ABSTRACT
It is widely accepted that the glucocorticoid receptor (GR), a ligand-regulated transcription factor that triggers anti-inflammatory responses, binds specific response elements as a homodimer. Here, we will discuss the original primary data that established this model and contrast it with a recent report characterizing the GR–DNA complex as a tetramer.
KEYWORDS: DNA binding, dimer, glucocorticoid receptor, monomer, steroid receptors, tetramer
The glucocorticoid receptor (GR) is a modular transcription factor, organized in three distinct structural and functional domains: the N-terminal domain (NTD), a central DNA-binding (DBD) domain, and a C-terminal Ligand-binding domain (LBD).1 The GR is expressed in most cell tissues, and is involved in critical biological processes including homeostasis and metabolism. Moreover, pharmacological activation of the GR triggers powerful anti-inflammatory and immunosuppressive actions, making the receptor one of the most targeted proteins for therapy. GR oligomeric manipulation is considered a key aspect in the search of synthetic ligands that would eliminate the side-effects associated with chronic glucocorticoid treatment.2
In current dogma, GR is considered to bind to specific glucocorticoid response elements (GREs) as a homodimer. However, primary data references for this model are largely absent in current review articles, or in research articles. This is expected for long-standing, widely accepted models. We recently reported that DNA binding at response elements induces tetramerization of the GR,3 suggesting that tetramers maybe the final, active chromatin bound form of the receptor. The report3 presents the first in vivo experiments that directly address the oligomeric state of GR when bound to chromatin. In this brief comment, we will argue that the dimerization model exclusively relies on in vitro data. Furthermore, the recent observations in living cells are, to some extent, consistent with early in vitro data, indicating that new efforts are needed to resolve the long-standing issue regarding the active form of the DNA bound GR.
GR as a dimer: In vitro studies
We refer here to in vitro studies as any experiment that has been performed in a cell-free environment. The first description of GR's oligomeric status was presented in 1983 by the Yamamoto lab. The authors purified GR from rat liver and analyzed by electron microscopy GR “particles” either bound or unbound to a DNA fragment containing a GRE sequence from the mouse mammary tumor virus (MMTV) promoter region.4 Based on the size of the particles, these investigators concluded: “…the 94 kD receptor subunits seem to form homotetramers in a DNA-independent manner under these conditions (…) but this has not been confirmed by independent methods.” Later, the Gustafsson group evaluated the relative stoichiometry between GR and DNA.5 Purified receptor bound to 3H-triamcinolone acetonide (a GR agonist) is incubated with 32P-labed DNA. After glycerol centrifugation, each fraction is analyzed and the relative radioactive signals are obtained. The authors concluded that only one GR molecule binds a single GRE, suggesting that GR binds to DNA as a monomer. However, the same technique applied on a “stronger” GRE sequence indicated that the activated GR exists as a homodimer when unbound as well as when bound to DNA.6
Another commonly used technique to study DNA binding is the electro-mobility shift assay (EMSA). In this case, in vitro translated, GST-purified (usually only the DBD fragment) or endogenous purified receptor is mixed with labeled DNA and the binding products analyzed by polyacrylamide gel. Early efforts identified two distinct shifted bands. These were assigned as monomeric and dimeric complexes, a conclusion perhaps also biased from the dyad symmetry of the GRE sequence.7 Therefore, results from EMSA experiments suggested that the functional entity that binds to a GRE is a dimer.7,8 Although some studies reported that the monomer binds first to DNA,7,9,10 other reports provided evidence for DNA-independent dimer formation.6,8,11–14 In general, groups that worked with the entire receptor argued for a DNA-independent pathway,8,11,12,14 whereas those working with the DBD fragment found that it was the monomer that first binds DNA, which in turn favors the binding of the second monomer, a concept known as positive cooperative effect.7,9,10 The use of other in vitro approaches did not help to solve the controversy. Although DNA-independent positive interactions between in vitro translated GR and cell-extracted immunoprecipitated GR have been observed in solution,15 other investigators argue that GR exists almost exclusively in a monomeric state.16 Finally, several early papers suggested that either adjacent GREs17–19 or unusual GREs20 could lead to the formation of homo-tetramers. However, when the solution and crystal structures of GR's DBD were elucidated,21,22 the community rapidly adopted the idea that the activated GR was in fact a homodimer.23
From the structural perspective, only the DBD22 and LBD24 domains have been crystalized, although separately. The NTD has eluded crystallization and high-resolution structure, most likely due to its intrinsically disordered domain.25 The first crystal structure of GR's DBD bound to DNA revealed a clear dimerization region between the two receptor monomers.22 Although several studies in the 90s suggested a region outside the DBD could be involved in GR dimerization,10,11,26,27 the predominant view portrayed the DBD as the exclusive domain responsible for dimer formation. This dogma developed largely because a point mutation in that region (known as the GRdim mutant) was allegedly sufficient to generate a monomeric GR,28,29 although no direct evidence was provided at the time.30,31 When the crystal structure of the LBD was reported and a second dimerization region discovered,24 concerns about the physiological relevance and functional contribution of both domains arose.32 Some investigators still argue that the LBD dimers are an artifact of crystallization, and that the LBD dimerization of GR is “unlikely.”33
Taken together, the evidence from in vitro data appears to indicate that GR is a dimer, although no clear consensus exists as to whether GR dimerizes before or after DNA binding, nor which domains are involved. The clearest results come, however, from studies performed on the DBD fragment alone and not the entire GR protein.
The in vivo perspective on GR dimerization
We refer here to in vivo studies as any experiment that has been performed inside living cells, or experiments wherein the biological parameter measured occurred in intact cells but was revealed with an in vitro technique. Inside the cell, in the absence of ligand, the receptor is mostly retained in the cytoplasmic compartment, as a monomer,34 by being part of a heterocomplex with Hsp90, Hsp70, p23, and immunophilins, among others.35 However, GR overexpression has been reported to induce ligand-independent cytoplasmic dimerization.36 Since the heterocomplex is necessary for proper folding that allows GR to bind hormone,37 it is not clear how the GR dimers can still remain associated with the heterocomplex, as they are able to bind ligand and translocate into the nucleus.36
Once GR is activated by ligand, it translocates almost completely to the nuclear compartment. Using a nuclear-import deficient receptor mutant in the context of its wild-type counterpart, the Hache lab demonstrated that GR can interact with itself before and/or during retrograde transport.15 This experiment constitutes the first demonstration that the GR can actually interact with itself in vivo (i.e., inside living cells), many years after the community had already adopted the dimerization paradigm. The Hache group noted at the time: “At present we cannot exclude the potential formation of higher-order complexes of GR.” Co-immunoprecipitations experiments from cell extracts also confirmed the presence of GR–GR interactions inside the cell,30,38 however, the interaction was always detected both in the presence and absence of hormone. Finally, using Förster resonance energy transfer (FRET) in vivo,39 GR–GR interactions have also been detected,40 although the assay cannot discriminate between different oligomerization states or accurately quantify the proportion of monomers against dimers in the population.
The first in vivo quantification of the quaternary structure of GR came from the use of the Number and Brightness assay (N&B), a microscopy technique that measures the oligomerization state of fluorescent proteins with high spatial resolution.41 Unexpectedly, results revealed that most of the ligand–GR complexes were dimeric in the nucleus,34 although a small proportion of monomers or higher oligomerization states could not been ruled out as well. Because the fraction of GR specifically bound to DNA at any given time in vivo is very low (3–5%),42–44 the virtually complete population of dimers observed in the nucleoplasmic N&B assay must arise from mostly unbound receptors, strongly suggesting a DNA-independent model for GR dimerization.31
Collectively, from the in vivo perspective, the activated GR appears to be mostly dimeric in the nucleoplasm. However, until recently, no in vivo experiment has measured directly the quaternary structure of the GR–chromatin complex. Theoretical models based on in vivo experimental kinetic studies predict that it is the GR monomer the entity that first binds to DNA.45 Alternatively, chromatin immunoprecipitation coupled with exonuclease digestion (ChIP-exo) “footprints” have been interpreted as in vivo evidence for dimeric binding of GR,46,47 based on the protection signature of the exonuclease from the DBD–DNA contacts. Since ChIP assays neither measure directly nor indirectly oligomerization states, the conclusion that GR is a dimer was drawn by assuming that (i) one-half GRE footprint always represents a monomeric GR event and (ii) GR dimerization exclusively relies on DBD–DBD contacts. However, direct measurement of the quaternary structure by N&B demonstrates that LBD–LBD contacts are more relevant in stabilizing the dimers than DBD–DBD contacts.3,31 Hence, it is conceivable that GR dimers can still bind a half-GRE wherein LBD–LBD contacts held the dimer together. In fact, when directly measured at a tandem array of GR-binding sites, the receptor appears tetrameric at full GREs.3
In vitro versus in vivo data: Any room for reconciliation?
In a recent report,3 we concluded that GR is mostly a tetramer when bound to DNA. This conclusion is based on four independent findings: (i) N&B measurements at an array of response elements show the presence of tetramers; (ii) A mutation that mimics the DNA-bound conformation of GR (P493R) triggers tetramerization in the whole nucleoplasm; (iii) Homo-FRET studies show higher oligomerization states in both nucleoplasm and the array; and (iv) Single-molecule photobleaching experiments detect the presence of greater-than-two-subunit oligomers randomly in the nucleus. Using several mutations, we also reported that tetramer formation depends on the presence of the LBD, is independent of dimerization surfaces, and requires DNA-induced conformational changes in the DBD.
How can these results be reconciled with the previous literature? One of the strongest evidence for GR dimerization is the crystallographic X-ray structure.22 However, lack of tetramerization of the DBD fragment is not surprising since tetramer formation depends on the LBD. Moreover, the monomer-to-dimer transition observed in EMSA assays7,9,10 is completely consistent with the N&B data once the LBD domain is removed. The NTD–DBD fragment behaves in vivo as the DBD behaves in vitro: fully monomeric in the nucleoplasm and fully dimeric at GREs.3 It is worth mention one study, by the Miguel Beato's group,8 where they mixed full-length and DBD fragments to form whole-GR/DBD-only heterodimers, strongly suggesting the presence of dimeric forms of the receptor. The in vivo evidence, on the contrary, only points to tetramerization3 as there are no other direct measurements reported.
If GR binds to DNA as a dimer in vitro and as a tetramer in vivo, then something must be missing in the in vitro system. They are several key elements present in vivo that are absent in the controlled in vitro environment: post-translational modifications, cofactor interactions, nucleosomes, the interphase chromatin landscape, to name a few. Some of these elements could be proven essential for this “next regulatory step” GR seems to have within live cells.
Toward a new model for GR oligomerization
After review of more than 30 years of research, we suggest that the evidence for GR dimerization is not as solid as originally thought. Based on the new N&B findings, we propose a new model that attempts to resolve all available data (Fig. 1).
Figure 1.
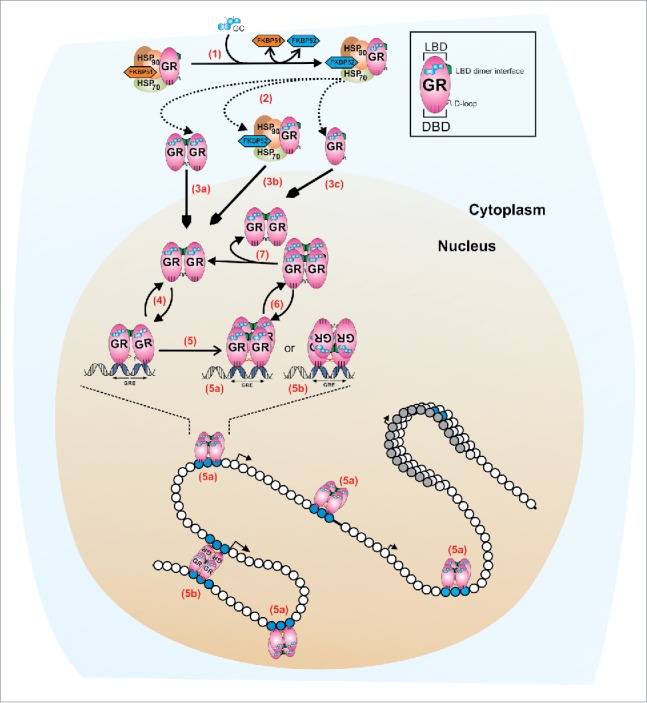
A revised model for GR quaternary structure dynamics. In the absence of ligand, the monomeric cytoplasmic GR forms a heterocomplex with Hsp90, Hsp70, FKBP51, and other proteins (not shown). (1) Ligand binding (GC) induces a conformational change in the GR that leads to either FKBP51-FKBP52 exchange within the heterocomplex, or the complete dissociation from the complex (2). At least two mechanisms appear to regulate the influx of GR molecules to the nuclear compartment: a microtubule/Hsp90 dependent pathway (3b, microtubule not shown) and a microtubule/Hsp90 independent pathway (3a, 3c).35 Early in vivo studies suggests that GR is a dimer before translocation15 (3a) but a monomeric population cannot be rule out (3c). Nucleoplasmic GR appears to be mostly dimeric,3,31,34 with at least two contact surfaces localized in the LBD (shown in green) and the DBD (D-loop, shown in black).31 The interaction with specify response elements (GREs) is very dynamic (4), in the order of seconds.42 DNA induces an allosteric change in the receptor's DBD domain51 (D-loop shown in red) which triggers a conformational change in the LBD, allowing the formation of tetramers, either in a head-to-head (5a) or head-to-tail (5b) configuration.3 GR tetramers also exchange dynamically with DNA (6) and, if it is the tetramer the quaternary structure that detaches from DNA, then the dissociation into dimers should occur at a much faster temporal scale (7), to account for the mostly complete population of dimers observed in the nucleoplasm.3 This highly dynamic regulation occurs in the context of a chromatin landscape. We speculate that a head-to-tail configuration (5b) may assist in bridging different points in the genome, thus favoring a looping mechanism between distant regulatory sites.
In the absence of ligand, the inactivated GR appears to be fully monomeric inside the nuclear compartment,31, 34 while their behavior in the cytoplasm could depend on its concentration: At endogenous levels, the GR is most likely a monomer but overexpression of the receptor could lead to ligand-independent dimerization.30,36,38,40
When the GR is activated by dexamethasone or its natural ligand corticosterone, dimer formation is initiated before or during the nuclear translocation process.15 The reported inability of the non-steroid GR ligand Compound A to efficiently promote GR nuclear translocation48 may be explained by its incapacity to induce GR dimer complexes.31 Once in the nucleus, virtually all agonist-bound GR molecules are in the dimeric form31,34 through LBD–LBD24 and DBD–DBD22 interactions, although mutational analyses indicate that these dimeric surfaces are not functionally equivalent. In fact, dimerization through the DBD is dependent upon the presence of the LBD.3 It has also been documented that after specific DNA binding, the DBD changes conformation,49 potentially favoring DBD–DBD interactions.50 Hence, the positive cooperative binding between monomers observed in vitro when only the DBD fragment is used10,50 may not be a key factor in vivo, and reflects further stabilization of the pre-formed dimers after engaging chromatin. Since GR molecules are already dimeric before binding to DNA, it is the dimer and not the monomer that is the hormone-activated GR entity (Fig. 1).
The GR is allosterically modulated not only by ligand binding, but also by DNA itself.51 This suggests that GREs do not merely serve as GR docking points, but may also modulate GR activity by altering its conformation.52 In fact, thermodynamic studies using GR's DBD have shown an induce-fit binding mode to DNA,53 therefore suggesting at least a two-step, and possibly a multi-step mechanism. We propose that this new conformation triggers a structural re-arrangement in the LBD, promoting the formation of higher order oligomers, predominantly tetramers, through LBD surfaces that are yet to be identified.3 This phenomenon may be more common than previously thought, as STAT3 has been recently described as transitioning from dimer to tetramers in a DNA-dependent manner.54 A deeper understanding on the intricacies of GR quaternary structure may help find new strategies in the search for safer glucocorticoids, or at least finally close some roads taken in the past2 that have led us nowhere. Finally, as combinatorial long-range interactions between regulatory elements play an important role in gene regulation,55 we speculate the tetrameric nature of some transcription factors such as GR or STAT3 can serve as a platform to bridge different points in the genome.
Abbreviations
- ChIP
chromatin immunoprecipitation
- DBD
DNA-binding domain
- EMSA
electro-mobility shift assay
- GC
glucocorticoids
- GR
glucocorticoid receptor
- GRE
glucocorticoid response element
- LBD
ligand-binding domain
- MMTV
mouse mammary tumor virus
- N&B
number and brightness
- NTD
N-terminal domain
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank S. Stoney Simons Jr., Ido Goldstein, and Ville Paakinaho for critical reading of the manuscript.
Funding
This work was supported by the Intramural Research Program of the National Institutes of Health (NIH) and the Center for Cancer Research (CCR) at the National Cancer Institute (NCI).
References
- [1].Moore JT, Collins JL, Pearce KH. The nuclear receptor superfamily and drug discovery. Chem Med Chem 2006; 1:504-523; PMID:16892386; http://dx.doi.org/ 10.1002/cmdc.200600006 [DOI] [PubMed] [Google Scholar]
- [2].Clark AR, Belvisi MG. Maps and legends: the quest for dissociated ligands of the glucocorticoid receptor. Pharmacol Ther 2012; 134:54-67; PMID 22212616; http://dx.doi.org/ 10.1016/j.pharmthera.2011.12.004 [DOI] [PubMed] [Google Scholar]
- [3].Presman DM, Ganguly S, Schiltz RL, Johnson TA, Karpova TS, Hager GL. DNA binding triggers tetramerization of the glucocorticoid receptor in live cells. Proc Natl Acad Sci U S A 2016; 113:8236-8241; PMID:27382178; http://dx.doi.org/ 10.1073/pnas.1606774113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Payvar F, DeFranco DB, Firestone GL, Edgar B, Wrange O, Okret S, Gustafsson JA, Yamamoto KR. Sequence-specific binding of glucocorticoid receptor to MTV DNA at sites within and upstream of the transcribed region. Cell 1983; 35:381-392; PMID:6317184; http://dx.doi.org/ 10.1016/0092-8674(83)90171-X [DOI] [PubMed] [Google Scholar]
- [5].Wrange O, Carlstedt-Duke J, Gustafsson JA. Stoichiometric analysis of the specific interaction of the glucocorticoid receptor with DNA. J Biol Chem 1986; 261:11770-11778; PMID:3017946 [PubMed] [Google Scholar]
- [6].Wrange O, Eriksson P, Perlmann T. The purified activated glucocorticoid receptor is a homodimer. J Biol Chem 1989; 264:5253-5259; PMID:2494184 [PubMed] [Google Scholar]
- [7].Tsai SY, Carlstedt-Duke J, Weigel NL, Dahlman K, Gustafsson JA, Tsai MJ, O'Malley BW. Molecular interactions of steroid hormone receptor with its enhancer element: evidence for receptor dimer formation. Cell 1988; 55:361-369; PMID:3167984; http://dx.doi.org/ 10.1016/0092-8674(88)90059-1 [DOI] [PubMed] [Google Scholar]
- [8].Chalepakis G, Schauer M, Cao X, Beato M. Efficient binding of glucocorticoid receptor to its responsive element requires a dimer and DNA flanking sequences. DNA Cell Biol 1990; 9:355-368; PMID:2372377; http://dx.doi.org/ 10.1089/dna.1990.9.355 [DOI] [PubMed] [Google Scholar]
- [9].Dahlman-Wright K, Siltala-Roos H, Carlstedt-Duke J, Gustafsson JA. Protein-protein interactions facilitate DNA binding by the glucocorticoid receptor DNA-binding domain. J Biol Chem 1990; 265:14030-14035; PMID:1974254 [PubMed] [Google Scholar]
- [10].Dahlman-Wright K, Wright A, Gustafsson JA, Carlstedt-Duke J. Interaction of the glucocorticoid receptor DNA-binding domain with DNA as a dimer is mediated by a short segment of five amino acids. J Biol Chem 1991; 266:3107-3112; PMID:1993683 [PubMed] [Google Scholar]
- [11].Cairns W, Cairns C, Pongratz I, Poellinger L, Okret S. Assembly of a glucocorticoid receptor complex prior to DNA binding enhances its specific interaction with a glucocorticoid response element. J Biol Chem 1991; 266:11221-11226; PMID:2040629 [PubMed] [Google Scholar]
- [12].Drouin J, Sun YL, Tremblay S, Lavender P, Schmidt TJ, de LA, Nemer M. Homodimer formation is rate-limiting for high affinity DNA binding by glucocorticoid receptor. Mol Endocrinol 1992; 6:1299-1309; PMID:1406707 [DOI] [PubMed] [Google Scholar]
- [13].Eriksson P, Wrange O. Protein-protein contacts in the glucocorticoid receptor homodimer influence its DNA binding properties. J Biol Chem 1990; 265:3535-3542; PMID:2303460 [PubMed] [Google Scholar]
- [14].Perlmann T, Eriksson P, Wrange O. Quantitative analysis of the glucocorticoid receptor-DNA interaction at the mouse mammary tumor virus glucocorticoid response element. J Biol Chem 1990; 265:17222-17229; PMID:2170368 [PubMed] [Google Scholar]
- [15].Savory JG, Prefontaine GG, Lamprecht C, Liao M, Walther RF, Lefebvre YA, Hache RJ. Glucocorticoid receptor homodimers and glucocorticoid-mineralocorticoid receptor heterodimers form in the cytoplasm through alternative dimerization interfaces. MolCell Biol 2001; 21:781-793; PMID 11154266; http://dx.doi.org/ 10.1128/MCB.21.3.781-793.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Robblee JP, Miura MT, Bain DL. Glucocorticoid receptor-promoter interactions: energetic dissection suggests a framework for the specificity of steroid receptor-mediated gene regulation. Biochemistry 2012; 51:4463-4472; PMID:22587663; http://dx.doi.org/ 10.1021/bi3003956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Schmid W, Strahle U, Schutz G, Schmitt J, Stunnenberg H. Glucocorticoid receptor binds cooperatively to adjacent recognition sites. EMBO J 1989; 8:2257-2263; PMID:2792086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wright AP, Gustafsson JA. Mechanism of synergistic transcriptional transactivation by the human glucocorticoid receptor. Proc Natl Acad Sci USA 1991; 88:8283-8287; PMID:1924286; http://dx.doi.org/ 10.1073/pnas.88.19.8283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Liu W, Wang J, Yu G, Pearce D. Steroid receptor transcriptional synergy is potentiated by disruption of the DNA-binding domain dimer interface. Mol Endocrinol 1996; 10:1399-1406; PMID:8923466 [DOI] [PubMed] [Google Scholar]
- [20].Garlatti M, Daheshia M, Slater E, Bouguet J, Hanoune J, Beato M, Barouki R. A functional glucocorticoid-responsive unit composed of two overlapping inactive receptor-binding sites: evidence for formation of a receptor tetramer. Mol Cell Biol 1994; 14:8007-8017; PMID:7969140; http://dx.doi.org/ 10.1128/MCB.14.12.8007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hard T, Kellenbach E, Boelens R, Maler BA, Dahlman K, Freedman LP, Carlstedt-Duke J, Yamamoto KR, Gustafsson JA, Kaptein R. Solution structure of the glucocorticoid receptor DNA-binding domain. Science 1990; 249:157-160; PMID:2115209; http://dx.doi.org/ 10.1126/science.2115209 [DOI] [PubMed] [Google Scholar]
- [22].Luisi BF, Xu WX, Otwinowski Z, Freedman LP, Yamamoto KR, Sigler PB. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA [see comments]. Nature 1991; 352:497-505; PMID:1865905; http://dx.doi.org/ 10.1038/352497a0 [DOI] [PubMed] [Google Scholar]
- [23].Beato M, Herrlich P, Schutz G. Steroid hormone receptors: many actors in search of a plot. Cell 1995; 83:851-857; PMID:8521509; http://dx.doi.org/ 10.1016/0092-8674(95)90201-5 [DOI] [PubMed] [Google Scholar]
- [24].Bledsoe RK, Montana VG, Stanley TB, Delves CJ, Apolito CJ, McKee DD, Consler TG, Parks DJ, Stewart EL, Willson TM et al.. Crystal structure of the glucocorticoid receptor ligand binding domain reveals a novel mode of receptor dimerization and coactivator recognition. Cell 2002; 110:93-105; PMID:12151000; http://dx.doi.org/ 10.1016/S0092-8674(02)00817-6 [DOI] [PubMed] [Google Scholar]
- [25].Simons SS Jr, Edwards DP, Kumar R. Minireview: dynamic structures of nuclear hormone receptors: new promises and challenges. Mol Endocrinol 2014; 28:173-182; PMID:24284822; http://dx.doi.org/ 10.1210/me.2013-1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Dahlman-Wright K, Wright AP, Gustafsson JA. Determinants of high-affinity DNA binding by the glucocorticoid receptor: evaluation of receptor domains outside the DNA-binding domain. Biochemistry 1992; 31:9040-9044; PMID:1390690; http://dx.doi.org/ 10.1021/bi00152a047 [DOI] [PubMed] [Google Scholar]
- [27].Segard-Maurel I, Rajkowski K, Jibard N, Schweizer-Groyer G, Baulieu EE, Cadepond F. Glucocorticosteroid receptor dimerization investigated by analysis of receptor binding to glucocorticosteroid responsive elements using a monomer-dimer equilibrium model. Biochemistry 1996; 35:1634-1642; PMID:8634295; http://dx.doi.org/ 10.1021/bi951369h [DOI] [PubMed] [Google Scholar]
- [28].Heck S, Kullmann M, Gast A, Ponta H, Rahmsdorf HJ, Herrlich P, Cato AC. A distinct modulating domain in glucocorticoid receptor monomers in the repression of activity of the transcription factor AP-1. EMBO J 1994; 13:4087-4095; PMID:8076604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Reichardt HM, Kaestner KH, Tuckermann J, Kretz O, Wessely O, Bock R, Gass P, Schmid W, Herrlich P, Angel P et al.. DNA binding of the glucocorticoid receptor is not essential for survival [see comments]. Cell 1998; 93:531-541; PMID:9604929; http://dx.doi.org/ 10.1016/S0092-8674(00)81183-6 [DOI] [PubMed] [Google Scholar]
- [30].Jewell CM, Scoltock AB, Hamel BL, Yudt MR, Cidlowski JA. Complex human glucocorticoid receptor dim mutations define glucocorticoid induced apoptotic resistance in bone cells. Mol Endocrinol 2012; 26:244-256; PMID:22174376; http://dx.doi.org/ 10.1210/me.2011-1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Presman DM, Ogara MF, Stortz M, Alvarez LD, Pooley JR, Schiltz RL, Grontved L, Johnson TA, Mittelstadt PR, Ashwell JD et al.. Live cell imaging unveils multiple domain requirements for in vivo dimerization of the Glucocorticoid Receptor. PLoS Biol 2014; 12:e1001813; PMID:24642507; http://dx.doi.org/ 10.1371/journal.pbio.1001813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Bledsoe RK, Stewart EL, Pearce KH. Structure and function of the glucocorticoid receptor ligand binding domain. VitamHorm 2004; 68:49-91; PMID 15193451; http://dx.doi.org/ 10.1016/S0083-6729(04)68002-2 [DOI] [PubMed] [Google Scholar]
- [33].Helsen C, Claessens F. Looking at nuclear receptors from a new angle. Mol Cell Endocrinol 2014; 382:97-106; PMID:24055275; http://dx.doi.org/ 10.1016/j.mce.2013.09.009 [DOI] [PubMed] [Google Scholar]
- [34].Presman DM, Alvarez LD, Levi V, Eduardo S, Digman MA, Marti MA, Veleiro AS, Burton G, Pecci A. Insights on glucocorticoid receptor activity modulation through the binding of rigid steroids. PLoS One 2010; 5:e13279; PMID:20949009; http://dx.doi.org/ 10.1371/journal.pone.0013279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Echeverria PC, Picard D. Molecular chaperones, essential partners of steroid hormone receptors for activity and mobility. BiochimBiophysActa 2010; 1803:641-649; PMID 20006655; http://dx.doi.org/ 10.1016/j.bbamcr.2009.11.012 [DOI] [PubMed] [Google Scholar]
- [36].Robertson S, Rohwer JM, Hapgood JP, Louw A. Impact of glucocorticoid receptor density on ligand-independent dimerization, cooperative ligand-binding and Basal priming of transactivation: a cell culture model. PLoS One 2013; 8:e64831; PMID:23717665; http://dx.doi.org/ 10.1371/journal.pone.0064831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Bresnick EH, Dalman FC, Sanchez ER, Pratt WB. Evidence that the 90-kDa heat shock protein is necessary for the steroid binding conformation of the L cell glucocorticoid receptor. J Biol Chem 1989; 264:4992-4997; PMID:2647745 [PubMed] [Google Scholar]
- [38].Dewint P, Gossye V, De BK, Vanden BW, Van BK, Deforce D, Van CS, Muller-Ladner U, Vander CB, Verbruggen G et al.. A plant-derived ligand favoring monomeric glucocorticoid receptor conformation with impaired transactivation potential attenuates collagen-induced arthritis. J Immunol 2008; 180:2608-2615; PMID:18250472; http://dx.doi.org/ 10.4049/jimmunol.180.4.2608 [DOI] [PubMed] [Google Scholar]
- [39].Jares-Erijman EA, Jovin TM. Imaging molecular interactions in living cells by FRET microscopy. Curr Opin Chem Biol 2006; 10:409-416; PMID: 16949332; http://dx.doi.org/ 10.1016/j.cbpa.2006.08.021 [DOI] [PubMed] [Google Scholar]
- [40].Robertson S, lie-Reid F, Vanden BW, Visser K, Binder A, Africander D, Vismer M, De BK, Hapgood J, Haegeman G et al.. Abrogation of glucocorticoid receptor dimerization correlates with dissociated glucocorticoid behavior of compound A. J Biol Chem 2010; 285:8061-8075; PMID:20037160; http://dx.doi.org/ 10.1074/jbc.M109.087866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Digman MA, Dalal R, Horwitz AF, Gratton E. Mapping the number of molecules and brightness in the laser scanning microscope. BiophysJ 2008; 94:2320-2332; PMID: 18096627; http://dx.doi.org/25034201 10.1529/biophysj.107.114645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Morisaki T, Muller WG, Golob N, Mazza D, McNally JG. Single-molecule analysis of transcription factor binding at transcription sites in live cells. Nat Commun 2014; 5:4456; PMID:25034201; http://dx.doi.org/ 10.1038/ncomms5456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Swinstead Erin E, Miranda Tina B, Paakinaho V, Baek S, Goldstein I, Hawkins M, Karpova Tatiana S, Ball D, Mazza D, Lavis Luke D et al.. Steroid receptors reprogram FoxA1 occupancy through dynamic chromatin transitions. Cell 2016; 165:593-605; PMID:27062924; http://dx.doi.org/ 10.1016/j.cell.2016.02.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Voss TC, Schiltz RL, Sung MH, Yen PM, Stamatoyannopoulos JA, Biddie SC, Johnson TA, Miranda TB, John S, Hager GL. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell 2011; 146:544-554; PMID:21835447; http://dx.doi.org/ 10.1016/j.cell.2011.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Ong KM, Blackford JA Jr, Kagan BL, Simons SS Jr, Chow CC. A theoretical framework for gene induction and experimental comparisons. Proc Natl Acad Sci USA 2010; 107:7107-7112; PMID: 20351279; http://dx.doi.org/25957148 10.1073/pnas.0911095107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Lim HW, Uhlenhaut NH, Rauch A, Weiner J, Hubner S, Hubner N, Won KJ, Lazar MA, Tuckermann J, Steger DJ. Genomic redistribution of GR monomers and dimers mediates transcriptional response to exogenous glucocorticoid in vivo. Genome Res 2015; 25:836-844; PMID:25957148; http://dx.doi.org/ 10.1101/gr.188581.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Starick SR, Ibn-Salem J, Jurk M, Hernandez C, Love MI, Chung HR, Vingron M, Thomas-Chollier M, Meijsing SH. ChIP-exo signal associated with DNA-binding motifs provide insights into the genomic binding of the glucocorticoid receptor and cooperating transcription factors. Genome Res 2015; 25(6):825-35; PMID:25720775; http://dx.doi.org/ 10.1101/gr.185157.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Sundahl N, Bridelance J, Libert C, De Bosscher K, Beck IM. Selective glucocorticoid receptor modulation: New directions with non-steroidal scaffolds. Pharmacol Therapeutics 2015; 152:28-41; PMID:25958032; http://dx.doi.org/ 10.1016/j.pharmthera.2015.05.001 [DOI] [PubMed] [Google Scholar]
- [49].Lefstin JA, Thomas JR, Yamamoto KR. Influence of a steroid receptor DNA-binding domain on transcriptional regulatory functions. Genes Dev 1994; 8:2842-2856; PMID:7995522; http://dx.doi.org/ 10.1101/gad.8.23.2842 [DOI] [PubMed] [Google Scholar]
- [50].Watson LC, Kuchenbecker KM, Schiller BJ, Gross JD, Pufall MA, Yamamoto KR. The glucocorticoid receptor dimer interface allosterically transmits sequence-specific DNA signals. Nat Struct Mol Biol 2013; 20:876-883; PMID: 23728292; http://dx.doi.org/19372434 10.1038/nsmb.2595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Meijsing SH, Pufall MA, So AY, Bates DL, Chen L, Yamamoto KR. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science 2009; 324:407-410; PMID:19372434; http://dx.doi.org/ 10.1126/science.1164265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Gronemeyer H, Bourguet W. Allosteric effects govern nuclear receptor action: DNA appears as a player. Sci Signal 2009; 2:pe34; PMID:19491383; http://dx.doi.org/ 10.1126/scisignal.273pe34 [DOI] [PubMed] [Google Scholar]
- [53].Spolar RS, Record MT Jr. Coupling of local folding to site-specific binding of proteins to DNA. Science 1994; 263:777-784; PMID:8303294; http://dx.doi.org/ 10.1126/science.8303294 [DOI] [PubMed] [Google Scholar]
- [54].Hinde E, Pandzic E, Yang Z, Ng IH, Jans DA, Bogoyevitch MA, Gratton E, Gaus K. Quantifying the dynamics of the oligomeric transcription factor STAT3 by pair correlation of molecular brightness. Nat Commun 2016; 7:11047; PMID:27009358; http://dx.doi.org/ 10.1038/ncomms11047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Stavreva DA, Hager GL. Chromatin structure and gene regulation: a dynamic view of enhancer function. Nucleus 2015; 6:442-448; PMID:26765055; http://dx.doi.org/ 10.1080/19491034.2015.1107689 [DOI] [PMC free article] [PubMed] [Google Scholar]


