Abstract
Recently, an association between Merkel cell polyomavirus (MCPyV) and epidermal growth factor receptor (EGFR) mutations in nonsmall cell lung cancer (NSCLC) was reported. However, the underlying carcinogenic effects and the prognosis related to MCPyV are still unclear. The aim of this study was to clarify the incidence and prognosis related to MCPyV infections in NSCLC.
Tissue samples from 167 NSCLC patients (92 with squamous cell carcinomas [SCCs] and 75 with adenocarcinomas) were analyzed for the presence of MCPyV and EGFR mutations. Clinicopathological characteristics, disease-free survival rate, and overall survival rate were assessed with respect to MCPyV.
MCPyV DNA was detected in 30 patients (18.0%) out of 167 patients, and EGFR mutations were found in 31 out of 127 patients (24.4%). EGFR mutations were more frequently detected in MCPyV-positive patients than in MCPyV-negative patients; however, this did not reach statistical significance (P = 0.075). There was no difference in overall survival between patients with and without MCPyV infections. The disease-free survival rate of patients with pN0 stage, SCC, or EGFR mutations was lower for patients with MCPyV than without MCPyV (P = 0.036, 0.042, and 0.050, respectively).
Although the prevalence of MCPyV infection was relatively low, the presence of MCPyV DNA was significantly correlated with cancer prognosis in subgroups of NSCLC patients. These results suggest that MCPyV may be partly associated with pathogenesis and prognosis in some cases of NSCLC.
Keywords: EGFR mutations, Merkel cell polyomavirus, nonsmall cell lung cancer, prognosis
1. Introduction
Lung cancer is the leading cause of cancer-related deaths worldwide, with only 16.8% of lung cancer patients alive 5 years after diagnosis.[1] Many causative factors for lung cancer have been identified, including active smoking, secondhand smoke, occupational agents, radiation, and environmental pollutants. In addition, genetic and hormonal factors and infections (eg, tuberculosis, human papillomavirus [HPV], or human immunodeficiency virus) may play a role in the development of lung cancer.[2–4]
Approximately 80% of Merkel cell carcinomas of the skin are infected with Merkel cell polyomavirus (MCPyV).[5] Since this discovery in 2008, MCPyV infections have been investigated in lung cancer, especially in small cell lung cancer. Presence of the virus is correlated with hypermethylation of the tumor suppressor gene RASSF1A in small cell lung cancers.[6] MCPyV DNA has been detected in the lower respiratory tract of adults admitted to the hospital, although the mode of transmission and pathogenic role of MCPyV in the respiratory system has not been established.[7] A possible association between HPV and MCPyV infections in lung cancer, especially in nonsmall cell lung cancer (NSCLC), has been reported.[8] The infection rate of MCPyV in NSCLCs ranges from 4.7% to 17.9% in different cohorts.[9–12]
Xu et al[12] first reported an association between MCPyV infections and epidermal growth factor receptor (EGFR) mutations in NSCLC. Several studies have suggested that EGFR mutation and expression in NSCLC are associated with poor survival, frequent lymph node metastasis, and reduced chemosensitivity.[13] We hypothesized that MCPyV infections might be related to the prognosis of NSCLC. The aim of this study was to clarify the incidence and prognosis related to MCPyV infections in NSCLC.
2. Materials and methods
2.1. Patients and clinical characteristics
Tumor specimens and corresponding nonmalignant lung tissue specimens (n = 167) were provided by the National Biobank of Korea, Kyungpook National University Hospital (KNUH), supported by the Ministry of Health, Welfare, and Family Affairs. All materials from the National Biobank of Korea, KNUH, were obtained under institutional review board-approved protocols. We collected basic clinical data including age, gender, disease stage, and smoking status. The pathologic staging of lung cancer was based on the 7th AJCC staging system.[14]
2.2. Identification of MCPyV
Genomic DNA was extracted using a QIAamp DNA Mini Kit (QIAGEN [New York, NY]). The polymerase chain reaction (PCR) was conducted with 200 ng of DNA using the AmpliTaq Gold 360 Master Mix (Life Technologies, Tokyo, Japan) and 0.4 mM of each primer in a total volume of 50 mL. The PCR amplification of MCPyV was performed as described previously with minor modification.[5,8] PCR was performed using AmpliTaq Gold DNA polymerase (Applied Biosystems [Foster City, California]). To detect the MCPyV large T antigen (LT) and viral protein 1 (VP1) genes, 3 primer sets (LT1, LT3, and VP1) were used as described previously.[5] The b-globin gene (HBB) was amplified to confirm the presence of PCR-amplifiable DNA. The PCR products were electrophoresed on a 1.5% agarose gel and stained with ethidium bromide to confirm the size of the bands.
2.3. Analysis of EGFR mutations
Exons 19 and 21 of the EGFR gene were amplified by PCR as described previously with minor modification.[12] PCR was performed using AmpliTaq Gold DNA polymerase (Applied Biosystems [Foster City, California]). The PCR products were electrophoresed on a 1.5% agarose gel and stained with ethidium bromide to confirm the size of the bands. Direct DNA sequencing for EGFR mutations was then performed using an ABI 3730 DNA sequencer by Bionics Inc, Korea. All sequencing reactions were performed in both the sense and antisense directions. Once EGFR mutations were identified, DNA isolated from matched blood or normal lung tissue was also amplified and sequenced to verify the EGFR alteration as a somatic mutation.
2.4. Statistical analysis
The SPSS statistical package (version 22.0 for Windows; Chicago, IL) was used for all statistical analyses. Chi-square and Fisher exact tests were used to analyze the relationship between variables. Survival curves, estimated with the Kaplan–Meier method (univariate analysis), were compared by log-rank test. Overall survival was defined as the time between diagnosis and either death from any cause or last follow-up. Disease-free survival was defined as the time between diagnosis and the development of local or distant recurrence. All P-values ≤0.05 were considered statistically significant.
3. Results
3.1. Clinicopathological characteristics of Merkel cell polyomavirus and EGFR mutations in NSCLC
Tissue from 167 NSCLC patients (92 with squamous cell carcinomas [SCCs] and 75 with adenocarcinomas [ADCs]) was analyzed in this study. There were 124 men (74.3%) and 43 women (25.7%), with a mean age of 64.7 ± 8.1 years. The mean follow-up time was 34.8 ± 22.3 months. MCPyV DNA was detected in 30 patients (18.0%). There were no statistically significant differences in demographic and histopathological variables according to the presence or absence of MCPyV DNA (Table 1).
Table 1.
Demographic and histopathological characteristics according to the presence or absence of MCPyV DNA in patients with NSCLC.
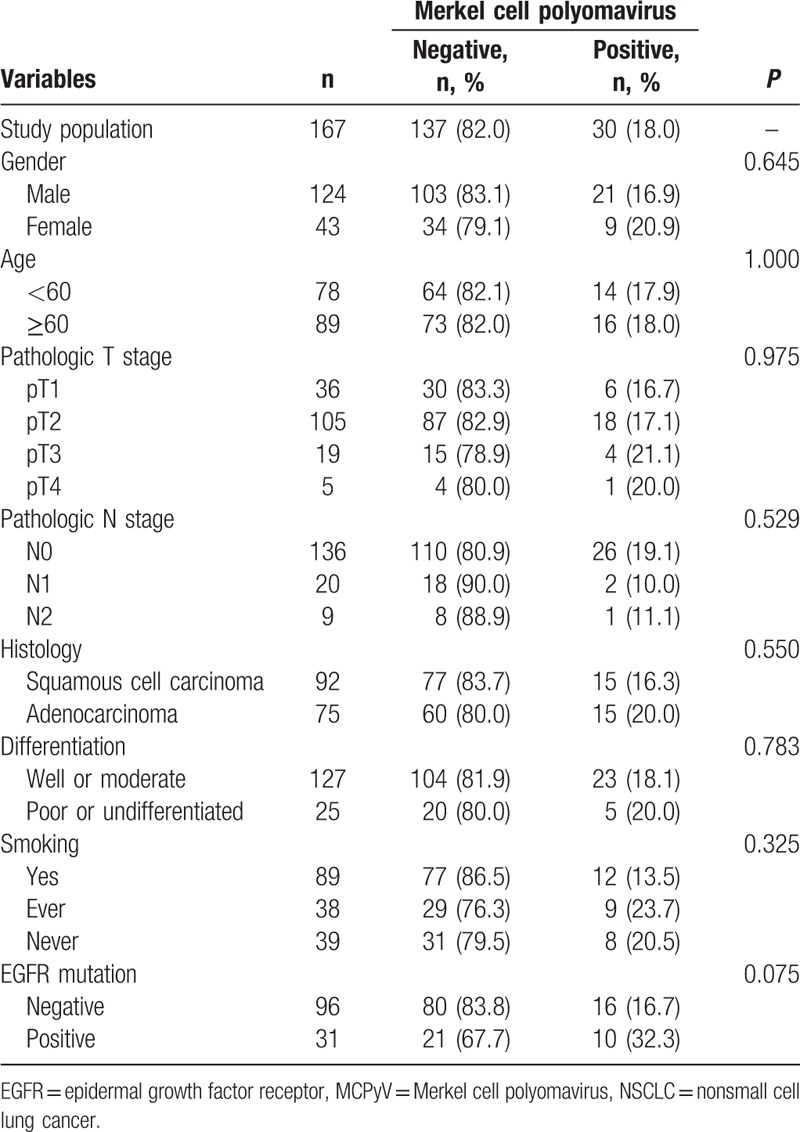
EGFR mutations were analyzed in a randomly selected subset of 127 patients and found in 31 of these patients (24.4%). EGFR mutations were more common in women, ADCs, and never smokers (Table 2). The infection rate of MCPyV was higher in NSCLCs with EGFR mutations (32.3%) than without EGFR mutations (16.7%); however, this difference was not statistically significant (P = 0.075; Table 1).
Table 2.
Clinicopathological characteristics of EGFR mutation in the study population.

3.2. Prognostic value of Merkel cell polyomavirus in NSCLC
Of the 167 patients in the study population, 59 had recurrent disease and 47 died after surgery during the follow-up period. The 5-year disease-free and overall survival rates for all patients in the cohort were 52.2% and 63.7%, respectively. The following variables were evaluated by the Kaplan–Meier method as potential prognostic factors associated with disease-free and overall survival: age (<60 vs ≥60), gender, pT stage, pN stage, histology (SCC vs ADC), tumor cell differentiation (well or moderate vs poor or undifferentiated), presence or absence of EGFR mutations, and presence or absence of MCPyV (Table 3). The disease-free survival rate was higher for patients with lower pN stage (P < 0.001), SCC (P = 0.040), or without EGFR mutations (P = 0.007). The overall survival rate was higher for patients with lower pT stage (P < 0.001), lower pN stage (P < 0.001), or well or moderate differentiation (P = 0.003). Median disease-free survival was shorter for MCPyV-positive patients (38.2 months) than MCPyV-negative patients (74.0 months, χ2 = 2.36); however, this difference was not statistically significant (P = 0.125) (Fig. 1). Other variables including MCPyV infections with respect to overall survival did not show a prognostic value (Fig. 1).
Table 3.
Univariate analysis of prognostic factors in NSCLC.
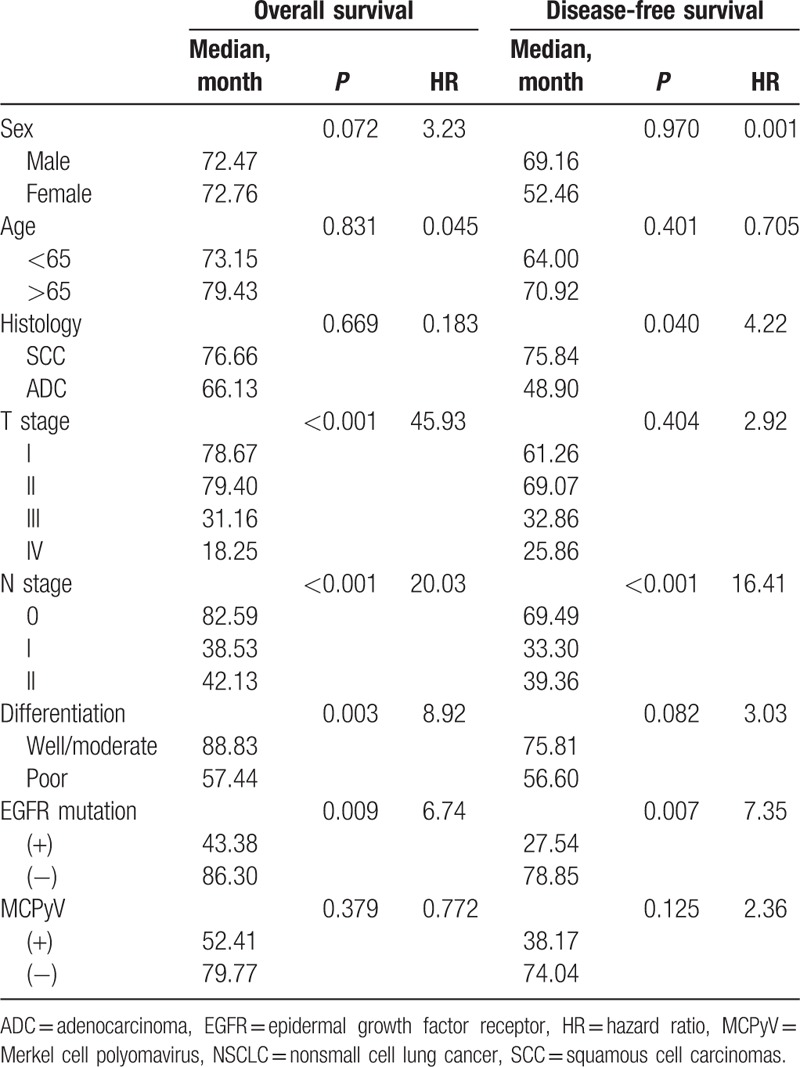
Figure 1.
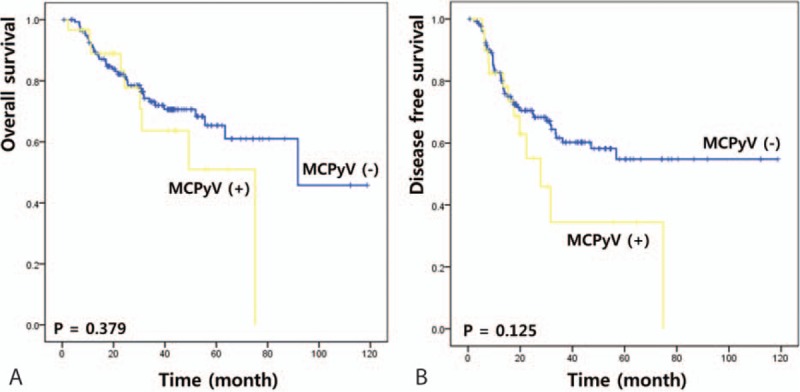
Prognostic impact of MCPyV infection in NSCLC patients. (A) Overall survival, (B) disease-free survival. MCPyV = Merkel cell polyomavirus, NSCLC = nonsmall cell lung cancer.
Disease-free survival and overall survival were analyzed in various patient subgroups with respect to MCPyV. There was no difference in overall survival rates for any of the parameters between MCPyV-positive and negative patients. However, the disease-free survival rate of patients with pN0 stage, SCC, or EGFR mutations was lower for patients with MCPyV than without MCPyV (P = 0.036, 0.042, and 0.050, respectively; Fig. 2).
Figure 2.
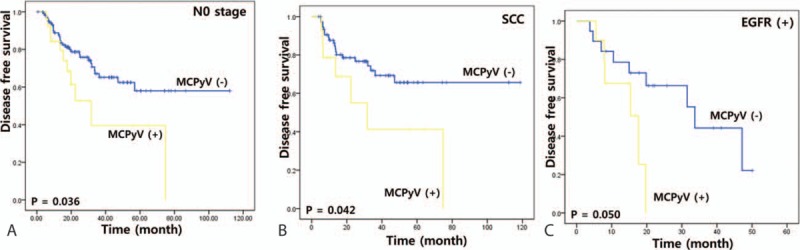
Stratifying analysis of disease-free survival of MCPyV infection in NSCLC patients with pN0 stage (A), squamous cell carcinoma (B), and positive EGFR mutation (C). EGFR = epidermal growth factor receptor, MCPyV = Merkel cell polyomavirus, NSCLC = nonsmall cell lung cancer.
4. Discussion
Several viral agents have been identified that cause or contribute to approximately 20% of the global cancer incidence (eg, hepatitis B and C viruses for hepatocellular carcinoma, Epstein–Barr virus for lymphoma, and HPV for cervical cancer).[15] Several other cancers have potential links to infections, including basal cell carcinomas of the skin, breast cancer, colorectal cancer, and some lung cancers.[16]
Polyomaviruses are nonenveloped, icosahedral viruses containing small circular, double-stranded DNA of approximately 5000 base pairs.[17] MCPyV is the first polyomavirus identified as oncogenic for humans.[5,18] The genome of MCPyV has 5387 base pairs, divided into 3 functional regions including a large T antigen, small T antigen, and the genes for capsid proteins VP1, VP2, and VP3.[5]
Since MCPyV was found in most Merkel cell carcinomas, possible associations with squamous carcinomas of the skin, cervical carcinomas, Bowen disease, basal cell skin carcinomas, and extrapulmonary small cell carcinomas have been reported.[19–23] MCPyV infections have been investigated for lung cancer, especially small cell lung cancer, which is histologically similar to Merkel cell carcinoma.[6,24] The prevalence of MCPyV in NSCLC was reported as 16.7%, 4.7%, and 9.1% in 3 different cohorts of Americans and Europeans.[8,9,11] In Asia, its infection rate in NSCLCs was recently found to be 17.9% in a Japanese cohort and 16.9% in a Chinese cohort.[10,12] In the present study, MCPyV DNA was detected in 18.0% of patients.
Previous studies have focused on detecting the presence of newly identified MCPyV in NSCLCs but not on understanding whether MCPyV contributes to the pathogenesis or the prognosis of NSCLCs. Xu et al[12] reported an association between MCPyV and EGFR mutation-driven NSCLC and suggested the possible role of MCPyV in the development of NSCLC. Several studies have suggested that EGFR mutations in NSCLC are associated with poor survival, frequent lymph node metastasis, and reduced chemosensitivity.[13] Thus, we were interested in the association between MCPyV infections and EGFR mutations, and cancer prognosis in MCPyV-infected NSCLCs. In this study, the MCPyV rate was higher in NSCLC patients with EGFR mutations (32.3%) compared to those without EGFR mutations (16.7%). This difference was not statistically significant due to the small number of EGFR-positive NSCLC patients, but it suggests a possible role for MCPyV in inducing EGFR mutations in lung tissue.
We investigated the prognosis related to MCPyV infections. There were no statistically significant differences between the presence of MCPyV and overall survival in our study. The disease-free survival rate of patients with MCPyV was lower than for patients without MCPyV in subgroup analyses based on pN0 stage, SCC, and EGFR mutations. It indicated that MCPyV infection and EGFR mutation have a synergistic effect on NSCLC prognosis. The prognostic value of MCPyV infections only in EGFR mutation-positive patients is likely explained by the deep association between MCPyV infection and EGFR mutation. The infection rate of MCPyV was similar in SCC and ADC; however, our analysis suggests that MCPyV has a more significant effect during SCC progression. Interestingly, MCPyV infection had a powerful prognostic value in the patients without lymph node spread suggesting their independent role in the progression of NSCLC.
This study has several limitations. First, it was a retrospective analysis. Second, the statistical power of the analysis may be weak as the number of patients in each subgroup was small. In particular, only 30 patients had MCPyV DNA and 31 patients had an EGFR mutation. Finally, we did not determine how MCPyV affects the prognosis of NSCLC in certain patients. A subsequent study with a larger patient cohort should be performed, and the molecular changes associated with MCPyV infections should be investigated.
5. Conclusions
To our knowledge, this is the first study to report a relationship between MCPyV infections and NSCLC prognosis. The results suggest that MCPyV may be associated with pathogenesis and progression in SCC NSCLC and EGFR-positive patients. This discovery may lead to new therapeutic strategies aimed at the molecular pathway of MCPyV and an improved classification of NSCLC based on this infection.
Acknowledgments
The authors thank Biomedical Research Institute grant, Kyungpook National University Hospital (2015) for the support.
Footnotes
Abbreviations: ADC = adenocarcinoma, EGFR = epidermal growth factor receptor, HPV = human papillomavirus, MCPyV = Merkel cell polyomavirus, NSCLC = nonsmall cell lung cancer, SCC = squamous cell carcinomas.
GJK and JHL contributed equally to this work.
Funding/support: This work was supported by a Biomedical Research Institute grant, Kyungpook National University Hospital (2015).
The authors have no conflicts of interest to disclose.
References
- [1].Howlader N, Noone A, Krapcho M, et al. SEER Cancer Statistics Review, 1975–2012. Bethesda: National Cancer Institute; 2015. [Google Scholar]
- [2].Alberg AJ, Brock MV, Ford JG, et al. Epidemiology of lung cancer: diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest 2013;143:e1S–29S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Rezazadeh A, Laber DA, Ghim SJ, et al. The role of human papilloma virus in lung cancer: a review of the evidence. Am J Med Sci 2009;338:64–7. [DOI] [PubMed] [Google Scholar]
- [4].Wu CY, Hu HY, Pu CY, et al. Pulmonary tuberculosis increases the risk of lung cancer: a population-based cohort study. Cancer 2011;117:618–24. [DOI] [PubMed] [Google Scholar]
- [5].Feng H, Shuda M, Chang Y, et al. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 2008;319:1096–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Helmbold P, Lahtz C, Herpel E, et al. Frequent hypermethylation of RASSF1A tumour suppressor gene promoter and presence of Merkel cell polyomavirus in small cell lung cancer. Eur J Cancer 2009;45:2207–11. [DOI] [PubMed] [Google Scholar]
- [7].Babakir-Mina M, Ciccozzi M, Lo Presti A, et al. Identification of Merkel cell polyomavirus in the lower respiratory tract of Italian patients. J Med Virol 2010;82:505–9. [DOI] [PubMed] [Google Scholar]
- [8].Joh J, Jenson AB, Moore GD, et al. Human papillomavirus (HPV) and Merkel cell polyomavirus (MCPyV) in non-small cell lung cancer. Exp Mol Pathol 2010;89:222–6. [DOI] [PubMed] [Google Scholar]
- [9].Gheit T, Munoz JP, Levican J, et al. Merkel cell polyomavirus in non-small cell lung carcinomas from Chile. Exp Mol Pathol 2012;93:162–6. [DOI] [PubMed] [Google Scholar]
- [10].Hashida Y, Imajoh M, Nemoto Y, et al. Detection of Merkel cell polyomavirus with a tumour-specific signature in non-small cell lung cancer. Br J Cancer 2013;108:629–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lasithiotaki I, Antoniou K, Derdas S, et al. The presence of Merkel cell polyomavirus is associated with deregulated expression of BRAF and Bcl-2 genes in non-small cell lung cancer. Int J Cancer 2013;133:604–11. [DOI] [PubMed] [Google Scholar]
- [12].Xu S, Jiang J, Yu X, et al. Association of Merkel cell polyomavirus infection with EGFR mutation status in Chinese non-small cell lung cancer patients. Lung Cancer 2014;83:341–6. [DOI] [PubMed] [Google Scholar]
- [13].Bethune G, Bethune D, Ridgway N, et al. Epidermal growth factor receptor (EGFR) in lung cancer: an overview and update. J Thoracic Dis 2011;2:48–51. [PMC free article] [PubMed] [Google Scholar]
- [14].Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol 2010;17:1471–4. [DOI] [PubMed] [Google Scholar]
- [15].Zur Hausen H, Fox JG, Wang TC, et al. Infections Causing Human Cancer. Weinheim: Wiley-VCH; 2006. [Google Scholar]
- [16].Zur Hausen H. The search for infectious causes of human cancers: where and why. Virology 2009;392:1–0. [DOI] [PubMed] [Google Scholar]
- [17].Johnson EM. Structural evaluation of new human polyomaviruses provides clues to pathobiology. Trends Microbiol 2010;18:215–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Chang Y, Moore PS. Merkel cell carcinoma: a virus-induced human cancer. Ann Rev Pathol 2012;7:123–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Dworkin AM, Tseng SY, Allain DC, et al. Merkel cell polyomavirus in cutaneous squamous cell carcinoma of immunocompetent individuals. J Invest Dermatol 2009;129:2868–74. [DOI] [PubMed] [Google Scholar]
- [20].Zur Hausen A. Merkel cell polyomavirus in the pathogenesis of non-melanoma skin cancer. Pathologe 2009;30(Suppl 2):217–20. [DOI] [PubMed] [Google Scholar]
- [21].Murakami M, Imajoh M, Ikawa T, et al. Presence of Merkel cell polyomavirus in Japanese cutaneous squamous cell carcinoma. J Clin Virol 2011;50:37–41. [DOI] [PubMed] [Google Scholar]
- [22].Imajoh M, Hashida Y, Nemoto Y, et al. Detection of Merkel cell polyomavirus in cervical squamous cell carcinomas and adenocarcinomas from Japanese patients. Virol J 2012;9:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hourdequin KC, Lefferts JA, Brennick JB, et al. Merkel cell polyomavirus and extrapulmonary small cell carcinoma. Oncol Lett 2013;6:1049–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wetzels CT, Hoefnagel JG, Bakkers JM, et al. Ultrastructural proof of polyomavirus in Merkel cell carcinoma tumour cells and its absence in small cell carcinoma of the lung. PloS One 2009;4:e4958. [DOI] [PMC free article] [PubMed] [Google Scholar]


