Abstract
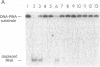
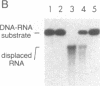
The RAD3 gene of Saccharomyces cerevisiae is required for excision repair of UV-damaged DNA and is essential for cell viability. The RAD3 protein exhibits a remarkable degree of sequence homology to the human excision repair protein ERCC2. The RAD3 protein is a single-stranded DNA-dependent ATPase and a DNA helicase capable of denaturing long regions of duplex DNA. Here, we demonstrate that RAD3 also possesses a potent DNA.RNA helicase activity similar in efficiency to its DNA helicase activity. The rad3 Arg-48 mutant protein, which binds but does not hydrolyze ATP, lacks the DNA.RNA unwinding activity, indicating a dependence on ATP hydrolysis. RAD3 does not show any RNA-dependent NTPase activity and, as expected, does not unwind duplex RNA. This observation suggests that RAD3 translocates on DNA in unwinding DNA.RNA duplexes. That the rad3 Arg-48 mutation inactivates the DNA and DNA.RNA helicase activities and confers a substantial reduction in the incision of UV-damaged DNA suggests a role for these activities in incision. We discuss how RAD3 helicase activities could function in tracking of DNA in search of damage sites and effect enhanced excision repair of actively transcribed genes.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bedinger P., Hochstrasser M., Jongeneel C. V., Alberts B. M. Properties of the T4 bacteriophage DNA replication apparatus: the T4 dda DNA helicase is required to pass a bound RNA polymerase molecule. Cell. 1983 Aug;34(1):115–123. doi: 10.1016/0092-8674(83)90141-1. [DOI] [PubMed] [Google Scholar]
- Bohr V. A., Smith C. A., Okumoto D. S., Hanawalt P. C. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985 Feb;40(2):359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- Bootsma D., Keijzer W., Jung E. G., Bohnert E. Xeroderma pigmentosum complementation group XP-I withdrawn. Mutat Res. 1989 Sep;218(2):149–151. doi: 10.1016/0921-8777(89)90021-9. [DOI] [PubMed] [Google Scholar]
- Caron P. R., Kushner S. R., Grossman L. Involvement of helicase II (uvrD gene product) and DNA polymerase I in excision mediated by the uvrABC protein complex. Proc Natl Acad Sci U S A. 1985 Aug;82(15):4925–4929. doi: 10.1073/pnas.82.15.4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins D. R., Prakash S., Reynolds P., Polakowska R., Weber S., Prakash L. Isolation and characterization of the RAD3 gene of Saccharomyces cerevisiae and inviability of rad3 deletion mutants. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5680–5684. doi: 10.1073/pnas.80.18.5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain I., Van Houten B., Thomas D. C., Abdel-Monem M., Sancar A. Effect of DNA polymerase I and DNA helicase II on the turnover rate of UvrABC excision nuclease. Proc Natl Acad Sci U S A. 1985 Oct;82(20):6774–6778. doi: 10.1073/pnas.82.20.6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. T., Elliott G. C., Squires S., Joysey V. C. Lack of complementation between xeroderma pigmentosum complementation groups D and H. Hum Genet. 1989 Feb;81(3):203–210. doi: 10.1007/BF00278989. [DOI] [PubMed] [Google Scholar]
- Koo H. S., Claassen L., Grossman L., Liu L. F. ATP-dependent partitioning of the DNA template into supercoiled domains by Escherichia coli UvrAB. Proc Natl Acad Sci U S A. 1991 Feb 15;88(4):1212–1216. doi: 10.1073/pnas.88.4.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn B., Abdel-Monem M., Krell H., Hoffmann-Berling H. Evidence for two mechanisms for DNA unwinding catalyzed by DNA helicases. J Biol Chem. 1979 Nov 25;254(22):11343–11350. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Madhani H. D., Bohr V. A., Hanawalt P. C. Differential DNA repair in transcriptionally active and inactive proto-oncogenes: c-abl and c-mos. Cell. 1986 May 9;45(3):417–423. doi: 10.1016/0092-8674(86)90327-2. [DOI] [PubMed] [Google Scholar]
- Matson S. W. Escherichia coli DNA helicase II (uvrD gene product) catalyzes the unwinding of DNA.RNA hybrids in vitro. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4430–4434. doi: 10.1073/pnas.86.12.4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson S. W., Kaiser-Rogers K. A. DNA helicases. Annu Rev Biochem. 1990;59:289–329. doi: 10.1146/annurev.bi.59.070190.001445. [DOI] [PubMed] [Google Scholar]
- Mellon I., Hanawalt P. C. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature. 1989 Nov 2;342(6245):95–98. doi: 10.1038/342095a0. [DOI] [PubMed] [Google Scholar]
- Mellon I., Spivak G., Hanawalt P. C. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987 Oct 23;51(2):241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- Miller R. D., Prakash L., Prakash S. Genetic control of excision of Saccharomyces cerevisiae interstrand DNA cross-links induced by psoralen plus near-UV light. Mol Cell Biol. 1982 Aug;2(8):939–948. doi: 10.1128/mcb.2.8.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumovski L., Friedberg E. C. A DNA repair gene required for the incision of damaged DNA is essential for viability in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4818–4821. doi: 10.1073/pnas.80.15.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E. Y., Grossman L. Characterization of the helicase activity of the Escherichia coli UvrAB protein complex. J Biol Chem. 1989 Jan 15;264(2):1336–1343. [PubMed] [Google Scholar]
- Oh E. Y., Grossman L. Helicase properties of the Escherichia coli UvrAB protein complex. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3638–3642. doi: 10.1073/pnas.84.11.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds P., Higgins D. R., Prakash L., Prakash S. The nucleotide sequence of the RAD3 gene of Saccharomyces cerevisiae: a potential adenine nucleotide binding amino acid sequence and a nonessential acidic carboxyl terminal region. Nucleic Acids Res. 1985 Apr 11;13(7):2357–2372. doi: 10.1093/nar/13.7.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A., Rupp W. D. A novel repair enzyme: UVRABC excision nuclease of Escherichia coli cuts a DNA strand on both sides of the damaged region. Cell. 1983 May;33(1):249–260. doi: 10.1016/0092-8674(83)90354-9. [DOI] [PubMed] [Google Scholar]
- Sauerbier W., Hercules K. Gene and transcription unit mapping by radiation effects. Annu Rev Genet. 1978;12:329–363. doi: 10.1146/annurev.ge.12.120178.001553. [DOI] [PubMed] [Google Scholar]
- Scheffner M., Knippers R., Stahl H. RNA unwinding activity of SV40 large T antigen. Cell. 1989 Jun 16;57(6):955–963. doi: 10.1016/0092-8674(89)90334-6. [DOI] [PubMed] [Google Scholar]
- Selby C. P., Sancar A. Transcription preferentially inhibits nucleotide excision repair of the template DNA strand in vitro. J Biol Chem. 1990 Dec 5;265(34):21330–21336. [PubMed] [Google Scholar]
- Stahl H., Dröge P., Knippers R. DNA helicase activity of SV40 large tumor antigen. EMBO J. 1986 Aug;5(8):1939–1944. doi: 10.1002/j.1460-2075.1986.tb04447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P., Higgins D., Prakash L., Prakash S. Mutation of lysine-48 to arginine in the yeast RAD3 protein abolishes its ATPase and DNA helicase activities but not the ability to bind ATP. EMBO J. 1988 Oct;7(10):3263–3269. doi: 10.1002/j.1460-2075.1988.tb03193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P., Prakash L., Matson S. W., Prakash S. RAD3 protein of Saccharomyces cerevisiae is a DNA helicase. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8951–8955. doi: 10.1073/pnas.84.24.8951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P., Prakash L., Weber S., Prakash S. The RAD3 gene of Saccharomyces cerevisiae encodes a DNA-dependent ATPase. Proc Natl Acad Sci U S A. 1987 Sep;84(17):6045–6049. doi: 10.1073/pnas.84.17.6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terleth C., van Sluis C. A., van de Putte P. Differential repair of UV damage in Saccharomyces cerevisiae. Nucleic Acids Res. 1989 Jun 26;17(12):4433–4439. doi: 10.1093/nar/17.12.4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson L. H., Brookman K. W., Dillehay L. E., Mooney C. L., Carrano A. V. Hypersensitivity to mutation and sister-chromatid-exchange induction in CHO cell mutants defective in incising DNA containing UV lesions. Somatic Cell Genet. 1982 Nov;8(6):759–773. doi: 10.1007/BF01543017. [DOI] [PubMed] [Google Scholar]
- Thompson L. H., Shiomi T., Salazar E. P., Stewart S. A. An eighth complementation group of rodent cells hypersensitive to ultraviolet radiation. Somat Cell Mol Genet. 1988 Nov;14(6):605–612. doi: 10.1007/BF01535314. [DOI] [PubMed] [Google Scholar]
- Weber C. A., Salazar E. P., Stewart S. A., Thompson L. H. ERCC2: cDNA cloning and molecular characterization of a human nucleotide excision repair gene with high homology to yeast RAD3. EMBO J. 1990 May;9(5):1437–1447. doi: 10.1002/j.1460-2075.1990.tb08260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeda G., van Ham R. C., Vermeulen W., Bootsma D., van der Eb A. J., Hoeijmakers J. H. A presumed DNA helicase encoded by ERCC-3 is involved in the human repair disorders xeroderma pigmentosum and Cockayne's syndrome. Cell. 1990 Aug 24;62(4):777–791. doi: 10.1016/0092-8674(90)90122-u. [DOI] [PubMed] [Google Scholar]
- Wilcox D. R., Prakash L. Incision and postincision steps of pyrimidine dimer removal in excision-defective mutants of Saccharomyces cerevisiae. J Bacteriol. 1981 Nov;148(2):618–623. doi: 10.1128/jb.148.2.618-623.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung A. T., Mattes W. B., Oh E. Y., Grossman L. Enzymatic properties of purified Escherichia coli uvrABC proteins. Proc Natl Acad Sci U S A. 1983 Oct;80(20):6157–6161. doi: 10.1073/pnas.80.20.6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Duin M., de Wit J., Odijk H., Westerveld A., Yasui A., Koken M. H., Hoeijmakers J. H., Bootsma D. Molecular characterization of the human excision repair gene ERCC-1: cDNA cloning and amino acid homology with the yeast DNA repair gene RAD10. Cell. 1986 Mar 28;44(6):913–923. doi: 10.1016/0092-8674(86)90014-0. [DOI] [PubMed] [Google Scholar]