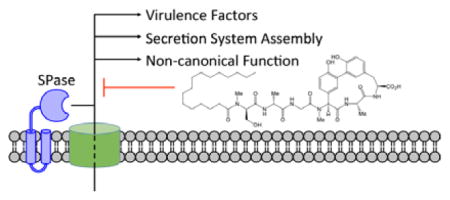
Abstract
The looming antibiotic crisis has prompted the development of new strategies towards fighting infection. Traditional antibiotics target bacterial processes essential for viability, whereas proposed antivirulence approaches rely on the inhibition of factors that are required only for the initiation and propagation of infection within a host. Although antivirulence compounds have yet to prove their efficacy in the clinic, bacterial signal peptidase I (SPase) represents an attractive target in that SPase inhibitors exhibit broad-spectrum antibiotic activity, but even at sub-MIC doses also impair the secretion of essential virulence factors. The potential consequences of SPase inhibition on bacterial virulence have not been thoroughly examined, and are explored within this review. In addition, we review growing evidence that SPase has relevant biological functions outside of mediating secretion, and discuss how the inhibition of these functions may be clinically significant.
Keywords: Secretion, Arylomycin, Antivirulence, SPase, Protein Translocation
Graphical Abstract
1. Introduction
We are at risk of losing effective antibiotic therapies as resistance continues to rise at an alarming rate.1 Bacteria resistant to one or more antibiotics are currently responsible for at least 50,000 deaths per year in Europe and the USA alone.2 However, the real threat of antibiotic resistance lies ahead. Unless the current rates of resistance are checked, that mortality rate could rise to 10 million worldwide per year by 2050.3 In addition to these direct consequences, antibiotics are fundamental to all aspects of modern healthcare. Prophylactic antibiotics are prescribed for most surgeries, without which such procedures might be too risky to perform, and for treatments such as chemotherapy, which reduces immune system defenses. Due to all of these factors, the World Health Organization has identified antibiotic resistance as one of the three greatest threats to human health.4
Given the rise in antibiotic resistance, one might expect research into novel antibiotics to grow in response, but just the opposite has occurred. Most major pharmaceutical companies have reduced early development efforts or abandoned the field completely. According to a recent Pew Charitable Trust analysis, only 5 of the top 50 pharmaceutical companies are pursuing new antibiotics.5 This can be attributed to the difficulty in simply developing a new antibiotic and the lure of more profitable ventures. Antibiotics are only prescribed for a short period of time, as opposed to medicines for chronic conditions such as diabetes or arthritis. It costs around $2.6 billion and takes eight to fifteen years to fully develop a new antibiotic.6–8 Only about one new antibiotic gains approval for every 16 candidates that enter phase I clinical trials,9 and even after approval it is possible that the clinical use of any new antibiotic will be reserved for last-resort cases in an effort to stave off resistance, further limiting potential returns on investment. As a result of reduced pharmaceutical interest, the antibiotic pipeline has all but run dry. Only four new classes of antibiotics have been introduced into the clinic in the last forty years (daptomycin, linezolid, fidaxomicin, bedaquiline), and none of them have activity against Gram-negative pathogens.
It would appear that our system for generating new antibiotics is broken.10 This has sparked a variety of proposals for alternative antibiotic strategies, including antivirulence strategies.11 Antivirulence compounds attenuate virulence without directly killing bacteria, and instead prevent processes that are essential for infection such as adhesion to host cells or the production of toxins. The immune system can then clear these emasculated bacteria. Some proponents of antivirulence strategies hypothesize that antivirulence compounds would put less selective pressure on bacteria and mitigate the emergence of resistance that has plagued traditional antibiotic therapy. Since there are no antivirulence drugs currently approved to treat bacterial infections in the clinic, this hypothesis remains to be tested.
The cited advantages of antivirulence strategies are also their greatest flaws. Inherently, antivirulence compounds will have a narrow spectrum of activity as virulence factors are poorly conserved across bacterial species. Their use in the clinic would be extremely limited in comparison to traditional broad-spectrum antibiotics, and thus in most cases are even less attractive to the pharmaceutical industry for development.
Type I signal peptidase (SPase) is an attractive target for therapeutic intervention because it is essential, supporting the possibility that inhibitors would have broad spectrum antibacterial activity, and because it is required for virulence. SPase is an essential part of the secretion apparatus; its proteolytic activity is required to release proteins from their N-terminal leader sequence, which remain membrane bound after the preprotein translocates across the cytoplasmic membrane. The antivirulence effects of inhibiting SPase are expected due to the many proteinaceous virulence factors that rely on SPase for processing into functional forms. SPase inhibition results in the accumulation of unprocessed proteins in the cytoplasmic membrane, which eventually causes it to lose its integrity and leads to cell death.12,13 This suggests that secretion is significantly impaired before the concentration and duration of exposure sufficient to kill cells is achieved. Indeed, in vitro studies have confirmed that even sub-MIC doses of arylomycin, a natural product SPase inhibitor, drastically reduce the secretion of proteins including, as discussed below, many virulence factors.14,15 Thus, treatment of infections with an SPase inhibitor may be able to kill bacterial cells and, even at lower doses, reduce virulence. In this review, we will summarize the current understanding of the role of SPase in virulence, as well as the potential antivirulence consequences of its inhibition. Due to the overwhelming number of effectors dependent on SPase for secretion, we will focus on those that highlight the diverse functions of SPase and which are likely the most relevant to human health.
2. SPase and the arylomycin family of natural products
By far, the most common route for protein secretion is the general secretory (Sec) pathway. Proteins designated for this route of secretion are translated with a specific motif at their N-terminus known as the signal (or leader) peptide.16 Typically conserved features of signal peptides include several positively charged residues at the N-terminus, a stretch of lipophilic residues, and a downstream polar cleavage site, which is commonly A-X-A. Before the protein achieves its mature form, the Sec machinery recognizes the signal peptide and translocates the pre-protein across the cytoplasmic membrane, during which time the lipophilic region of the signal peptide becomes embedded in the cytoplasmic membrane (Figure 1). SPase then cleaves the signal peptide to release the protein. SPase also functions at the terminal step of the Twin-Arginine Translocase (Tat) pathway. The Tat pathway is functionally similar to the Sec pathway, but it recognizes signal peptides containing a highly conserved R-R motif, and its pre-protein cargo fold prior to translocation across the cytoplasmic membrane. It is worth noting that two other signal peptidases exist: type II signal peptidase processes lipoproteins and type III signal peptidase (also known as type IV prepilin peptidase) specifically processes type IV prepilins and type IV prepilin-like proteins. Unless required for clarity, we will refer to type I signal peptides that are processed by SPase simply as “signal peptides.”
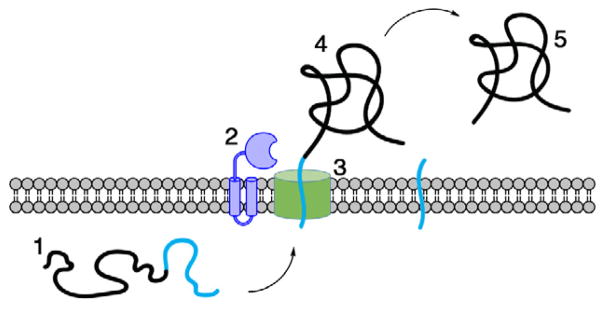
Figure 1.
Cleavage of the signal peptide (shown in cyan) from the encoded pre-protein completes secretion across the cytoplasmic membrane. Components shown are the (1) unfolded pre-protein, (2) SPase, (3) Sec translocon, (4) translocated pre-protein, and (5) secreted mature protein.
Several classes of inhibitors exist for SPase.17 Krisynomycin18 and the arylomycin family19–21 represent natural product inhibitors, whereas 5S penems,22,23 peptide substrate mimics,24–26 and a β-aminoketone27 are synthetic inhibitors. Of these, the arylomycins have been the most developed as potential therapeutics.28 While arylomycins have activity against a variety of Gram-positive and Gram-negative bacteria, mutations within SPase that ablate a hydrogen bond limit their spectrum. Interestingly, it has been demonstrated that restoring the hydrogen bond sensitizes an even wider spectrum of bacterial species to the arylomycins, including the Gram-negative pathogens Escherichia coli and Pseudomonas aeruginosa.29 This suggests that the arylomycin scaffold is a small increase in affinity away from becoming broad-spectrum SPase inhibitors. Derivatization efforts to impart them with this affinity have already led to compounds with broader spectrum and more potent activity.30
3. Virulence factors of Gram-positive bacteria
Gram-positive bacteria rely on Sec for the secretion of most virulence factors. Some Gram-positive bacteria also possess an accessory Sec system consisting of at least an additional SecA homolog (SecA2),31,32 as well as the Tat pathway.33–37 While each of these constitute different pathways across the cytoplasmic membrane, SPase functions unilaterally at their terminal step. As a result, the role of SPase in secretion of Gram-positive virulence factors is relatively well-defined. Only a few exceptions to Sec-dependent secretion exist, including type IV,38–43 type VII,32–34,36,44 and flagella biosynthesis, the substrates of which lack N-terminal signal peptides.33,45 The central importance of SPase is highlighted in in vivo studies of Listeria monocytogenes infection. As is common with Gram-positive bacteria, the genome of L. monocytogenes includes three separate SPase genes (SipX, SipY, and SipZ) that each play distinct roles in virulence. Deletion of sipX results in a 100-fold reduction of virulence, whereas deletion of sipZ results in an almost complete loss of infectivity in a mouse model.46 Other Gram-positive bacteria may encode one (e.g. Staphylococcus aureus and Mycobacterium tuberculosis) to as many as 7 (Bacillus cereus) SPases. When only a single SPase is encoded, it is essential for viability.47
After signal peptide cleavage, SPase substrates in Gram-positive bacteria are either covalently anchored to the cell wall via sortases or released into the environment.44,48–50 While no cell wall anchored proteins have been identified that are exported via Tat, the contribution of Tat-secreted, free virulence factors has been demonstrated in a variety of bacteria.32,35,37,51 For example, bioinformatics studies predict that M. tuberculosis encodes 31 Tat substrates, of which 18 have been experimentally confirmed and a number of which are known to contribute directly to virulence, including the phospholipase C enzymes, PlcA and PlcB, and the β-lactamase BlaC.52,53 SecA2 contributes virulence factors to both the cell wall and external environment.35,54,55
As opposed to SecA2 and Tat, the analysis of the general Sec-mediated secretome has been complicated due to the essentiality of SPase. Substrates have primarily been identified simply by analyzing proteins in the media of bacterial cultures. To reduce the potentially confounding contribution of contamination due to cell lysis, the identified proteins may be further delimited to those with in silico-predicted SPase cleavage sites.56 This approach has been applied extensively to staphylococcal and streptococcal species.57 One analysis of the secretome of S. aureus USA300 identified 174 proteins, ~52% of which were predicted to be authentic SPase substrates.58 Analysis of the secretomes of Streptococcus agalactiae, Streptococcus pneumoniae and Streptococcus gordonii revealed 8, 42 and 72 SPase substrates, respectively.59–61 However, the presence of a predicted SPase cleavage site does not demonstrate actual processing, and thus Ravipaty and Reilly identified 59 proteins in the media fraction of growing S. aureus COL, which by MS were shown to have processed N-termini, suggesting that they had been processed at predicted SPase cleavage sites.62
We have employed arylomycins to more directly identify proteins whose secretion into the media is dependent on SPase. Importantly, this approach can unambiguously identify SPase substrates because only authentic substrates will show a dose-dependent decrease with addition of the SPase inhibitor. With this approach we have characterized the SPase-dependent secretome of two staphylococci pathogens, Staphylococcus aureus15 and Staphylococcus epidermidis.14 We identified 11 S. epidermidis proteins and 47 S. aureus proteins whose presence in the media fraction was inversely correlated with the arylomycin dose. Among the SPase secretome are many known virulence factors such as membrane damaging toxins, cell wall attached proteins for immune evasion, proteases that cleave host factors, and coagulases that promote prothrombin activation and may lead to protection from phagocytosis.
Surface-associated SPase substrates also play a vital role in the pathogenesis of Gram-positive bacteria. Gram-positive pathogens such as Actinomyces spp.,63 B. cereus,64 Corneybacterium diphtheriae,65 Enterococcus spp.,66 Lactobacillus rhamnosus,67 S. agalactiae,68 Streptococcus gallolyticus,69 S. pneumoniae,70 and Streptococcus pyogenes71 all secrete pilin components that have an N-terminal signal peptides with a predicted SPase cleavage site (as well as a C-terminal sortase signal for processing and attachment to the cell wall).42,72 Pili in general are important for adherence to host cells, although they serve other functions in specific bacteria. For example, deletion mutants of Enterococcus faecalis that are unable to form pili show attenuated virulence in a rat model due to the resulting reduction in biofilm formation.66 Biofilms in general can have severe consequences in treating pathogenic bacteria as they can facilitate surface adherence and provide protection from the host immune system, and SPase is also directly involved in this process. In S. aureus, which along with S. epidermidis is responsible for the majority of biofilm-associated infections involving indwelling medical devices,73–76 biofilm formation relies heavily on cell wall anchored proteins including biofilm-associated protein (Bap), fibronectin-binding proteins A/B (FnbpA/B), clumping factor B (ClfB), protein A (Spa), serine-aspartate repeat protein C (SdrC), and surface proteins C and G (SasC and SasG). Unlike S. aureus, the pathogenicity of S. epidermidis relies almost solely on its ability to form biofilms.73,77 In S. epidermidis, autolysin E (AtlE), accumulation-associated protein (AAP), Bap, extracellular matrix protein (Ebh), and the surface protein SSP1, have all been implicated in biofilm formation, and each are predicted to be SPase substrates. Finally, SPase is involved in the formation of the S-layer,54,55 which is a crystalline-like array of proteins, glycoprotein, or both that coat the surface of the cell. It is perhaps best characterized in Bacillus anthracis and Clostridium difficile. Deletion of bslA, which encodes a component of the B. anthracis S-layer results in at least a 100-fold increase in the LD50 in a guinea pig model.78
While type VII secretion systems likely function independently of SPase,33,45 SPase may influence flagellar assembly and type IV secretion systems (T4SSs), as components of the translocation machinery itself are predicted to require SPase processing.79,80 For example, the T4SS mediates the direct transfer of proteins into target cells, but is perhaps best known for its role in the direct transfer of DNA, as this has been implicated as a primary means by which bacteria acquire foreign DNA leading to antibiotic resistance. Unlike Gram-negative bacteria, which use pili to establish the required cell-to-cell contact, Gram-positive bacteria mediate cell-to-cell contact via cell surface adhesions. One of the best studied Gram-positive T4SSs is encoded on the plasmid pCF10 and includes three cell wall proteins, PrgA, PrgB and PrgC, which are all encoded with SPase cleavage sites and sortase signal sequences.81 PrgB has been demonstrated to mediate attachment to bacteria and host cells in E. faecalis, and mutants of prgB show reduced plasmid transfer. It should be noted that many plasmid-based T4SSs do not appear to encode dedicated cell wall proteins, and it is unclear how surface contact is achieved. One possibility is that genome-encoded cell wall proteins play a role in mediating contact, although this has yet to be explored
4. A direct role for SPase in Gram-negative virulence
Unlike Gram-positive bacteria, Gram-negative bacteria typically encode only a single SPase. In addition, its substrates must transverse two membranes to exert their effects on their environment. Several widespread types of secretion systems have evolved to execute this task, and as a result the secretion systems of Gram-negative bacteria are both more diverse and more elaborate than their Gram-positive counterparts (Table 1). Secretion is either a one-step or two-step process. Sec-independent systems transport substrates directly from the cytoplasm to the external environment in a single step. Sec-dependent systems rely on Sec or Tat to translocate substrates into the periplasm, after which additional machinery is necessary for secretion past the outer membrane. Type II, V, VIII, and the chaperone/usher secretion systems are Sec-dependent (Figure 2). As such, pre-proteins secreted via these pathways invariably contain an N-terminal signal peptide, and signal peptidase processing is necessary to complete the process of secretion. It is through these secretion systems that SPase directly facilitates virulence in Gram-negative pathogens.
Table 1.
Summary of secretion systems in Gram-negative bacteria and the role of SPase in their biogenesis and function
| System | Sec-dependent substrates? | Machinery requires SPase processing? |
|---|---|---|
| T1SS | No | Yes |
| T2SS | Yes | Yes |
| T3SS | No | Yes |
| T4SS | Rarely | Yes |
| T5SS | Yes | Yes |
| T6SS | No | No |
| T8SS | Yes | Yes |
| C/U | Yes | Yes |
| T4PS | No | Yes |
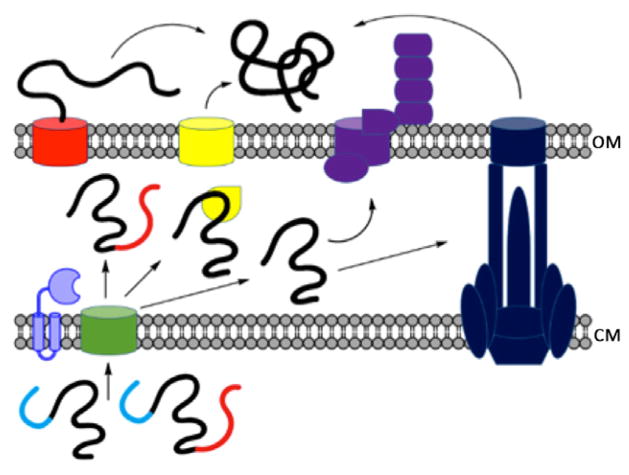
Figure 2.
Gram-negative SPase-dependent secretion systems: type II (blue), Type V (substrate’s β-barrel domain is shown in red before and after insertion into the OM), type VIII (purple) chaperone/usher (yellow). The Sec translocon and SPase are shown in green and light purple, respectively. Signal peptides are shown in cyan. OM: outer membrane; CM: cytoplasmic membrane
4.1. A variety of functions for type II secretion
The type II secretion system (T2SS) is referred to as the main terminal branch of the Sec pathway in Gram-negative bacteria, and has been reviewed extensively.82,83 Its machinery, or secreton, is composed of at least twelve proteins that form a pilus-like structure spanning both membranes.84 After SPase processing, T2SS substrates in the periplasm enter the secreton and are then transported across the outer membrane. The T2SS was first discovered in Klebsiella pneumoniae,85 although its role in virulence was not well explored until subsequent work with model organisms such as E. coli,86 Legionella pneumophila,87 P. aeruginosa,88,89 and Vibrio cholera90 elucidated the varied and vital roles of T2SS-dependent virulence factors. The genes encoding a complete or nearly complete T2SS are present in a wide variety of bacteria,82 suggesting that the lessons learned from these model organisms are applicable to a broader understanding of bacterial virulence.
T2SS-dependent virulence factors generally function as toxins and degradative enzymes. Prominent examples include the heat-labile enterotoxins of V. cholera and enterotoxigenic E. coli (ETEC).91,92 These toxins are responsible for the potentially lethal diarrhea associated with cholera93,94 and ETEC infection, of which there are millions of reported cases and hundreds of thousands of deaths annually.95,96 P. aeruginosa also produces an ADP-ribosylating toxin, exotoxin A, and it is this pathogen’s most toxic compound by weight, the LD50 for which is only 1 μg/ml in mice.97 P. aeruginosa provides an example of the variety of T2SS-dependent virulence factors that can be found within a single organism such as protease,98 phospholipase,99 lipase,100,101 DNAse,102 elastase,103,104 and phosphatase88 enzymes.
In vivo studies of mutants deficient in type II secretion have demonstrated a direct role for this secretion system in virulence. Mutation of the prepilin peptidase of V. cholera, which is necessary for the assembly of the secreton, abolished the production of cholera toxin and resulted in a 100-fold increase in the inoculum LD50 in an infant mouse cholera model.91 Introducing a similar mutation in L. pneumophila resulted in a severe defect in its ability to survive and proliferate in the lungs of mice.105 Subsequent studies determined specific virulence factors secreted by L. pneumophila and established a role for the T2SS in intracellular infection and immune system evasion.106–108 P. aeruginosa requires mutations in both type II and type III secretion to be rendered avirulent; either secretion system is sufficient to cause bacterial cell death in a murine model of lung infection.109
Mutational analyses of the T2SS has also led to surprises, such as the identification of the Shiga toxin of enterohemorrhagic E. coli (EHEC) as a Tat substrate despite the lack of the characteristic R-R motif of Tat substrates,110 and evidence that pyoverdine-mediated iron uptake in P. aeruginosa is dependent on SPase processing of pyoverdine biosynthetic genes.111 Although the T2SS mutants of P. aeruginosa are still virulent in a murine lung infection model, pyoverdine was shown to be absolutely essential for infection using a burned mouse model.112 This demonstrates that the T2SS works in tandem with other systems to promote virulence, and that the importance of specific virulence factors may be dependent on the context of infection. Many T2SS-dependent virulence factors have been identified outside of these model organisms, and the established role for type II secretion in the diverse and sometimes essential aspects of virulence merits further exploration.
4.2. Other SPase-dependent secretion systems
Not all virulence factors in Gram-negative bacteria require complicated machinery for translocation across the outer membrane, and the type V secretion systems (T5SSs) are the simplest. Type Va secretion systems (autotransporters) encode a passenger domain, a β-barrel domain, and typically a type I signal peptide, all within a single polypeptide. After signal peptide cleavage, the β-barrel forms a pore in the outer membrane and translocates the attached passenger domain.113,114 Other subcategories of type V secretion, such as type Vb (two-partner), vary in the organization of these domains and do not necessarily encode all components on a single polypeptide chain.115–117 Genetic analysis has identified over 700 autotransporters in Gram-negative bacteria and shows that they are nearly ubiquitous among Proteobacteria.118 Because of the mechanism of the T5SS, it is not possible to totally inhibit type V secretion in an organism by altering a single gene, as it is with secretion systems that use conserved machinery to translocate their substrates. Therefore, the study of T5SSs has been limited to the functions of specific virulence factors. A well-studied example is the IgA1 protease,119 which inhibits human immunoglobulin A1 and has been implicated in colonization at mucosal surfaces.119,120 Other prominent proteins secreted via the T5SS include IcsA (VirG),121,122 vacuolating cytotoxin VacA,123 the Haemophilus influenzae Hap adhesin,124 pertactin,125 and members of the large group of serine protease autotransporters of the Enterobacteriacea (SPATEs).126–132 These effectors play clear roles in processes such as adhesion, proteolysis, biofilm formation, toxicity, and immune system evasion.
The chaperone-usher (C/U) pathway, which relies on SPase processing for all of its substrates, is responsible for the assembly of at least 30 different types of surface fibers that are vital for the initial stage of infection for pathogens such as E. coli,133,134 Acinetobacter baumannii,135 Salmonella enterica serovars,136–138 Yersinia species,139,140 H. influenzae,141 P. aeruginosa,142 Bordetella pertussis,143 K. pneumoniae,144 and Proteus mirabilis.145,146 These surface fibers function to provide adhesion to host cells, evade the immune response, and contribute to biofilm formation. E. coli express P and type I pili, perhaps the best studied structures of this class.147 The C/U pathway also assembles the F1 antigen of Y. pestis and the haemagglutinating pilus of H. influenzae, which aid in resistance to phagocytic cells and colonization of the respiratory tract, respectively.148,149 Curli fibers are another surface fiber involved in virulence functions such as biofilm formation, immune modulation, and colonization. They are secreted independently of the C/U system, and instead secreted via the Sec-dependent type VIII secretion system (T8SS).150,151
The T4SS of Gram-positive bacteria have only been shown to o function in the context of conjugation, but Gram-negative T4SSs possess a number of structural differences that also allow them to function in effector translocation and release/uptake systems.152 The substrates of the T4SS generally do not contain a signal peptide, and are processed independently of SPase. The only exception with clear clinical relevance is the pertussis toxin of B. pertussis, for which signal peptide processing is required for secretion.153–155 This bacterium is well known as the causative agent of whooping cough, and the pertussis toxin is responsible for most of the symptoms.156 At the genetic level, mutation of the pertussis toxin gene has been shown to increase the LD50 three orders of magnitude compared to wild-type B. pertussis in a mouse model.157 Clearly, it is an essential virulence factor for this bacterium.
5. SPase processes components of multimeric secretion systems
The substrates of type I, III, (usually) IV, and VI secretion systems, as well as the type IV pilin system (T4PS), do not contain N-terminal signal peptides. Instead, their substrates possess other secretion signals to direct them to the appropriate secretion machinery and out of the cell in a single step. Because Sec components do not directly process the substrates of these secretion systems, they are commonly referred to as “Sec-Independent.” However, from our perspective this is misleading. While these one-step secretion systems have adapted to bypass the periplasm, integral components of the secretion machinery itself must be exported into the periplasm for proper localization (Table 1). As a result, these secretion systems rely on the Sec machinery for biogenesis. Specifically, many of the protein complexes that form outer membrane channels, as well as other components that at some point enter the periplasm, contain signal peptides, highlighting a greater role for SPase in virulence than is often appreciated (Figure 3).
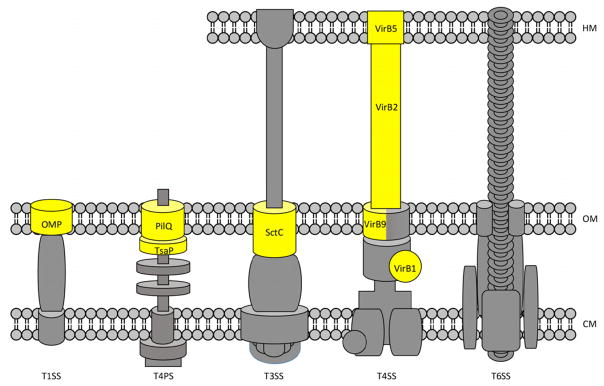
Figure 3.
Sec-independent secretion systems. Components of the secretion machinery that require SPase for processing are highhlighted in yellow and labeled. Grey components lack identifiable type I signal peptides. VirB9 forms a complex with VirB7 (not labeled). The T6SS is the only identified system without a component that depends on SPase. Detailed discussion of the structure and biogenesis of these secretion systems can be found elsewhere.39,158–160 HM: host membrane; OM: outer membrane; CM: cytoplasmic membrane.
Type I secretion systems (ABC transporters) are the simplest of the Sec-ndependent secretion systems. They are composed of three proteins: an inner membrane ATPase, a membrane fusion protein (MFP) spanning the periplasmic space, and an outer membrane protein (OMP). These OMPs form a β-barrel structure and are structurally similar to the outer membrane components of T5SSs. The proper localization of OMPs depends on their encoded signal peptide.161–163 TolC is the prototypical example of an OMP that functions as part of the T1SS, and contains a signal peptide.164,165 As is typical of OMPs, TolC can be recruited by multiple ATPase/MFP complexes to form functional type I secretion machines including HlyA and CvaC in E. coli alone. Virulence factors exported via T1SSs include α-hemolysin and others in the repeats-in-toxin (RTX) family, proteases, lipases, adhesins, S-layer proteins, and siderophores.166,167 In addition to the secretion of virulence factors, OMPs such as porins and outer membrane components of efflux pumps play a major role in resistance to most antibiotics.168 Inhibition of porin localization to the OM could actually decrease antibiotic susceptibility.169 Conversely, efflux pump inhibitors are being explored as antibiotic adjuvants. The AcrAB-TolC and MexAB-OprM from E. coli and P. aeruginosa, respectively, are the best studied of these efflux pumps. Unlike other mechanisms of antibiotic resistance, these pumps are polyspecific, meaning that they confer resistance to multiple classes of antibiotics. The role of efflux pumps in multidrug resistance has been reviewed recently.168,170
SPase plays an essential role in the assembly of multiple secretion systems through the processing of secretins, which are large, multimeric proteins that localize to the OM. Secretins are found within T2SSs, T3SSs, and T4PSs. Depending on the organism and the specific secretion system, they are synthesized with either a type I or type II signal peptide. The structure and function of secretins within the context of these complex secretion systems has been reviewed recently.171
The T3SS is a multimeric structure known as the injectisome that allows pathogens to inject virulence factors directly into host cells. Its assembly, which is covered in an excellent review,158 begins with a Sec-mediated phase in which the export apparatus, membrane ring, and supramembrane ring localize to the cytoplasmic membrane, and the outer-membrane ring localizes to the outer membrane. SPase processing is not required for IM components, but the secretin SctC does have a signal peptide that is necessary for localization.172 The closely related flagellar type III secretion system (fT3SS) shares many similar components, including the proteins of the P and L rings, FlgI and FlgH, which localize to the peptidoglycan and outer membrane, respectively. Although FlgH is likely processed by type II signal peptidase, SPase does process FlgI.173,174 Interestingly, SPase processing is also required for fully functional FliP, which is an essential component of the fT3SS that contains several transmembrane regions after its signal peptide and localizes to the IM.79,80 The T2SS and T4PS, which are structurally similar and share a common evolutionary origin,175–177 assemble in a similar fashion to the T3SS and the fT3SS.84 The secretin PilQ localizes to the outer membrane early in biogenesis and acts as a scaffold for further assembly, although some components of the IM localize independently.178 In the case of type IV pili, TsaP is another component of the OM pore that is likely dependent on SPase processing.179 Depending on the specific organism, the secretin of the T2SS may rely on the type II signal peptidase for processing.180–182
The T4PS, non-flagellar T3SS, and the fT3SS all play an integral role in pathogenicity. Their study in model organisms such as E. coli, P. aeruginosa, and species of Yersinia, Salmonella, Shigella, Bordetella, Vibrio, and Chlamydia have elucidated a variety of functions including toxicity, adherence, immune system evasion, biofilm formation, cellular invasion, motility, and DNA uptake.183,184 The contributions to virulence of the fT3SS, which secretes its own set of effectors in addition to assembling flagella, have been reviewed elsewhere.185 In several cases, T3SS-deficient bacterial strains show a defect or a complete absence of infectivity.183 The same is true of bacteria lacking Type IV pili.93,186,187
In addition to virulence, SPase inhibition has implications for the spread of antibiotic resistance. The T4SS secretes virulence factors in diverse pathogens such as L. pneumophila,188,189 H. pylori,190,191 Bartonella spp,192,193 and and is responsible for the spread of antibiotic resistance genes within clinical settings.194 Therefore, inhibiting type IV secretion could have profound implications from a therapeutic standpoint. With the exception of BPE123 and pertussin toxin, the T4SS secretes substrates that lack a signal peptide. The structure and nomenclature of T4SS subgroups have been reviewed recently.39 Typically, the nomenclature is borrowed from the Agrobacterium tumefaciencs Ti-plasmid encoded VirB/D4 T4SS, which is composed of VirB1 through VirB11 and VirD4. Both the VirB/D4 system and the E. coli plasmid pKM101- or R388-encoded T4SS are well-studied models of the T4SS in general. From these model systems, it is evident that VirB7, VirB9, and VirB10 associate at the outer membrane. Although VirB7 is processed by signal peptidase II, VirB9 contains a type I signal peptide and shows modest similarity to secretins of the T3SS, T2SS, and T4PS.195,196 For T4SSs, the VirB7/B9/B10 complex serves as a scaffold to initiate biogenesis of the secretion machinery, suggesting that inhibition of SPase would ablate formation of the T4SS entirely. Interestingly, the hydrolase VirB1 and the pilus components VirB2 and VirB5 are predicted SPase substrates.197 Multiple components of the F-like T4SS also contain putative signal peptides.198
6. Implications of non-canonical SPase substrates
SPase was originally discovered as part of Sec, and its study has historically focused on the characterization of this function, leading to the implicit assumption, pervasive in the literature, that SPase only processes proteins with N-terminal signal peptides. However, recent evidence indicates that SPase also processes proteins at internal (non-canonical) sites that otherwise retain many of the features of a canonical signal peptide (Figure 4).
Figure 4.
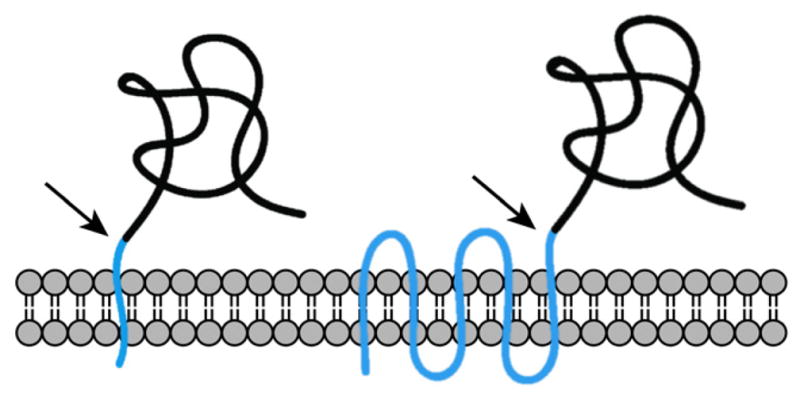
Canonical (left) and non-canonical (right) signal peptides (cyan). Cleavage sites are indicated with an arrow.
Increasing doses of arylomycin added to cultures of S. aureus resulted in a decrease in the levels of the extracellular domains of LtaS and OatA in the media.14 LtaS and OatA are proteins with five transmembrane segments that function in the synthesis of lipoteichoic acid and lysozyme resistance, respectively. The mRNA levels of these two genes were unaffected by arylomycin treatment, but MS analysis revealed that the proteins had been cleaved at an internal site downstream of the transmembrane domain. When the requirement for an N-terminal location is relaxed, the SignalP program predicts a viable signal-peptide-like sequence within this region of the protein, suggesting that SPase processes these proteins at a non-canonical internal site, which has been independently confirmed.56,199 LtaS and BlaR1 were shown to be SPase substrates in S. aureus in a similar manner, and the internal processing of LtaS has likewise been confirmed.199 Importantly, BlaR1 is essential for sensing β-lactam antibiotics and inducing resistance via β-lactamase production. This is highly suggestive that SPase plays a role in this clinically relevant process.
It is likely that SPase functions in this non-canonical manner in bacteria outside of the Staphylococci. Proteolysis at or within the cytoplasmic membrane has been observed in many different processes, and SPase appears ideally suited for this activity.200–202 For instance, the phosphoglycerol transferase of E. coli has been shown to contain a putative internal SPase cleavage site.201 Bioinformatics analysis of E. coli (TMHMM and SignalP programs) predicts that 27 polytopic membrane proteins are encoded with such sites.203 Clearly, the role of SPase in processing non-canonical substrates remains underexplored. Further studies may reveal additional therapeutically interesting roles, adding to the possible effects of SPase inhibition.
7. Conclusions
Targeting virulence is an attractive concept, but one that is in practice unlikely to be pursued due to the limited spectrum of activity expected for the resulting therapeutics. This is because the limited spectrum would result in fewer opportunities for use, and since antivirulence therapies are unlikely to be less expensive to develop than traditional antibiotics, they provide even less financial incentive to the pharmaceutical industry. However, SPase inhibitors are fundamentally unlike antivirulence compounds that target a single virulence factor. SPase is essential for viability and simultaneously functions as a global virulence determinant. As a result, it is likely that compounds that inhibit it should have both broad-spectrum antibacterial and broad-spectrum antivirulence activity. This strategy, inhibiting an essential protein that is also required for virulence, likely represents the only feasible route to the development of compounds with broad-spectrum antivirulence activity. As discussed in this review, SPase is required to directly or indirectly process a wide variety of virulence factors involved in processes such as biofilm formation, motility, adherence, immune evasion, pili formation, host degradation, and cellular invasion in a number of the most notorious human pathogens. Because SPase is required for the proper localization of antibiotic efflux pumps, biogenesis of the cell wall (a common target of other antibiotics),204–207 secretion of antibiotic-degrading enzymes, and processing of machinery involved in lateral gene transfer, SPase inhibition could also have a profound impact on the efficacy of other antibiotics and the spread of resistance. Finally, the recent discovery that SPase processes non-canonical substrates suggest that the full consequences of its inhibition remain underexplored. There may be much more to the inhibition of SPase than meets the eye.
Acknowledgments
We acknowledge financial support from the National Institutes of Health (AI-109809 to F.E.R.) and the Natural Sciences and Engineering Research Council of Canada (postdoctoral fellowship to A.C.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.World Health Organization. Antimicrobial Resistance: Global Report on Surveillance. Geneva, Switzerland: WHO Document Production Services; 2014. [Google Scholar]
- 2.Kostyanev T, Bonten MJ, O'Brien S, Steel H, Ross S, Francois B, Tacconelli E, Winterhalter M, Stavenger RA, Karlén A, Harbarth S, Hackett J, Jafri HS, Vuong C, MacGowan A, Witschi A, Angyalosi G, Elborn JS, deWinter R, Goossens H. J Antimicrob Chemother. 2016;71:290. doi: 10.1093/jac/dkv339. [DOI] [PubMed] [Google Scholar]
- 3.O'Neill J. Antimicrobial Resistance: Tackling a crisis for the Health and Wealth of Nations. London, UK: Review on Antimicrobial Resistance; 2014. [Google Scholar]
- 4.Velkov T, Roberts KD, Nation RL, Thompson PE, Li J. Future Microbiol. 2013;8:711. doi: 10.2217/fmb.13.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antibiotics Currently in Clinical Development. Philadelphia, PA: The Pew Charitable Trusts; 2015. [Google Scholar]
- 6.DiMasi JA, Grabowski HG. In: The Oxford Handbook of the Economics of the Biopharmaceutical Industry. Danzon PM, Nicholson S, editors. Oxford University Press; Oxford, UK: 2012. [Google Scholar]
- 7.DiMasi JA, Hansen RW, Grabowski HG. J Health Econ. 2003;22:151. doi: 10.1016/S0167-6296(02)00126-1. [DOI] [PubMed] [Google Scholar]
- 8.Mullard A. Nat Rev Drug Discov. 2014;13:877. doi: 10.1038/nrd4218. [DOI] [PubMed] [Google Scholar]
- 9.Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Nat Rev Drug Discov. 2007;6:29. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 10.Cooper MA, Shlaes D. Nature. 2011;472:32. doi: 10.1038/472032a. [DOI] [PubMed] [Google Scholar]
- 11.Anthouard R, DiRita VJ. Infect Immun. 2015;83:456. doi: 10.1128/IAI.02021-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith PA, Romesberg FE. Antimicrob Agents Chemother. 2012;56:5054. doi: 10.1128/AAC.00785-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steed DB, Liu J, Wasbrough E, Miller L, Halasohoris S, Miller J, Somerville B, Hershfield JR, Romesberg FE. Antimicrob Agents Chemother. 2015;59:3887. doi: 10.1128/AAC.00181-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Powers ME, Smith PA, Roberts TC, Fowler BJ, King CC, Trauger SA, Siuzdak G, Romesberg FE. J Bacteriol. 2011;193:340. doi: 10.1128/JB.01052-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schallenberger MA, Niessen S, Shao C, Fowler BJ, Romesberg FE. J Bacteriol. 2012;194:2677. doi: 10.1128/JB.00064-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paetzel M, Karla A, Strynadka NC, Dalbey RE. Chem Rev. 2002;102:4549. doi: 10.1021/cr010166y. [DOI] [PubMed] [Google Scholar]
- 17.Rao CVS, De Waelheyns E, Economou A, Anné J. Biochim Biophys Acta. 2014;1843:1762. doi: 10.1016/j.bbamcr.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 18.Therien AG, Huber JL, Wilson KE, Beaulieu P, Caron A, Claveau D, Deschamps K, Donald RG, Galgoci AM, Gallant M, Gu X, Kevin NJ, Lafleur J, Leavitt PS, Lebeau-Jacob C, Lee SS, Lin MM, Michels AA, Ogawa AM, Painter RE, Parish CA, Park YW, Benton-Perdomo L, Petcu M, Phillips JW, Powles MA, Skorey KI, Tam J, Tan CM, Young K, Wong S, Waddell ST, Miesel L. Antimicrob Agents Chemother. 2012;56:4662. doi: 10.1128/AAC.00726-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kulanthaivel P, Kreuzman AJ, Strege MA, Belvo MD, Smitka TA, Clemens M, Swartling JR, Minton KL, Zheng F, Angelton EL, Mullen D, Jungheim LN, Klimkowski VJ, Nicas TI, Thompson RC, Peng SB. J Biol Chem. 2004;279:36250. doi: 10.1074/jbc.M405884200. [DOI] [PubMed] [Google Scholar]
- 20.Schimana J, Gebhardt K, Höltzel A, Schmid DG, Süssmuth R, Müller J, Pukall R, Fiedler HP. J Antibiot (Tokyo) 2002;55:565. doi: 10.7164/antibiotics.55.565. [DOI] [PubMed] [Google Scholar]
- 21.Tan YX, Romesberg FE. MedChemComm. 2012;3:916. [Google Scholar]
- 22.Allsop AE, Brooks G, Edwards PD, Kaura AC, Southgate R. J Antibiot (Tokyo) 1996;49:921. doi: 10.7164/antibiotics.49.921. [DOI] [PubMed] [Google Scholar]
- 23.Harris DA, Powers ME, Romesberg FE. Bioorg Med Chem Lett. 2009;19:3787. doi: 10.1016/j.bmcl.2009.04.034. [DOI] [PubMed] [Google Scholar]
- 24.Barkocy-Gallagher GA, Bassford PJJ. J Biol Chem. 1992;267:1231. [PubMed] [Google Scholar]
- 25.Bruton G, Huxley A, O'Hanlon PJ, Oriek B, Eggleston D, Humphries J, Readshaw S, West A, Ashman S, Brown MH, Moore K, Pope A, O'Dwyer K, Wang L. Eur J Med Chem. 2003;38:351. doi: 10.1016/s0223-5234(03)00040-0. [DOI] [PubMed] [Google Scholar]
- 26.Nilsson I, von Heijne G. FEBS Lett. 1992;299:243. doi: 10.1016/0014-5793(92)80124-y. [DOI] [PubMed] [Google Scholar]
- 27.Ollinger J, O'Malley T, Ahn J, Odingo J, Parish T. J Bacteriol. 2012;194:2614. doi: 10.1128/JB.00224-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Craney A, Romesberg FE. Bioorg Med Chem Lett. 2015;25:4761. doi: 10.1016/j.bmcl.2015.07.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith PA, Roberts TC, Romesberg FE. Chem Biol. 2010;17:1223. doi: 10.1016/j.chembiol.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J, Smith PA, Steed DB, Romesberg F. Bioorg Med Chem Lett. 2013;23:5654. doi: 10.1016/j.bmcl.2013.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bensing BA, Seepersaud R, Yen YT, Sullam PM. Biochim Biophys Acta. 2014;1843:1674. doi: 10.1016/j.bbamcr.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feltcher ME, Sullivan JT, Braunstein M. Future Microbiol. 2010;5:1581. doi: 10.2217/fmb.10.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Desvaux M, Hébraud M. FEMS Microbiol Rev. 2006;30:774. doi: 10.1111/j.1574-6976.2006.00035.x. [DOI] [PubMed] [Google Scholar]
- 34.Freudl R. Res Microbiol. 2013;164:664. doi: 10.1016/j.resmic.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 35.Goosens VJ, Monteferrante CG, van Dijl JM. Biochim Biophys Acta. 2014;1843:1698. doi: 10.1016/j.bbamcr.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 36.Ligon LS, Hayden JD, Braunstein M. Tuberculosis (Edinb) 2012;92:121. doi: 10.1016/j.tube.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vasil ML, Tomaras AP, Pritchard AE. Antimicrob Agents Chemother. 2012;56:6223. doi: 10.1128/AAC.01575-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alvarez-Martinez CE, Christie PJ. Microbiol Mol Biol Rev. 2009;73:775. doi: 10.1128/MMBR.00023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chandran Darbari V, Waksman G. Annu Rev Biochem. 2015;84:603. doi: 10.1146/annurev-biochem-062911-102821. [DOI] [PubMed] [Google Scholar]
- 40.Christie PJ, Whitaker N, González-Rivera C. Biochim Biophys Acta. 2014;1843:1578. doi: 10.1016/j.bbamcr.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goessweiner-Mohr N, Arends K, Keller W, Grohmann E. Plasmid. 2013;70:289. doi: 10.1016/j.plasmid.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guglielmini J, Néron B, Abby SS, Garcillán-Barcia MP, de la Cruz F, Rocha EP. Nucleic Acids Res. 2014;42:5715. doi: 10.1093/nar/gku194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zechner EL, Lang S, Schildbach JF. Philos Trans R Soc Lond B Biol Sci. 2012;367:1073. doi: 10.1098/rstb.2011.0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneewind O, Missiakas DM. Philos Trans R Soc Lond B Biol Sci. 2012;367:1123. doi: 10.1098/rstb.2011.0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Solomonson M, Setiaputra D, Makepeace KA, Lameignere E, Petrotchenko EV, Conrady DG, Bergeron JR, Vuckovic M, DiMaio F, Borchers CH, Yip CK, Strynadka NC. Science. 2015;23:571. doi: 10.1016/j.str.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 46.Bonnemain C, Raynaud C, Reglier-Poupet H, Dubail I, Frehel C, Lety MA, Berche P, Charbit A. Mol Microbiol. 2004;51:1251. doi: 10.1111/j.1365-2958.2004.03916.x. [DOI] [PubMed] [Google Scholar]
- 47.van Roosmalen ML, Geukens N, Jongbloed JD, Tjalsma H, Dubois JY, Bron S, van Dijl JM, Anné J. Biochim Biophys Acta. 2004;1694:279. doi: 10.1016/j.bbamcr.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 48.Bierne H, Dramsi S. Curr Opin Microbiol. 2012;15:715. doi: 10.1016/j.mib.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 49.Desvaux M, Dumas E, Chafsey I, Hébraud M. FEMS Microbiol Lett. 2006;256:1. doi: 10.1111/j.1574-6968.2006.00122.x. [DOI] [PubMed] [Google Scholar]
- 50.Dramsi S, Magnet S, Davison S, Arthur M. FEMS Microbiol Rev. 2008;32:307. doi: 10.1111/j.1574-6976.2008.00102.x. [DOI] [PubMed] [Google Scholar]
- 51.Bhuwan M, Arora N, Sharma A, Khubaib M, Pandey S, Chaudhuri TK, Hasnain SE, Ehtesham NZ. MBio. 2016;7:e02259. doi: 10.1128/mBio.02259-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McDonough JA, Hacker KE, Flores AR, Pavelka MS, Jr, Braunstein M. J Bacteriol. 2005;187:7667. doi: 10.1128/JB.187.22.7667-7679.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raynaud C, Guilhot C, Rauzier J, Bordat Y, Pelicic V, Manganelli R, Smith I, Gicquel B, Jackson M. Mol Microbiol. 2002;45:203. doi: 10.1046/j.1365-2958.2002.03009.x. [DOI] [PubMed] [Google Scholar]
- 54.Fagan RP, Fairweather NF. J Biol Chem. 2011;286:27483. doi: 10.1074/jbc.M111.263889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nguyen-Mau SM, Oh SY, Kern VJ, Missiakas DM, Schneewind O. J Bacteriol. 2012;194:3841. doi: 10.1128/JB.00384-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Petersen TN, Brunak S, von Heijne G, Nielsen H. Nat Methods. 2011;8:785. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 57.Sibbald MJJB, Ziebandt AK, Engelmann S, Hecker M, de Jong A, Harmsen HJM, Raangs GC, Stokroos I, Arends JP, Dubois JYF, Van Dijl JM. Microbiol Mol Biol Rev. 2006;70:755. doi: 10.1128/MMBR.00008-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Enany S, Yoshida Y, Magdeldin S, Zhang Y, Bo X, Yamamoto T. Peptides. 2012;37:128. doi: 10.1016/j.peptides.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 59.Choi CW, Lee YG, Kwon SO, Kim HY, Lee JC, Chung YH, Yun CY, Kim SI. Diagn Microbiol Infect Dis. 2012;72:318. doi: 10.1016/j.diagmicrobio.2011.12.013. [DOI] [PubMed] [Google Scholar]
- 60.Maddi A, Haase E, Scannapieco F. J Proteomics Bioinform. 2014;7:287. doi: 10.4172/jpb.1000331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Papasergi S, Galbo R, Lanza-Cariccio V, Domina M, Signorino G, Biondo C, Pernice I, Poyart C, Trieu-Cuot P, Teti G, Beninati C. J Proteomics. 2013;89:154. doi: 10.1016/j.jprot.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 62.Ravipaty S, Reilly JP. Mol Cell Proteomics. 2010;9:1898. doi: 10.1074/mcp.M900494-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mishra A, Das A, Cisar JO, Ton-That H. J Bacteriol. 2007;189:3156. doi: 10.1128/JB.01952-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Budzik JM, Marraffini LA, Schneewind O. Mol Microbiol. 2007;66:495. doi: 10.1111/j.1365-2958.2007.05939.x. [DOI] [PubMed] [Google Scholar]
- 65.Ton-That H, Schneewind O. Mol Microbiol. 2003;50:1429. doi: 10.1046/j.1365-2958.2003.03782.x. [DOI] [PubMed] [Google Scholar]
- 66.Nallapareddy SR, Singh KV, Sillanpää J, Garsin DA, Höök M, Erlandsen SL, Murray BE. J Clin Invest. 2006;116:2799. doi: 10.1172/JCI29021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.von Ossowski I, Reunanen J, Satokari R, Vesterlund S, Kankainen M, Huhtinen H, Tynkkynen S, Salminen S, de Vos WM, Palva A. Appl Environ Microbiol. 2010;76:2049. doi: 10.1128/AEM.01958-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lauer P, Rinaudo CD, Soriani M, Margarit I, Maione D, Rosini R, Taddei AR, Mora M, Rappuoli R, Grandi G, Telford JL. Science. 2005;309:105. doi: 10.1126/science.1111563. [DOI] [PubMed] [Google Scholar]
- 69.Vanrobaeys M, De Herdt P, Charlier G, Ducatelle R, Haesebrouck F. Microbiology. 1999;145:335. doi: 10.1099/13500872-145-2-335. [DOI] [PubMed] [Google Scholar]
- 70.Barocchi MA, Ries J, Zogaj X, Hemsley C, Albiger B, Kanth A, Dahlberg S, Fernebro J, Moschioni M, Masignani V, Hultenby K, Taddei AR, Beiter K, Wartha F, von Euler A, Covacci A, Holden DW, Normark S, Rappuoli R, Henriques-Normark B. Proc Natl Acad Sci USA. 2006;103:2857. doi: 10.1073/pnas.0511017103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mora M, Bensi G, Capo S, Falugi F, Zingaretti C, Manetti AG, Maggi T, Taddei AR, Grandi G, Telford JL. Proc Natl Acad Sci USA. 2005;102:15641. doi: 10.1073/pnas.0507808102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mandlik A, Swierczynski A, Das A, Ton-That H. Trends Microbiol. 2008;16:33. doi: 10.1016/j.tim.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Büttner H, Mack D, Rohde H. Front Cell Infect Microbiol. 2015;5:14. doi: 10.3389/fcimb.2015.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Götz F. Mol Microbiol. 2002;43:1367. doi: 10.1046/j.1365-2958.2002.02827.x. [DOI] [PubMed] [Google Scholar]
- 75.Hobley L, Harkins C, MacPhee CE, Stanley-Wall NR. FEMS Microbiol Rev. 2015;39:649. doi: 10.1093/femsre/fuv015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Speziale P, Pietrocola G, Foster TJ, Geoghegan JA. Front Cell Infect Microbiol. 2014;4:171. doi: 10.3389/fcimb.2014.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fey PD, Olson ME. Future Microbiol. 2010;5:917. doi: 10.2217/fmb.10.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kern J, Schneewind O. Mol Microbiol. 2010;75:324. doi: 10.1111/j.1365-2958.2009.06958.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ohnishi K, Fan F, Schoenhals GJ, Kihara M, Macnab RM. J Bacteriol. 1997;179:6092. doi: 10.1128/jb.179.19.6092-6099.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pradel N, Ye C, Wu LF. Biochem Biophys Res Commun. 2004;319:1276. doi: 10.1016/j.bbrc.2004.05.123. [DOI] [PubMed] [Google Scholar]
- 81.Bhatty M, Cruz MR, Frank KL, Gomez JAL, Andrade F, Garsin DA, Dunny GM, Kaplan HB, Christie PJ. Molecular Microbiology. 2015;95:660. doi: 10.1111/mmi.12893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cianciotto NP. Trends Microbiol. 2005;13:581. doi: 10.1016/j.tim.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 83.Sandkvist M. Infect Immun. 2001;69:3523. doi: 10.1128/IAI.69.6.3523-3535.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Korotkov KV, Sandkvist M, Hol WG. Nat Rev Microbiol. 2012;10:336. doi: 10.1038/nrmicro2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.d'Enfert C, Ryter A, Pugsley AP. EMBO J. 1987;6:3531. doi: 10.1002/j.1460-2075.1987.tb02679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Francetic O, Belin D, Badaut C, Pugsley AP. EMBO J. 2000;19:6697. doi: 10.1093/emboj/19.24.6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cianciotto NP. Future Microbiol. 2009;4:797. doi: 10.2217/FMB.09.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ball G, Durand E, Lazdunski A, Filloux A. Mol Microbiol. 2002;43:475. doi: 10.1046/j.1365-2958.2002.02759.x. [DOI] [PubMed] [Google Scholar]
- 89.Wretlind B, Pavlovskis OR. J Bacteriol. 1984;158:801. doi: 10.1128/jb.158.3.801-808.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sandkvist M, Michel LO, Hough LP, Morales VM, Bagdasarian M, Koomey M, DiRita VJ, Bagdasarian M. J Bacteriol. 1997;179:6994. doi: 10.1128/jb.179.22.6994-7003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Marsh JW, Taylor RK. Mol Microbiol. 1998;29:1481. doi: 10.1046/j.1365-2958.1998.01031.x. [DOI] [PubMed] [Google Scholar]
- 92.Tauschek M, Gorrell RJ, Strugnell RA, Robins-Browne RM. Proc Natl Acad Sci USA. 2002;99:7066. doi: 10.1073/pnas.092152899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Herrington DA, Hall RH, Losonsky G, Mekalanos JJ, Taylor RK, Levine MM. J Exp Med. 1988;168:1487. doi: 10.1084/jem.168.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Levine MM, Kaper JB, Black RE, Clements ML. Microbiol Rev. 1983;47:510. doi: 10.1128/mr.47.4.510-550.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Turner SM, Scott-Tucker A, Cooper LM, Henderson IR. FEMS Microbiol Lett. 2006;263:10. doi: 10.1111/j.1574-6968.2006.00401.x. [DOI] [PubMed] [Google Scholar]
- 96.Verma R, Khanna P, Chawla S. Hum Vaccin Immunother. 2012;8:682. doi: 10.4161/hv.19083. [DOI] [PubMed] [Google Scholar]
- 97.Essam F, Saleh AJBH, Hussain SM. IOSR J Pharm Biol Sci. 2013;7:49. [Google Scholar]
- 98.Passmore IJ, Nishikawa K, Lilley KS, Bowden SD, Chung JC, Welch M. J Bacteriol. 2015;197:762. doi: 10.1128/JB.02404-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Martínez A, Ostrovsky P, Nunn DN. Mol Microbiol. 1998;28:1235. doi: 10.1046/j.1365-2958.1998.00888.x. [DOI] [PubMed] [Google Scholar]
- 100.Martinez A, Ostrovsky P, Nunn DN. Mol Microbiol. 1999;34:317. doi: 10.1046/j.1365-2958.1999.01601.x. [DOI] [PubMed] [Google Scholar]
- 101.Rosenau F, Jaeger K. Biochimie. 2000;82:1023. doi: 10.1016/s0300-9084(00)01182-2. [DOI] [PubMed] [Google Scholar]
- 102.Mulcahy H, Charron-Mazenod L, Lewenza S. Environ Microbiol. 2010;12:1621. doi: 10.1111/j.1462-2920.2010.02208.x. [DOI] [PubMed] [Google Scholar]
- 103.Braun P, de Groot A, Bitter W, Tommassen J. J Bacteriol. 1998;180:3467. doi: 10.1128/jb.180.13.3467-3469.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kessler E, Safrin M. J Bacteriol. 1988;170:5241. doi: 10.1128/jb.170.11.5241-5247.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rossier O, Starkenburg SR, Cianciotto NP. Infect Immun. 2004;72:310. doi: 10.1128/IAI.72.1.310-321.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.McCoy-Simandle K, Stewart CR, Dao J, DebRoy S, Rossier O, Bryce PJ, Cianciotto NP. Infect Immun. 2011;79:1984. doi: 10.1128/IAI.01077-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rossier O, Cianciotto NP. Infect Immun. 2005;73:2020. doi: 10.1128/IAI.73.4.2020-2032.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rossier O, Dao J, Cianciotto NP. Appl Environ Microbiol. 2008;74:753. doi: 10.1128/AEM.01944-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jyot J, Balloy V, Jouvion G, Verma A, Touqui L, Huerre M, Chignard M, Ramphal R. J Infect Dis. 2011;203:1369. doi: 10.1093/infdis/jir045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pradel N, Ye C, Livrelli V, Xu J, Joly B, Wu LF. Infect Immun. 2003;71:4908. doi: 10.1128/IAI.71.9.4908-4916.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Voulhoux R, Filloux A, Schalk IJ. J Bacteriol. 2006;188:3317. doi: 10.1128/JB.188.9.3317-3323.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Meyer JM, Neely A, Stintzi A, Georges C, Holder IA. Infect Immun. 1996;64:518. doi: 10.1128/iai.64.2.518-523.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Junker M, Besingi RN, Clark PL. Mol Microbiol. 2009;71:1323. doi: 10.1111/j.1365-2958.2009.06607.x. [DOI] [PubMed] [Google Scholar]
- 114.Peterson JH, Tian P, Ieva R, Dautin N, Bernstein HD. Proc Natl Acad Sci USA. 2010;107:17739. doi: 10.1073/pnas.1009491107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Henderson IR, Nataro JP. Infect Immun. 2001;69:1231. doi: 10.1128/IAI.69.3.1231-1243.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Henderson IR, Navarro-Garcia F, Desvaux M, Fernandez RC, Ala'Aldeen D. Microbiol Mol Biol Rev. 2004;68:692. doi: 10.1128/MMBR.68.4.692-744.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Leo JC, Grin I, Linke D. Philos Trans R Soc Lond B Biol Sci. 2012;367:1088. doi: 10.1098/rstb.2011.0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pallen MJ, Chaudhuri RR, Henderson IR. Curr Opin Microbiol. 2003;6:519. doi: 10.1016/j.mib.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 119.Mistry D, Stockley RA. Int J Biochem Cell Biol. 2006;38:1244. doi: 10.1016/j.biocel.2005.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kilian M, Reinholdt J, Lomholt H, Poulsen K, Frandsen EV. Acta Pathol Microbiol Immmunol Scand. 1996;104:321. doi: 10.1111/j.1699-0463.1996.tb00724.x. [DOI] [PubMed] [Google Scholar]
- 121.Brotcke Zumsteg A, Goosmann C, Brinkmann V, Morona R, Zychlinsky A. Cell Host Microbe. 2014;15:435. doi: 10.1016/j.chom.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 122.Phalipon A, Sansonetti P. Biologicals. 1995;23:125. doi: 10.1006/biol.1995.0023. [DOI] [PubMed] [Google Scholar]
- 123.Palframan SL, Kwok T, Gabriel K. Front Cell Infect Microbiol. 2012;2:92. doi: 10.3389/fcimb.2012.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Spahich NA, St Geme JW., 3rd Front Cell Infect Microbiol. 2011;1:5. doi: 10.3389/fcimb.2011.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Leininger E, Roberts M, Kenimer JG, Charles IG, Fairweather N, Novotny P, Brennan MJ. Proc Natl Acad Sci USA. 1991;88:345. doi: 10.1073/pnas.88.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Benjelloun-Touimi Z, Sansonetti PJ, Parsot C. Mol Microbiol. 1995;17:123. doi: 10.1111/j.1365-2958.1995.mmi_17010123.x. [DOI] [PubMed] [Google Scholar]
- 127.Brunder W, Schmidt H, Karch H. Mol Microbiol. 1997;24:767. doi: 10.1046/j.1365-2958.1997.3871751.x. [DOI] [PubMed] [Google Scholar]
- 128.Eslava C, Navarro-Garcia F, Czeczulin JR, Henderson IR, Cravioto A, Nataro JP. Infect Immun. 1998;66:3155. doi: 10.1128/iai.66.7.3155-3163.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Guyer DM, Henderson IR, Nataro JP, Mobley HL. Mol Microbiol. 2000;38:53. doi: 10.1046/j.1365-2958.2000.02110.x. [DOI] [PubMed] [Google Scholar]
- 130.Henderson IR, Czeczulin J, Eslava C, Noriega F, Nataro JP. Infect Immun. 1999;67:5587. doi: 10.1128/iai.67.11.5587-5596.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kostakioti M, Stathopoulos C. Infect Immun. 2004;72:5548. doi: 10.1128/IAI.72.10.5548-5554.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Stein M, Kenny B, Stein MA, Finlay BB. J Bacteriol. 1996;178:6546. doi: 10.1128/jb.178.22.6546-6554.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hull RA, Gill RE, Hsu P, Minshew BH, Falkow S. Infect Immun. 1981;33:933. doi: 10.1128/iai.33.3.933-938.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Madhavan TP, Sakellaris H. Adv Appl Microbiol. 2015;90:155. doi: 10.1016/bs.aambs.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 135.Gaddy JA, Actis LA. Future Microbiol. 2009;4:273. doi: 10.2217/fmb.09.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Althouse C, Patterson S, Fedorka-Cray P, Isaacson RE. Infect Immun. 2003;71:6446. doi: 10.1128/IAI.71.11.6446-6452.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ewen SW, Naughton PJ, Grant G, Sojka M, Allen-Vercoe E, Bardocz S, Thorns CJ, Pusztai A. FEMS Immunol Med Microbiol. 1997;18:185. doi: 10.1111/j.1574-695X.1997.tb01044.x. [DOI] [PubMed] [Google Scholar]
- 138.Naughton PJ, Grant G, Bardocz S, Allen-Vercoe E, Woodward MJ, Pusztai A. J Med Microbiol. 2001;50:191. doi: 10.1099/0022-1317-50-2-191. [DOI] [PubMed] [Google Scholar]
- 139.Felek S, Jeong JJ, Runco LM, Murray S, Thanassi DG, Krukonis ES. Microbiology. 2011;157:805. doi: 10.1099/mic.0.044826-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hatkoff M, Runco LM, Pujol C, Jayatilaka I, Furie MB, Bliska JB, Thanassi DG. Infect Immun. 2012;80:3490. doi: 10.1128/IAI.00434-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gilsdorf JR, McCrea KW, Marrs CF. Infect Immun. 1997;65:2997. doi: 10.1128/iai.65.8.2997-3002.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Vallet I, Olson JW, Lory S, Lazdunski A, Filloux A. Proc Natl Acad Sci USA. 2001;98:6911. doi: 10.1073/pnas.111551898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Scheller EV, Melvin JA, Sheets AJ, Cotter PA. MBio. 2015;6:e00500. doi: 10.1128/mBio.00500-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Fader RC, Davis CP. Infect Immun. 1980;30:554. doi: 10.1128/iai.30.2.554-561.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sauer FG, Remaut H, Hultgren SJ, Waksman G. Biochim Biophys Acta. 2004;1694:259. doi: 10.1016/j.bbamcr.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 146.Massad G, Fulkerson JF, Jr, Watson DC, Mobley HL. Infect Immun. 1996;64:4390. doi: 10.1128/iai.64.10.4390-4395.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lillington J, Geibel S, Waksman G. Biochim Biophys Acta. 2014;1840:2783. doi: 10.1016/j.bbagen.2014.04.021. [DOI] [PubMed] [Google Scholar]
- 148.Barsum W, Wilson R, Read RC, Rutman A, Todd HC, Houdret N, Roussel P, Cole PJ. Eur Respir J. 1995;8:709. [PubMed] [Google Scholar]
- 149.Du Y, Rosqvist R, Forsberg A. Infect Immun. 2002;70:1453. doi: 10.1128/IAI.70.3.1453-1460.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Cao B, Zhao Y, Kou Y, Ni D, Zhang XC, Huang Y. Proc Natl Acad Sci USA. 2014;111:E5439. doi: 10.1073/pnas.1411942111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Evans ML, Chapman MR. Biochim Biophys Acta. 2014;1843:1551. doi: 10.1016/j.bbamcr.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Bhatty M, Gomez JAL, Christie PJ. Res Microbiol. 2013;164:620. doi: 10.1016/j.resmic.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Nicosia A, Perugini M, Franzini C, Casagli MC, Borri MG, Antoni G, Almoni M, Neri P, Ratti G, Rappuoli R. Proc Natl Acad Sci USA. 1986;83:4631. doi: 10.1073/pnas.83.13.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Smith AM, Yan H, Groves N, Dalla Pozza T, Walker MJ. FEMS Microbiol Lett. 2000;191:177. doi: 10.1111/j.1574-6968.2000.tb09336.x. [DOI] [PubMed] [Google Scholar]
- 155.Weiss AA, Johnson FD, Burns DL. Proc Natl Acad Sci USA. 1993;90:2970. doi: 10.1073/pnas.90.7.2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Carbonetti NH. Future Microbiol. 2010;5:455. doi: 10.2217/fmb.09.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Weiss AA, Hewlett EL, Myers GA, Falkow S. J Infect Dis. 1984;150:219. doi: 10.1093/infdis/150.2.219. [DOI] [PubMed] [Google Scholar]
- 158.Portaliou AG, Tsolis KC, Loos MS, Zorzini V, Economou A. Trends Biochem Sci. 2016;41:175. doi: 10.1016/j.tibs.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 159.Chang YW, Rettberg LA, Treuner-Lange A, Iwasa J, Sogaard-Andersen L, Jensen GJ. Science. 2016;351:aad2001. doi: 10.1126/science.aad2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zoued A, Brunet YR, Durand E, Aschtgen MS, Logger L, Douzi B, Journet L, Cambillau C, Cascales E. Biochim Biophys Acta. 2014;1843:1664. doi: 10.1016/j.bbamcr.2014.03.018. [DOI] [PubMed] [Google Scholar]
- 161.Danese PN, Silhavy TJ. Annu Rev Genet. 1998;32:59. doi: 10.1146/annurev.genet.32.1.59. [DOI] [PubMed] [Google Scholar]
- 162.Osborn MJ, Rick PD, Lehmann V, Rupprecht E, Singh M. Ann N Y Acad Sci. 1974;235:52. doi: 10.1111/j.1749-6632.1974.tb43256.x. [DOI] [PubMed] [Google Scholar]
- 163.Tokuda H. Biosci Biotechnol Biochem. 2009;73:465. doi: 10.1271/bbb.80778. [DOI] [PubMed] [Google Scholar]
- 164.Hackett J, Misra R, Reeves P. FEBS Lett. 1983;156:307. doi: 10.1016/0014-5793(83)80518-3. [DOI] [PubMed] [Google Scholar]
- 165.Masi M, Duret G, Delcour AH, Misra R. Microbiology. 2009;155:1847. doi: 10.1099/mic.0.027219-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Delepelaire P. Biochim Biophys Acta. 2004;1694:149. doi: 10.1016/j.bbamcr.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 167.Thomas S, Holland IB, Schmitt L. Biochim Biophys Acta. 2014;1843:1629. doi: 10.1016/j.bbamcr.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 168.Li XZ, Plesiat P, Nikaido H. Clin Microbiol Rev. 2015;28:337. doi: 10.1128/CMR.00117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Fernández L, Hancock RE. Clin Microbiol Rev. 2012;25:661. doi: 10.1128/CMR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Nikaido H, Pagès JM. FEMS Microbiol Rev. 2012;36:340. doi: 10.1111/j.1574-6976.2011.00290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Korotkov KV, Gonen T, Hol WG. Trends Biochem Sci. 2011;36:433. doi: 10.1016/j.tibs.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, Galan JE, Aizawa SI. Science. 1998;280:602. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- 173.De Buck E, Lammertyn E, Anné J. Trends Microbiol. 2008;16:442. doi: 10.1016/j.tim.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 174.Jones CJ, Homma M, Macnab RM. J Bacteriol. 1989;171:3890. doi: 10.1128/jb.171.7.3890-3900.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Ayers M, Howell PL, Burrows LL. Future Microbiol. 2010;5:1203. doi: 10.2217/fmb.10.76. [DOI] [PubMed] [Google Scholar]
- 176.Nunn D. Trends Cell Biol. 1999;9:402. doi: 10.1016/s0962-8924(99)01634-7. [DOI] [PubMed] [Google Scholar]
- 177.Peabody CR, Chung YJ, Yen MR, Vidal-Ingigliardi D, Pugsley AP, Saier MH., Jr Microbiology. 2003;149:3051. doi: 10.1099/mic.0.26364-0. [DOI] [PubMed] [Google Scholar]
- 178.Friedrich C, Bulyha I, Søgaard-Andersen L. J Bacteriol. 2014;196:378. doi: 10.1128/JB.01094-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Siewering K, Jain S, Friedrich C, Webber-Birungi MT, Semchonok DA, Binzen I, Wagner A, Huntley S, Kahnt J, Klingl A, Boekema EJ, Søgaard-Andersen L, van der Does C. Proc Natl Acad Sci USA. 2014;111:E953. doi: 10.1073/pnas.1322889111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Bose N, Taylor RK. J Bacteriol. 2005;187:2225. doi: 10.1128/JB.187.7.2225-2232.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Schmidt SA, Bieber D, Ramer SW, Hwang J, Wu CY, Schoolnik G. J Bacteriol. 2001;183:4848. doi: 10.1128/JB.183.16.4848-4859.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Viarre V, Cascales E, Ball G, Michel GP, Filloux A, Voulhoux R. J Biol Chem. 2009;284:33815. doi: 10.1074/jbc.M109.065938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Coburn B, Sekirov I, Finlay BB. Clin Microbiol Rev. 2007;20:535. doi: 10.1128/CMR.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Giltner CL, Nguyen Y, Burrows LL. Microbiol Mol Biol Rev. 2012;76:740. doi: 10.1128/MMBR.00035-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zhou M, Yang Y, Chen P, Hu H, Hardwidge PR, Zhu G. Appl Microbiol Biotechnol. 2015;99:8883. doi: 10.1007/s00253-015-6946-x. [DOI] [PubMed] [Google Scholar]
- 186.Bieber D, Ramer SW, Wu CY, Murray WJ, Tobe T, Fernandez R, Schoolnik GK. Science. 1998;280:2114. doi: 10.1126/science.280.5372.2114. [DOI] [PubMed] [Google Scholar]
- 187.Tacket CO, Taylor RK, Losonsky G, Lim Y, Nataro JP, Kaper JB, Levine MM. Infect Immun. 1998;66:692. doi: 10.1128/iai.66.2.692-695.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ninio S, Roy CR. Trends Microbiol. 2007;15:372. doi: 10.1016/j.tim.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 189.Marra A, Blander SJ, Horwitz MA, Shuman HA. Proc Natl Acad Sci USA. 1992;89:9607. doi: 10.1073/pnas.89.20.9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Fischer W. FEBS J. 2011;278:1203. doi: 10.1111/j.1742-4658.2011.08036.x. [DOI] [PubMed] [Google Scholar]
- 191.Odenbreit S, Püls J, Sedlmaier B, Gerland E, Fischer W, Haas R. Science. 2000;287:1497. doi: 10.1126/science.287.5457.1497. [DOI] [PubMed] [Google Scholar]
- 192.Schulein R, Dehio C. Mol Microbiol. 2002;46:1053. doi: 10.1046/j.1365-2958.2002.03208.x. [DOI] [PubMed] [Google Scholar]
- 193.Seubert A, Hiestand R, de la Cruz F, Dehio C. Mol Microbiol. 2003;49:1253. doi: 10.1046/j.1365-2958.2003.03650.x. [DOI] [PubMed] [Google Scholar]
- 194.Juhas M, Crook DW, Dimopoulou ID, Lunter G, Harding RM, Ferguson DJ, Hood DW. J Bacteriol. 2007;189:761. doi: 10.1128/JB.01327-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Bayliss R, Harris R, Coutte L, Monier A, Fronzes R, Christie PJ, Driscoll PC, Waksman G. Proc Natl Acad Sci USA. 2007;104:1673. doi: 10.1073/pnas.0609535104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Christie PJ. Biochim Biophys Acta. 2004;1694:219. doi: 10.1016/j.bbamcr.2004.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Lai EM, Eisenbrandt R, Kalkum M, Lanka E, Kado CI. J Bacteriol. 2002;184:327. doi: 10.1128/JB.184.1.327-330.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Lawley TD, Klimke WA, Gubbins MJ, Frost LS. FEMS Microbiol Lett. 2003;224:1. doi: 10.1016/S0378-1097(03)00430-0. [DOI] [PubMed] [Google Scholar]
- 199.Wörmann ME, Reichmann NT, Malone CL, Horswill AR, Gründling A. J Bacteriol. 2011;193:5279. doi: 10.1128/JB.00369-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Brown MS, Ye J, Rawson RB, Goldstein JL. Cell. 2000;100:391. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- 201.Lequette Y, Lanfroy E, Cogez V, Bohin JP, Lacroix JM. Microbiology. 2008;154:476. doi: 10.1099/mic.0.2007/013169-0. [DOI] [PubMed] [Google Scholar]
- 202.Thöny-Meyer L, James P, Hennecke H. Proc Natl Acad Sci USA. 1991;88:5001. doi: 10.1073/pnas.88.11.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. J Mol Biol. 2001;305:567. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 204.Asoh S, Matsuzawa H, Ishino F, Strominger JL, Matsuhashi M, Ohta T. Eur J Biochem. 1986;160:231. doi: 10.1111/j.1432-1033.1986.tb09961.x. [DOI] [PubMed] [Google Scholar]
- 205.Broome-Smith JK, Edelman A, Yousif S, Spratt BG. Eur J Biochem. 1985;147:437. doi: 10.1111/j.1432-1033.1985.tb08768.x. [DOI] [PubMed] [Google Scholar]
- 206.Duez C, Piron-Fraipont C, Joris B, Dusart J, Urdea MS, Martial JA, Frere JM, Ghuysen JM. Eur J Biochem. 1987;162:509. doi: 10.1111/j.1432-1033.1987.tb10669.x. [DOI] [PubMed] [Google Scholar]
- 207.Pratt JM, Holland IB, Spratt BG. Nature. 1981;293:307. doi: 10.1038/293307a0. [DOI] [PubMed] [Google Scholar]