Abstract
Safe and effective delivery is required for siRNA and mRNA-based therapeutics to reach their potential. Here, we report on the development of poly(glycoamidoamine) brush nanoparticles as delivery vehicles for siRNA and mRNA. These polymers were capable of significant delivery of siRNA against FVII and mRNA-encoding erythropoietin (EPO) in mice. Importantly, these nanoparticles were well-tolerated at their effective dose based on analysis of tissue histology, systemic cytokine levels, and liver enzyme chemistry. The polymer brush nanoparticles reported here are promising for therapeutic applications.
Keywords: siRNA, mRNA, polymer brush, erythropoietin, nanomaterial
Graphical Abstract
There is a growing interest in use of RNA, including small interfering RNA (siRNA) and mRNA (mRNA), as therapeutics.1–8 siRNA can be used to downregulate target genes, while mRNA can elicit the expression of an encoded protein.9–13 siRNA and mRNA can therefore also serve as useful tools to study signaling pathways,14–16 in addition to their therapeutic applications.7,17 The efficient delivery of siRNA or mRNA remains the key challenge for broad applications of RNA based therapeutics in vivo.6 Lipid and polymer nanoparticles have previously demonstrated efficient delivery of siRNA and mRNA,18–20 and some successful examples for siRNA delivery have advanced to clinical trials to treat diverse diseases.2,7,8,17 However, delivery of siRNA and mRNA using the same materials has not been reported to date, and the combination of siRNA and mRNA may provide more effective tools for biological studies and clinical applications.
Here we report on the development of a new class of polymer-brush materials (Figure 1a) for siRNA and mRNA delivery, with subsequent evaluation of their delivery efficiency in vivo as well as a preliminary assessment of their safety profile. To facilitate interaction with siRNA or mRNA, amino groups were incorporated into the polymer for electrostatic complexation and entrapment of negatively charged siRNA or mRNA. In addition, multiple hydroxyl groups on the polymer increase hydrophilicity, while alkyl tail brushes added along the polymer backbone enable incorporation of the polymer brush materials into lipid-based nanoparticle formulations. This modular design offers the ability to tune the RNA delivery system through modification of a number of chemical and structural properties.
Figure 1.
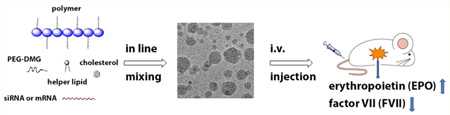
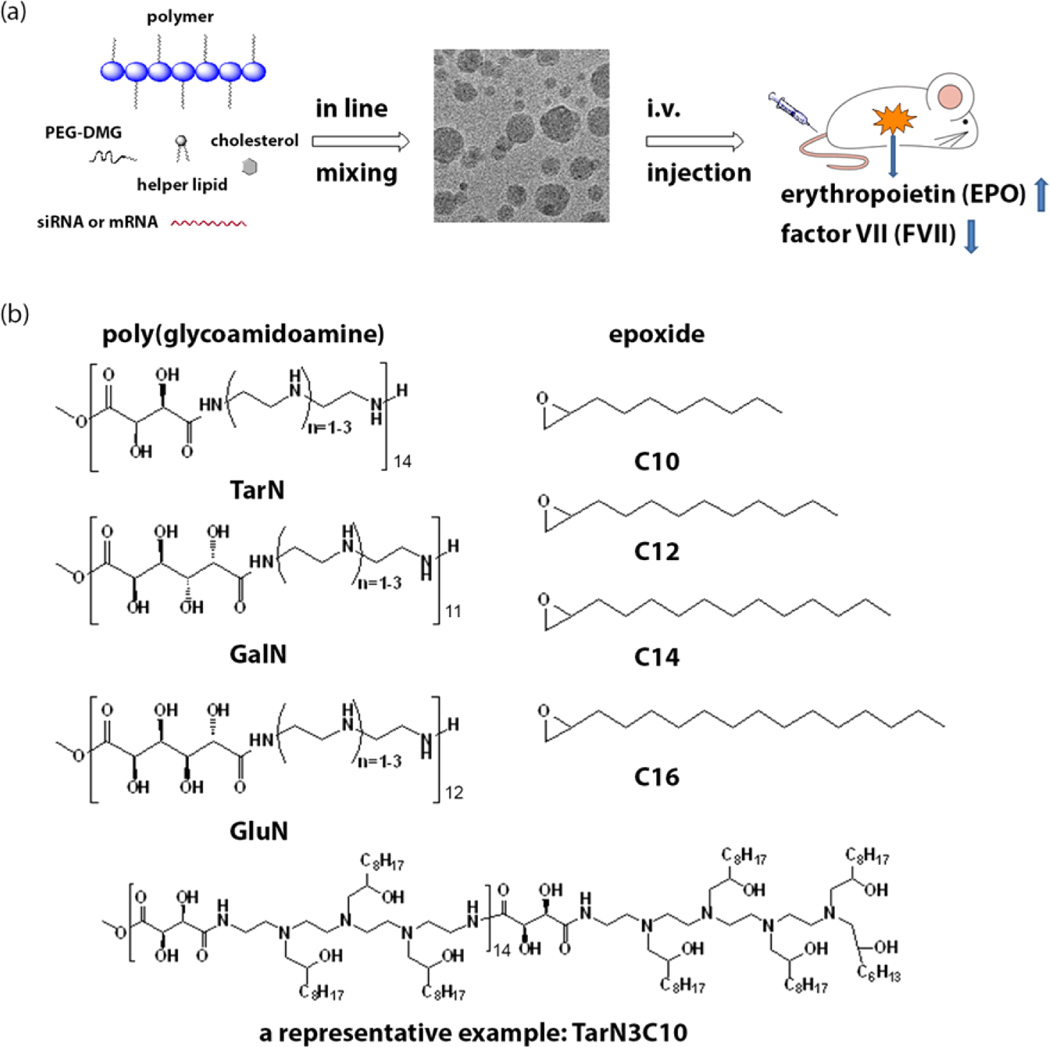
Polymer-brush materials for siRNA and mRNA delivery. (a) Illustration of particle formulation with cholesterol, helper lipid, mPEG2000-DMG, and siRNA/mRNA via a microfluidic based mixing device and evaluation through intravenous delivery. (b) Synthesis of polymer-brush materials through ring opening reactions between poly(glycoamidoamine) (PGAAs) and epoxides, along with a representative structure (TarN3C10). TarN, GalN, and GluN were synthesized using the methods reported by Reineke (TarN1, m = 12; TarN2, m = 11; TarN3, m = 11; GalN1, m = 11; GalN2, m = 14; GalN3, m = 14; GluN1, m = 11; GluN2, m = 11; GluN3, m = 11).21–24 1HNMR of PGAA polymers is consistent with reported data.21,24
Reineke and co-workers previously reported on the development of poly(glycoamidoamines) (PGAAs), which contain amines and multiple hydroxyl groups along their polymer backbone.21–23,25,26 These polymers previously demonstrated efficient delivery of both DNA and siRNA in different cell types.21–23,25,26 Beginning with the PGAA polymer backbone,18,26,27 we prepared modified PGAAs to create new polymer-brush materials (Figure 1b) for incorporation into lipid nanoparticle formulations. First, we synthesized three different PGAA polymers based on tartarate, galactarate, or glucarate sugars combined with three different amine-containing monomers using the synthetic methods reported by Reineke.21–24 1H NMR of PGAA polymers is consistent with reported data.21–23,25,26 Next, alkyl tails were added to amines on the PGAA backbone using ring-opening reactions with epoxides to afford modified polymer-brush materials.27–30 In total, 31 new polymers were synthesized. Structures of polymers were confirmed by 1H NMR and their molecular weight was calculated based on the results reported by Reineke and 1H NMR of final products.22 The nomenclature for polymer identification signifies the combination of these three structural building blocks; a three letter code (Tar, tartarate; Gal, galactarate; Glu, glucarate) denoting the sugar used to prepare the PGAA backbone followed by the number of amines in the amine-containing monomer (N1, N2, or N3), and finally the number of carbons (C10, C12, C14, or C16) on the epoxides used for modification.
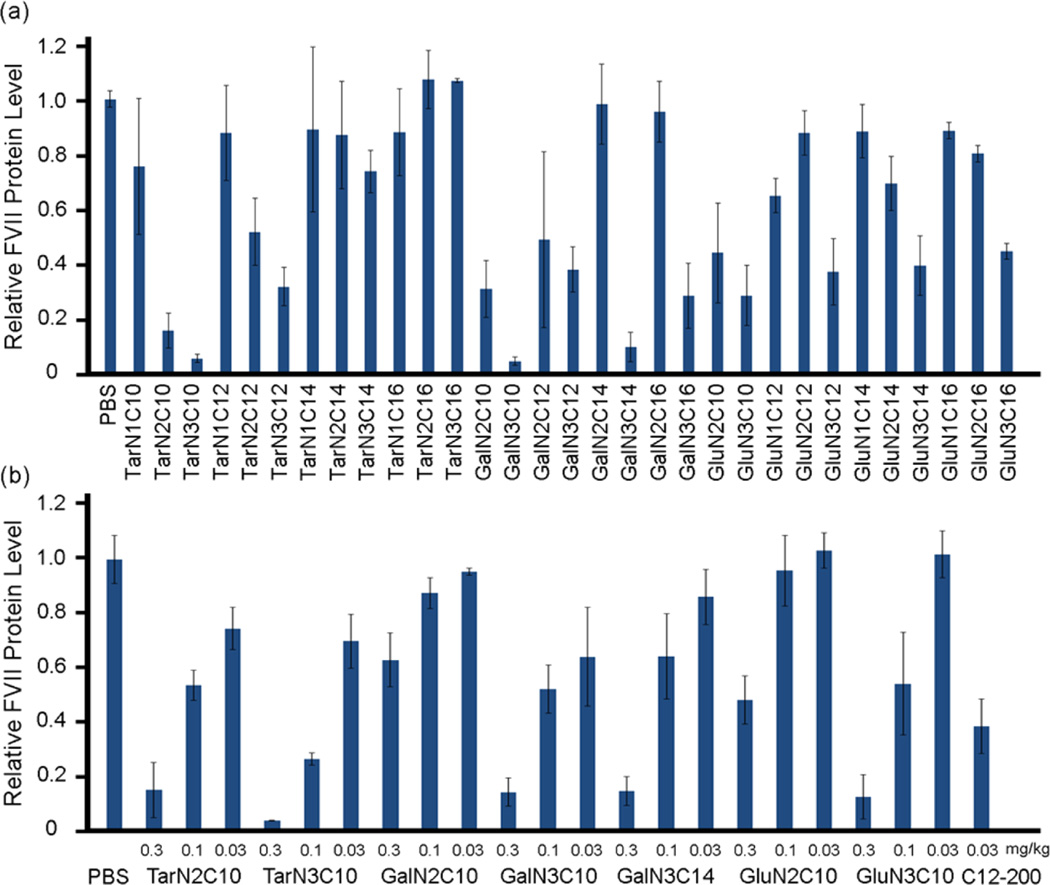
To formulate polymer-siRNA nanoparticles, we first mixed polymers with siRNA without adding additional components. However, the resulting complexation produces particles that are too large to be suitable for in vivo evaluation. For example, the formulated mixture of TarN3C1 with siRNA produces particles 831 nm in diameter (Table S1 in Supporting Information). In order reduce particle size and improve polydispersity, we incorporated additional formulation components based on previous experience in siRNA delivery.29 The polymer brush materials were subsequently formulated into nanoparticles through combination with cholesterol, DSPC (1,2-distearoyl-sn-glycero-3-phosphocholine), mPEG2000-DMG (1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy-(polyethylene glycol)-2000]), and siRNA using a microfluidic mixing device.31 To evaluate the delivery efficiency of these polymer-brush nanoparticles, siRNA against factor VII (FVII) was incorporated into the formulations. FVII is a blood clotting factor, which is a commonly used reporter for evaluation of siRNA delivery due to ease of sampling. We measured the particle size using the dynamic light scattering, which demonstrated particles between 68 and 137 nm in diameter (Table S1 in Supporting Information). Additionally, the siRNA loading efficiency was measured using the RiboGreen assay (Table S1 in Supporting Information).18 Polymer-brush nanoparticles were administered intravenously via tail vein in mice using a FVII siRNA dose of 0.3 mg/kg. Twenty-four hours following injection, blood was collected and FVII levels were measured by a chromogenic assay. In sum, 14 out of the 31 polymers evaluated showed FVII silencing of 50% or more. We then performed a dose-dependency study of the best polymer nanoparticles at the siRNA dose of 0.3, 0.1, and 0.03 mg/kg (Figure 2a). As shown in Figure 2b, all of these polymer nanoparticles displayed dose-dependent silencing of FVII. TarN3C10 nanoparticles silenced over 70% of FVII at a dose of 0.1 mg/kg.
Figure 2.
siRNA delivery efficiency of polymer-brush nanoparticles. (a) Silencing of FVII in mouse serum for nanoparticles at siRNA dose of 0.3 mg/kg. Control: free mRNA. (b) A dose-dependence study of lead materials. Lead materials showed dose-dependent silencing of FVII. C12–200 as a positive control. Data shown is mean ± s.d. (n = 3).
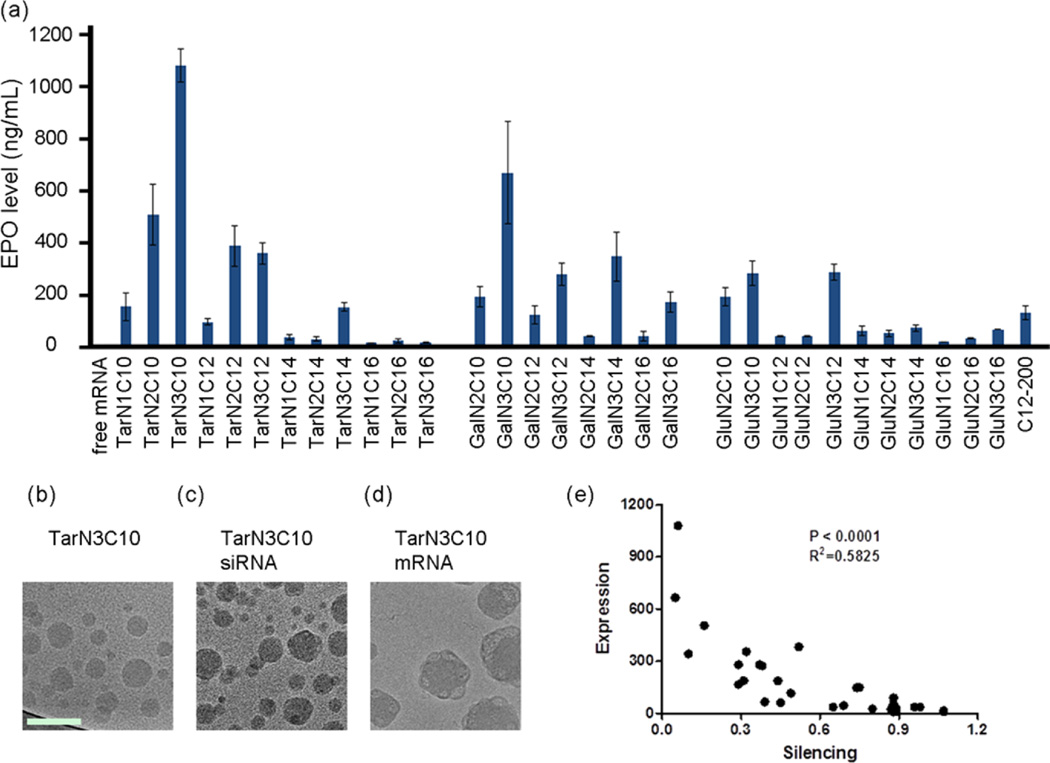
To evaluate the mRNA delivery efficiency of these polymer-brush nanoparticles, mRNA for human erythropoietin (EPO) was incorporated into the formulations. EPO functions to regulate red blood cell production13 and is used therapeutically by patients with anemia and myelodysplasia.32 The polymer-brush materials were subsequently formulated into nanoparticles as previously described.31 The mRNA loading efficiency, measured by the RiboGreen assay,18 was as high as 81% for these formulations. Polymer-brush nanoparticles were administered intravenously via tail vein in mice using an EPO mRNA dose of 0.3 mg/kg, with free mRNA as a control. Protein expression with mRNA delivery is known to peak around 5 to 7 h.11 Therefore, 6 h following injection, blood was collected and EPO levels were measured by ELISA, with several polymer-brush nanoparticles demonstrating efficacy in the delivery of functional EPO mRNA (Figure 3a). TarN3C10 nanoparticles were further characterized using cryogenic transmission electron microscopy.33–35 The TarN3C10 and TarN3C10-siRNA nanoparticles form round spherical particles. The addition of mRNA (Figure 3b–d) results in the formation of more complex structures. The molecular weight difference between siRNA and mRNA could contribute to changing the particle formulation and structure.
Figure 3.
mRNA delivery efficiency of polymer-brush nanoparticles. (a) Expression of EPO in mouse serum for nanoparticles at mRNA dose of 0.3 mg/kg. C12–200 as a positive control. Data shown is mean ± s.d. (n = 3). TarN3C10 nanoparticles demonstrated EPO expression over 1000-fold higher than free mRNA. Data shown is mean ± s.d. (n = 3). (b) Cryo-TEM of TarN3C10 without RNA. (c) siRNA formulated TarN3C10 and (d) mRNA formulated TarN3C10. The TarN3C10 nanoparticles form round spherical objects with/without siRNA, and the addition of mRNA leads to formation of more complex structures. Scale bar is 100 nm for all cryo-TEM images. (e) Correlation analysis for siRNA and mRNA delivery (p < 0.0001).
In sum, the results presented in Figures 2 and 3 confirm that members of each of the three polymer building blocks were able to facilitate at least some delivery of siRNA and mRNA. No correlation was found between delivery efficiency and particle size/RNA entrapment (Figure S1 in Supporting Information), while significant correlation between siRNA and mRNA delivery was observed (Figure 3e, p < 0.0001). We also summarized several structure–activity relationship from these data. First, if the sugar unit and alkyl tails remain constant, efficiency increases as the number of amino groups is increased (N1 < N2 < N3; e.g., TarN1C10 vs TarN2C10 vs TarN3C10; GalN2C10 vs GalN3C10; GluN2C10 vs GluN3C10, Figures 2 and 3). Second, when evaluating the significance of the sugar alone, the tartarate series was generally more efficient than either the galactarate or glucarate series (e.g., TarN2C10 vs GalN2C10 vs GluN2C10; TarN3C10 vs GalN3C10 vs TarN3C10, Figures 2 and 3), while the galactarate series demonstrated similar or better efficiency when compared to the glucarate series (e.g., GalN2C10 vs GluN2C10; GalN3C10 vs TarN3C10, Figures 2 and 3). Taken together, the results for the importance of sugar indicate that the number of hydroxyl groups is important to delivery efficiency. Third, when comparing polymer-brush materials on the basis of alkyl tail length, in general materials with shorter tails showed better efficiency than those with longer tails (C10 > C12 > C14 > C16,; e.g., TarN1C10 vs TarN1C12 vs TarN1C14 vs TarN1C16; TarN2C10 vs TarN2C12 vs TarN2C14 vs TarN2C16; TarN3C10 vs TarN3C12 vs TarN3C14 vs TarN3C16). In summation, under the current formulation methods, we can arrive at several guidelines for structure–activity relationships: (1) efficiency is increased as then number of amino groups is increased (N3 > N2 > N1); (2) the tartarate sugar improves efficiency relative to the galactarate or glucarate sugar; and (3) shorter alkyl tails improve efficiency (C10 > C12 > C14 > C16). Consistent with these guidelines, TarN3C10 was therefore found to be the best-performing material. TarN3C10 induced EPO expression that resulted in serum EPO concentrations of 1080 ng/mL at a dose of 0.3 mg/kg, which is much higher than C12–200 LLNs, a previously reported lipid-like nanoparticle. This was over 1000-fold higher than expression following delivery of free EPO mRNA.
We next evaluated the biodistribution of the best-performing material by formulating TarN3C10 with luciferase mRNA. In these studies, TarN3C10 nanoparticles were injected intravenously at an mRNA dose of 1 mg/kg. Through luminescence imaging, we measured signal arising from the pancreas, liver, spleen, kidneys, uterus/ovaries, fat, muscle, lungs, heart, and thymus (Figure S2 in Supporting Information). Luciferase expression following mRNA delivery with TarN3C10 nanoparticles was over 1000-fold higher in the liver and spleen compared to the administration of free mRNA, normalized by tissue weight. Additionally, over 10-fold higher luciferase expression was detected in the pancreas, uterus/ovaries, fat, kidneys, muscle, lungs, heart, and thymus for TarN3C10-treated animals relative to the free mRNA control.
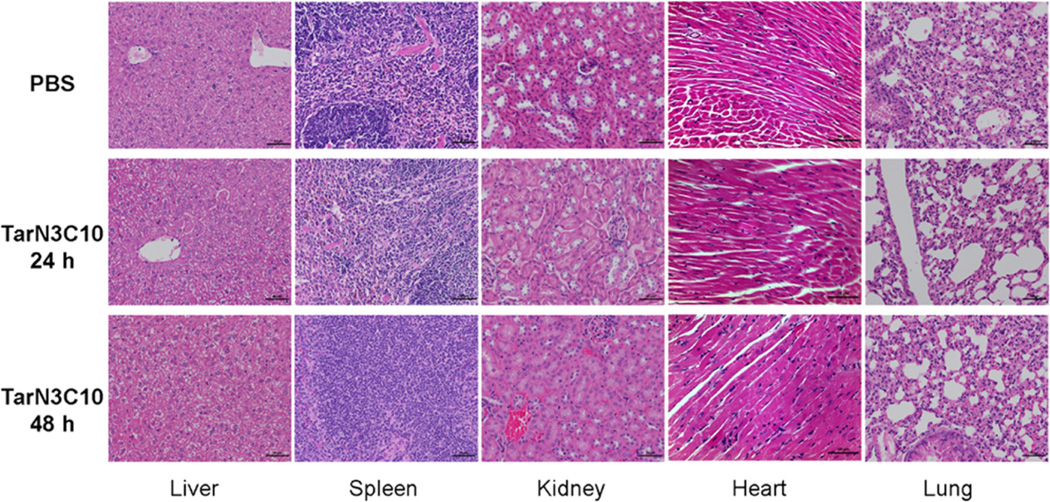
In addition to delivery efficiency, the clinical use of RNA therapeutics requires an acceptable safety profile.36 A preliminary single dose toxicity study of TarN3C10 nanoparticles was conducted, followed by histopathology, a serum cytokine panel, and liver blood chemistry in mice dosed at 1 mg/kg siRNA. Tissue and blood samples were collected at 24 and 48 h after injection. No histological abnormalities were observed in liver, spleen, kidney, heart, or lung at either time point (Figure 4). We also analyzed a panel of 31 different cytokines in serum. We detected a transient increase in expression of G-CSF and MIG at 24 h in the treated groups compared with control groups, and this increase returned to baseline at 48 h (Figure S3 in Supporting Information). Finally, we analyzed serum chemistry for alanine transaminase (ALT), aspartate transaminase (AST), and total bilirubin of TarN3C10-treated animals in comparison to controls (Table S2 in Supporting Information), and no significant differences were observed. Taken together, this preliminary single dose toxicity study supports a conclusion that TarN3C10 nanoparticles are well tolerated in mice at these doses. Detailed toxicity studies are ongoing and will be reported in due course.
Figure 4.
Histology images of liver, spleen, kidney, heart, and lung (scale bar = 50 um). PBS control, TarN3C10-treated group at 24 h, and TarN3C10-treated group at 48 h. No histological abnormalities were observed in the treated group at either time.
We have described the synthesis and evaluation of novel polymer-brush nanoparticles for siRNA and mRNA delivery. A modular design strategy enabled the creation of 31 new polymers with building blocks consisting of amino groups, multiple hydroxyl groups, and alkyl tails. Analyzing structure–activity relationships indicates that all three building blocks contribute to efficient mRNA encapsulation and delivery. The key structural features of top performing formulations revealed that more amino groups are favorable, the tartarate series is in general more potent than the galactarate or glucarate series, and shorter alkyl tails improve performance. These guidelines can be used to inform the design of next-generation RNA delivery systems. Importantly, we found significant correlation for siRNA and mRNA delivery. Of interest, we note that human patients with chemotherapy-related anemia receive EPO at 40,000 units/week, translating to approximately 308 µg.37 Our lead polymeric nanoparticle reported here, TarN3C10, induced EPO expression of 1080 ng/mL with an mRNA dose of 0.3 mg/kg. This would translate to a dose roughly 10-fold higher than that used clinically for EPO. Finally, a single dose toxicity study revealed TarN3C10 nanoparticles are well tolerated based on histopathology, broad panel cytokine screening, and liver blood chemistry profiles. As such, strategies based on the platform of polymer-brush nanoparticles reported here may have promise for use in RNA-based therapy.
Methods
Materials
Cholesterol was purchased from Sigma-Aldrich, DSPC (1,2-distearoyl-sn-glycero-3-phosphocholine) was purchased from Avanti Polar Lipids, and mPEG2000-DMG (1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000] was provided by Alnylam. Slide-A-Lyzer dialysis cassettes were obtained from Pierce Thermo Scientific. The microfluidics devices were made as previously published.31 The RiboGreen was ordered from Invitrogen Life Technologies and used as per the manufacturer guidelines. The EPO and luciferase mRNA used were generously provided by Shire. Serum separator tubes were purchased from BD Biosciences. The EPO ELISA kits were from R&D Systems Inc.
General Procedures for Synthesis of Polymer Brush Materials
Poly(glycoamidoamine) (PGAA) was synthesized according to synthetic methods reported by Reineke.21 Polymer structures were confirmed with 1HNMR. PGAAs undergo ring opening reactions with diverse epoxides to afford the desired polymer brush materials. A mixture of PGAA and epoxides (a ratio of 1.5:1 epoxides/amine) in EtOH was irradiated in a microwave oven at 140 °C for 5 h. The reaction mixture was purified by flash chromatography using a solvent system CH2Cl2/MeOH/NH4OH(aq) 87.5:11:1.5.
Formulation Procedure
Liposomes were formed using a microfluidics devices, as previously described.31 Briefly, the polymer brush, DSPC, cholesterol, and mPEG2000-DMG were dissolved in ethanol and combined in a 5:2:2:1 weight ratio. The siRNA/mRNA was dissolved in a 10 mM citrate buffer, pH = 3.0. The polymer/siRNA–mRNA weight ratio was 5:1 and 10:1, respectively. The ethanol and aqueous solutions were combined in a 1:1 ratio using the microfluidics device and immediately diluted 2-fold in PBS. Formulations were then dialyzed against PBS dialysis cassettes. The relative mRNA entrapment was determined using a RiboGreen fluorescent assay, and the volume mean particle diameter was determined via dynamic light scattering (ZetaPALS, Brookhaven Instruments).
Cryo-Transmission Electron Microscopy (cryo-TEM)
Cryo-TEM samples are prepared in a controlled environment vitrification system (CEVS) or using the commercial environmentally controlled automated Vitrobot (FEI, Netherlands), always at a controlled temperature (25 °C) and at saturation. A 6 µL drop of the suspension is placed on a 200-mesh TEM copper grid covered with a perforated carbon film. To remove excess solution and produce a thin liquid film the drop is blotted, manually in the CEVS and automatically in the Vitrobot. The blotted sample is then plunged into liquid ethane (−183 °C) to form a vitrified specimen and transferred to liquid nitrogen (−196 °C) for storage. Vitrified specimens are examined at temperatures below −175 °C using a Gatan 626 cryo holder either in a Tecnai T12 G2 TEM (FEI, Netherlands) or a Philips CM120 TEM operating at 120 kV. Images are recorded on a Gatan MultiScan 791 camera or Gatan UltraScan 1000 using the DigitalMicrograph software (Gatan, U.K.) in the low-dose imaging mode to minimize beam exposure and electron-beam radiation damage, as described.35,38
Preparation of FFL and Human EPO mRNA
Firefly luciferase (FFL) and human erythropoietin (hEPO) mRNA were synthesized via in vitro transcription from a plasmid DNA template encoding the respective gene. The subsequent transcript was further reacted by the enzymatic addition of a 5′ cap structure (Cap 1) and a 3′ poly(A) tail of approximately 300 nucleotides in length as determined by gel electrophoresis.39 The mRNA was purified using commercially available silica-based spin column technology.
In Vivo Factor VII Silencing
All procedures used in animal studies conducted at MIT and Alnylam were approved by the Institutional Animal Care and Use Committee (IACUC) and were also consistent with local, state, and federal regulations as applicable. C57BL/6 mice were administered intravenously via tail vein injection for siRNA silencing experiments. After 24 h, animals were anaesthetized by isofluorane inhalation for blood sample collection by retro-orbital eye bleed using serum separation tubes. Protein levels of Factor VII were calculated by chromogenic assay (Biophen FVII, Aniara Corporation) with a standard curve obtained from control mice.
In Vivo EPO mRNA Delivery in Mice
All procedures used in animal studies conducted at MIT were approved by the Institutional Animal Care and Use Committee (IACUC) and were also consistent with local, state, and federal regulations as applicable. Formulated mRNA was administered intravenously via tail vein injection using C57BL/6 mice (Charles River Laboratories, 6 to 8 weeks old, 18–22 g) were for mRNA expression experiments. After 6 h, blood was collected from the mice via the tail vein, and serum was obtained using serum separation tubes. EPO levels were measured by an ELISA assay using standard EPO protein.
Biodistribution of TarN3C10-mRNA Nanopartilces in Mice
C57BL/6 mice were administered intravenously via tail vein injection for luciferase mRNA expression experiments. The mice were sacrificed 24 h postinjection; the pancreas, spleen, liver, kidneys, ovaries/uterus, heart, lungs, and thymus as well as a section of the adipose tissue and muscle tissue were then dissected. The tissues were examined with an IVIS imaging system from Caliper. Signal strength of the individual tissue was normalized against tissue weight.
Toxicity Study in Mice
C57Bl/6 mice were administered intravenously TarN3C10 nanoparticles via tail vein injection. Blood and tissue samples were collected 24 and 48 h after injection from the animals. Histopathology on liver, spleen, kidneys, heart, and lungs were processed and evaluated by the histology core facility of Koch Institute. Immunoassays were used to measure the levels of cytokines in a 96-well plate using Bio-Plex Pro assays formatted on magnetic beads. Thirty different cytokines were analyzed: IL-1, IL-2, IL-3, IL-4, IL-5, IL-6, IL-9, IL-10, IL-12 (p40), IL-12 (p70), IL-13, IL-17, Exotaxin, G-CSF, GM-CSF, IFN-γ, KC, MCP-1, MIP-1a, MIP-1b, RANTES, TNG-α, IL-18, FGF-basic, LIF, M-CSF, MIG, MIP-2, PDGF-bb, and VEGF. Clinical chemistry of ALT, AST, and total bilirubin were measured by IDEXX Laboratories.
Supplementary Material
Acknowledgments
The authors thank the Koch Institute Swanson Biotechnology Center for technical support, specifically Animal Imaging and Histology Core. This work was supported by the National Cancer Institute Center of Cancer Nanotechnology Excellence at MIT-Harvard (U54-CA151884), the National Heart, Lung, and Blood Institute, National Institutes of Health (NIH), as a Program of Excellence in Nanotechnology (PEN) Award, Contract #HHSN268201000045C, as well as by Shire, Alnylam Pharmaceuticals, and the NIH Grants R01-EB000244-27, 5-R01-CA132091-04, and R01-DE016516-03. Y.D. acknowledges support from the National Institute of Biomedical Imaging and Bioengineering for his postdoctoral fellowship 1F32EB017625.
Footnotes
ASSOCIATED CONTENT
Supporting Information
- Experimental methods and supplementary figures (PDF)
The authors declare no competing financial interest.
REFERENCES
- 1.Tavernier G, Andries O, Demeester J, Sanders NN, De Smedt SC, Rejman J. J. Controlled Release. 2011;150(3):238–247. doi: 10.1016/j.jconrel.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 2.Pascolo S. Handb. Exp. Pharmacol. 2008;183:221–235. doi: 10.1007/978-3-540-72167-3_11. [DOI] [PubMed] [Google Scholar]
- 3.Sahin U, Kariko K, Tureci O. Nat. Rev. Drug Discovery. 2014;13(10):759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 4.McIvor RS. Mol. Ther. 2011;19(5):822–823. doi: 10.1038/mt.2011.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weissman D, Kariko K. Mol. Ther. 2015;23(9):1416–1417. doi: 10.1038/mt.2015.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whitehead KA, Langer R, Anderson DG. Nat. Rev. Drug Discovery. 2009;8(2):129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burnett JC, Rossi JJ, Tiemann K. Biotechnol. J. 2011;6(9):1130–1146. doi: 10.1002/biot.201100054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Fougerolles A, Vornlocher HP, Maraganore J, Lieberman J. Nat. Rev. Drug Discovery. 2007;6(6):443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kariko K, Muramatsu H, Keller JM, Weissman D. Mol. Ther. 2012;20(5):948–953. doi: 10.1038/mt.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kariko K, Muramatsu H, Welsh FA, Ludwig J, Kato H, Akira S, Weissman D. Mol. Ther. 2008;16(11):1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phua KK, Leong KW, Nair SK. J. Controlled Release. 2013;166(3):227–233. doi: 10.1016/j.jconrel.2012.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zangi L, Lui KO, von Gise A, Ma Q, Ebina W, Ptaszek LM, Spater D, Xu H, Tabebordbar M, Gorbatov R, Sena B, Nahrendorf M, Briscoe DM, Li RA, Wagers AJ, Rossi DJ, Pu WT, Chien KR. Nat. Biotechnol. 2013;31:898–907. doi: 10.1038/nbt.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kormann MS, Hasenpusch G, Aneja MK, Nica G, Flemmer AW, Herber-Jonat S, Huppmann M, Mays LE, Illenyi M, Schams A, Griese M, Bittmann I, Handgretinger R, Hartl D, Rosenecker J, Rudolph C. Nat. Biotechnol. 2011;29(2):154–157. doi: 10.1038/nbt.1733. [DOI] [PubMed] [Google Scholar]
- 14.Kariko K, Ni HP, Capodici J, Lamphier M, Weissman D. J. Biol. Chem. 2004;279(13):12542–12550. doi: 10.1074/jbc.M310175200. [DOI] [PubMed] [Google Scholar]
- 15.Kariko K, Buckstein M, Ni H, Weissman D. Immunity. 2005;23(2):165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 16.Sahay G, Querbes W, Alabi C, Eltoukhy A, Sarkar S, Zurenko C, Karagiannis E, Love K, Chen D, Zoncu R, Buganim Y, Schroeder A, Langer R, Anderson DG. Nat. Biotechnol. 2013;31(7):653–658. doi: 10.1038/nbt.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weide B, Carralot JP, Reese A, Scheel B, Eigentler TK, Hoerr I, Rammensee HG, Garbe C, Pascolo S. J. Immunother. 2008;31(2):180–188. doi: 10.1097/CJI.0b013e31815ce501. [DOI] [PubMed] [Google Scholar]
- 18.Akinc A, Zumbuehl A, Goldberg M, Leshchiner ES, Busini V, Hossain N, Bacallado SA, Nguyen DN, Fuller J, Alvarez R, Borodovsky A, Borland T, Constien R, de Fougerolles A, Dorkin JR, Narayanannair Jayaprakash K, Jayaraman M, John M, Koteliansky V, Manoharan M, Nechev L, Qin J, Racie T, Raitcheva D, Rajeev KG, Sah DW, Soutschek J, Toudjarska I, Vornlocher HP, Zimmermann TS, Langer R, Anderson DG. Nat. Biotechnol. 2008;26(5):561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Semple SC, Akinc A, Chen J, Sandhu AP, Mui BL, Cho CK, Sah DW, Stebbing D, Crosley EJ, Yaworski E, Hafez IM, Dorkin JR, Qin J, Lam K, Rajeev KG, Wong KF, Jeffs LB, Nechev L, Eisenhardt ML, Jayaraman M, Kazem M, Maier MA, Srinivasulu M, Weinstein MJ, Chen Q, Alvarez R, Barros SA, De S, Klimuk SK, Borland T, Kosovrasti V, Cantley WL, Tam YK, Manoharan M, Ciufolini MA, Tracy MA, de Fougerolles A, MacLachlan I, Cullis PR, Madden TD, Hope MJ. Nat. Biotechnol. 2010;28(2):172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 20.Cheng C, Convertine AJ, Stayton PS, Bryers JD. Biomaterials. 2012;33(28):6868–6876. doi: 10.1016/j.biomaterials.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y, Wenning L, Lynch M, Reineke TM. J. Am. Chem. Soc. 2004;126(24):7422–7423. doi: 10.1021/ja049831l. [DOI] [PubMed] [Google Scholar]
- 22.Liu Y, Reineke TM. Biomacromolecules. 2010;11(2):316–325. doi: 10.1021/bm9008233. [DOI] [PubMed] [Google Scholar]
- 23.McLendon PM, Fichter KM, Reineke TM. Mol. Pharmaceutics. 2010;7(3):738–750. doi: 10.1021/mp900282e. [DOI] [PubMed] [Google Scholar]
- 24.Liu Y, Wenning L, Lynch M, Reineke TM. Polymeric Drug Delivery I. ACS Symp. Ser. 2006;923:217–227. [Google Scholar]
- 25.Ingle NP, Malone B, Reineke TM. Trends Biotechnol. 2011;29(9):443–453. doi: 10.1016/j.tibtech.2011.04.012. [DOI] [PubMed] [Google Scholar]
- 26.Tranter M, Liu Y, He S, Gulick J, Ren X, Robbins J, Jones WK, Reineke TM. Mol. Ther. 2012;20(3):601–608. doi: 10.1038/mt.2011.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Love Kevin T, Mahon Kerry P, Levins Christopher G, Whitehead Kathryn A, Querbes W, Dorkin JR, Qin J, Cantley W, Qin Liu L, Racie T, Frank-Kamenetsky M, Yip KN, Alvarez R, Sah Dinah WY, de Fougerolles A, Fitzgerald K, Koteliansky V, Akinc A, Langer R, Anderson Daniel G. Proc. Natl. Acad. Sci. U. S. A. 2010;107(5):1864–1869. doi: 10.1073/pnas.0910603106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siegwart DJ, Whitehead KA, Nuhn L, Sahay G, Cheng H, Jiang S, Ma M, Lytton-Jean A, Vegas A, Fenton P, Levins CG, Love KT, Lee H, Cortez C, Collins SP, Li YF, Jang J, Querbes W, Zurenko C, Novobrantseva T, Langer R, Anderson DG. Proc. Natl. Acad. Sci. U. S. A. 2011;108(32):12996–13001. doi: 10.1073/pnas.1106379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong Y, Love KT, Dorkin JR, Sirirungruang S, Zhang Y, Chen D, Bogorad RL, Yin H, Chen Y, Vegas AJ, Alabi CA, Sahay G, Olejnik KT, Wang W, Schroeder A, Lytton-Jean AK, Siegwart DJ, Akinc A, Barnes C, Barros SA, Carioto M, Fitzgerald K, Hettinger J, Kumar V, Novobrantseva TI, Qin J, Querbes W, Koteliansky V, Langer R, Anderson DG. Proc. Natl. Acad. Sci. U. S. A. 2014;111(11):3955–3960. doi: 10.1073/pnas.1322937111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, Pelet JM, Heller DA, Dong Y, Chen D, Gu Z, Joseph BJ, Wallas J, Anderson DG. Adv. Mater. 2013;25(33):4641–4645. doi: 10.1002/adma.201301917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen D, Love KT, Chen Y, Eltoukhy AA, Kastrup C, Sahay G, Jeon A, Dong Y, Whitehead KA, Anderson DG. J. Am. Chem. Soc. 2012;134(16):6948–6951. doi: 10.1021/ja301621z. [DOI] [PubMed] [Google Scholar]
- 32.Coleman T, Brines M. Crit Care. 2004;8(5):337–341. doi: 10.1186/cc2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dahlman JE, Barnes C, Khan OF, Thiriot A, Jhunjunwala S, Shaw TE, Xing Y, Sager HB, Sahay G, Speciner L, Bader A, Bogorad RL, Yin H, Racie T, Dong Y, Jiang S, Seedorf D, Dave A, Singh Sandhu K, Webber MJ, Novobrantseva T, Ruda VM, Lytton-Jean AK, Levins CG, Kalish B, Mudge DK, Perez M, Abezgauz L, Dutta P, Smith L, Charisse K, Kieran MW, Fitzgerald K, Nahrendorf M, Danino D, Tuder RM, von Andrian UH, Akinc A, Panigrahy D, Schroeder A, Koteliansky V, Langer R, Anderson DG. Nat. Nanotechnol. 2014;9(8):648–655. doi: 10.1038/nnano.2014.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michel R, Kesselman E, Plostica T, Danino D, Gradzielski M. Angew. Chem., Int. Ed. 2014;53:12441–12445. doi: 10.1002/anie.201406927. [DOI] [PubMed] [Google Scholar]
- 35.Danino D. Curr. Opin. Colloid Interface Sci. 2012;17(6):316–329. doi: 10.1016/j.cocis.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barros SA, Gollob JA. Adv. Drug Delivery Rev. 2012;64(15):1730–1737. doi: 10.1016/j.addr.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 37.Jelkmann W. Nephrol., Dial., Transplant. 2009;24(5):1366–1368. doi: 10.1093/ndt/gfp058. [DOI] [PubMed] [Google Scholar]
- 38.Danino D, Bernheim-Groswasser A, Talmon Y. Colloids Surf., A. 2001;183:113–122. [Google Scholar]
- 39.Fechter P, Brownlee GG. J. Gen. Virol. 2005;86:1239–1249. doi: 10.1099/vir.0.80755-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.