duration from the date the patient was first scanned, to the date of tumor-related death for the patients. Subjects with irrelevant death reasons (e.g., suicide) were excluded. A threshold of 22 months was used to divide the patients into 2 groups: (1) short-term survival and (2) long-term survival, with 34 and 35 subjects in each group, respectively. This threshold was defined based on the OS rates in the literature [6], as approximately 50% of the high-grade glioma patients died within this time period.
3 The Proposed Method
Deep learning models can learn a hierarchy of features, in which high-level features are built upon low-level image features layer-by-layer. CNN [4,5] is a useful deep learning tool, when trained with appropriate regularizations, CNN has been shown with superior performance on both visual object recognition and image classification tasks (e.g., [5]).
In this paper, we first employ CNN architecture to train one survival time prediction model with T1 MRI, fMRI and DTI modalities, respectively. With such trained deep models, we can extract features from the respective image modalities in a supervised manner. Then, a binary classifier (e.g., SVM) is trained to predict OS time. In the following, we first introduce our supervised feature extraction strategies, followed by a classifier training step. Note that the feature extraction strategy for T1 MRI data is different from the feature extraction strategies for fMRI and DTI. This is because fMRI and DTI have multiple channels of data from each subject, while T1 MRI has a single channel of data. Thus, we first introduce our 3D CNN architecture for single-channel data (T1 MRI), and then extend it for multi-channel data (fMRI and DTI).
Single-Channel Feature Extraction
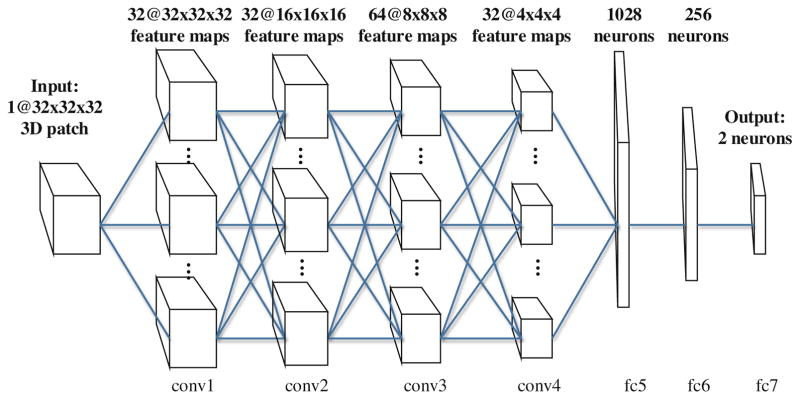
As described earlier, T1 MRI data is in 3D, and therefore we propose a 3D CNN model with a set of 3D trainable filters. The architecture used in our paper is depicted in Fig. 3, in which we define four convolutional layer groups (conv1 to conv4, note both conv1 and conv2 are composed of two convolutional layers), and three fully-connected layers (fc5 to fc7). The convolutional layers will associate their outputs to the local 3D regions in their inputs, each computing a convolutional operation with a 3D filter of size 3 × 3 × 3 (i.e., between their weights and the 3D regions they are operating on). These results are 3D volumes of the same size as their inputs, which are followed by a max-pooling procedure to perform a downsampling operation along the 3D volume dimensions. The fully-connected layers include neurons connected to all activations in their previous layer, as in the conventional neural networks. The last layer (fc7) would have 2 neurons, whose outputs are associated with the class scores. The supervision on the class scores would lead to a back-propagation procedure for learning the most relevant features in the fc layers. We use the outputs from the last two layers of this CNN (fc6 and fc7) as our fully supervised learned features, and also compare their efficiency and effectiveness in the experiments.
Fig. 3.
The CNN architecture for single-channel feature extraction from 3D patches
Multi-channel Feature Extraction
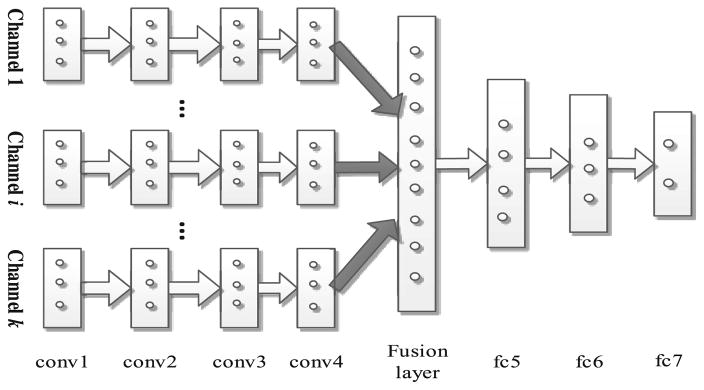
As mentioned, both fMRI and DTI images are composed of multiple channels of data (as in Fig. 2). To effectively employ all multi-channel data, we propose a new multi-channel-CNN (mCNN) architecture to train one mCNN for one modality by considering multi-channel data that can provide different information for the brain tumor. On the other hand, different channels may have little direct complementary information in their original image spaces, due to different acquisition techniques. Inspired by the work in [10], in which a multi-modal deep Boltzmann machine was introduced, we modify our 3D CNN architecture to deal with multi-channel data. Specifically, in our proposed mCNN, the same convolution layers are applied to each channel of data separately, but a fusion layer is added to fuse the outputs of conv4 layers from all different channels. Then, three fully connected layers are further incorporated to finally extract the features. This new mCNN architecture is illustrated in Fig. 4. In this architecture, the networks in the lower layers (i.e., the conv layers) for each pathway are different, accounting for different input distributions from each channel. The fusion layer combines the outputs from these different streams. Note that all these layer groups are identical to those described for the single-channel data.
Fig. 4.
Architecture of mCNN for feature extraction from multi-channel data
It is very important to note that the statistical properties of different channels of the data are often different, which makes it difficult for a single-channel model (as illustrated in Fig. 3) to directly find correlations across channels. In contrast, on our proposed mCNNs model, the differences of multi-channel data can be largely bridged by fusing their respective higher-layer features.
Classification
Once we train a CNN (Fig. 3) for T1 MRI images and two mCNNs (Fig. 4) for fMRI and DTI, respectively, we can map each raw (3D) image from all three modalities to its high-level representation. This is actually accomplished by feeding each image patch to the corresponding CNN or mCNN model, according to the modality where the current patch is extracted. The outputs of the last fully-connected layers are regarded as the learned features for the input image patch. Finally, each subject will be represented by a same-length feature vector. Together with survival labels, we train a SVM classifier for survival classification.
4 Experiments and Results
As described earlier, we learn features from multi-modal brain tumor data in a supervised manner, and then these extracted features to train a SVM classifier for survival prediction. For the training of deep networks, we adopt a back-propagation algorithm to update the network parameters. The network weights are initialized by Xavier algorithm [4]. The initial learning rate and weight decay parameter are determined by conducting a coarse line search, followed by decreasing the learning rate during training. To evaluate our proposed method, we incorporate different sets of features for training our method. In particular, the two commonly-used sets of features from CNNs are the outputs of the fc6 and fc7 layers, as can be seen in Figs. 3 (for single-channel CNN) and 4 (for mCNN). The network for each modality is trained on the respective modality data of training set. Then, each testing patch is fed into the trained network to obtain its respective features. Note that the layer before the output layer, denoted as fc6, has 256 neurons, while the output layer (fc7) is composed of 2 neurons. In addition to these features, the handcrafted features (HF) are also included in our experiments. These HF features consist of generic brain tumor features, including gender, age at diagnosis, tumor location, size of tumor, and the WHO grade. We also take advantage of scale-invariant transform (SIFT) [7] as a comparison feature extraction approach. To show the advantage of the 3D filters and architecture of the proposed method, we also provide the results obtained using the conventional 2D-CNN with the same settings.
As stated in Sect. 2, each patient has 3 modalities of images (i.e., T1 MRI, fMRI and DTI). From each of these three modalities (of each subject), 8 different patches are extracted. This leads to 8 × 256 = 2048 learned fc6 features, and 8×2 = 16 learned fc7 features, for each modality, while totally, 2048×3 = 6144 and 16 × 3 = 48 learned features in fc6 and fc7 layers, for three modalities. Due to the large number of features in fc6, we conducted a feature selection/reduction procedure on them. Specifically, we used Principal Component Analysis (PCA) and Sparse Representation (SR).
Results
We used 10-fold cross-validation, in which for each testing fold, 9 other folds are used to train both the CNN and mCNNs, and then the SVM with the learned features. The performance measures averaged for the 10 folds are reported in Table 1, including accuracy (ACC), sensitivity (SEN), specificity (SPE), positive predictive rate (PPR) and negative predictive rate (NPR), which are defined in the following (TP: true positive; TN: true negative; FP: false positive; FN: false negative):
Table 1.
Performance evaluation of different features and selection/reduction methods.
| ACC (%) | SEN (%) | SPE (%) | PPR (%) | NPR (%) | |
|---|---|---|---|---|---|
| HF | 62.96 | 66.39 | 58.53 | 63.18 | 65.28 |
| HF + SIFT | 78.35 | 80.00 | 77.28 | 67.59 | 87.09 |
| HF + 2D-CNN | 81.25 | 81.82 | 80.95 | 74.23 | 88.35 |
| fc7 | 80.12 | 85.60 | 77.64 | 71.71 | 87.50 |
| fc6-PCA | 80.55 | 84.85 | 76.92 | 75.68 | 85.71 |
| fc6-SR | 76.39 | 86.67 | 69.05 | 66.67 | 87.88 |
| HF + fc7 | 89.58 | 92.19 | 88.22 | 84.44 | 95.57 |
| HF + fc6-PCA | 89.85 | 96.87 | 83.90 | 84.94 | 93.93 |
| HF + fc6-SR | 85.42 | 92.60 | 80.39 | 75.36 | 96.83 |
As could be seen, incorporating both deeply-learned (with the proposed 3D CNN and 3D mCNN models) and hand crafted (HF) features results in the best classification accuracy of 89.85 %. In contrast, with HF alone, or in combination with unsupervised learned features (SIFT), we obtain just an accuracy of 62.96% or 78.35 %, respectively. Furthermore, the 3D architecture outperforms the conventional 2D architecture (i.e., 2D-CNN), which suggests that the 3D filters can lead to better feature learning. Regarding sensitivity and specificity, we know that the higher the sensitivity, the lower the chance of misclassifying the short survival patients; on the other hand, the higher the specificity, the lower the chance of misclassifying the long survival patients. The proposed feature extraction method resulted in an approximate 30% higher sensitivity and specificity, compared to the traditional handcrafted features. Interestingly, our model predicts the short survival patients with more confidence than the long survival patients. Furthermore, the features from different layers of the CNN and mCNN models (fc6 and fc7) exhibit roughly similar power in predicting the OS time, on this dataset.
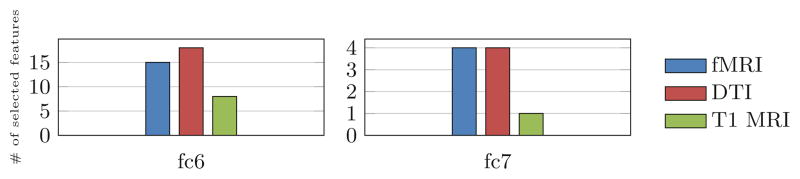
To analyze the importance of the features for predicting OS, we also visualize the number of features selected from each modality. To do this, for the features from fc6, we count the features selected by sparse representation, and for the fc7 layer, we use ℓ1-regularized SVM for classification for the features from fc7 layer, to enforce selection of the most discriminative features. We average the number of the selected features over all cross-validation folds. The results are depicted in Fig. 5. As it is obvious, the fMRI data have contributions for the prediction model as well as the DTI and T1 MRI.
Fig. 5.
The average number of selected features from each modality, using our model.
5 Conclusions
In this study, we proposed a 3D deep learning model to predict the (long or short) OS time for brain gliomas patients. We trained 3D CNN and mCNN models for learning features from single-channel (T1 MRI) and multi-channel (fMRI and DTI) data in a supervised manner, respectively. The extracted features were then fed into a binary SVM classifier. Experimental results showed that our supervised-learned features significantly improved the predictive accuracy of gliomas patients’ OS time. This indicates that our proposed 3D deep learning frameworks can provoke computational models to extract useful features for such neuro-oncological applications. In addition, the analysis on the selected features further shows that DTI data can contribute slightly more than fMRI, but both fMRI and DTI play more significant roles compared to T1 MRI, in building such successful prediction models.
References
- 1.Bisdas S, et al. Cerebral blood volume measurements by perfusion-weighted MR imaging in gliomas: ready for prime time in predicting short-term outcome and recurrent disease? Am J Neuroradiol. 2009;30(4):681–688. doi: 10.3174/ajnr.A1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeAngelis LM. Brain tumors. N Engl J Med. 2001;344(2):114–123. doi: 10.1056/NEJM200101113440207. [DOI] [PubMed] [Google Scholar]
- 3.Fan RE, et al. Liblinear: a library for large linear classification. JMLR. 2008;9:1871–1874. [Google Scholar]
- 4.Glorot X, Bengio Y. Understanding the difficulty of training deep feedforward neural networks. AISTATS. 2010:249–256. [Google Scholar]
- 5.Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. NIPS. 2012:1097–1105. [Google Scholar]
- 6.Lacroix M, et al. A multivariate analysis of 416 patients with glioblastoma multiforme: prognosis, extent of resection, and survival. J Neurosurg. 2001;95(2):190–198. doi: 10.3171/jns.2001.95.2.0190. [DOI] [PubMed] [Google Scholar]
- 7.Lowe DG. Distinctive image features from scale-invariant keypoints. Int J Comput Vis. 2004;60(2):91–110. [Google Scholar]
- 8.Pillai JJ, Zacá D. Clinical utility of cerebrovascular reactivity mapping in patients with low grade gliomas. 2011 doi: 10.5306/wjco.v2.i12.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pope WB, et al. MR imaging correlates of survival in patients with high-grade gliomas. Am J Neuroradiol. 2005;26(10):2466–2474. [PMC free article] [PubMed] [Google Scholar]
- 10.Srivastava N, Salakhutdinov RR. Multimodal learning with deep boltzmann machines. NIPS. 2012:2222–2230. [Google Scholar]
- 11.Zacharaki EI, et al. Survival analysis of patients with high-grade gliomas based on data mining of imaging variables. Am J Neuroradiol. 2012;33(6):1065–1071. doi: 10.3174/ajnr.A2939. [DOI] [PMC free article] [PubMed] [Google Scholar]