Summary
DNA double-strand breaks (DSBs) are highly cytotoxic DNA lesions, whose accurate repair by non-homologous end-joining (NHEJ) or homologous recombination (HR) is crucial for genome integrity and is strongly influenced by the local chromatin environment. Here, we identify SCAI (Suppressor of Cancer Cell Invasion) as a 53BP1-interacting chromatin-associated protein that promotes the functionality of several DSB repair pathways in mammalian cells. SCAI undergoes prominent enrichment at DSB sites through dual mechanisms involving 53BP1-dependent recruitment to DSB-surrounding chromatin and 53BP1-independent accumulation at resected DSBs. Cells lacking SCAI display reduced DSB repair capacity, hypersensitivity to DSB-inflicting agents and genome instability. We demonstrate that SCAI is a mediator of 53BP1-dependent repair of heterochromatin-associated DSBs, facilitating ATM kinase signaling at DSBs in repressive chromatin environments. Moreover, we establish an important role of SCAI in meiotic recombination, as SCAI deficiency in mice leads to germ cell loss and subfertility associated with impaired retention of the DMC1 recombinase on meiotic chromosomes. Collectively, our findings uncover SCAI as a physiologically important component of both NHEJ- and HR-mediated pathways that potentiates DSB repair efficiency in specific chromatin contexts.
In response to genotoxic insults such as DNA double-strand breaks (DSBs), eukaryotic cells mount a coordinated DNA damage response (DDR) that activates DNA repair pathways to mitigate the deleterious consequences of DNA lesions1, 2. DSBs can be repaired by non-homologous end-joining (NHEJ) or homologous recombination (HR)3. Dysfunctions in DSB repair pathways cause severe hereditary disorders with symptoms including cancer predisposition, neurodegeneration, subfertility, and immunodeficiency4.
The state and organization of chromatin, the natural environment of cellular DNA, fundamentally influences DSB repair efficiency and pathway choice, and major compositional and structural changes are imposed onto chromatin during the course of DSB formation and repair5, 6. DNA damage-induced modifications of chromatin-associated proteins near the lesions serve as direct recognition marks for numerous factors involved in DSB repair, enabling their local accumulation at the damage sites3, 5. The ATM kinase is a master organizer of this response, phosphorylating numerous substrates including the histone H2A variant H2AX, whose phosphorylation product (referred to as γ-H2AX) triggers events that lead to recruitment of the E3 ubiquitin ligases RNF8 and RNF168. Ubiquitin-dependent modification of histones at DSB sites by these ligases then promotes accumulation of critical DSB repair factors such as BRCA1 and 53BP1 to the DSB-surrounding chromatin areas7. However, the structure of chromatin can present a substantial barrier to efficient DSB repair. A spectrum of chromatin states exists, ranging from open, transcriptionally active euchromatin to highly compacted, transcriptionally inert heterochromatin. The latter state interferes with the accessibility of repair factors to DNA lesions, and heterochromatin-associated DSBs are generally repaired with slower kinetics than euchromatic breaks6. Cells therefore possess multiple factors that remodel chromatin structure to enhance the targeting of DNA repair factors to lesions in heterochromatic regions6.
The chromatin-associated protein 53BP1 is a key mediator of DSB repair in mammalian cells. 53BP1 is an HR-inhibitory factor that mediates end-joining of unprotected telomeres and other toxic DNA repair reactions8. 53BP1 is also crucial for long-range end-joining during V(D)J recombination and immunoglobulin heavy-chain (IgH) class-switch recombination (CSR) in developing lymphocytes; consequently 53BP1−/− B cells are severely impaired for CSR 9, 10. These functions of 53BP1 are, to a large extent, mediated by the 53BP1-binding factors RIF1 and PTIP11–15. Finally, 53BP1 has an established, but less well understood, role in promoting ATM-dependent repair of DSBs in heterochromatin. This impinges on 53BP1-mediated, localized phosphorylation of the transcriptional co-repressor KAP1 at S824 in heterochromatin by ATM, triggering the release of the chromatin remodeler CHD3.1 to enable chromatin relaxation and efficient lesion repair16–18. While dedicated effectors of 53BP1-dependent repair of heterochromatin-associated DSBs have remained unknown, we identified here the poorly characterized protein SCAI (Suppressor of Cancer Cell Invasion) as a mediator of this 53BP1 function.
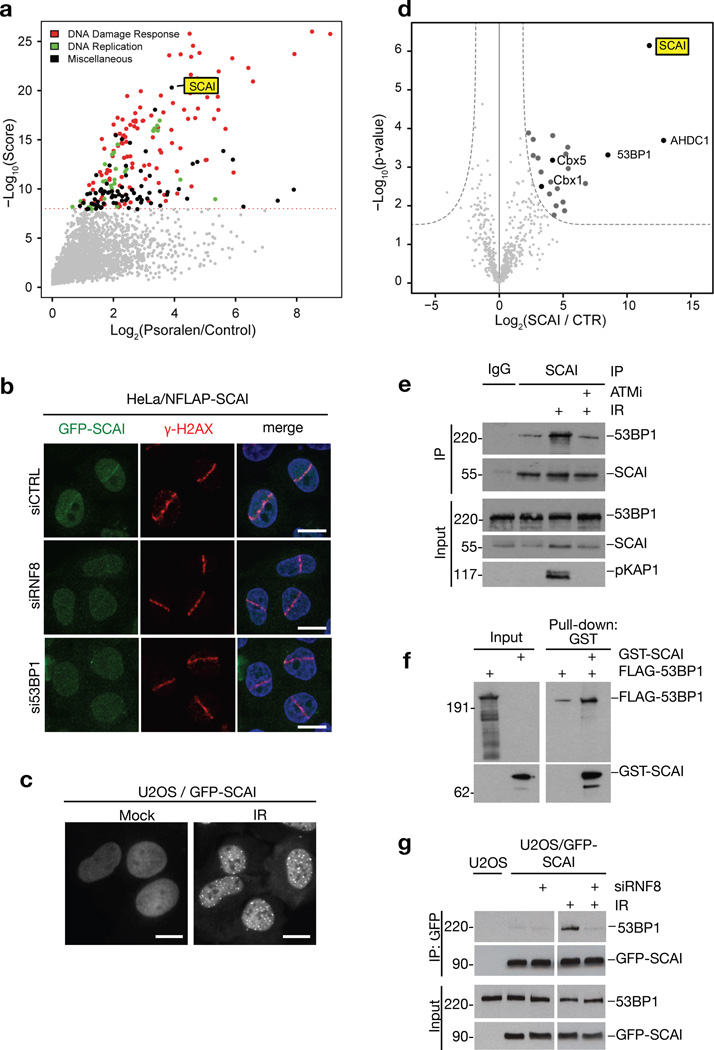
Using the CHROMASS method for systems-wide profiling of protein recruitment to chromatin templates incubated in Xenopus egg extracts that we recently described19, we observed prominent enrichment of SCAI at DNA damage-containing chromatin along with multiple known DDR components (Figure 1a;S1a,b). SCAI is highly conserved among vertebrates and has been implicated in transcriptional regulation20, 21, but has no annotated domains and shares little sequence homology with other proteins. Using cells expressing GFP-tagged human SCAI at near-physiological levels, we found that SCAI is recruited to microlaser- and ionizing radiation (IR)-generated DSB sites (Figure 1b,c), suggesting it is involved in DSB repair processes. To gain insight into this function, we used quantitative mass spectrometry to identify SCAI-interacting proteins, revealing 53BP1 as well as heterochromatin-associated factors (including the HP1 proteins HP1β (Cbx1) and HP1α (Cbx5)) among prominently enriched, prospective SCAI-binding proteins (Figure 1d). Consistently, biochemical fractionation experiments showed that SCAI is predominantly associated with chromatin (Figure S1c). In co-immunoprecipitation assays, SCAI interacted with 53BP1 in an IR- and ATM-stimulated manner, and purified SCAI and 53BP1 interacted in vitro (Figure 1e,f;S1d), suggesting their interaction is direct and functionally relevant in the context of DSB repair. Knockdown of 53BP1 or its upstream recruitment factor RNF87 strongly attenuated SCAI accumulation at microlaser-generated DSBs, but not vice versa (Figure 1b;S1e–i), suggesting that SCAI is recruited to DSB-surrounding chromatin via direct binding to 53BP1, downstream of RNF8/RNF168-mediated histone ubiquitylation. Like ATM inhibition, RNF8 depletion suppressed IR-induced SCAI-53BP1 interaction (Figure 1g), suggesting that the SCAI-53BP1 complex is stabilized once recruited to DSBs.
Figure 1. SCAI is recruited to DSB-surrounding chromatin via interaction with 53BP1.
a. Analysis of protein recruitment to psoralen crosslinked chromatin (PSO) compared to undamaged control. Chromatin templates were replicated in repair-proficient Xenopus egg extracts. After chromatin re-isolation, associated proteins were analyzed by mass spectrometry. Maximal protein intensity is plotted against an overall score determined from several independent experiments. The dashed line indicates the significance threshold (q-value < 0.01). Data was replotted from19. b. HeLa cells stably expressing GFP-tagged human SCAI at endogenous levels from a BAC (NFLAP-SCAI) were transfected with control (CTRL), RNF8 or 53BP1 siRNAs. Cells were subsequently subjected to laser micro-irradiation, fixed 1 h later, immunostained with γ-H2AX antibody and counter-stained with DAPI. c. U2OS cells stably expressing GFP-SCAI were exposed to ionizing radiation (IR, 5 Gy) and fixed 4 h later. d. GFP-SCAI was affinity-purified on GFP-Trap beads from HeLa/NFLAP-SCAI cells, and co-purifying proteins were analyzed by QUBIC mass spectrometry. Intensities and p-values for interacting proteins are shown in a volcano plot. e. Chromatin-enriched fractions of U2OS cells exposed to IR and/or ATM inhibitor (ATMi) were subjected to SCAI immunoprecipitation (IP) followed by immunoblotting with antibodies against 53BP1, SCAI and phospho-KAP1. f. Interaction between recombinant full-length FLAG-tagged 53BP1 and GST-tagged SCAI was analyzed by GST pulldown followed by immunoblotting with antibodies against FLAG and GST. g. U2OS cells stably expressing GFP-SCAI were transfected with control or RNF8 siRNA and exposed to IR where indicated. Chromatin-enriched fractions were subjected to GFP immunoprecipitation followed by immunoblotting with antibodies against 53BP1 and GFP. All scale bars, 10 µm. Uncropped blots (e,f,g) are shown in Figure S6.
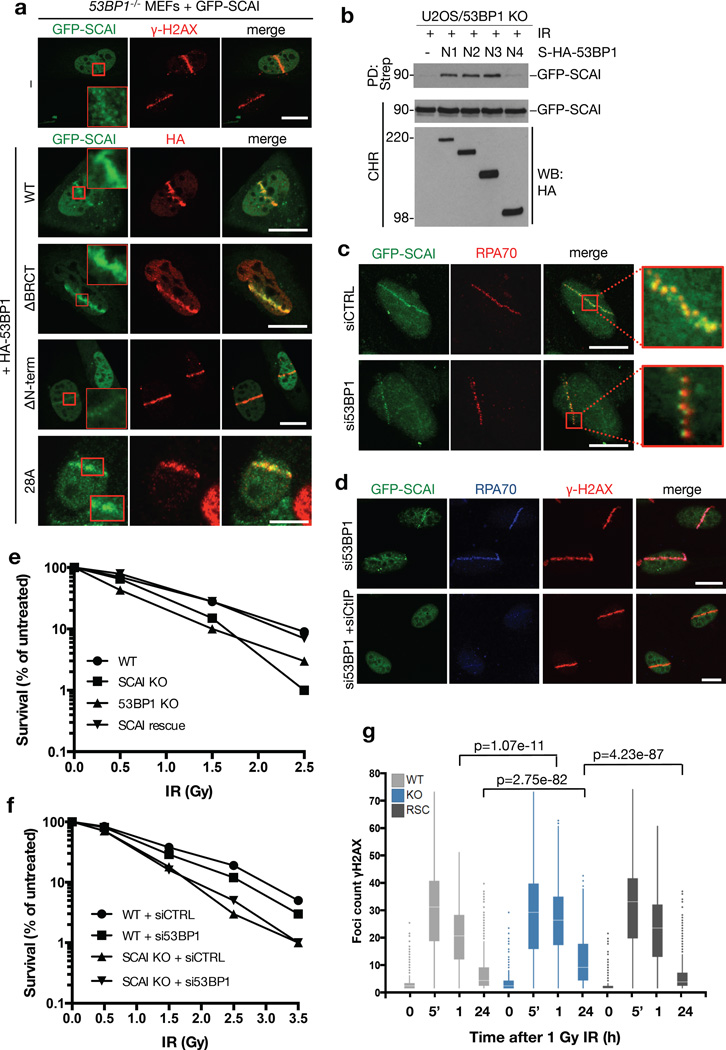
Reconstitution of 53BP1−/− MEFs with different 53BP1 constructs showed that its N-terminus, which undergoes multi-site phosphorylation by ATM to provide binding sites for RIF1 and PTIP11–15, was required for SCAI recruitment to DSB sites (Figure 2a;S2a). Within this 53BP1 region we mapped the SCAI-binding site to amino acids 900-1230, which form part of its ATM phosphorylation domain (Figure 2b). However, unlike RIF1 and PTIP, SCAI was recruited to damaged DNA independently of ATM-dependent 53BP1 phosphorylation, as expression of a 53BP1 28A mutant refractory to phosphorylation by ATM22 in 53BP1−/− cells restored SCAI recruitment to DSBs as efficiently as wild-type (WT) 53BP1 (Figure 2a;S2a). Also, downstream effectors of 53BP1, such as RIF1, accumulated at DSB sites independently of SCAI (Figure S2b). Interestingly, while 53BP1 depletion markedly impaired SCAI retention at DSB sites, we noted that a subset of cells displayed residual SCAI recruitment to punctate foci along microlaser-generated DNA damage tracks, which colocalized with RPA (Figure 2a,c;S2c). Based on our previous findings on compartmentalization of nuclear areas flanking DSBs3, 23, we surmised that the 53BP1-independent SCAI microfoci might colocalize with RPA-coated single-stranded DNA (ssDNA) regions generated by DSB end resection (Figure S2c). Indeed, these SCAI microfoci in 53BP1 knockdown cells were eliminated upon co-depletion of the key resection factor CtIP, but not by downstream HR factors including BRCA1 and BRCA2 (Figure 2d;S2d). The mechanism underlying SCAI recruitment to resected DSBs may involve its direct binding to ssDNA stretches, as SCAI interacted with ssDNA oligos but not RPA (Figure 1d;Figure S2e). We conclude from these findings that SCAI undergoes enrichment at both the chromatin and ssDNA regions surrounding DSBs, an unusual recruitment pattern observed so far only for BRCA1 and the MRE11-NBS1-RAD50 (MRN) complex23, key factors in HR.
Figure 2. SCAI is required for optimal DSB repair.
a. 53BP1−/− MEFs were co-transfected with GFP-SCAI and indicated WT or deletion constructs of HA-tagged 53BP1 (Figure S2a), subjected to laser micro-irradiation, fixed 1 h later, and immunostained with γ-H2AX or HA antibody. Inserts show larger magnifications of the highlighted regions. b. U2OS cells with targeted knockout of 53BP1 were co-transfected with GFP-SCAI construct and plasmids encoding indicated Strep-HA-tagged fragments of 53BP1. To analyze SCAI-53BP1 interaction, chromatin-enriched fractions were subjected to Strep pull-down followed by immunoblotting with GFP antibody. N1, 53BP1 residues 250-1972; N2, 600-1972; N3, 900-1972; N4, 1230-1972. c. U2OS cells stably expressing GFP-SCAI were transfected with control (CTRL) or siRNA targeting 53BP1 and treated as in (a). 1 h later, cells were pre-extracted and immunostained with RPA antibody. Inserts show larger magnifications of the highlighted regions. d. Cells from (c) transfected with indicated siRNAs were processed as in (c) and co-immunostained with RPA70 and γ-H2AX antibodies. All scale bars, 10 µm. e. U2OS WT, 53BP1 KO and SCAI KO cells (Figure S3b, c) were, exposed to increasing doses of ionizing radiation (IR) and plated for clonogenic survival assays. After 14 days, colonies were fixed, stained and counted. Data points indicate the mean from three observations. f. Indicated U2OS cell lines (Figure S3b) were transfected with control (CTRL) or 53BP1 siRNAs and were treated and analyzed as in (e). g. U2OS cells and derivative cell lines in (e) were fixed at indicated times after exposure to IR (1 Gy) and stained with γ-H2AX antibody. The number of foci per cell was measured by high content microscopy. Centre indicates the median and whiskers the borders of the 95% quantiles. 1000 cells (n=1000 independent measurements) were measured per condition and p-values were calculated from a non-parametric two-tailed Mann-Whitney U test.
To understand how SCAI functions in DSB repair, we employed CRISPR/Cas9 technology to generate human cell lines with targeted SCAI knockout (KO). While deletion of SCAI did not significantly impact cell cycle distribution, SCAI KO cells showed reduced cell survival following exposure to IR (Figure 2e,f;S3a,b), consistent with a role for SCAI in promoting DSB repair. Reconstitution of SCAI KO cells with full-length ectopic SCAI at near-physiological levels fully rescued this defect (Figure 2e;S3b), demonstrating that it was a specific consequence of SCAI ablation. Notably, while 53BP1 loss also sensitized cells to IR as expected, we observed no additive effect of co-depleting SCAI and 53BP1 (Figure 2f;S3c), suggesting that they operate in a common DSB repair pathway. Using quantitative image analysis to monitor DSB repair kinetics through enumeration of γ-H2AX and 53BP1 foci, we observed a significant increase in persistent γ-H2AX and 53BP1 foci in SCAI KO cells, which was restored to WT levels by reintroduction of ectopic SCAI (Figure 2g;S3b,d). Using established reporter assays for NHEJ- and HR-mediated repair of DSBs24, we found that SCAI deficiency in human cells led to a pronounced reduction in NHEJ efficiency, while overall HR activity as measured by this system, as well as RAD51 foci formation in response to IR, were not significantly impaired (Figure S3e–i). However, as described below, we obtained evidence that SCAI is important for HR in specific chromosomal contexts.
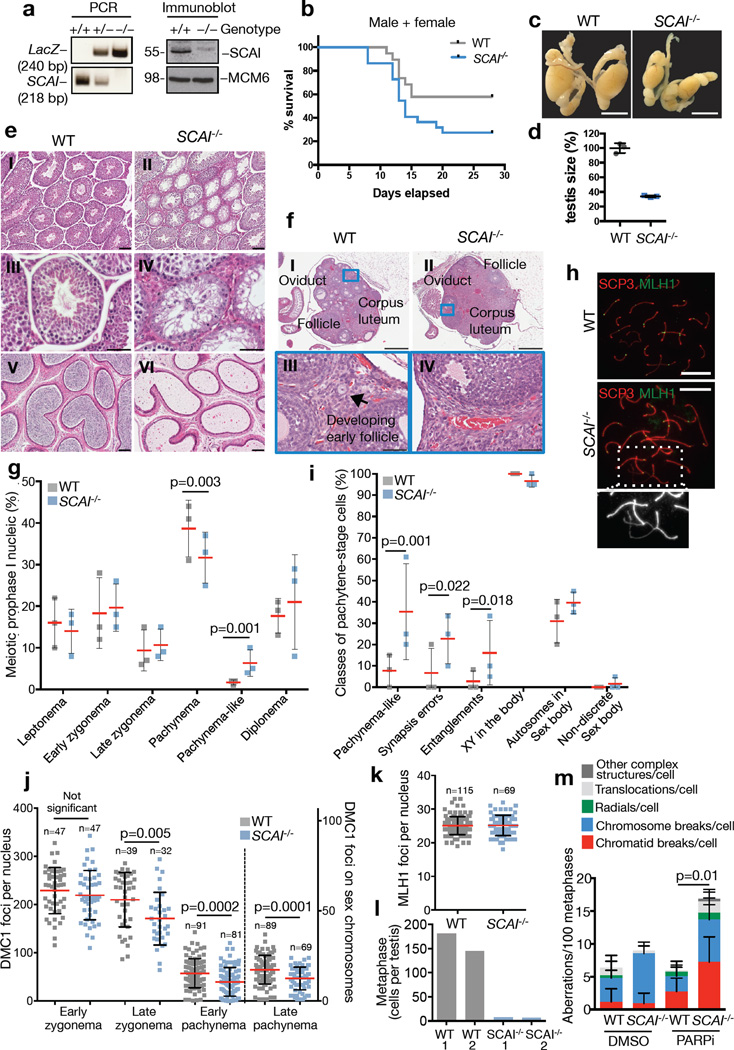
To characterize the physiological consequences of SCAI loss, we generated SCAI knockout mice and verified complete loss of SCAI protein expression in MEFs fromSCAI−/− animals (Figure 3a). SCAI−/− mice were born at the expected Mendelian frequency (Table S1) and showed no overt developmental or survival defects, demonstrating that SCAI is not an essential gene. Moreover, SCAI−/− primary MEFs proliferated similarly to WT littermate controls (Figure S4a). To test whether loss of SCAI compromises DSB repair capacity in murine cells, we exposed G2 phase WT and SCAI−/− MEFs to low doses of IR and monitored 53BP1 foci clearance over time. Similar to human SCAI KO cells, we observed a pronounced persistence of 53BP1 foci at late time points across independent SCAI−/−primary MEF lines compared to WT lines (Figure S4b,c). Upon exposure of WT and SCAI−/− mice to whole-body IR, we found that SCAI−/− animals died more quickly, ultimately showing an approx. 2-fold survival decrease in both males and females, compared to controls (Figure 3b;S4d,e). Together, these data suggest that SCAI has a physiologically important role in promoting DSB repair efficiency and survival after DNA damage in mammals.
Figure 3. SCAI deficiency leads to meiotic recombination defects, germ cell loss and subfertility in mice.
a. Confirmation of SCAI gene disruption by PCR on mouse tails (WT=218 bp, KO=240 bp) and immunoblotting of MEFs with the indicated antibodies. Uncropped blots are shown in Figure S6. b. 8 Gy whole-body gamma-irradiation of 19 age-matched WT and 22 SCAI−/− mice. Sex-separated data in (Figure S4d,e). c. Testes from 8-week old WT and SCAI−/− mice. Scale bar, 10 mm. d. Sizes of n=3 independent testes from (c). e. Hematoxylin and eosin-stained sections of testes (I-IV) and caudal epidydimis (V-VI) from 8-week-old mice. Scale bars I, II, V, and VI, 100 µm; III – IV, 50 µm. f. Hematoxylin and eosin-stained sections of ovaries from 14-week-old mice. Bars I and II, 500 µm; III and IV, 50 µm. g. Spermatocyte spreads from WT and SCAI−/− mice stained with SYCP1 and SYCP3 antibodies. 100 cells each from n=3 independent animals were scored and the percentage of cells at each stage plotted. Data were analyzed by Fisher’s exact test, two-tailed. h. Representative images of spermatocytes stained for MLH1 and SYCP3 showing a pachynema WT cell and a pachynema-like SCAI−/− cell. Inset: magnification of the boxed area with entangled chromosomes and loss of synapsis indicated by weaker SYCP3 staining. Scale bars, 10 µm i. Spermatocytes from (h) were stained for SYCP3 and γ-H2AX to identify chromosome entanglements and sex body. n=3 independent animals were examined and a total of 52 and 58 cells analyzed for WT and SCAI−/−, respectively. Statistical analysis was done as in (g). j. DMC1 foci were counted at the indicated stages of meiotic prophase I. Pooled cells from three independent animals. P-values were calculated from a Mann-Whitney test. k. MLH1 foci counts were plotted as in (j). Pooled cells from three independent animals. l. Total number of metaphase cells from one testis. m. Metaphase spreads of primary B cells from WT and SCAI−/− mice treated with DMSO or PARP inhibitor (PARPi) for 16 h were FISH-stained for telomeric DNA and analyzed for chromosomal aberrations. See Table S3 and Figure S4m. P-value was calculated as in (j) (n=6 independent mice of each genotype). All data points are represented as mean ± SD.
While 53BP1 is a key DSB repair factor promoting class-switch recombination (CSR) in B cells9,10, we found that SCAI has no obvious role in facilitating this function of 53BP1. Specifically, SCAI−/− and control mice displayed comparable splenic B cell numbers and frequencies; moreover, proliferation and class-switching to IgG1 and IgG3 was indistinguishable between SCAI−/− and control B cells stimulated ex vivo (Figure S4f–i). In addition, SCAI−/− mice showed no differences in levels of IgG1, IgG3 or IgM in blood serum compared to control mice (Figure S4j). Instead, full-body necropsies showed that male SCAI−/− mice had markedly reduced testis size (Figure 3c,d), suggesting that, unlike 53BP1 knockout25,26, ablation of SCAI might result in defective spermatogenesis and subfertility. Indeed, while histological examination of SCAI−/− testes showed normal distributions of seminiferous tubules at different stages of spermatogenesis, the lumen of the seminiferous tubules were largely devoid of maturing sperm, with some tubules displaying a Sertoli-cell-only (SCO) phenotype and concomitant expansion of extra-tubular Leydig cells (Figure 3e (I-IV)). As a consequence of these defects, the caudal epididymis of SCAI−/− males contained few if any mature spermatids (Figure 3e (V-VI)). Ovaries from SCAI−/− and control females were similar in size and shape and both contained fully developed corpora lutea (Figure 3f (I-II)), indicating that overall development of ovary structure and hormonal signaling per se were not affected by SCAI loss. However, ovaries from SCAI−/− mice contained few or no developing primary follicles (Figure 3f (I-IV)). Consistent with these germ cell maturation defects, we observed substantial reductions in fertility rates of both male and female SCAI−/− mice (nearly 3- and 7-fold, respectively) compared to controls (Table S2). Moreover, the few litters generated by SCAI−/− female breeding cages were smaller compared to control cages (Figure S4k). We conclude that, unlike loss of 53BP1, SCAI deficiency leads to germ cell development defects and subfertility in both males and females.
To investigate the underlying cause of defective germ cell development associated with SCAI deficiency, we analyzed spermatocyte spreads from SCAI−/− and control testes stained for meiosis-specific synaptonemal complex markers (SYCP1 and SYCP3). The frequency of meiotic prophase I spermatocytes in leptonema, early zygonema, late zygonema, and diplonema were indistinguishable from controls (Figure 3g). However, SCAI−/− testes showed reduced levels of spermatocytes in pachynema and a concomitant increase in aberrant pachynema-like cells characterized by irregular synaptic behavior including gaps, breaks and entangled chromosomes (Figure 3g–i). These data suggest that loss of SCAI leads to impaired meiotic recombination of DNA breaks. Consistently, while the meiosis-specific recombinase DMC1 was loaded normally onto meiotic chromosomes at early stages, DMC1 foci were reduced in late zygonema, early pachynema and on the sex chromosomes of pachynema SCAI−/− spermatocytes (Figure 3j;S4l). Nevertheless, SCAI−/−spermatocytes form a proper sex body and show normal numbers of diplonema spermatocytes (Figure 3g, i). This suggests that while SCAI−/− spermatocytes have a reduced ability to synapse homologs, most are capable of progressing through the mid-pachynema checkpoint. Additionally, late pachynema SCAI−/− spermatocytes show a normal frequency of MLH1 foci (Figure 3k), marking sites of future crossovers. Intriguingly, we observed a dramatic reduction in the number of metaphase I cells in SCAI−/− testis (Figure 3l). Metaphase I spermatocytes are lost through apoptosis as a consequence of lagging chromosomes, which are primarily caused by the absence of crossing over between homologs27. Our observations suggest that, while crossover designation may be normal in the absence of SCAI, crossing-over itself is disrupted. Thus, loss of SCAI may cause impaired accumulation and/or retention of the HR recombinase DMC1 on meiotic chromosomes and aberrant progression through pachynema, ultimately leading to loss of spermatocytes at metaphase I. These results demonstrate that the germ cell developmental defects and subfertility of SCAI−/− mice are at least partially due to aberrant meiotic recombination although relatively mild compared to fully HR-deficient spermatocytes (SPO11−/− or DMC1−/−) showing severe synapsis and/or pairing defects28. Further supporting a role of SCAI in HR, we observed an increase in chromosomal aberrations in primary SCAI−/− B cells compared to controls treated with Olaparib, an established sensitizer of HR-compromised cells (Figure 3m;S4m; Table S3)29–31. We conclude that SCAI deficiency gives rise to common features of compromised HR-mediated DSB repair in both meiotic and mitotically growing cells.
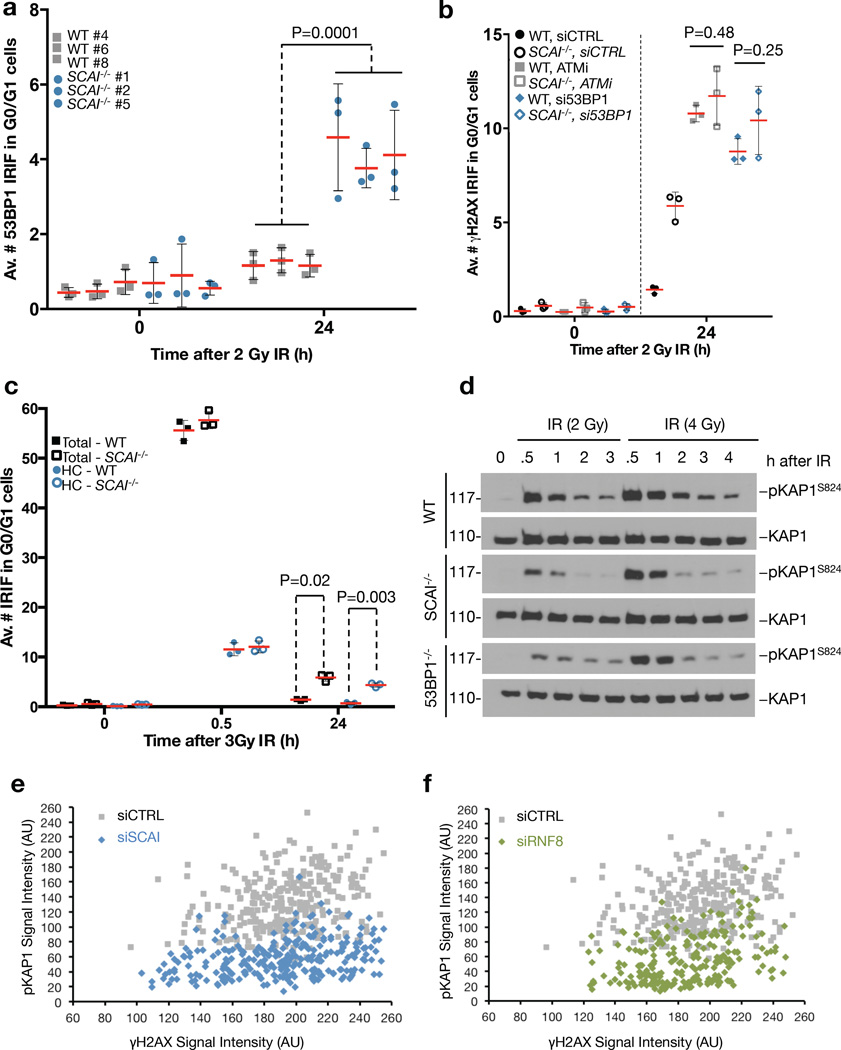
While SCAI is dispensable for 53BP1-dependent CSR, we reasoned that it might mediate other 53BP1 functions in DSB repair. The SCAI interactome (Figure 1d) revealed an enrichment of heterochromatin-associated factors including HP1 proteins, which we confirmed biochemically (Figure S5a,b). This raised the possibility that SCAI promotes the function of 53BP1 in repair of heterochromatin-associated DSBs relying on localized, ATM-dependent phosphorylation of KAP1 (pKAP1) in heterochromatin6, 16. Indicative of a heterochromatin-associated NHEJ defect16, SCAI deficiency in MEFs arrested in G0/G1 phase gave rise to an increase in persistent 53BP1 foci after IR (Figure 4a;S5c). Moreover, epistasis experiments using WT and SCAI−/− MEFs treated with 53BP1 siRNA or ATM inhibitor showed that loss of SCAI did not exacerbate the DSB repair defect observed upon impaired 53BP1 or ATM function (Figure 4b;S5d), suggesting that SCAI and 53BP1 operate in a common pathway to mediate ATM-dependent repair of heterochromatin-associated DSBs. SCAI was recently found to be specifically enriched in pull-downs with histone H3 tail peptides containing trimethylated K9 (H3K9me3)32, the main repressive histone mark in heterochromatin. Indeed, a heterochromatin correlation analysis confirmed that most unrepaired DSBs at late time points in SCAI−/− MEFs were associated with H3K9me3-positive chromocenters (Figure 4c;S5e). Similar to 53BP1−/− MEFs, immediate IR-induced pKAP1, a marker of productive DSB repair in heterochromatin16, was markedly reduced after low IR doses in quiescent SCAI−/− MEFs (Figure 4d). This effect was partly masked upon increasing IR doses (Figure 4d), as seen also in 53BP1-deficient cells16. The pKAP1 defect was also evident at persistent heterochromatin-associated breaks, as SCAI knockdown in quiescent 48BR primary human fibroblasts strongly reduced the decoration of γ-H2AX foci with pKAP1 after IR (Figure 4e;S5f). Knockdown of RNF8, an essential mediator of 53BP1 accumulation at DSB sites7, phenocopied the effect of SCAI depletion (Figure 4f; S5f). Collectively, these data suggest that SCAI functions downstream of 53BP1 in heterochromatin-associated DSB repair to mediate ATM-dependent KAP1 phosphorylation in repressive chromatin environments.
Figure 4. ATM, 53BP1 and SCAI operate in a common pathway to mediate repair of heterochromatin-associated DSBs.
a. Independent, immortalized WT and SCAI−/− MEF cell lines were arrested in G0/G1 by growing to full confluency. Cultures were mock-treated or exposed to IR (2 Gy), fixed 24 h later and stained with 53BP1 antibody. Images were acquired as Z-stacks and the number of 53BP1 foci per cell was counted through the entire nuclear volume. P-value was calculated from a one-tailed t-test using Welch correction (n = 9 independent measurements across 3 MEF lines). Bars indicate mean ± SD. See Figure S5c for full data set including a 0.5 h time point. IRIF: Ionizing Radiation Induced Foci b. Immortalized WT and SCAI−/− MEFs were grown to full confluency while transfecting with 53BP1 siRNA for 72 h or incubating with ATM inhibitor (ATMi) for 1 h prior to irradiation. Cells were treated and analyzed as in (a), except that they were immunostained for γ-H2AX as a marker of unrepaired DSB (n = 3 biologically independent samples). See Figure S5d for full data set including a 0.5 h time point. c. Immortalized WT and SCAI−/− MEFs were treated as in (a), except that cells were co-stained with antibodies to γ-H2AX and the heterochromatin marker H3K9me3 to determine chromatin context (n = 3 biologically independent samples). HC (heterochromatin). See Figure S5e for analysis of ATM inhibitor treated samples. d. Immortalized WT, SCAI−/− and 53BP1−/− MEFs were grown to confluency, exposed to IR (2 or 4 Gy) and harvested at the indicated time points. Lysates were analyzed by immunoblotting with antibodies against total and phosphorylated KAP1. Uncropped blots are shown in Figure S6. e. Quiescent 48BR primary human fibroblasts were transfected with control (CTRL) or SCAI siRNAs, irradiated with IR and fixed after 24 h. Cells were immunostained with antibodies against γ-H2AX and phosphorylated KAP1 (pKAP1), and the relative fluorescence intensities were measured by high content microscopy. Each data point represents one individual IRIF. See Figure S5f for representative images. f. As in (e), except that cells were transfected with control (CTRL) or RNF8 siRNAs. See Figure S5f for representative images.
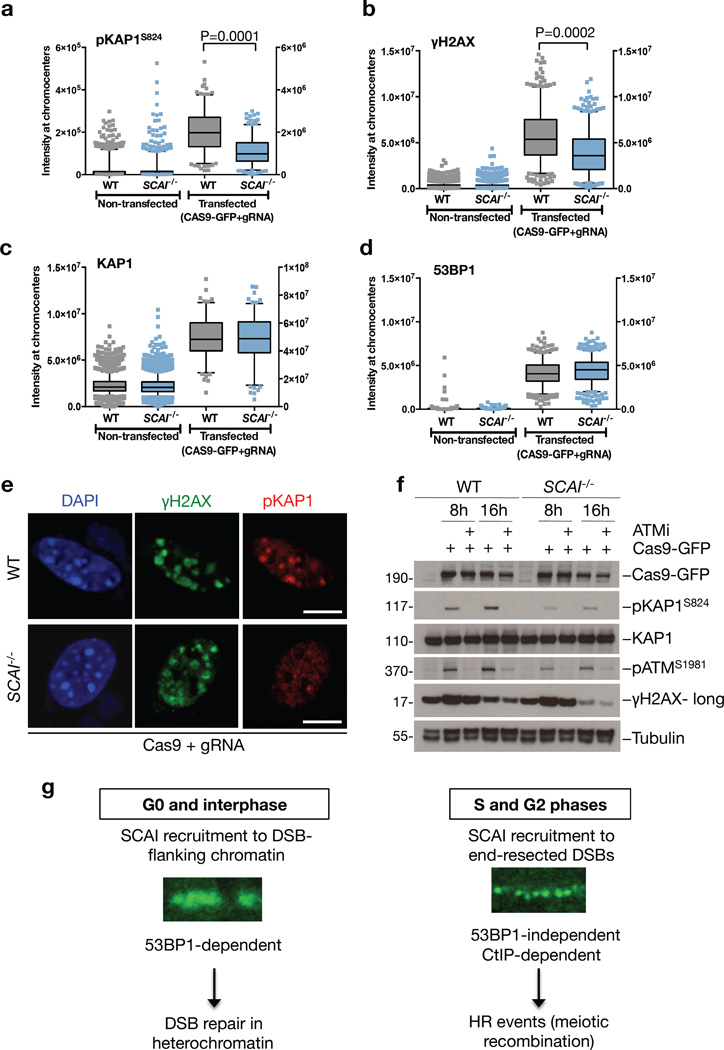
To further characterize the DSB repair function of SCAI in heterochromatin, we employed a CRISPR/Cas9-based system using gRNAs targeting major satellite repeats to induce heterochromatin-specific DSBs in murine cells33. The resulting breaks caused rapid accumulation of 53BP1 and GFP-SCAI in DAPI-rich chromocenters corresponding to heterochromatin (Figure S5g). Employing this system to assay signaling from heterochromatin-associated DSBs in WT and SCAI−/− MEFs, we found that SCAI deficiency specifically compromised ATM-dependent phosphorylation of KAP1 and H2AX upon DSB formation in heterochromatin, while it had no effect on total levels of KAP1 or 53BP1 accumulation at these structures (Figure 5a–f). Moreover, loss of SCAI did not significantly impact the size and composition of DSB-containing chromocenters, as evidenced by markers such as DAPI, HP1 and H3K9me3 (Figure S5h–k). Consistent with a role for SCAI in promoting overall ATM signaling at heterochromatin-associated DSBs, overexpression of SCAI enhanced ATM-mediated phosphorylations upon formation of such breaks (Figure S5l). Importantly, SCAI−/− MEFs did not display obvious ATM signaling defects after IR-induced DSBs, which mostly target euchromatic regions of the genome34 (Figure S5m). Together, these results demonstrate that SCAI functions downstream of 53BP1 to mediate ATM-dependent signaling after DSBs specifically in heterochromatin. 53BP1 promotes repair of heterochromatin-associated DSBs via both NHEJ (in G0/G1 phase cells) and HR (in G2 phase cells)16, 35, 36. Because SCAI deficiency gives rise to a DSB repair defect in both G0 and G2 phase cells, it is possible that SCAI mediates productive DSB repair in compacted heterochromatin via either of these major DSB repair pathways through chromatin remodeling events that facilitate the access of the repair machinery to the lesions.
Figure 5. SCAI mediates ATM signaling from DSBs in heterochromatin.
a. Immortalized WT and SCAI−/− MEFs were transfected with Cas9-GFP and gRNAs targeting the major satellite repeats to induce CRISPR-mediated DSBs in heterochromatin-containing chromocenters. After 8 h cells were fixed and immunostained with antibodies against phosphorylated KAP1 (pKAP1). Cells were analyzed by high content microscopy using DAPI signal as a mask for chromocenters. P-values were calculated from two-tailed t-tests using Welch correction. Centre indicates the median and whiskers the borders of the 95% quantiles. Y-axis on the left side corresponds to the non-transfected conditions, while y-axis on the right side corresponds to the transfected conditions. (n=200 independent measurements) b. As in (a), except cells were immunostained with γ-H2AX antibodies. (n=430 independent measurements) c. As in (a), except cells were immunostained with KAP1 antibodies. (n=125 independent measurements) d. As in (a), except cells were immunostained with 53BP1 antibodies. (n=550 independent measurements) e. Representative images from the experiments in (a) and (b). Scale bars, 10 µm. f. Immortalized WT and SCAI−/− MEFs were transfected as in (a) while treated with ATM inhibitor (ATMi) where indicated. Cell extracts were analyzed by immunoblotting with indicated antibodies. Uncropped blots are shown in Figure S6. g. Model of SCAI function in DSB repair. SCAI is recruited to DSB-proximal chromatin throughout interphase through direct interaction with 53BP1, promoting 53BP1- and ATM-mediated repair of heterochromatic DSBs. Notably, SCAI is dispensable for other 53BP1-dependent functions, such as immunoglobulin class-switching. During the S and G2 phases of the cell cycle, SCAI also accumulates at CtIP-resected ssDNA regions in a 53BP1-independent manner. From this locale, SCAI supports a subset of HR events, and its deficiency is associated with defects in meiotic recombination and germ cell development.
Collectively, our data establish SCAI as a physiologically important chromatin-associated component of the cellular machinery that mediates DSB repair in different chromosomal contexts. This involvement minimally includes roles of SCAI in promoting 53BP1-dependent DSB repair in heterochromatin and 53BP1-independent crossover/DSB repair reactions on resected DNA ends during meiotic recombination, likely reflecting its unusual, dual presence at chromatin and end resection-dependent ssDNA regions flanking DSBs, respectively (Figure 5g). Whether SCAI promotes these processes via common or distinct mechanisms, and precisely how it exerts its DSB repair functions at the molecular level, are important future areas of study.
Methods
Plasmids and siRNAs
Full-length SCAI cDNA was amplified by PCR and inserted into pEGFP-C1 (Clontech) and pcDNA4/TO (Life Technologies) containing an N-terminal Strep-HA-tag to generate mammalian expression plasmids for GFP-tagged and Strep-HA-tagged SCAI, respectively. The CMK6-HA-53BP1 plasmid was described previously1. 53BP1 N-terminal deletion constructs (N1-N4) were amplified by PCR and inserted into pcDNA4/TO-Strep-HA. Plasmid transfections were performed using GeneJuice (Novagen) or FuGene 6 (Promega) according to the manufacturer’s instructions. siRNA transfections were done using RNAiMAX (Life Technologies) according to the manufacturer’s instructions. siRNA target sequences used in this study were: Control (5’-GGGAUACCUAGACGUUCUA-3’), SCAI(#9) (5’-GAGGCGGAUCCUGUAAUGGUA-3’); SCAI(#10) (5’-GGACAGACCUGAAUUGGUA-3’); 53BP1 (5’-GGACUCCAGUGUUGUCAUUUU-3’), RNF8 (5’-UGCGGAGUAUGAAUAUGAA-3’); CtIP (5’-GCUAAAACAGGAACGAAUCTT-3’); BRCA1 (5’-GGAACCUGUCUCCACAAAGTT-3’); RNF8 (5’-UGCGGAGUAUGAAUAUGAATT-3’); and RNF168 (5’-GGCGAAGAGCGAUGGAGGATT-3’). BRCA2 siRNA was an siGENOME SMARTpool from Dharmacon (M-003462-01).
Plasmids for generation of SCAI knock-out cells by CRISPR/Cas9 were generated as described 2. Briefly, SCAI gRNA sequences were introduced into pEsgRNA by PCR-based insertion mutagenesis. gRNA sequences used were: SCAI#2: GTCTAATAGTGTTGCGTATAAGG (chr9:127757212-127757234); SCAI#4: GGCTTGAAGCGCTGGCAAATAGG (chr9:127790713-127790735); 53BP1#1: GCCAGCTCCTGCTCGAAGCTGGG (chr15:43701875-43701897); and 53BP1#2: GTTGACTCTGCCTGATTGTATGG (chr15:43724790-43724812). gRNA targeting major satellite repeats was cloned into vector containing U6 promoter plus followed by a gRNA scaffold. Sequence: Ma-sat#3: GAAATGTCCACTGTAGGACG. Cas9 cDNA was amplified from pX330-U6-Chimeric_BB-CBh-hSpCas9 (kind gift from Feng Zhang) and cloned using golden gate cloning into pCX5-CMVp-Cas9-EGFP-SV40p-Puro-pA and pX-86-U6p-gRNA(Ma-sat#3)-CMVp-Cas9-mCherry-SV40p-HygroR-pA plasmids to generate EGFP-tagged and mCherry-tagged Cas9 expression constructs, respectively.
Cell culture and reagents
All standard cell lines were obtained from ATCC and regularly tested for mycoplasma infection. The cell lines were not further authenticated and are not found in the database of commonly misidentified cell lines that is maintained by ICLAC and NCBI Biosample. Human U2OS, HeLa and 48BR cells were cultured in DMEM (GIBCO) containing 10% fetal bovine serum. Mouse NIH-3T3 cells were cultured in DMEM containing 10 % Newborn calf serum. To generate cell lines stably expressing GFP-tagged SCAI, U2OS cells were co-transfected with pEGFP-C1-SCAI and pBabe.puro plasmids and positive single cell clones expanded in the presence of puromycin (1 µg/ml, Sigma). Doxycycline-induced Strep-HA-tagged SCAI cell lines were obtained by co-transfection of pcDNA4/TO-Strep-HA-SCAI and pcDNA6/TR (Life Technologies) and expansion of single cell clones under Zeocin (0.2 µg/ml, Life Technologies) and Blasticidin S (5 µg/ml, InVivoGen) selection. The HeLa/NFLAP-SCAI BAC cell line was a kind gift from Dr. Anthony Hyman (Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany). 53BP1−/− MEFs and reconstituted cell lines were a kind gift from Dr. Andre Nussenzweig (National Institutes of Health, Bethesda, USA). For B cell cultures, resting splenic B cells were isolated from 8–14-week-old WT or SLA1−/− mice with anti-CD43 microbeads (anti-Ly48; Miltenyi Biotech #130-049-801) and stimulated to undergo class switching with either LPS (25 µg/ml), α-IgD-dextran (2.5 ng/ml) and RP105 (0.5 µg/ml) for CSR to IgG3 or LPS (25 µg/ml), IL4 (5 ng/ml) and RP105 (0.5 µg/ml) for CSR to IgG1, as described previously3 . B cell proliferation was analyzed by CFSE-like labeling using CellTrace Violet proliferation kit (#C34557, Lifetechnologies) according to the manufacturer’s instructions. Primary MEFs derived from E13.5 were obtained by intercrossing mice following standard procedures. For immortalization, MEFs were subjected to retroviral infections with SV40LT at passage 2 and cultured in DMEM supplemented with 15% fetal bovine serum (GIBCO), 100 U/ml penicillin and 0.1 mg/ml streptomycin (Sigma). Fibroblast proliferative capacities were assayed by plating passage 2 primary MEF lines (P2). Every 2 days, cells from each dish were trypsinized, counted and replated. Cells were treated with inhibitors targeting ATM kinase (KU60019 (10 µM, Selleckchem) or KU55933 (10 µM, Selleckchem)), proteasome (MG132 (20 µM, AH Diagnostics)) and PARP-1 (Olaparib (1 µM, AZD2281, Selleckchem)). To induce DSBs, cells were exposed to the indicated doses of x-rays using a Y.SMART tube (YXLON A/S, Denmark) at 6 mA and 160 kV through a 3-mm aluminium filter. For high content imaging of RAD51 foci, cells were exposed to IR from a cesium irradiator.
CRISPR/Cas9 genome editing
SCAI or 53BP1 CRISPR knock-out cell lines were generated as described2 . Briefly, gRNA plasmids were co-transfected with pBabe.Puro in Cas9-FLAG U2OS SEC-C cells (a kind gift from Dr. John Rouse) 4 . Cells were grown in DMEM in the presence of doxycycline to induce Cas9-FLAG expression. Subsequently, cells were grown in the presence of puromycin during clonal selection for 7–10 days. Knockdown efficiency was validated by qPCR and immunoblotting. Generation of heterochromatin-specific DSBs by Cas9 was achieved by transfecting cells with major satellite-specific gRNA and GFP-mCherry-Cas9 for 8 or 16 h, before pre-extraction in 0.1% Triton/PBS for 30 s followed by fixation in 4% paraformaldehyde/PBS for 10 min.
Mass spectrometry
Analysis of replication-dependent recruitment of proteins to damaged chromatin by means of the CHROMASS method was done as described 1. In brief, psoralen-crosslinked chromatin was incubated in repair-proficient Xenopus egg extracts. Chromatin was isolated by sedimentation through a sucrose cushion and analyzed by mass spectrometry.
SCAI-interacting proteins were identified by QUBIC, as described previously 2. HeLa BAC cells expressing GFP-tagged SCAI (NFLAP-SCAI) under the control of the endogenous promoter were cultured in DMEM. Pellets from ∼107 cells were resuspended in 1 ml lysis buffer (50 mM Tris, pH 7.5; 150 mM NaCl; 5% Glycerol; 1% NP-40; 1 mM MgCl2) containing 200 U Benzonase (Merck) and EDTA-free complete protease inhibitor cocktail (Roche) and incubated for 30 min on ice. Cell lysates were cleared by centrifugation and GFP-tagged proteins were bound to 50 µl magnetic beads coupled to monoclonal mouse GFP antibody (Miltenyi Biotec, #130-091-125) for 15 min on ice. Bound proteins were washed three times with 800 µl ice cold wash buffer I (50 mM Tris, pH 7.5; 150 mM NaCl; 5% Glycerol; 0.05% NP-40) and two times with 500 µl wash buffer II (50 mM Tris, pH 7.5; 150 mM NaCl; 5% Glycerol). Purified proteins were digested on beads at room temperature by adding 25 µl digestion buffer (50 mM Tris, pH 7.5; 2 M urea) containing 150 ng Trypsin (Promega) and 1 mM DTT. After 30 min, peptides were eluted by adding twice 50 µl digestion buffer containing 5 mM chloracetamid. After overnight digestion at room temperature, peptides were acidified by addition of 1 µl trifluoroacetic acid and purified on C18 material. Peptides were separated on RP ReproSil-Pur C18-AQ 3µm resin (Dr. Maisch) columns (15 cm) and directly injected into a LTQ-Orbitrap mass spectrometer (Q Exactive, Thermo Scientific, Germany) 2. Raw data was analyzed with MaxQuant using the label-free algorithm 5. ProteinGroups were filtered to have at least three valid values in the LFQ intensities of the SCAI replicates and to be identified by at least two peptides. Missing values in the control pull-downs were imputed by values simulating noise around the detection limit. SCAI interactors were identified by comparing the LFQ intensities in the SCAI and mock pull-downs using a modified two-sided t-test (FDR < 0.01, S0=1, see www.maxquant.org for details).
Immunoblotting, immunoprecipitation and antibodies
For whole cell extracts, cells were lysed in EBC buffer (50 mM Tris, pH 7.5; 150 mM NaCl; 1 mM EDTA; 0.5% NP-40) or RIPA buffer (1% NP40, 0.5% sodium deoxycholate, 0.1% SDS, 150 mM NaCl, 50 mM TRIS pH 8.0) supplemented with protease and phosphatase inhibitors. To obtain chromatin-enriched fractions, cells were lysed in low-salt buffer (10 mM HEPES, pH 7.4; 10 mM KCl; 0.05% NP-40) supplemented with protease and phosphatase inhibitors, and chromatin-associated proteins were released from the pellet by treatment with micrococcal nuclease. Strep pull-downs were done with Strep-Tactin sepharose (IBA) and GFP immunoprecipitation was performed with GFP-Trap agarose (Chromotek). Bound material was resolved on SDS-PAGE and transferred to nitrocellulose membranes. Antibodies used in this study included: rabbit polyclonals to 53BP1 (sc-22760, Santa Cruz, 1:5000 (WB)/1:1000 (IF)), RAD51 (sc-8349, Santa Cruz, 1:150 (IF)), DMC1 (sc-22768, Santa Cruz, 1:200 (IF)), RIF1 (A300-569A, Bethyl, 1:200 (IF)), Tubulin (ab6046, Abcam, 1:10.000 (WB)), Histone H3K9me3 (ab8898, Abcam, 1:500 (IF)), 53BP1 (ab21083 and ab36823, Abcam, 1:1000 (IF); NB100–304, Novus Biologicals, 1:1000 (IF)), SYCP3 (sc-33195, Santa Cruz, 1:200 (IF)), SYCP1 (ab15090, Abcam, 1:200 (IF)), KAP1 (A300-274A, Bethyl, 1:500 (WB)) and phospho-KAP1 (S824) (A300-767A, Bethyl, 1:1000 (WB & IF)); mouse monoclonals to GFP (sc-9996 (clone B-2), Santa Cruz, 1:500 (WB)), FLAG (F-1804 (clone M2), Sigma, 1:100 (IF)/1:500 (WB)), HA (sc-7392 (clone F-7), Santa Cruz, 1:500 (WB)/1:1000 IF)), γ-H2AX (S139) (05–636, (clone JBW301) Millipore, 1:1000 (IF); ab22551 (clone 3F2) Abcam, 1:1000 (IF & WB)) , SYCP3 (sc-74569 (clone D-1) Santa Cruz, 1:200 (IF)), MLH1 (51-1327GR, (clone G168-15) BD Pharmingen, 1:20 (IF)), BRCA1 (sc-6954 (clone D-9), Santa Cruz, 1:100 (IF)), HP-1γ (MAB3450 (clone 2MOD-1G6), Millipore, 1:1000 (WB)), HP-1β (MAB3448 (clone 1MOD-1A9), Millipore, 1:1000 (WB)), H3K9me2+3 (ab71604 (clone 6F12-H4), Abcam, 1:1000 (IF)), ATM pS1981 (200-301-400 (clone 10H11.E12), Rockland, 1:500 (IF)/1:1000 (WB)) and phospho-H3 (S13) (ab14955 (clone mAbcam 14955), Abcam, 1:1000 (IF)); rabbit monoclonal to RPA70 (ab79398 (clone EPR3472), Abcam, 1:1000 (IF)); goat polyclonal to MCM6 (sc-9843, Santa Cruz, 1:500 (WB)). A sheep polyclonal antibody to SCAI was generated by immunization with a full-length GST-fusion protein produced in bacteria (1µg/µl for IP). Rat monoclonal antibody to SCAI (IH2) was described (1:50 (WB)) 6.
Immunofluorescence, confocal microscopy and laser microirradiation
Cells were fixed in 4% formaldehyde, permeabilized or pre-extracted prior to fixation with PBS containing 0.2% Triton X-100 for 5 min or 1 min, respectively, and incubated with primary antibodies diluted in DMEM for 1 h at room temperature. Following staining with secondary antibodies (Alexa Fluor 488 and 568; Life Technologies) for 30 min, coverslips were mounted in Vectashield mounting medium (Vector Laboratories) containing the nuclear stain DAPI. For detection of nucleotide incorporation during DNA replication, an EdU labeling kit (Life Technologies) was used according to the manufacturer’s instructions. Confocal images were acquired on an LSM-780 (Carl Zeiss) mounted on a Zeiss-AxioObserver Z1 equipped with a Plan-Neofluar 40x/1.3 oil immersion objective. Dual and triple color confocal images were acquired with standard settings for excitation of DAPI, Alexa Fluor 488, Alexa Fluor 568, and Alexa Fluor 647 dyes (Molecular Probes, Life Technologies), respectively. Image acquisition and analysis was carried out with LSM-ZEN software. Laser microirradiation of cells was performed essentially as described 7. Imaging of Cas9-induced heterochromatin damage at chromocenters was acquired on Confocal Laser Scanning Microscope TCS SP8 (Leica), using a 63X objective. Spermatocyte spreads were prepared, stained, and scored as previously reported 8. Images were acquired on a Zeiss Axio Imager M2 with a Plan-Apochromat 100X/1.4 oil immersion objective. Regarding animals used in spermatocyte spread analyses, age-matched animals were between 18–27 weeks of age, no statistical method was used to predetermine sample size, experiments were not randomized, nor were the investigators blinded to allocation during the experiments or outcome assessment.
Flow Cytometry and ELISA
Cells were stained with antibodies and measured with an LSR Fortessa cell analyzer (BD Pharmingen) using a DAPI negative live lymphocyte gate. Data were analyzed using FlowJo X 10 software. Antibodies used for flow cytometric analysis included B220 (RA3-6B2), CD19 (1D3), IgM (II/41), IgG1 (A85-1), and IgG3 (R40-82) (BD and eBiosciences). To measure Ig in the blood serum by ELISA, plates were coated with anti-mouse IgM (#406501) or IgG (#1030-01) (Southern Biotechnology Associates, Inc.), and Ig was detected with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG1 (#1070-05), IgG3 (#1100-05) or IgM (#1020-05) (Southern Biotechnology Associates, Inc.). In all cases, wells were developed with the Ultra TMB peroxidase substrate system (Thermo Scientific) and OD was measured at 450 nm using a Fluostar Omega microplate reader (BMG-Labtech). Regarding animals used in FACS and ELISA experiments, animals were between 8–12 weeks of age, no statistical method was used to predetermine sample size, experiments were not randomized, nor were the investigators blinded to allocation during the experiments or outcome assessment.
Chromosome metaphase spreads
For genome instability analysis, B cells isolated from animals between 8–12 weeks of age were harvested after 3 days in culture stimulated to undergo class switching to IgG1. Metaphase spreads were prepared and processed for FISH analysis as previously described 9–12. PARP inhibitor Olaparib (2µM, AZD2281, Selleckchem) was added to cells stimulated ex vivo for 16 h and Colcemid (100 ng/ml, Roche) added 1 h before preparation of metaphase spreads, and imaging as described below using a high content microscope. Experiments were performed with the investigator blinded to the group allocation. An assistant labeled the slides and/or dissected spleen/cultured cells before analysis by the investigator, and the data were subsequently related to the identity of the specimens. A total of 165 (WT) and 189 (SCAI−/−) metaphase spreads from DMSO-treated cells and 452 (WT) and 453 (SCAI−/−) spreads from PARPi-treated cells were analyzed, across multiple mice, and detailed in the Table S3. Spermatocyte metaphase spreads were prepared as previously described 13, except a 2.9% isotonic sodium citrate dihydrate solution was used and the slides were stained with Giemsa. Spermatocyte metaphase spread images were acquired on a Zeiss Axio Scope.A1 LED with a Plan-Apochromat 100X/1.4 oil immersion objective.
Generation of SCAI KO mice and histology
To generate SCAI−/− mice, ES cells carrying a targeted allele of the Scai gene were obtained from EUCOMM (allele name: SCAI (tm1a(EUCOMM)Hmgu); clone ID: HEPD0516_1_G04). Correct targeting was verified by PCR using primers spanning the homology arms. PCR fragments from the 5’ and 3’ end of the targeting construct were cloned into pCR4-TOPO (Invitrogen) and sequenced. Following blastocyst injection, chimeras were mated with C57Bl/6 WT mice and germline transmission of the targeted allele was achieved. The resulting mouse line was crossed with E2A-Cre to remove floxed sequences. Cycling conditions for genotyping PCR were: 94 °C (60 s), 60 °C (90 s), 72 °C (120 s), 32 cycles, 72 °C (10 m). Primers LacZ_EUCOMM_for03 (5’-ccagttcaacatcagccgctacagtc-3’) and SV40_EUCOMM_rev01 (5’-ctagagcttagatcccccctgcc-3’) yield a 240-bp product specific for the targeted allele and primers mSCAI_for01 (5’-ccagcacttgggaggcagagac-3’) and mSCAI_rev01 (5’-gcagctaaggatagacgatcatagcag-3’) yield a 218-bp product for the WT allele.
All animal experiments were approved by the Department of Experimental Medicine (University of Copenhagen), the Danish Working Environment Authority, the Danish Animal Experiment Inspectorate, and the MDACC Institutional Animal Care and Use Committee (IACUC).
Testes and ovaries from WT and SCAI−/− mice were fixed in 10% formalin, and paraffin sections were stained with hematoxylin and eosin. Images were acquired with an inverted microscope (Axiovert 200M; Carl Zeiss, Inc.) equipped with a 10x NA 0.45 objective lens (Plan-Apochromat; Nikon) and a color charge-coupled device camera (Axiocam MRc5; Carl Zeiss, Inc.) using AxioVision software (version 4.6.3.0; Carl Zeiss, Inc.). Male animals used for histology were 8 weeks old and female animals were 14 weeks old. Similar results were obtained from at least 3 mice of each genotype. For histological analyses, no statistical method was used to predetermine sample size, experiments were not randomized, nor were the investigators blinded to allocation during the experiments or outcome assessment.
Whole-body irradiation of mice
Age-matched male and female WT and SCAI−/− mice were subjected to whole-body gamma-irradiation with a one-time dose of 8 Gy of gamma-rays from a Gammacell 40 Exactor Cs137 source and were carefully monitored every day to assess survival. Post-irradiation, the mice were put on antibiotic water for the duration of the study (0.1mg/ml Ciproxin). The experiment with male mice was performed twice with a reproducible result (Figure. S4d) and the experiment with females was performed once (Figure. S4e). Figure. 3b represents the total data of all 3 experiments. A scoring sheet used by the animal care-taker was generated to carefully monitor weight-loss, abnormal posture, and lack of movement/lethargy on a daily basis. Animals were euthanized by the care-taker before severe distress/suffering was observed, as determined by the scoring system. As such, the care-takers were blinded to allocation of the genotypes during the experiments and informed the investigator of the data. All remaining animals in the experiment were euthanized by day 28 post-irradiation. WT and SCAI−/− mice were age-matched (male exp. 1: 17–27 weeks of age, male exp. 2: 14–42 weeks of age, female exp.: 17–64 weeks of age), no statistical method was used to predetermine sample size, and experiments were not randomized.
High content microscopy and image analysis
Quantitative image-based cytometry (QIBC) for measurement of fluorescence intensities was done as described previously 14,15. The images were obtained with a 40× 0.95 NA, FN 26.5 (UPLSAPO40x) dry objective, a quadruple-band filter set for DAPI, FITC, Cy3 and Cy5 fluorescent dyes, a MT20 Illumination system and a digital monochrome Hamamatsu C9100 EM-CCD (electron-multiplying charge-coupled device) camera. Camera resolution is 200 nm x 200 nm per pixel (binning 1, 40x). Image analysis was performed with Olympus ScanR automated image and data analysis software using standard algorithms for detection of nuclei and sub-objects within nuclei. Typically, 49 images (corresponding to 1500–3000 sub-objects) were acquired under non-saturating conditions for each data point, allowing robust measurements of experimental parameters such as intensities. Automated unbiased image acquisition was carried out with the ScanR acquisition software. Automated detection and imaging of high-resolution images of metaphase spreads were obtained using Olympus ScanR image analysis and Xcellence software. Images for quantification of 53BP1 and yH2AX foci were acquired with Olympus ScanR image analysis and Xcellence software. Twenty-five images were acquired and at least 2500 cells were analyzed per sample. High-throughput analysis of chromocenters and heterochromatin marker intensities after Cas9 damage induction, were obtained using the IN Cell Analyzer 1000 Cellular Imaging System, followed by analysis using Cellomics Cell-Insight software. Briefly, cells were selected based on DAPI-dense regions and cells expressing Cas9-EGFP-gRNA yielding damage-induced 53BP1/γH2AX pattern at chromocenters were chosen for intensity analysis.
DSB repair by IRIF enumeration
Immunofluorescence and DSB repair analysis was carried out as described 16–18; briefly, cells were fixed in 3% Paraformaldehyde containing 2% sucrose for 10 min, permeabilized for 3 min in 0.2% Triton X-100 in PBS and immunostained for 1 h with primary antibody (diluted in PBS containing 2% BSA), then 30 min with 1:200 dilutions of secondary antibodies (in PBS containing 2% BSA). Cells were counterstained with 0.1 µg/ml DAPI to visualize nuclei and were mounted using Polymount G. Samples were imaged with a Zeiss Axio Observer Z1 platform microscope, with a Plan-Apochromat 20x/0.8, an EC Plan-Neofluar 40x/0.75 or a Plan-Apochromatin 63x/1.4 (oil immersion) objective and an AxioCam MRm Rev.3 camera. Acquisition and analysis was done with Zen Pro (Zeiss) software. All error bars on DSB repair graphs indicate the standard deviation. DSB repair analysis within regions of heterochromatin was performed as described 18.
Clonogenic survival assays
Between 250 and 3000 cells were seeded in 6 cm dishes followed by X-ray irradiation the next day as indicated. After 10–14 days cells were stained with crystal violet solution (0.5% crystal violet, 25% methanol) and colonies containing >100 cells were scored. The experiments were carried out in triplicates and the fraction of surviving cells was normalized to the untreated control.
HR and NHEJ reporter assays
NHEJ or HR reporter constructs (kind gift from Dr. Vera Gorbunova, University of Rochester) were digested in vitro with HindIII endonuclease. SCAI CRISPR WT or KO cells were co-transfected with RFP and either circular (negative control) or linearized reporter plasmids. Cells were collected 3 days after transfection and analyzed by FACS as described previously 19.
Statistics and Reproducibility
All western blots and microscopy experiments shown in figures were successfully repeated at least 3 times. For statistical testing of parameters where normal distributions and equal variance could be assumed we calculated p-values by the standard students t-test (Figure S3e,f; Figure S4k). In cases where equal variance could not be assumed, we used t-test with Welch correction (Figure 4a,b,c; Figure 5a,b,c,d; Figure S4b, S5c,d,e,h,i,j,k). For data sets where normal distribution could not be assumed, we employed the non-parametrical Mann-Whitney U test (Figure 2g; Figure 3j,k,m; Figure S3d) or Fisher’s exact test (Figure 3g,i).
Data availability
The entire CHROMASS mass spectrometry data set has been deposited to the ProteomeXchange Consortium via the PRIDE 20 partner repository with the dataset identifier PXD000490 (http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD000490), and was previously published 1. SCAI interactome recorded by Label Free Quantification (Figure 1d) has been deposited with the dataset identifier PXD004912 (http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD004912). All other data supporting the findings of this study are available from the corresponding authors upon request.
Supplementary Material
Acknowledgments
We thank Drs. Chikashi Obuse (University of Hokkaido, Japan) and Hiroshi Kimura (University of Osaka, Japan) for sharing unpublished data, Andre Nussenzweig, John Rouse, Vera Gorbunova, Anthony Hyman and Ina Poser for providing reagents, Gopal Karemore for statistical analysis support and Jutta Bulkesher for technical assistance with imaging. Work in the Mailand lab was supported by grants from the Novo Nordisk Foundation (Grants no. NNF14CC0001 and NNF15OC0016926), European Research Council (ERC), The Danish Cancer Society, Danish Medical Research Council, and the Lundbeck Foundation. Work in the Mann lab was supported by Center for Integrated Protein Research Munich (CIPSM). Work in the Daniel lab was supported by grants from the Novo Nordisk Foundation (Grant no. NNF14CC0001) and the Danish Medical Research Council, and AM is supported by a Marie Curie Intra-European Fellowship for Career Development (Project #627187). The Goodarzi laboratory is supported by the Canadian Institutes of Health Research. Dr. Goodarzi is currently the Canada Research Chair for Genome Damage and Instability Disease and this work was undertaken, in part, thanks to funding from the Canada Research Chairs program. Work in the Cole lab is supported by the Cancer Prevention and Research Institute of Texas (R1213), the Jeanne F. Shelby Scholarship Fund for the R. Lee Clark Fellowship, and the National Institutes of Health (DP2HD087943). Work in the Grosse lab is supported by the German Research Foundation (Grant GR 2111/2-2). All experiments were performed in full compliance with the ethical guidelines for biological research in Denmark. Work with all animals has been approved by the Department of Experimental Medicine (University of Copenhagen), the Danish Working Environment Authority, the Danish Animal Experiment Inspectorate, the MDACC Institutional Animal Care and Use Committee (IACUC) and the Regierungspräsidia Karlsruhe and Darmstadt.
Footnotes
Author contributions
R.K.H., A.M., S.L.P., K.T., and K.K. performed the biochemical and cell biological experiments. R.S., A.M. and M.S. carried out mouse experiments. B.R. performed and analyzed the mouse histology experiments. M.R. performed and analyzed the proteomics experiments. R.K.H and M.T. designed and generated CRISPR-based knock-out cell lines. S.O., T.W., R.G. and D.B. generated the SCAI knockout mouse. M.R., M.M., F.C., E.S., A.G., J.D., N.M., and S.B.-J. designed the experiments and N.M., and S.B.-J. conceived the project and wrote the manuscript. All authors discussed the results and commented on the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 2.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bekker-Jensen S, Mailand N. Assembly and function of DNA double-strand break repair foci in mammalian cells. DNA Repair (Amst) 2010;9:1219–1228. doi: 10.1016/j.dnarep.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 4.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lukas J, Lukas C, Bartek J. More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance. Nat Cell Biol. 2011;13:1161–1169. doi: 10.1038/ncb2344. [DOI] [PubMed] [Google Scholar]
- 6.Lemaitre C, Soutoglou E. Double strand break (DSB) repair in heterochromatin and heterochromatin proteins in DSB repair. DNA Repair (Amst) 2014;19:163–168. doi: 10.1016/j.dnarep.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 7.Schwertman P, Bekker-Jensen S, Mailand N. Regulation of DNA double-strand break repair by ubiquitin and ubiquitin-like modifiers. Nat Rev Mol Cell Biol. 2016;17:379–394. doi: 10.1038/nrm.2016.58. [DOI] [PubMed] [Google Scholar]
- 8.Bunting SF, Nussenzweig A. End-joining, translocations and cancer. Nat Rev Cancer. 2013;13:443–454. doi: 10.1038/nrc3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manis JP, et al. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat Immunol. 2004;5:481–487. doi: 10.1038/ni1067. [DOI] [PubMed] [Google Scholar]
- 10.Ward IM, et al. 53BP1 is required for class switch recombination. J Cell Biol. 2004;165:459–464. doi: 10.1083/jcb.200403021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Callen E, et al. 53BP1 mediates productive and mutagenic DNA repair through distinct phosphoprotein interactions. Cell. 2013;153:1266–1280. doi: 10.1016/j.cell.2013.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Virgilio M, et al. Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science. 2013;339:711–715. doi: 10.1126/science.1230624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Escribano-Diaz C, et al. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol Cell. 2013;49:872–883. doi: 10.1016/j.molcel.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 14.Chapman JR, et al. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol Cell. 2013;49:858–871. doi: 10.1016/j.molcel.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zimmermann M, Lottersberger F, Buonomo SB, Sfeir A, de Lange T. 53BP1 regulates DSB repair using Rif1 to control 5’ end resection. Science. 2013;339:700–704. doi: 10.1126/science.1231573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noon AT, et al. 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat Cell Biol. 2010;12:177–184. doi: 10.1038/ncb2017. [DOI] [PubMed] [Google Scholar]
- 17.Goodarzi AA, Kurka T, Jeggo PA. KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nat Struct Mol Biol. 2011;18:831–839. doi: 10.1038/nsmb.2077. [DOI] [PubMed] [Google Scholar]
- 18.Klement K, et al. Opposing ISWI- and CHD-class chromatin remodeling activities orchestrate heterochromatic DNA repair. J Cell Biol. 2014;207:717–733. doi: 10.1083/jcb.201405077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raschle M, et al. Proteomics reveals dynamic assembly of repair complexes during bypass of DNA cross-links. Science. 2015;348 doi: 10.1126/science.1253671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brandt DT, Xu J, Steinbeisser H, Grosse R. Regulation of myocardin-related transcriptional coactivators through cofactor interactions in differentiation and cancer. Cell Cycle. 2009;8:2523–2527. doi: 10.4161/cc.8.16.9398. [DOI] [PubMed] [Google Scholar]
- 21.Brandt DT, et al. SCAI acts as a suppressor of cancer cell invasion through the transcriptional control of beta1-integrin. Nat Cell Biol. 2009;11:557–568. doi: 10.1038/ncb1862. [DOI] [PubMed] [Google Scholar]
- 22.Bothmer A, et al. Regulation of DNA end joining, resection, and immunoglobulin class switch recombination by 53BP1. Mol Cell. 2011;42:319–329. doi: 10.1016/j.molcel.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bekker-Jensen S, et al. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J Cell Biol. 2006;173:195–206. doi: 10.1083/jcb.200510130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gunn A, Stark JM. I-SceI-based assays to examine distinct repair outcomes of mammalian chromosomal double strand breaks. Methods Mol Biol. 2012;920:379–391. doi: 10.1007/978-1-61779-998-3_27. [DOI] [PubMed] [Google Scholar]
- 25.Ward IM, Minn K, van Deursen J, Chen J. p53 Binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol Cell Biol. 2003;23:2556–2563. doi: 10.1128/MCB.23.7.2556-2563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Broering TJ, et al. BRCA1 establishes DNA damage signaling and pericentric heterochromatin of the X chromosome in male meiosis. J Cell Biol. 2014;205:663–675. doi: 10.1083/jcb.201311050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Svetlanov A, Cohen PE. Mismatch repair proteins, meiosis, and mice: understanding the complexities of mammalian meiosis. Exp Cell Res. 2004;296:71–79. doi: 10.1016/j.yexcr.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 28.Hunter N. Meiotic Recombination: The Essence of Heredity. Cold Spring Harbor perspectives in biology. 2015;7 doi: 10.1101/cshperspect.a016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jackson SP. The DNA-damage response: new molecular insights and new approaches to cancer therapy. Biochem Soc Trans. 2009;37:483–494. doi: 10.1042/BST0370483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 31.Bryant HE, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 32.Eberl HC, Spruijt CG, Kelstrup CD, Vermeulen M, Mann M. A map of general and specialized chromatin readers in mouse tissues generated by label-free interaction proteomics. Mol Cell. 2013;49:368–378. doi: 10.1016/j.molcel.2012.10.026. [DOI] [PubMed] [Google Scholar]
- 33.Tsouroula K, et al. Temporal and Spatial Uncoupling of DNA Double Strand Break Repair Pathways within Mammalian Heterochromatin. Mol Cell. 2016;63:293–305. doi: 10.1016/j.molcel.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 34.Cowell IG, et al. gammaH2AX foci form preferentially in euchromatin after ionising-radiation. PLoS One. 2007;2:e1057. doi: 10.1371/journal.pone.0001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goodarzi AA, et al. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell. 2008;31:167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 36.Kakarougkas A, et al. Opposing roles for 53BP1 during homologous recombination. Nucleic Acids Res. 2013;41:9719–9731. doi: 10.1093/nar/gkt729. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 1.Raschle M, et al. DNA repair. Proteomics reveals dynamic assembly of repair complexes during bypass of DNA cross-links. Science. 2015;348:1253671. doi: 10.1126/science.1253671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hubner NC, et al. Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J Cell Biol. 2010;189:739–754. doi: 10.1083/jcb.200911091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daniel JA, et al. PTIP promotes chromatin changes critical for immunoglobulin class switch recombination. Science. 2010;329:917–923. doi: 10.1126/science.1187942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Munoz IM, Szyniarowski P, Toth R, Rouse J, Lachaud C. Improved genome editing in human cell lines using the CRISPR method. PLoS One. 2014;9:e109752. doi: 10.1371/journal.pone.0109752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox J, et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandt DT, et al. SCAI acts as a suppressor of cancer cell invasion through the transcriptional control of beta1-integrin. Nat Cell Biol. 2009;11:557–568. doi: 10.1038/ncb1862. [DOI] [PubMed] [Google Scholar]
- 7.Mosbech A, Lukas C, Bekker-Jensen S, Mailand N. The deubiquitylating enzyme USP44 counteracts the DNA double-strand break response mediated by the RNF8 and RNF168 ubiquitin ligases. J Biol Chem. 2013;288:16579–16587. doi: 10.1074/jbc.M113.459917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cole F, et al. Homeostatic control of recombination is implemented progressively in mouse meiosis. Nat Cell Biol. 2012;14:424–430. doi: 10.1038/ncb2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bunting SF, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Callen E, et al. 53BP1 mediates productive and mutagenic DNA repair through distinct phosphoprotein interactions. Cell. 2013;153:1266–1280. doi: 10.1016/j.cell.2013.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Callen E, et al. ATM prevents the persistence and propagation of chromosome breaks in lymphocytes. Cell. 2007;130:63–75. doi: 10.1016/j.cell.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 12.Daniel JA, et al. Loss of ATM kinase activity leads to embryonic lethality in mice. J Cell Biol. 2012;198:295–304. doi: 10.1083/jcb.201204035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans EP, Breckon G, Ford CE. An Air-Drying Method for Meiotic Preparations from Mammalian Testes. Cytogenetics. 1964;3:289–294. doi: 10.1159/000129818. [DOI] [PubMed] [Google Scholar]
- 14.Gudjonsson T, et al. TRIP12 and UBR5 suppress spreading of chromatin ubiquitylation at damaged chromosomes. Cell. 2012;150:697–709. doi: 10.1016/j.cell.2012.06.039. [DOI] [PubMed] [Google Scholar]
- 15.Toledo LI, et al. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell. 2013;155:1088–1103. doi: 10.1016/j.cell.2013.10.043. [DOI] [PubMed] [Google Scholar]
- 16.Goodarzi AA, Kurka T, Jeggo PA. KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nat Struct Mol Biol. 2011;18:831–839. doi: 10.1038/nsmb.2077. [DOI] [PubMed] [Google Scholar]
- 17.Goodarzi AA, et al. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell. 2008;31:167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 18.Noon AT, et al. 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat Cell Biol. 2010;12:177–184. doi: 10.1038/ncb2017. [DOI] [PubMed] [Google Scholar]
- 19.Seluanov A, Mao Z, Gorbunova V. Analysis of DNA double-strand break (DSB) repair in mammalian cells. J Vis Exp. 2010 doi: 10.3791/2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vizcaino JA, et al. 2016 update of the PRIDE database and its related tools. rNucleic Acids Res. 2016;44:D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The entire CHROMASS mass spectrometry data set has been deposited to the ProteomeXchange Consortium via the PRIDE 20 partner repository with the dataset identifier PXD000490 (http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD000490), and was previously published 1. SCAI interactome recorded by Label Free Quantification (Figure 1d) has been deposited with the dataset identifier PXD004912 (http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD004912). All other data supporting the findings of this study are available from the corresponding authors upon request.