Abstract
Background
In monocentric studies, patients with mild cognitive impairment (MCI) and Alzheimer's disease (AD) dementia exhibited alterations of functional cortical connectivity in resting-state functional MRI (rs-fMRI) analyses. Multicenter studies provide access to large sample sizes, but rs-fMRI may be particularly sensitive to multiscanner effects.
Methods
We used data from five centers of the “German resting-state initiative for diagnostic biomarkers” (psymri.org), comprising 367 cases, including AD patients, MCI patients and healthy older controls, to assess the influence of the distributed acquisition on the group effects. We calculated accuracy of group discrimination based on whole brain functional connectivity of the posterior cingulate cortex (PCC) using pooled samples as well as second-level analyses across site-specific group contrast maps.
Results
We found decreased functional connectivity in AD patients vs. controls, including clusters in the precuneus, inferior parietal cortex, lateral temporal cortex and medial prefrontal cortex. MCI subjects showed spatially similar, but less pronounced, differences in PCC connectivity when compared to controls. Group discrimination accuracy for AD vs. controls (MCI vs. controls) in the test data was below 76% (72%) based on the pooled analysis, and even lower based on the second level analysis stratified according to scanner. Only a subset of quality measures was useful to detect relevant scanner effects.
Conclusions
Multicenter rs-fMRI analysis needs to employ strict quality measures, including visual inspection of all the data, to avoid seriously confounded group effects. While pending further confirmation in biomarker stratified samples, these findings suggest that multicenter acquisition limits the use of rs-fMRI in AD and MCI diagnosis.
Highlights
-
•
Diagnostic accuracy of multicenter rs-fMRI in AD and MCI
-
•
Quality metrics for multicenter rs-fMRI that should be used
-
•
Quality metrics for multicenter rs-fMRI that should not be used
-
•
Multicenter rs-fMRI will have limited diagnostic use in clinical routine diagnosis
1. Introduction
Criteria for prodromal Alzheimer’s disease (AD) (Albert et al., 2011, Dubois et al., 2010, Dubois et al., 2007, Dubois et al., 2014) and AD dementia (McKhann et al., 2011) diagnosis include structural imaging markers, such as MRI based hippocampus volumetry, molecular imaging markers, such as amyloid PET, and functional imaging markers, such as 18FDG-PET. All these imaging markers have already been evaluated in large multicenter cohorts, such as ADNI, EDSD, NEST-DD and others (Doraiswamy et al., 2014, Herholz, 2010, Kilimann et al., 2014, Risacher et al., 2009).
Particularly, FDG-PET has proven a precise predictor of imminent conversion from mild cognitive impairment (MCI) to AD dementia (Ito et al., 2015). At the same time, PET imaging is relatively expensive and availability of PET scanners is limited. Resting state fMRI (rs-fMRI) has been discussed as a functional imaging alternative for 18FDG-PET (Teipel et al., 2015). Decline of default mode network connectivity, a brain network encompassing key regions of AD pathology such as posterior cingulate, precuneus, inferior parietal lobes, prefrontal cortex and medial temporal lobes (Fox et al., 2005), has been shown in AD dementia and MCI patients compared to age matched controls in a range of studies (Chhatwal et al., 2013, Greicius et al., 2004, Thomas et al., 2014). Results on diagnostic accuracies are mixed, ranging from 62% to > 90% group separation of MCI or AD dementia cases from healthy control cases in monocenter studies (Dyrba et al., 2015b, Koch et al., 2012). Such variation across studies likely not only reflects differences in the cohorts, but also variation in acquisition parameters of rs-fMRI sequences between studies. High variability of group discrimination across sites, however, would severely diminish the value of rs-fMRI as an imaging biomarker of AD.
Multicenter studies in healthy people revealed high variability of task related functional MRI image properties, such as transient signals, smoothness and the shape of the hemodynamic response function, even when multicenter data stemmed from the same brand and model of scanners (Zou et al., 2005). Consistent with these findings, test-retest reliability studies of rs-fMRI suggest high intra-individual variability of resting state connectivity even in healthy people repeatedly scanned at the same scanner (Chen et al., 2015, Jovicich et al., 2016, Lin et al., 2015, Meindl et al., 2010, Orban et al., 2015, Shirer et al., 2015), including long-term evaluation after more than 12 months (Blautzik et al., 2013, Chou et al., 2012, Guo et al., 2012). Multiscanner evaluation suggests high variability of signal-to-noise and contrast-to-noise ratios, particularly when using field strengths of 3T and higher (Jovicich et al., 2016, Lin et al., 2015, Magnotta et al., 2006). In an explicit linear model, center accounted for a large amount of variance across voxel-wise resting state connectivity (Suckling et al., 2012).
Several rs-fMRI multicenter studies have investigated alterations of functional connectivity in AD and other neuropsychiatric conditions, but without consideration of multiscanner effects (Esslinger et al., 2011) even though protocols differed between sites in some studies (Chhatwal et al., 2013, Demertzi et al., 2015, Martucci et al., 2015, Sripada et al., 2014, Thomas et al., 2014). Some of these studies used the same scanner type across sites (Demertzi et al., 2015, Esslinger et al., 2011, Thomas et al., 2014), but some did not (Chhatwal et al., 2013, Martucci et al., 2015, Sripada et al., 2014).
Several studies reported techniques to reduce inter-scanner variability, mostly, however, in data from healthy people. One study probed a wide range of processing steps to reduce test-retest variability (Shirer et al., 2015). Another study comparing different connectivity metrics found most stable results for cross-correlation as compared to cross-coherence and partial cross-correlation (Fiecas et al., 2013). Two studies in healthy adults and young people at risk of psychosis, respectively, used scanner as a covariate in a second-level linear ANOVA model (Anticevic et al., 2015, Biswal et al., 2010), another study in healthy adults used conjunction analysis across scanners (Long et al., 2008). Only one previous study explicitly modelled center effects in healthy older people and MCI cases using a meta-analysis of between group effects across four different cohorts (Tam et al., 2015).
Here, we used rs-fMRI data of people with AD dementia, MCI and healthy older controls from the “German resting-state initiative for diagnostic biomarkers” (www.psymri.org) collected at five sites to compare previously employed measures of scan quality across sites (Jenkinson et al., 2002, Yan et al., 2013a), determine the effect of multicenter acquisition on between group effects, and assess diagnostic accuracies from different univariate analysis approaches. We expected to find large heterogeneity of between group effects that would likely impair the use of multicenter rs-fMRI data as diagnostic biomarker for AD. We used the widely established structural measure of hippocampus volume that has been found to be stable against multicenter effects (Ewers et al., 2006) as an internal benchmark for the functional connectivity metric.
2. Material and methods
The original data set consisted of 367 rs-fMRI scans that have been retrieved retrospectively from five sites within the framework of the “German resting-state initiative for diagnostic biomarkers” (www.psymri.org). After a first round of visual quality check, 350 rs-fMRI data were retained, whereas 17 scans were dropped due to severe problems with scan quality, incomplete scans or scans covering only parts of the brain. From the remaining 350 scans, all 100 scans from one site (site V) were rated as borderline quality due to severe susceptibility effects and subsequent analyses were conducted both with and without the scans from this site. Distribution of demographic characteristics of participants across sites is summarized in Table 1, Table 2; the number of participants per scanner is shown in supplementary table 1.
Table 1.
Demographic characteristics, all sites
| AD | MCI | Controls | |
|---|---|---|---|
| No. cases (women)1 | 84 (46) | 115 (59) | 151 (82) |
| Age (SD) [years]2 | 72.0 (9.0) | 72.6 (8.0) | 69.0 (7.8) |
| MMSE (SD), number3 | 22.4 (4.4), 84 | 26.7 (1.8), 115 | 28.9 (1.0) 115 |
| MoCA (SD), number4 | – | 22.7 (3.0), 22 | 26.4 (2.1), 19 |
| education (SD) [years]5 | 10.9 (2.4) | 12.4 (3.3) | 12.9 (3.1) |
MMSE – Mini Mental State Examination (Folstein et al., 1975)
MoCA – Montreal Cognitive Assessment (Nasreddine et al., 2005)
Not significantly different between groups, χ2 = 0.315, 2 df, p = 0.85.
Significantly different between groups, F(2, 347) = 7.5, p < 0.001.
Significantly different between groups, Kruskal Wallis Test, p < 0.001.
Significantly different between groups, Kruskal Wallis Test, p < 0.001.
Significantly different between groups, F(2, 323) = 11.4, p < 0.001.
Table 2.
Demographic characteristics, one site excluded.
| AD | MCI | Controls | |
|---|---|---|---|
| No. cases (women)1 | 53 (31) | 79 (43) | 118 (61) |
| Age (SD) [years]2 | 72.4 (8.8) | 74.8 (6.0) | 70.4 (6.2) |
| MMSE (SD), number3 | 22.5 (4.4), 53 | 26.5 (1.8), 79 | 28.8 (1.0) 97 |
| MoCA (SD), number4 | – | 22.7 (3.0), 22 | 26.4 (2.1), 19 |
| education (SD) [years]5 | 11.4 (2.1) | 13.0 (3.4) | 13.6 (3.1) |
MMSE – Mini Mental State Examination (Folstein et al., 1975)
MoCA – Montreal Cognitive Assessment (Nasreddine et al., 2005)
Not significantly different between groups, χ2 = 0. 689, 2 df, p = 0.71.
Significantly different between groups, F(2, 247) = 9.8, p < 0.001.
Significantly different between groups, Kruskal Wallis Test, p < 0.001.
Significantly different between groups, Kruskal Wallis Test, p < 0.001.
Significantly different between groups, F(2, 246) = 9.73, p < 0.001.
The retained data included scans from 84 patients with clinically probable AD according to NINCDS-ADRCA criteria (McKhann et al., 1984), 115 individuals fulfilling Mayo criteria of amnestic MCI (Petersen et al., 1999) and 151 healthy elderly control individuals. All participants were free of any significant neurological, psychiatric, or medical condition (except for AD or MCI in patients), in particular cerebrovascular apoplexy, vascular dementia, depression, or subclinical hypothyroidism, as well as substance abuse. Healthy controls were required to have no cognitive complaints and scored within one standard deviation of the age and education adjusted norm in all subtests of the Consortium to Establish a Registry of Alzheimer’s Disease (CERAD) cognitive battery (Morris et al., 1989).
Written informed consent was provided by all subjects, or their representatives. The study was approved by local ethics committees at each of the participating centers, and has been conducted in accord with the Helsinki Declaration of 1975.
2.1. Imaging and data acquisition
Data were obtained from five different 3.0 Tesla MRI scanners. Acquisition parameters for the rs-fMRI sequences are given in Table 3. In one center (site I), the subjects were instructed to keep their eyes open, whereas in the remaining centers (sites II-V) all subjects were requested to close their eyes, relax, but not to fall asleep. Functional MRI was based on echo-planar imaging using scan durations between 6 and 9 min for the rs-fMRI sequence. The number of acquired time points was between 120 and 240 with a voxel size ranging from 2 × 2 × 2.6 up to 3.28 × 3.28 × 4.4 mm3 (Table 3). Anatomical scans were obtained from all scanners with an isotropic resolution of 1 mm3 during the same session.
Table 3.
Scanner characteristics.
| Center | Model | Manufacturer | TR [s] |
TE [s] |
Flip angle [°] | Matrix size | Field of view [mm3] | Number of volumes | Voxel size [mm3] |
Gap [mm] | Slice thickness [mm] | Spacing between slices [mm] | Slice acquisition order |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I | TrioTim | Siemens | 2.61 | 0.030 | 80 | 64 × 64 × 42 | 192 × 192 × 151 | 200 | 3 × 3 × 3.6 | 0.6 | 3 | 3.6 | Interleaved, ascending |
| II | Verio | Siemens | 3 | 0.030 | 90 | 96 × 96 × 45 | 192 × 192 × 117 | 120 | 2 × 2 × 2.6 | 0.6 | 2 | 2.6 | Contiguous, descending |
| III | Verio | Siemens | 2.58 | 0.030 | 80 | 64 × 64 × 47 | 224 × 224 × 165 | 180 | 3.5 × 3.5 × 3.5 | 0 | 3.5 | 3.5 | Interleaved, ascending |
| IV | Trio | Siemens | 3 | 0.030 | 80 | 64 × 64 × 28 | 210 × 210 × 123 | 120 | 3.28 × 3.28 × 4.4 | 0.4 | 4 | 4.4 | Interleaved, ascending |
| V | Achieva | Philips | 2 | 0.043 | 82 | 96 × 96 × 32 | 220 × 220 × 128 | 240 | 2.29 × 2.29 × 4 | 0 | 4 | 4 | Interleaved, descending |
All centers used an echo-planar imaging (EPI) sequence with axial slice orientation.
2.2. MR processing
The anatomical T1-weighted image for each participant was segmented into gray matter, white matter, and cerebrospinal fluid (CSF) partitions of 1.5 mm isotropic voxel-size using the tissue prior free segmentation routine of the VBM8-toolbox (Gaser et al., 1999) that extends Statistical Parametric Mapping (SPM8) (Friston et al., 2007). The Diffeomorphic Anatomical Registration Through Exponentiated Lie algebra (DARTEL) algorithm (Ashburner, 2007) was applied to normalize the T1-weighted gray matter and white matter partitions to the Montreal Neurological Institute (MNI) reference coordinate system using the default brain template included in VBM8. Individual flow-fields resulting from the DARTEL registration to the reference template were used to warp the gray matter segments. Voxel values of the warped gray matter segments were only modulated for the non-linear component of the deformation field, thus accounting at this step for differences in head size which are modeled by the affine component of the transformation.
Functional MRI data processing was carried out using Data Processing Assistant for Resting-State fMRI (DPARSF 3.2) (Chao-Gan and Yu-Feng, 2010), considering the recommendations from a recent systematic evaluation of processing alternatives (Shirer et al., 2015). After the removal of the first six images to account for gradient field stabilization, the rs-fMRI data was slice time corrected and realigned to the temporal mean image. Slice time correction addresses the problem that, for functional MRI, the 3D image of one time point is typically obtained by acquiring a series of 2D slices, with each slice being acquired one after another within the full period of one repetition time, for instance three seconds (Table 3). Thus, different slices of one 3D image measure the brain activity at a slightly different moment in time (Sladky et al., 2011). Slice time correction compensates for phase shifts in the time series signal using the cardinal sine interpolation based on the fast Fourier transform (Sladky et al., 2011). Sladky et al. found that this correction step improved the stability of estimates and magnitude of effects obtained from event-related and block design paradigms in task-based functional MRI (Sladky et al., 2011). It is also commonly applied to rs-fMRI data for both approaches seed-based functional connectivity and independent component analysis (Dyrba et al., 2015b, Koch et al., 2012, Meindl et al., 2010, Power et al., 2014, Yan et al., 2013b), which assess the correlation or homogeneity of the time series signal of remote brain regions or voxels. Controversially, previous studies only found minor, non-significant effects of applying slice time correction to rs-fMRI data (Shirer et al., 2015, Wu et al., 2011). These observations may be due to the subsequent step of bandpass filtering, which eliminates high-frequency components of the data with a wavelength of less than ten seconds and, thus, reduces the influence of slight short-term inaccuracies. The anatomical T1-weighted image for each participant was coregistered to the mean functional image. The deformation fields generated by DARTEL from the anatomical T1-weighted images were used to project the functional scans from each subjects’ native image space into the MNI reference space. We combined this step with the reslicing of all functional data to an isotropic resolution of 3 mm. The subsequent nuisance regression included covariates of head movement (rotation, translation, and derivatives) and the mean time courses for the global brain signal, the white matter segment signal, and the CSF segment signal. Although global signal regression was found to introduce negative correlations (Murphy et al., 2009, Shirer et al., 2015), studies consistently reported that it effectively reduces the signal-to-noise ratio (Power et al., 2014, Shirer et al., 2015, Yan et al., 2013a). Recently, Shirer et al. evaluated the influence of global signal regression on group separation but only found a minor, non-significant effect (Shirer et al., 2015). Subsequently, the images were band-pass filtered using the frequency band 0.1–0.01 Hz and smoothed using a 6 mm full-width-at-half-maximum (FWHM) Gaussian kernel. Ventral posterior cingulate cortex (PCC) functional connectivity maps were calculated using a spherical seed with 4 mm radius, which was set at MNI position 0,-53,26 (Hedden et al., 2009). Finally, Pearson correlation coefficients of the signal time courses were adjusted to be normally distributed using Fisher's Z-transform (Fisher, 1915): z = 0.5 ln [(1 + r)/(1 − r)].
2.3. Extraction of hippocampus volumes
A mask for the hippocampus was obtained by manual delineation of the hippocampus in the reference template (Grothe et al., 2012) using the interactive software package Display (McConnell Brain Imaging Centre at the Montreal Neurological Institute) and a previously described protocol for segmentation of the medial temporal lobe (Pruessner et al., 2000). Individual gray matter volumes of the hippocampus were extracted automatically from the warped gray matter segments by summing up the modulated gray matter voxel values within hippocampus ROI in the reference space.
3. Statistics
3.1. Quality control measures for scanner effects
We compared previously employed scan characteristics across scanners and diagnostic groups, including:
-
•
Framewise displacement (FD) – mean and percentage above threshold (0.5 mm) (Jenkinson et al., 2002, Power et al., 2012, Power et al., 2014, Yan et al., 2013a)
-
•
Temporal signal-to-noise ratio (tSNR) (Marcus et al., 2013, Welvaert and Rosseel, 2013)
-
•
Standardized DVARS – root mean square of change in signal intensity from one time point to the next (Power et al., 2012)
-
•
Percentage of outlier voxels (Zuo et al., 2014)
-
•
Foreground to background energy ratio (FBER) (Zuo et al., 2014)
-
•
Fractional amplitude of low frequency fluctuations (fALFF) (Yan et al., 2013b).
Additionally, we compared the regional correlations between PCC and anterior medial prefrontal cortex (aMPFC) time courses between scanners and groups, based on spherical seed regions with a radius of 4 mm at MNI coordinates 0, − 53, 26 (PCC) (Hedden et al., 2009) and − 6, 52, − 2 (aMPFC) (Andrews-Hanna et al., 2010).
To limit the number of measures, we decided not to use some previously employed measures (Zuo et al., 2014), such as entropy focus criterion (EFC) (Atkinson et al., 1997), image smoothness (IS) (Zuo et al., 2014), or ghost-to-signal ratio (GSR). The GSR needs manual interaction for the definition of the area of ghost artifacts in native subject space and was obsolete for the detection of poor scan quality as our scans underwent visual inspection. We excluded EFC and IS which target strong blurring, motion, and noise and become redundant when including tSNR, percentage of outlier voxels, FD, and FBER.
A description of the quality measures can be found in the supplementary material section.
3.2. Spatial pattern of group differences
We determined differences in voxel-wise correlations of PCC activity between AD patients and controls and between MCI patients and controls using two different univariate approaches to take scanner effects into account:
-
•
First, we determined group differences using a fixed effects linear model with diagnosis and scanner as independent factors, henceforth referred to as pooled analysis with scanner covariate. Significant clusters were identified with at least 10 voxels passing the uncorrected threshold of p < 0.01.
-
•
Secondly, we used a second level analysis with linear models of between group differences at the first level and a one-sample t-test of the between group effects across the 3 scanners for AD vs. control comparison and 5 scanners for the MCI vs. control comparison at the second level. Significant clusters were identified with at least 10 voxels passing the uncorrected threshold of p < 0.01.
Additionally, we assessed the spatial coherence of voxel-wise group differences between single scanners using conjunction analysis (Friston et al., 2005). Conjunction analysis resembles an ANOVA model for detecting group effects for more than two groups, but allows setting a threshold k to define the minimum number of effects, so that a second level group effect is considered to be present in a given voxel if a significant group difference had been found for at least k individual scanners (Friston et al., 2005). With our data, the value of k could range from 1, indicating an effect for at least one single scanner, to 5, indicating that a group effect must be present for each of the five scanners.
3.3. Accuracy of group discrimination
We defined regions of interest (ROI) as those brain regions that showed significant group differences in the voxel-based comparisons of AD or MCI and healthy control subjects. Specifically, we binarized the statistical maps thresholded at p < 0.01 as described above for each statistical approach (i.e., pooled analysis, and second level analysis) yielding 2 (statistical approach) × 2 (AD vs. controls and MCI vs. controls) = 4 different ROIs. For each of these ROIs, we extracted averaged Fisher's Z-transformed correlation coefficients. To this end, the individual voxel-wise correlation maps in MNI standard space were multiplied by the thresholded binary ROIs, and the voxel values within each ROI were averaged for each individual scan, yielding scalar markers as predictors in linear logistic regression analyses.
To obtain an estimate of the accuracy of group discrimination for each modality and analysis technique, we used block-wise cross validation with repeated random sampling, based on Gaussian-distributed random numbers generated in R. We repeatedly split the data set into 63.2% of training data and 36.8% of test data. For each of the repeatedly drawn training samples, the logistic regression parameters were estimated and subsequently applied to the remaining test data set. Classification accuracy, sensitivity, and specificity as well as area under the receiver operating characteristic curves were recorded for each test data set. The entire cross-validation process was iterated 1000 times to determine the variability of the estimates of accuracy across runs. We determined nonparametric bootstrap confidence intervals with the 2.5 and 97.5 percentiles defining the lower and upper limits of the confidence interval ((Efron and Tibshirani, 1993), Chapter 13). Logistic regression analysis was calculated in R, using function glm with the parameter 'family' = binary.
To define a benchmark for the effect size of group discrimination, we repeated the bootstrapped determination of the area under the receiver operating characteristic curves for the widely established measure of hippocampus volume, averaged across left and right hemispheres.
3.4. Scanner effects
We employed variance component analysis using libraries “nlme” and “ape” in R with the function “varcomp” to determine the effect of scanner on functional connectivity, with diagnosis as fixed effect covariate and scanner as random effect covariate. We determined the proportion of variance attributable to scanner relative to the variance attributable to error. Variances were scaled to sum to 1.
4. Results
4.1. Quality control measures for scanner effects
Framewise displacement showed comparable displacements across sites, both in mean values as well as in percentage of framewise displacement > 0.5 mm. Similarly, the foreground-to-background energy ratio, the fractional amplitude of low frequency fluctuations in PCC, and the mean functional connectivity between PCC and anterior medial prefrontal cortex indicated no outlying center (Supplementary Fig. 1 to 5). When looking at single sites, differences between diagnostic groups showed a general trend in the expected direction that only occasionally reached statistical significance. For instance, cognitively impaired patients showed slightly more head motion than controls (Supplementary Fig. 1) and lower temporal signal-to-noise ratio (supplementary figure 6). Mean whole brain temporal signal-to-noise ratio, the mean percentage of outlier voxels, and standardized DVARS identified an outlier in the site V data, with significantly decreased tSNR and standardized DVARS, and increased number of outlier voxels in the healthy control group compared to the MCI and AD group (Supplementary Figs. 6 to 8), one-sided Wilcoxon tests, p < 0.01. Site II showed a significantly reduced tSNR compared to the other sites (Supplementary Fig. 6), two-sided t-test, p < 0.001; but this systematic bias was evenly distributed across all subject groups.
4.2. Spatial pattern of group differences
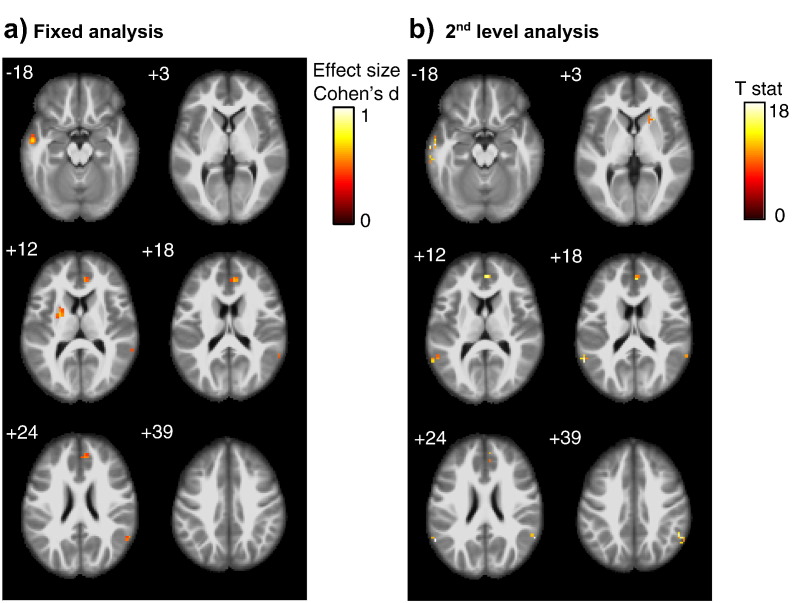
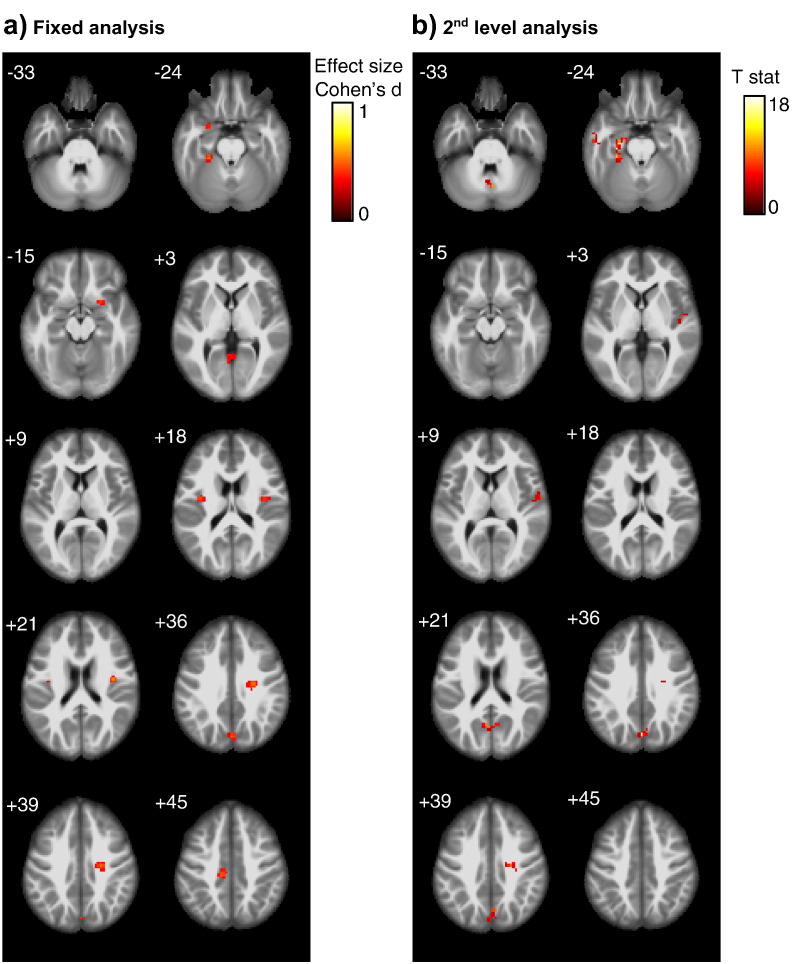
We found group differences between AD patients and controls and between MCI patients and controls both in the pooled data analysis as well as the second level analysis only at an uncorrected level of significance of p < 0.01, but no effects at an uncorrected p-value of 0.001. Functional connectivity of the PCC was smaller in AD and MCI cases compared to controls when the data of site V were removed from the analysis. Peak areas of group effects were located in the mid temporal cortex, anterior cingulum and inferior parietal cortex (including angular gyrus) for the AD vs. control comparison, and in the precuneus, middle cingulate cortex, insula cortex, fusiform gyrus and medial temporal lobes (including amygdala and parahippocampal cortex) for the MCI vs. controls comparison (Fig. 1, Fig. 2). The conjunction analysis revealed small clusters in only few regions when setting the minimum number of effects to k = 2, i.e. when group effects were significant in data from at least two scanners (data not shown). For the MCI vs. controls comparison, no cluster survived when the number of effects k was > 2. When the data of site V were included in the analysis, effects were in the opposite direction with larger functional connectivity in the AD and MCI cases compared to controls (data not shown).
Fig. 1.
AD vs. control comparison
Group effects of PCC functional connectivity differences between AD patients and controls, using (Panel a) a fixed effect analysis pooling all scans across scanners with scanner as covariate, and (Panel b) a second level analysis with scanner as second level factor. Significant cluster of at least 10 voxel passing an uncorrected threshold of significance of p < 0.01, are projected onto an anatomical MRI scan in MNI space. Numbers in the upper left corner of each image slice indicate the MNI z-coordinate, i.e. the axial section in MNI space.
Color bars represent color coding for Cohen's d effect size estimates (Cohen, 1977) for the pooled analysis, and T values for the second level analysis, respectively.
Fig. 2.
MCI vs. control comparison
Group effects of PCC functional connectivity differences between MCI patients and controls, using (Panel a) a fixed effect analysis pooling all scans across scanners with scanner as covariate, and (Panel b) a second level analysis with scanner as second level factor. Significant cluster of at least 10 voxel passing an uncorrected threshold of significance of p < 0.01, are projected onto an anatomical MRI scan in MNI space. Numbers in the upper left corner of each image slice indicate the MNI z-coordinate, i.e. the axial section in MNI space.
Color bars represent color coding for Cohen's d effect size estimates (Cohen, 1977) for the pooled analysis, and T values for the second level analysis, respectively.
4.3. Accuracy of group discrimination
These analyses were only conducted in the sample without including the data of site V. The distribution of MCI and control cases was relatively well balanced across the four sites. Since the distribution of AD cases was imbalanced across sites, the analyses for the AD vs. control comparisons were repeated across all sites and across the only two sites with a balanced number of AD cases and controls.
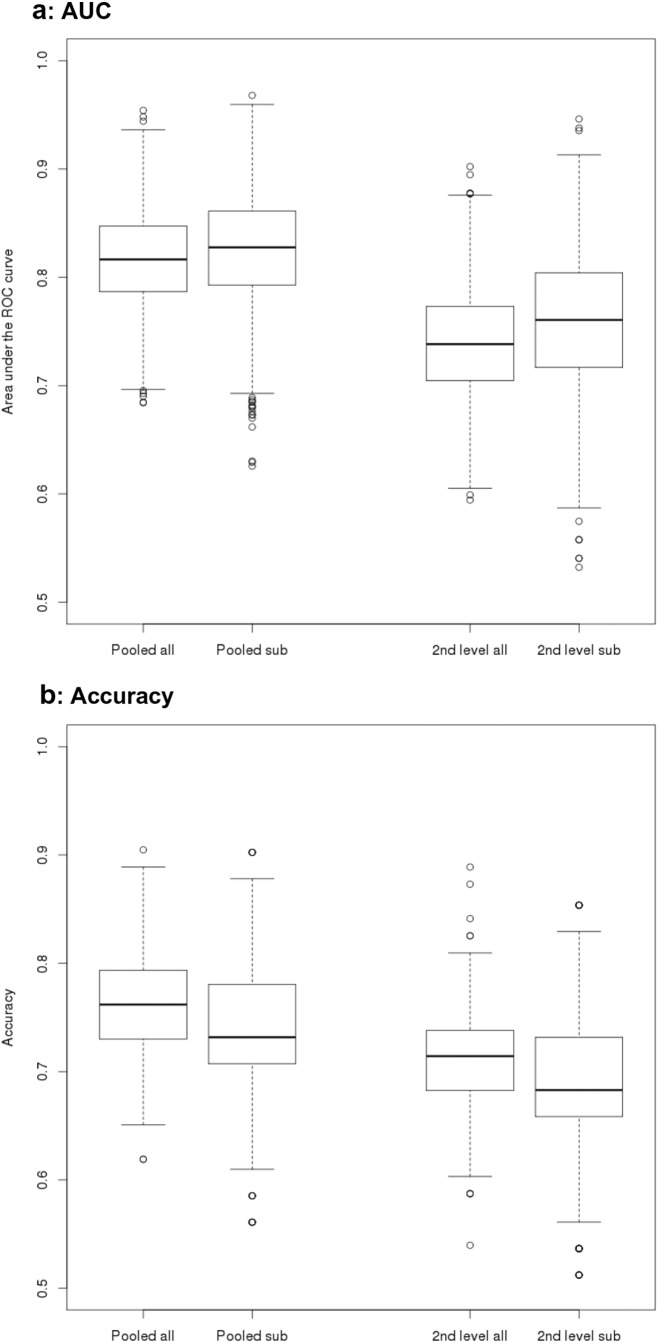
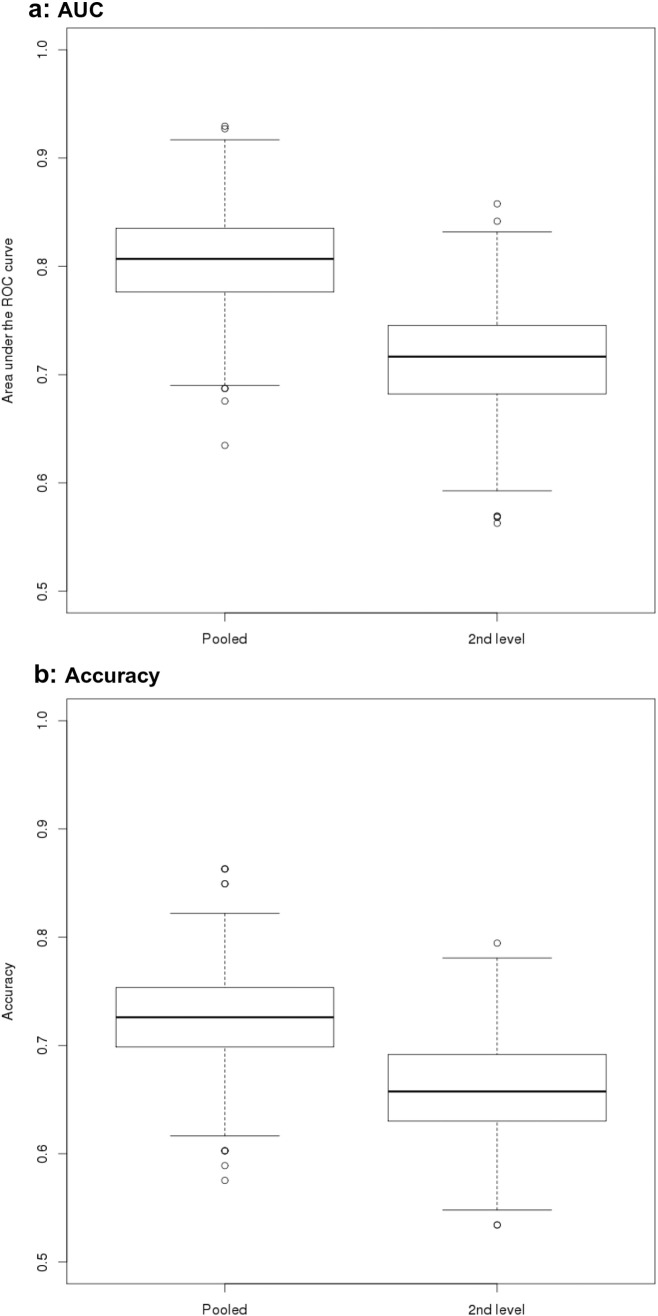
For the AD vs. controls comparison, mean AUCs in the test data ranged from 74% for the second level data to 82% for the pooled data, and accuracies ranged from 69% for the second level data to 76% for the pooled data (Table 4 and Figure 3). For the MCI vs. controls comparison, AUCs (accuracies) ranged from 71% (66%) for the second level data to 81% (72%) for the pooled data (Table 4 and Figure 4).
Table 4.
Group discrimination in the test sample
| AD vs. controls | ||||
|---|---|---|---|---|
| AUC pooled all | AUC pooled sub | AUC 2nd level all | AUC 2nd level sub | |
| Mean | 0.816 | 0.822 | 0.739 | 0.757 |
| Lower CI | 0.73 | 0.721 | 0.63 | 0.635 |
| Upper CI | 0.898 | 0.918 | 0.836 | 0.87 |
| Ac pooled all | Ac pooled sub | Ac 2nd level all | Ac 2nd level sub | |
| Mean | 0.761 | 0.738 | 0.708 | 0.688 |
| Lower CI | 0.667 | 0.634 | 0.619 | 0.585 |
| Upper CI | 0.841 | 0.854 | 0.794 | 0.805 |
| MCI vs. controls | ||||
| AUC pooled all | AUC 2nd level all | |||
| Mean | 0.805 | 0.713 | ||
| Lower CI | 0.719 | 0.617 | ||
| Upper CI | 0.885 | 0.803 | ||
| Ac pooled all | Ac 2nd level all | |||
| Mean | 0.72 | 0.662 | ||
| Lower CI | 0.644 | 0.575 | ||
| Upper CI | 0.808 | 0.74 | ||
Ac – Accuracy.
AUC – Area under the ROC curve.
sub - subsample from two scanners with matched numbers of AD patients and controls.
Fig. 3.
Areas under ROC and accuracy for AD vs. control comparisons
Box plots of AUC and accuracy levels from cross-validation logistic regression. Levels of AUC (Panel a) and accuracy (Panel b) were determined using bootstrapped logistic regression models on the discrimination between AD patients and controls following a pooled analysis with center covariate (“pooled”), and a second level analysis with center as second level factor (“2nd level”), respectively. Analyses were repeated, using all AD and control data (“all”) as well as only data from a subset of centers where number of AD cases and controls was matched between centers (“sub”).
Fig. 4.
Areas under ROC and accuracy for MCI vs. control comparisons
Box plots of AUC and accuracy levels from cross-validation logistic regression. Levels of AUC (Panel a) and accuracy (Panel b) were determined using bootstrapped logistic regression models on the discrimination between MCI patients and controls following a pooled analysis with center covariate (“pooled”), and a second level analysis with center as second level factor (“2nd level”), respectively.
For comparison, the AUCs for left and right averaged hippocampus volume were 86% [2.5/97.5th percentile confidence interval 77%/95%] for the AD vs. controls comparison, and 74% [2.5/97.5th percentile confidence interval 65%/84%] for the MCI vs. controls comparison.
To determine if levels of accuracy measures (AUC and overall accuracy) differed significantly between the values derived from the pooled vs. the 2nd level data, we used the degree of overlap between confidence intervals of the bootstrapped cross-validation data following Afshartous' rule (Afshartous and Preston, 2010). This rule considers the correlation of accuracy measures between samples and the ratio of the standard errors of the accuracy measures of both samples. Following this approach, neither AUCs nor overall accuracies were significantly different between the pooled and the 2nd level test data for the AD vs. controls and the MCI vs. controls comparisons, respectively, at a two tailed significance level of p < 0.05.
4.4. Scanner effects
Excluding site V, for the AD vs. control comparison, the proportion of variance attributable to scanner relative to the error variance was 6.6% across all sites and 6.3% for the two sites with balanced group distribution, and was 5.1% for the MCI vs. control comparison.
5. Discussion
In a relatively large multicenter data set of retrospectively pooled rs-fMRI data we found spatially restricted differences in PCC whole brain functional connectivity between AD patients and controls and MCI patients and controls, both in a pooled analysis and a second level analysis stratified according to scanner. The effects were in the expected direction with connectivity smaller in AD/MCI than in controls when removing the data of one site that had failed on visual data inspection, the quality assessments for tSNR and the standardized DVARS, but met all other quality assessments employed. Our findings lead to two major conclusions: Multicenter rs-fMRI using seed based functional connectivity has limited accuracy in the discrimination of AD and MCI cases from controls, and requires careful data quality checks beyond evaluation of global quality metrics, including visual inspection of all the data.
The regional distribution of diagnostic group effects in PCC connectivity found in the subset of data passing the visual quality check resembles the results in previous monocenter studies that reported reduced connectivity in MCI or AD within the posterior cingulate gyrus, inferior parietal lobes and medial temporal lobes (Balthazar et al., 2014, Binnewijzend et al., 2012, Chhatwal et al., 2013, Greicius et al., 2004, Koch et al., 2012, Thomas et al., 2014). Overall, the effect sizes of group differences were small, with regional effects passing only an uncorrected level of p < 0.01. This finding suggests that multiscanner variability decreases between group effects in functional connectivity. This interpretation is supported by the contribution of 5.2% to 6.6% of the overall variability by scanner related variance in the variance component analysis. In addition, the poor overlap of between group effects across scanners in the conjunction analysis indicates major confounding of group differences by multiscanner variability.
Levels of diagnostic accuracy ranged between 69% based on second level analysis and 76% based on pooled analysis for the AD vs. control comparisons and 66% and 72% for the MCI vs. control comparisons, respectively, in our study. These values are at the lower range of those previously reported from monocenter studies that involved small samples and failed to employ a cross-validation analysis (Balthazar et al., 2014, Koch et al., 2012).They are, however, close to previous estimates from the test data of cross-validated monocenter studies (Dyrba et al., 2015b). It is important to note that the benchmark for assessing performance of a technique is the cross-validated accuracy in the test data, not the accuracy in the training data. According to this benchmark, our multicenter study is at the level of accuracy of monocenter studies. Thus, although the use of multicenter data increases the degrees of freedom of the test statistics it did not increase the power of group discrimination due to confounding inter-scanner variance. One has to consider, however, that the identification of the peak areas that were included in the accuracy estimation was not part of the cross-validation so that effects may be slightly overestimated.
The levels of accuracy for functional connectivity in our study were below the levels of accuracy for hippocampus volume, one of the best established imaging markers of AD to date (for review see (Teipel et al., 2013)), reaching 86% and 74% AUC for the AD vs. controls comparison, respectively. For the MCI vs. controls comparison, the mean AUC for pooled data functional connectivity (81%) was numerically higher than the AUC for hippocampus (74%). The confidence interval of the hippocampus AUC, however, was largely contained within the confidence interval of the functional connectivity measures, suggesting that the functional connectivity measures were not significantly more accurate for the MCI vs. controls discrimination than the easily accessible hippocampus volumetry.
The results were clearly sensitive to scan quality. When we included the large data set of site V that had severe susceptibility artifacts, the direction of the group differences was inverted. When we considered the global scan quality measures, the tSNR (Marcus et al., 2013) and standardized DVARS (Power et al., 2012) suggested that insufficient signal in the healthy control group was driving this effect. Interestingly, other metrics employed in other multiscanner data pooling activities, including the intrinsic functional connectivity for two key areas of the DMN (Zuo et al., 2014), fractional ALFF (Yan et al., 2013b, Zuo et al., 2014), foreground to background energy ratio (Zuo et al., 2014), or subject head motion (Jenkinson et al., 2002, Power et al., 2012, Power et al., 2014, Yan et al., 2013a), were inconspicuous for these data, suggesting that determining tSNR (Marcus et al., 2013), standardized DVARS (Power et al., 2012), and visual inspection of all the data are indispensable for multiscanner data pooling. This is relevant since large scale data pooling efforts such as the PCP Quality Assessment Protocol (preprocessed-connectomes-project.org/quality-assessment-protocol/index.html) and the 1000 functional connectomes project (Yan et al., 2013b, Zuo et al., 2014) focus on the detection and correction of spatial displacements and head motion that were inconspicuous with the site V data.
Despite the high relevance of multiscanner variability of rs-fMRI data (Jovicich et al., 2016, Lin et al., 2015, Magnotta et al., 2006), the large majority of studies on multicenter rs-fMRI in neuropsychiatric diseases did not take multiscanner effects into account, even if protocols differed between sites (Chhatwal et al., 2013, Demertzi et al., 2015, Esslinger et al., 2011, Martucci et al., 2015, Sripada et al., 2014, Thomas et al., 2014). Regional effects of group differences strongly overlapped between a fixed effect analysis, including scanner as covariate, and a second level analysis stratified according to scanner, but were more extended for the pooled analysis. Numerically, group discrimination was smaller based on the second level analysis compared to the pooled analysis, albeit this difference was not statistically significant. A second level analysis of voxel-wise functional connectivity using Fisher's z-transformed correlation coefficients resembles a center-wise voxel-based meta-analysis (Teipel et al., 2012) that determines voxel-wise effect size within sites and then assesses the confidence level of the voxel-wise effect size estimates across sites. Such an approach has been used in one previous study across four cohorts of 129 MCI cases and 99 controls (Tam et al., 2015). The main outcome of this previous study were Cohen's d (Cohen, 1977) effect size estimates that ranged between 0.10 to 0.48, representing moderate effect sizes of MCI vs. control differences in regions of interests that were empirically derived from proximity metrics without a priori region selection. These moderate effect sizes agree with the effect sizes of group differences below Cohen's d = 1 in our peak voxel analysis (figures 1a and 2a).
An interesting question is the effect of multicenter acquisition on between subjects variability in trajectories of intra-individual change from longitudinal studies. Evidence here is still very limited. One recent study evaluated reproducibility of rs-fMRI connectivity across 13 different scanners at baseline and 7 to 60 days of follow-up in five healthy people per site (Jovicich et al., 2016), including different scanner types and vendors. In this study site differences in test-retest-variability of PCC connectivity were marginally not significant (p < 0.06). This finding suggests that multicenter acquisition not only introduces higher variability of between group differences as shown in our current study, but may introduce additional noise into the assessment of trajectories of intra-individual change.
We need to consider several limitations of our study. First, the scan protocols were different between scanners. This is not the case in a prospectively planned cohort study with a unified protocol. Still, even in absence of a harmonized protocol previous studies have pooled rs-fMRI data in studying neuropsychiatric diseases so that these findings are pertinent to the present state of research. More homogeneous acquisition parameters may amend some of the scanner effects but at the same time limit the usefulness of multicenter acquisition in routine care, where differences in scanner type and manufacturer will not allow perfect alignment of scanning parameters across sites. We carefully checked the image quality of each single scan by visual inspection. As a result, we excluded the data of site V. The remaining data had high image quality upon visual inspection, consistent with the results of the quality metrics employed. Still, the combination of data from different scanning protocols and scanner resulted in high inter-scanner variability despite sufficient intra-scanner scan quality. In future, we plan to determine the effects of multiscanner acquisition from an ongoing prospective multicenter study in MCI, AD, and healthy controls that employs a harmonized rs-fMRI protocol across sites. Although we expect that the multicenter effects may be smaller in such a harmonized study, we still anticipate that multiscanner effects will limit accuracy of group discrimination. Secondly, different preprocessing protocols may be useful to reduce multiscanner variation. Here, we employed a preprocessing protocol that was oriented on the recommendations from a systematic evaluation of processing steps (Shirer et al., 2015), and used cross-correlation as connectivity metric that has been found more stable than other connectivity metrics, such as cross-coherence and partial cross-correlation, in a previous study (Fiecas et al., 2013). We did, however, not systematically explore other processing steps and connectivity metrics. Thirdly, group discrimination accuracy can never perform better than the reference standard. The reference standard in our study for AD and MCI definition lacked CSF or PET biomarker evidence for most cases, but data came from expert centers experienced in the early diagnosis of AD and MCI. Still, a final judgment of the added value of rs-fMRI for AD diagnosis must await systematic evaluation of diagnostic accuracy in multicenter data from biomarker stratified cases.
In summary, we found spatially restricted group differences in resting state functional connectivity in AD patients and MCI patients compared to controls, limited by high multiscanner variability. The accuracy of group discrimination resembled findings from previous monocenter studies using a training/test data set approach, encouraging the conclusion that rs-fMRI at least when using seed based functional connectivity metrics may play a limited role in early diagnosis of AD or MCI. The discrimination accuracy in the test data did not reach the internal benchmark set by the established marker of hippocampus volumetry. This conclusion needs further corroboration in biomarker qualified multicenter cohorts. From a practical viewpoint, studies pooling multicenter rs-fMRI data should employ careful data quality checks that need to include tSNR, standardized DVARS, and visual inspection of all the data besides other established global metrics, and should use explicit modelling of scanner effects such as provided by second level models or center-based meta-analysis when focusing on univariate approaches. Potential usefulness of multivariate non-linear approaches such as provided by machine learning algorithms that were successfully employed in reducing multiscanner effects for structural connectivity data (Dyrba et al., 2015a, Dyrba et al., 2013) is another open area of research.
Acknowledgment
SJT received support by a grant of the Federal Ministry of Research (BMBF) (AgeGain, 1GQ1425B). ME received support by grants from the Alzheimer Forschung Initiative (AFI), LMUexcellent and the European Commission (PCIG12-GA-2012-334259).
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.nicl.2017.01.018.
Appendix A. Supplementary data
Supplementary material.
References
- Afshartous D., Preston R.A. Confidence intervals for dependent data: equating non-overlap with statistical significance. Comput. Stat. Data Anal. 2010;54:2296–2305. [Google Scholar]
- Albert M.S., DeKosky S.T., Dickson D., Dubois B., Feldman H.H., Fox N.C., Gamst A., Holtzman D.M., Jagust W.J., Petersen R.C., Snyder P.J., Carrillo M.C., Thies B., Phelps C.H. The diagnosis of mild cognitive impairment due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7:270–279. doi: 10.1016/j.jalz.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews-Hanna J.R., Reidler J.S., Sepulcre J., Poulin R., Buckner R.L. Functional-anatomic fractionation of the brain's default network. Neuron. 2010;65:550–562. doi: 10.1016/j.neuron.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anticevic A., Haut K., Murray J.D., Repovs G., Yang G.J., Diehl C., McEwen S.C., Bearden C.E., Addington J., Goodyear B., Cadenhead K.S., Mirzakhanian H., Cornblatt B.A., Olvet D., Mathalon D.H., McGlashan T.H., Perkins D.O., Belger A., Seidman L.J., Tsuang M.T., van Erp T.G., Walker E.F., Hamann S., Woods S.W., Qiu M., Cannon T.D. Association of thalamic dysconnectivity and conversion to psychosis in youth and young adults at elevated clinical risk. JAMA Psychiatry. 2015;72:882–891. doi: 10.1001/jamapsychiatry.2015.0566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner J. A fast diffeomorphic image registration algorithm. NeuroImage. 2007;38:95–113. doi: 10.1016/j.neuroimage.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Atkinson D., Hill D.L.G., Stoyle P.N.R., Summers P.E., Keevil S.F. Automatic correction of motion artifacts in magnetic resonance images using an entropy focus criterion. IEEE Trans. Med. Imaging. 1997;16:903–910. doi: 10.1109/42.650886. [DOI] [PubMed] [Google Scholar]
- Balthazar M.L., de Campos B.M., Franco A.R., Damasceno B.P., Cendes F. Whole cortical and default mode network mean functional connectivity as potential biomarkers for mild Alzheimer's disease. Psychiatry Res. 2014;221:37–42. doi: 10.1016/j.pscychresns.2013.10.010. [DOI] [PubMed] [Google Scholar]
- Binnewijzend M.A., Schoonheim M.M., Sanz-Arigita E., Wink A.M., van der Flier W.M., Tolboom N., Adriaanse S.M., Damoiseaux J.S., Scheltens P., van Berckel B.N., Barkhof F. Resting-state fMRI changes in Alzheimer's disease and mild cognitive impairment. Neurobiol. Aging. 2012;33:2018–2028. doi: 10.1016/j.neurobiolaging.2011.07.003. [DOI] [PubMed] [Google Scholar]
- Biswal B.B., Mennes M., Zuo X.N., Gohel S., Kelly C., Smith S.M., Beckmann C.F., Adelstein J.S., Buckner R.L., Colcombe S., Dogonowski A.M., Ernst M., Fair D., Hampson M., Hoptman M.J., Hyde J.S., Kiviniemi V.J., Kotter R., Li S.J., Lin C.P., Lowe M.J., Mackay C., Madden D.J., Madsen K.H., Margulies D.S., Mayberg H.S., McMahon K., Monk C.S., Mostofsky S.H., Nagel B.J., Pekar J.J., Peltier S.J., Petersen S.E., Riedl V., Rombouts S.A., Rypma B., Schlaggar B.L., Schmidt S., Seidler R.D., Siegle G.J., Sorg C., Teng G.J., Veijola J., Villringer A., Walter M., Wang L., Weng X.C., Whitfield-Gabrieli S., Williamson P., Windischberger C., Zang Y.F., Zhang H.Y., Castellanos F.X., Milham M.P. Toward discovery science of human brain function. Proc. Natl. Acad. Sci. U.S.A. 2010;107:4734–4739. doi: 10.1073/pnas.0911855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blautzik J., Keeser D., Berman A., Paolini M., Kirsch V., Mueller S., Coates U., Reiser M., Teipel S.J., Meindl T. Long-term test-retest reliability of resting-state networks in healthy elderly subjects and with amnestic mild cognitive impairment patients. J Alzheimers Dis. 2013;34:741–754. doi: 10.3233/JAD-111970. [DOI] [PubMed] [Google Scholar]
- Chao-Gan Y., Yu-Feng Z. DPARSF: A MATLAB Toolbox for “Pipeline” data analysis of resting-state fMRI. Front. Syst. Neurosci. 2010;4:13. doi: 10.3389/fnsys.2010.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Xu T., Zhou C., Wang L., Yang N., Wang Z., Dong H.M., Yang Z., Zang Y.F., Zuo X.N., Weng X.C. Individual variability and test-retest reliability revealed by ten repeated resting-state brain scans over one month. PLoS One. 2015;10 doi: 10.1371/journal.pone.0144963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhatwal J.P., Schultz A.P., Johnson K., Benzinger T.L., Jack C., Jr., Ances B.M., Sullivan C.A., Salloway S.P., Ringman J.M., Koeppe R.A., Marcus D.S., Thompson P., Saykin A.J., Correia S., Schofield P.R., Rowe C.C., Fox N.C., Brickman A.M., Mayeux R., McDade E., Bateman R., Fagan A.M., Goate A.M., Xiong C., Buckles V.D., Morris J.C., Sperling R.A. Impaired default network functional connectivity in autosomal dominant Alzheimer disease. Neurology. 2013;81:736–744. doi: 10.1212/WNL.0b013e3182a1aafe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou Y.H., Panych L.P., Dickey C.C., Petrella J.R., Chen N.K. Investigation of long-term reproducibility of intrinsic connectivity network mapping: a resting-state fMRI study. AJNR Am. J. Neuroradiol. 2012;33:833–838. doi: 10.3174/ajnr.A2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. Academic Press; New York: 1977. Statistical Power Analysis for the Behavioural Sciences. [Google Scholar]
- Demertzi A., Antonopoulos G., Heine L., Voss H.U., Crone J.S., de Los Angeles C., Bahri M.A., Di Perri C., Vanhaudenhuyse A., Charland-Verville V., Kronbichler M., Trinka E., Phillips C., Gomez F., Tshibanda L., Soddu A., Schiff N.D., Whitfield-Gabrieli S., Laureys S. Intrinsic functional connectivity differentiates minimally conscious from unresponsive patients. Brain. 2015;138:2619–2631. doi: 10.1093/brain/awv169. [DOI] [PubMed] [Google Scholar]
- Doraiswamy P.M., Sperling R.A., Johnson K., Reiman E.M., Wong T.Z., Sabbagh M.N., Sadowsky C.H., Fleisher A.S., Carpenter A., Joshi A.D., Lu M., Grundman M., Mintun M.A., Skovronsky D.M., Pontecorvo M.J., Group, A.A.S Florbetapir F 18 amyloid PET and 36-month cognitive decline: a prospective multicenter study. Mol. Psychiatry. 2014;19:1044–1051. doi: 10.1038/mp.2014.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois B., Feldman H.H., Jacova C., Dekosky S.T., Barberger-Gateau P., Cummings J., Delacourte A., Galasko D., Gauthier S., Jicha G., Meguro K., O'Brien J., Pasquier F., Robert P., Rossor M., Salloway S., Stern Y., Visser P.J., Scheltens P. Research criteria for the diagnosis of Alzheimer's disease: revising the NINCDS-ADRDA criteria. Lancet Neurol. 2007;6:734–746. doi: 10.1016/S1474-4422(07)70178-3. [DOI] [PubMed] [Google Scholar]
- Dubois B., Feldman H.H., Jacova C., Cummings J.L., Dekosky S.T., Barberger-Gateau P., Delacourte A., Frisoni G., Fox N.C., Galasko D., Gauthier S., Hampel H., Jicha G.A., Meguro K., O'Brien J., Pasquier F., Robert P., Rossor M., Salloway S., Sarazin M., de Souza L.C., Stern Y., Visser P.J., Scheltens P. Revising the definition of Alzheimer's disease: a new lexicon. Lancet Neurol. 2010;9:1118–1127. doi: 10.1016/S1474-4422(10)70223-4. [DOI] [PubMed] [Google Scholar]
- Dubois B., Feldman H.H., Jacova C., Hampel H., Molinuevo J.L., Blennow K., DeKosky S.T., Gauthier S., Selkoe D., Bateman R., Cappa S., Crutch S., Engelborghs S., Frisoni G.B., Fox N.C., Galasko D., Habert M.O., Jicha G.A., Nordberg A., Pasquier F., Rabinovici G., Robert P., Rowe C., Salloway S., Sarazin M., Epelbaum S., de Souza L.C., Vellas B., Visser P.J., Schneider L., Stern Y., Scheltens P., Cummings J.L. Advancing research diagnostic criteria for Alzheimer's disease: the IWG-2 criteria. Lancet Neurol. 2014;13:614–629. doi: 10.1016/S1474-4422(14)70090-0. [DOI] [PubMed] [Google Scholar]
- Dyrba M., Ewers M., Wegrzyn M., Kilimann I., Plant C., Oswald A., Meindl T., Pievani M., Bokde A.L., Fellgiebel A., Filippi M., Hampel H., Kloppel S., Hauenstein K., Kirste T., Teipel S.J., Group, E.S Robust automated detection of microstructural white matter degeneration in Alzheimer's disease using machine learning classification of multicenter DTI data. PLoS One. 2013;8 doi: 10.1371/journal.pone.0064925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyrba M., Barkhof F., Fellgiebel A., Filippi M., Hausner L., Hauenstein K., Kirste T., Teipel S.J. Predicting prodromal Alzheimer's disease in subjects with mild cognitive impairment using machine learning classification of multimodal multicenter diffusion-tensor and magnetic resonance imaging data. J. Neuroimaging. 2015 doi: 10.1111/jon.12214. [DOI] [PubMed] [Google Scholar]
- Dyrba M., Grothe M., Kirste T., Teipel S.J. Multimodal analysis of functional and structural disconnection in Alzheimer's disease using multiple kernel SVM. Hum. Brain Mapp. 2015;36:2118–2131. doi: 10.1002/hbm.22759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efron B., Tibshirani R.J. Chapman & Hall/CRC; Boca Raton: 1993. An Introduction to the Bootstrap. [Google Scholar]
- Esslinger C., Kirsch P., Haddad L., Mier D., Sauer C., Erk S., Schnell K., Arnold C., Witt S.H., Rietschel M., Cichon S., Walter H., Meyer-Lindenberg A. Cognitive state and connectivity effects of the genome-wide significant psychosis variant in ZNF804A. NeuroImage. 2011;54:2514–2523. doi: 10.1016/j.neuroimage.2010.10.012. [DOI] [PubMed] [Google Scholar]
- Ewers M., Teipel S.J., Dietrich O., Schonberg S.O., Jessen F., Heun R., Scheltens P., van de Pol L., Freymann N.R., Moeller H.J., Hampel H. Multicenter assessment of reliability of cranial MRI. Neurobiol. Aging. 2006;27:1051–1059. doi: 10.1016/j.neurobiolaging.2005.05.032. [DOI] [PubMed] [Google Scholar]
- Fiecas M., Ombao H., van Lunen D., Baumgartner R., Coimbra A., Feng D. Quantifying temporal correlations: a test-retest evaluation of functional connectivity in resting-state fMRI. NeuroImage. 2013;65:231–241. doi: 10.1016/j.neuroimage.2012.09.052. [DOI] [PubMed] [Google Scholar]
- Fisher R.A. Frequency distribution of the values of the correlation coefficient in samples from an indefinitely large population. Biometrika. 1915;10:507. [Google Scholar]
- Folstein M.F., Folstein S.E., McHugh P.R. Mini-mental-state: a practical method for grading the cognitive state of patients for the clinician. J. Psychiatr. Res. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- Fox M.D., Snyder A.Z., Vincent J.L., Corbetta M., Van Essen D.C., Raichle M.E. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc. Natl. Acad. Sci. U.S.A. 2005;102:9673–9678. doi: 10.1073/pnas.0504136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston K.J., Penny W.D., Glaser D.E. Conjunction revisited. NeuroImage. 2005;25:661–667. doi: 10.1016/j.neuroimage.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Friston K.J., Ashburner J., Kiebel S., Nichols T., Penny W.D. First edition ed. Elsevier/Academic Press; Amsterdam; Boston: 2007. Statistical Parametric Mapping: The Analysis of Funtional Brain Images. [Google Scholar]
- Gaser C., Volz H.-P., Kiebel S., Riehemann S., Sauer H. Detecting structural changes in whole brain based on nonlinear deformations—application to schizophrenia research. NeuroImage. 1999;10:107–113. doi: 10.1006/nimg.1999.0458. [DOI] [PubMed] [Google Scholar]
- Greicius M.D., Srivastava G., Reiss A.L., Menon V. Default-mode network activity distinguishes Alzheimer's disease from healthy aging: evidence from functional MRI. Proc. Natl. Acad. Sci. U.S.A. 2004;101:4637–4642. doi: 10.1073/pnas.0308627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grothe M., Heinsen H., Teipel S.J. Atrophy of the cholinergic Basal forebrain over the adult age range and in early stages of Alzheimer's disease. Biol. Psychiatry. 2012;71:805–813. doi: 10.1016/j.biopsych.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C.C., Kurth F., Zhou J., Mayer E.A., Eickhoff S.B., Kramer J.H., Seeley W.W. One-year test-retest reliability of intrinsic connectivity network fMRI in older adults. NeuroImage. 2012;61:1471–1483. doi: 10.1016/j.neuroimage.2012.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden T., Van Dijk K.R., Becker J.A., Mehta A., Sperling R.A., Johnson K.A., Buckner R.L. Disruption of functional connectivity in clinically normal older adults harboring amyloid burden. J. Neurosci. 2009;29:12686–12694. doi: 10.1523/JNEUROSCI.3189-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herholz K. Cerebral glucose metabolism in preclinical and prodromal Alzheimer's disease. Expert Rev. Neurother. 2010;10:1667–1673. doi: 10.1586/ern.10.136. [DOI] [PubMed] [Google Scholar]
- Ito K., Fukuyama H., Senda M., Ishii K., Maeda K., Yamamoto Y., Ouchi Y., Ishii K., Okumura A., Fujiwara K., Kato T., Arahata Y., Washimi Y., Mitsuyama Y., Meguro K., Ikeda M., Group, S.-J.S Prediction of outcomes in mild cognitive impairment by using 18F-FDG-PET: a multicenter study. J Alzheimers Dis. 2015;45:543–552. doi: 10.3233/JAD-141338. [DOI] [PubMed] [Google Scholar]
- Jenkinson M., Bannister P., Brady M., Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. NeuroImage. 2002;17:825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- Jovicich J., Minati L., Marizzoni M., Marchitelli R., Sala-Llonch R., Bartres-Faz D., Arnold J., Benninghoff J., Fiedler U., Roccatagliata L., Picco A., Nobili F., Blin O., Bombois S., Lopes R., Bordet R., Sein J., Ranjeva J.P., Didic M., Gros-Dagnac H., Payoux P., Zoccatelli G., Alessandrini F., Beltramello A., Bargallo N., Ferretti A., Caulo M., Aiello M., Cavaliere C., Soricelli A., Parnetti L., Tarducci R., Floridi P., Tsolaki M., Constantinidis M., Drevelegas A., Rossini P.M., Marra C., Schonknecht P., Hensch T., Hoffmann K.T., Kuijer J.P., Visser P.J., Barkhof F., Frisoni G.B., PharmaCog C. Longitudinal reproducibility of default-mode network connectivity in healthy elderly participants: a multicentric resting-state fMRI study. NeuroImage. 2016;124:442–454. doi: 10.1016/j.neuroimage.2015.07.010. [DOI] [PubMed] [Google Scholar]
- Kilimann I., Grothe M., Heinsen H., Alho E.J., Grinberg L., Amaro E., Jr., Dos Santos G.A., da Silva R.E., Mitchell A.J., Frisoni G.B., Bokde A.L., Fellgiebel A., Filippi M., Hampel H., Kloppel S., Teipel S.J. Subregional basal forebrain atrophy in Alzheimer's disease: a multicenter study. J Alzheimers Dis. 2014;40:687–700. doi: 10.3233/JAD-132345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch W., Teipel S., Mueller S., Benninghoff J., Wagner M., Bokde A.L., Hampel H., Coates U., Reiser M., Meindl T. Diagnostic power of default mode network resting state fMRI in the detection of Alzheimer's disease. Neurobiol. Aging. 2012;33:466–478. doi: 10.1016/j.neurobiolaging.2010.04.013. [DOI] [PubMed] [Google Scholar]
- Lin Q., Dai Z., Xia M., Han Z., Huang R., Gong G., Liu C., Bi Y., He Y. A connectivity-based test-retest dataset of multi-modal magnetic resonance imaging in young healthy adults. Sci. Data. 2015;2:150056. doi: 10.1038/sdata.2015.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long X.Y., Zuo X.N., Kiviniemi V., Yang Y., Zou Q.H., Zhu C.Z., Jiang T.Z., Yang H., Gong Q.Y., Wang L., Li K.C., Xie S., Zang Y.F. Default mode network as revealed with multiple methods for resting-state functional MRI analysis. J. Neurosci. Methods. 2008;171:349–355. doi: 10.1016/j.jneumeth.2008.03.021. [DOI] [PubMed] [Google Scholar]
- Magnotta V.A., Friedman L., First B. Measurement of signal-to-noise and contrast-to-noise in the fBIRN multicenter imaging study. J. Digit. Imaging. 2006;19:140–147. doi: 10.1007/s10278-006-0264-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus D.S., Harms M.P., Snyder A.Z., Jenkinson M., Wilson J.A., Glasser M.F., Barch D.M., Archie K.A., Burgess G.C., Ramaratnam M., Hodge M., Horton W., Herrick R., Olsen T., McKay M., House M., Hileman M., Reid E., Harwell J., Coalson T., Schindler J., Elam J.S., Curtiss S.W., Van Essen D.C., Consortium W.U.-M.H. Human Connectome Project informatics: quality control, database services, and data visualization. NeuroImage. 2013;80:202–219. doi: 10.1016/j.neuroimage.2013.05.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martucci K.T., Shirer W.R., Bagarinao E., Johnson K.A., Farmer M.A., Labus J.S., Apkarian A.V., Deutsch G., Harris R.E., Mayer E.A., Clauw D.J., Greicius M.D., Mackey S.C. The posterior medial cortex in urologic chronic pelvic pain syndrome: detachment from default mode network-a resting-state study from the MAPP Research Network. Pain. 2015;156:1755–1764. doi: 10.1097/j.pain.0000000000000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKhann G., Drachman D., Folstein M., Katzman R., Price D., Stadlan E.M. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- McKhann G.M., Knopman D.S., Chertkow H., Hyman B.T., Jack C.R., Jr., Kawas C.H., Klunk W.E., Koroshetz W.J., Manly J.J., Mayeux R., Mohs R.C., Morris J.C., Rossor M.N., Scheltens P., Carrillo M.C., Thies B., Weintraub S., Phelps C.H. The diagnosis of dementia due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7:263–269. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meindl T., Teipel S., Elmouden R., Mueller S., Koch W., Dietrich O., Coates U., Reiser M., Glaser C. Test-retest reproducibility of the default-mode network in healthy individuals. Hum. Brain Mapp. 2010;31:237–246. doi: 10.1002/hbm.20860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J.C., Heyman A., Mohs R.C., Hughes J.P., van Belle G., Fillenbaum G., Mellits E.D., Clark C. The Consortium to Establish a Registry for Alzheimer's Disease (CERAD). Part I. Clinical and neuropsychological assessment of Alzheimer's disease. Neurology. 1989;39:1159–1165. doi: 10.1212/wnl.39.9.1159. [DOI] [PubMed] [Google Scholar]
- Murphy K., Birn R.M., Handwerker D.A., Jones T.B., Bandettini P.A. The impact of global signal regression on resting state correlations: Are anti-correlated networks introduced? NeuroImage. 2009;44:893–905. doi: 10.1016/j.neuroimage.2008.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasreddine Z.S., Phillips N.A., Bedirian V., Charbonneau S., Whitehead V., Collin I., Cummings J.L., Chertkow H. The Montreal Cognitive Assessment, MoCA: a brief screening tool for mild cognitive impairment. J. Am. Geriatr. Soc. 2005;53:695–699. doi: 10.1111/j.1532-5415.2005.53221.x. [DOI] [PubMed] [Google Scholar]
- Orban P., Madjar C., Savard M., Dansereau C., Tam A., Das S., Evans A.C., Rosa-Neto P., Breitner J.C., Bellec P., Group, P.-A.R Test-retest resting-state fMRI in healthy elderly persons with a family history of Alzheimer's disease. Sci. Data. 2015;2:150043. doi: 10.1038/sdata.2015.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen R.C., Smith G.E., Waring S.C., Ivnik R.J., Tangalos E.G., Kokmen E. Mild cognitive impairment: clinical characterization and outcome. Arch. Neurol. 1999;56:303–308. doi: 10.1001/archneur.56.3.303. [DOI] [PubMed] [Google Scholar]
- Power J.D., Barnes K.A., Snyder A.Z., Schlaggar B.L., Petersen S.E. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. NeuroImage. 2012;59:2142–2154. doi: 10.1016/j.neuroimage.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power J.D., Mitra A., Laumann T.O., Snyder A.Z., Schlaggar B.L., Petersen S.E. Methods to detect, characterize, and remove motion artifact in resting state fMRI. NeuroImage. 2014;84:320–341. doi: 10.1016/j.neuroimage.2013.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruessner J.C., Li L.M., Serles W., Pruessner M., Collins D.L., Kabani N., Lupien S., Evans A.C. Volumetry of hippocampus and amygdala with high-resolution MRI and three-dimensional analysis software: minimizing the discrepancies between laboratories. Cereb. Cortex. 2000;10:433–442. doi: 10.1093/cercor/10.4.433. [DOI] [PubMed] [Google Scholar]
- Risacher S.L., Saykin A.J., West J.D., Shen L., Firpi H.A., McDonald B.C., Alzheimer's Disease Neuroimaging I. Baseline MRI predictors of conversion from MCI to probable AD in the ADNI cohort. Curr. Alzheimer Res. 2009;6:347–361. doi: 10.2174/156720509788929273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirer W.R., Jiang H., Price C.M., Ng B., Greicius M.D. Optimization of rs-fMRI Pre-processing for enhanced signal-noise separation, test-retest reliability, and group discrimination. NeuroImage. 2015;117:67–79. doi: 10.1016/j.neuroimage.2015.05.015. [DOI] [PubMed] [Google Scholar]
- Sladky R., Friston K.J., Tröstl J., Cunnington R., Moser E., Windischberger C. Slice-timing effects and their correction in functional MRI. NeuroImage. 2011;58:588–594. doi: 10.1016/j.neuroimage.2011.06.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sripada C., Kessler D., Fang Y., Welsh R.C., Prem Kumar K., Angstadt M. Disrupted network architecture of the resting brain in attention-deficit/hyperactivity disorder. Hum. Brain Mapp. 2014;35:4693–4705. doi: 10.1002/hbm.22504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suckling J., Barnes A., Job D., Brennan D., Lymer K., Dazzan P., Marques T.R., MacKay C., McKie S., Williams S.R., Williams S.C., Deakin B., Lawrie S. The Neuro/PsyGRID calibration experiment: identifying sources of variance and bias in multicenter MRI studies. Hum. Brain Mapp. 2012;33:373–386. doi: 10.1002/hbm.21210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam A., Dansereau C., Badhwar A., Orban P., Belleville S., Chertkow H., Dagher A., Hanganu A., Monchi O., Rosa-Neto P., Shmuel A., Wang S., Breitner J., Bellec P., Alzheimer's Disease Neuroimaging I. Common effects of amnestic mild cognitive impairment on resting-state connectivity across four independent studies. Front. Aging Neurosci. 2015;7:242. doi: 10.3389/fnagi.2015.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teipel S.J., Wegrzyn M., Meindl T., Frisoni G., Bokde A.L., Fellgiebel A., Filippi M., Hampel H., Kloppel S., Hauenstein K., Ewers M. Anatomical MRI and DTI in the diagnosis of Alzheimer's disease: a European multicenter study. J. Alzheimers Dis. 2012;31(Suppl. 3):S33–S47. doi: 10.3233/JAD-2012-112118. [DOI] [PubMed] [Google Scholar]
- Teipel S.J., Grothe M., Lista S., Toschi N., Garaci F.G., Hampel H. Relevance of magnetic resonance imaging for early detection and diagnosis of Alzheimer disease. Med. Clin. North Am. 2013;97:399–424. doi: 10.1016/j.mcna.2012.12.013. [DOI] [PubMed] [Google Scholar]
- Teipel S., Drzezga A., Grothe M.J., Barthel H., Chetelat G., Schuff N., Skudlarski P., Cavedo E., Frisoni G.B., Hoffmann W., Thyrian J.R., Fox C., Minoshima S., Sabri O., Fellgiebel A. Multimodal imaging in Alzheimer's disease: validity and usefulness for early detection. Lancet Neurol. 2015;14:1037–1053. doi: 10.1016/S1474-4422(15)00093-9. [DOI] [PubMed] [Google Scholar]
- Thomas J.B., Brier M.R., Bateman R.J., Snyder A.Z., Benzinger T.L., Xiong C., Raichle M., Holtzman D.M., Sperling R.A., Mayeux R., Ghetti B., Ringman J.M., Salloway S., McDade E., Rossor M.N., Ourselin S., Schofield P.R., Masters C.L., Martins R.N., Weiner M.W., Thompson P.M., Fox N.C., Koeppe R.A., Jack C.R., Jr., Mathis C.A., Oliver A., Blazey T.M., Moulder K., Buckles V., Hornbeck R., Chhatwal J., Schultz A.P., Goate A.M., Fagan A.M., Cairns N.J., Marcus D.S., Morris J.C., Ances B.M. Functional connectivity in autosomal dominant and late-onset Alzheimer disease. JAMA Neurol. 2014;71:1111–1122. doi: 10.1001/jamaneurol.2014.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welvaert M., Rosseel Y. On the definition of signal-to-noise ratio and contrast-to-noise ratio for fMRI data. PLoS One. 2013;8 doi: 10.1371/journal.pone.0077089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.W., Chen C.L., Liu P.Y., Chao Y.P., Biswal B.B., Lin C.P. Empirical evaluations of slice-timing, smoothing, and normalization effects in seed-based, resting-state functional magnetic resonance imaging analyses. Brain Connect. 2011;1:401–410. doi: 10.1089/brain.2011.0018. [DOI] [PubMed] [Google Scholar]
- Yan C.G., Cheung B., Kelly C., Colcombe S., Craddock R.C., Di Martino A., Li Q., Zuo X.N., Castellanos F.X., Milham M.P. A comprehensive assessment of regional variation in the impact of head micromovements on functional connectomics. NeuroImage. 2013;76:183–201. doi: 10.1016/j.neuroimage.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C.G., Craddock R.C., Zuo X.N., Zang Y.F., Milham M.P. Standardizing the intrinsic brain: towards robust measurement of inter-individual variation in 1000 functional connectomes. NeuroImage. 2013;80:246–262. doi: 10.1016/j.neuroimage.2013.04.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou K.H., Greve D.N., Wang M., Pieper S.D., Warfield S.K., White N.S., Manandhar S., Brown G.G., Vangel M.G., Kikinis R., Wells W.M., Group, F.B.R Reproducibility of functional MR imaging: preliminary results of prospective multi-institutional study performed by Biomedical Informatics Research Network. Radiology. 2005;237:781–789. doi: 10.1148/radiol.2373041630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X.-N., Anderson J.S., Bellec P., Birn R.M., Biswal B.B., Blautzik J., Breitner J.C.S., Buckner R.L., Calhoun V.D., Castellanos F.X., Chen A., Chen B., Chen J., Chen X., Colcombe S.J., Courtney W., Craddock R.C., Di Martino A., Dong H.-M., Fu X., Gong Q., Gorgolewski K.J., Han Y., He Y., He Y., Ho E., Holmes A., Hou X.-H., Huckins J., Jiang T., Jiang Y., Kelley W., Kelly C., King M., LaConte S.M., Lainhart J.E., Lei X., Li H.-J., Li K., Li K., Lin Q., Liu D., Liu J., Liu X., Liu Y., Lu G., Lu J., Luna B., Luo J., Lurie D., Mao Y., Margulies D.S., Mayer A.R., Meindl T., Meyerand M.E., Nan W., Nielsen J.A., O’Connor D., Paulsen D., Prabhakaran V., Qi Z., Qiu J., Shao C., Shehzad Z., Tang W., Villringer A., Wang H., Wang K., Wei D., Wei G.-X., Weng X.-C., Wu X., Xu T., Yang N., Yang Z., Zang Y.-F., Zhang L., Zhang Q., Zhang Z., Zhang Z., Zhao K., Zhen Z., Zhou Y., Zhu X.-T., Milham M.P. An open science resource for establishing reliability and reproducibility in functional connectomics. Sci. Data. 2014;1:140049. doi: 10.1038/sdata.2014.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.