Abstract
The growth in epigenetics continues to attract considerable cross-disciplinary interest, apparently representing an opportunity to move beyond genomics towards the goal of understanding phenotypic variability from molecular through organismal to the societal level. The epigenome may also harbour useful information about life-time exposures (measured or unmeasured) irrespective of their influence on health or disease, creating the potential for a person-specific biosocial archive. Furthermore such data may prove of use in providing identifying information, providing the possibility of a future forensic epigenome. The mechanisms involved in ensuring that environmentally induced epigenetic changes perpetuate across the life course remain unclear. Here we propose a potential role of adult stem cells in maintaining epigenetic states provides a useful basis for formulating such epidemiologically-relevant concepts.
Epigenetics encompasses different mechanisms of gene expression regulation, the most commonly discussed ones being DNA methylation, histone modifications and non-coding RNAs 1, 2. The epigenetic mechanism most studied to date is DNA methylation – addition of a methyl group (CH3) at the 5’ position of a cytosine base typically at CpG dinucleotides, which are often clustered in CpG-rich DNA segments called CpG islands3, 4. This chemical addition is made by a covalent bond and is stable over time.
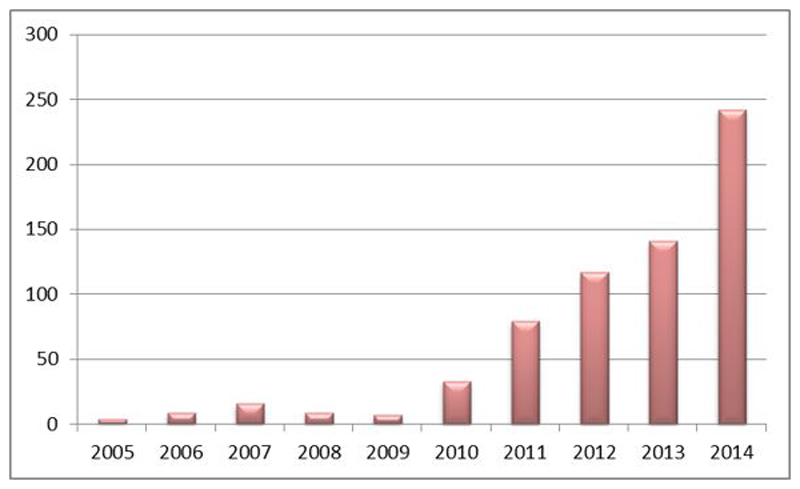
Since epigenetic processes are believed to be modifiable by environmental (i.e. non-genetic) factors, much of the current interest in epigenetics lies in understanding how the environment influences gene expression – even though as currently analyzable, epigenetics is unlikely to hold all of the answers for this broad and rather complex question 2,5. Technological advances now allow epigenome-wide investigations to be performed in large human populations at affordable costs – especially with respect to DNA methylation 6,7. Although other measures are technically possible they remain prohibitively expensive on a population scale. Epidemiological studies involving DNA methylation pertain to one of the most recent branches of epidemiology – epigenetic epidemiology 8. Large-scale profiling of DNA methylation levels has been applied to case-control studies 9,10, densely-phenotyped cohorts 11 and in some instances serial samples from the same longitudinal cohort 12. This allows investigation of the determinants of variation in DNA methylation and their importance in the context of different health outcomes and traits, as evidenced by the steady rise in epigenetic epidemiology publications (Figure 1) and the numerous studies reported in this issue of the IJE 13,14,15,16,17,18,19,20,21,22,23,24.
Figure 1.
Ten year publication trajectory in epigenetic epidemiology (2005-2014)
Long-term effects of environmental exposures on methylation
Methylation modifications are relatively stable. Indeed, such stability is important in the maintenance of methylation modifications that regulate developmental processes, such as cell differentiation 25,26,27,28. On the other hand, methylation modifications are sometimes reversible, and there is evidence that demethylation plays an important role in biological processes 29,30,31. Stochastic processes also contribute to some of the plasticity observed in the methylome.
The short term responsiveness of the epigenome to environmental challenges is an area that has not been widely studied. Experimental studies of short term exposure to particulate air matter have demonstrated that DNA methylation changes occur within hours of exposure 32. Inference can also be made with respect to responsiveness to exposures in utero detected at birth, given the specific time window of exposure 49. Critical windows of exposure can however be difficult to determine in many instances as the epigenetic measure inevitably captures a cross-sectional measure of epigenetic differences as opposed to being contemporaneous with the timing of the exposure itself.
Long-term effects of an early life exposure on methylation levels might mediate at least some of the associations between exposures and phenotypes at later life stages, although scepticism needs to be maintained in the breadth of claims regarding DNA cytosine methylation, given it plays no important role in the development of such important model organisms as Caenorhabditis elegans 33, Drosophila melanogaster and Saccharomyces cerevisiae 34 (a conclusion not altered by detection of very low levels of cytosine methylation in Drosophila 35and adenine methylation in both Drosophila and C. elegans 36). It is commonplace in epidemiological studies to see claims that epigenetic processes may provide biological plausibility to potentially causal associations, which then represent potential targets for intervention 2,5,8,37,38,39. Epigenetic mediation through persistent changes induced in early life has indeed become a widely accepted hypothetical model for developmental programming 38,39,40, particularly in generic reviews of this field 39. This has motivated many studies, including examples published in this issue of the IJE involving: early life determinants of methylation levels shortly after birth 16, 17, 20 and in adulthood 14; longitudinal associations of methylation levels with neurocognitive function and behavior in children 18 and with physical and cognitive fitness in the elderly 23; and the potential mediating role of DNA methylation in the association between maternal smoking and birth weight 15. It should be noted, however, that epigenetic persistence is not a pre-requisite for a programmed effect, since a transient change in DNA methylation could, in theory, set other biological processes in train which then precipitate long term effects.
Outside of the developmental programming literature there are also examples of long term health effects of an exposure potentially mediated by DNA methylation. Lung cancer risk is higher among past smokers compared to never smokers, and the relative risk is maintained over time after quitting smoking 41. It is possible that long-term epigenetic modifications are mediators of this association, since past smoking has recently been associated with DNA methylation levels even decades after cessation 42 (and could be considered a very useful biomarker of exposure at the tissue level). Nevertheless, disentangling mediation from other association-driving mechanisms – such as confounding and reverse causation – requires careful consideration 2,5,8, 37 and statistical approaches to such mediation analysis are severely compromised by measurement error43, 44, 45, 46. Furthermore the tissue from which methylation data are generated – blood, in the case of the above-referenced paper on dynamic methylation changes in relation to quitting smoking 42 – is not a plausible candidate for a mediator between smoking and long-term, post-cessation, risk of lung cancer. These serious problems are unlikely to restrain over-confident claims of causality and mediation in the reporting of studies incorporating methylation measures, however.
Perusal of existing literature indicates that evidence of long term effects of exposures on DNA methylation is largely limited to assumptions of persistence made in studies where methylation is measured at a single time point and related to historical exposure data. This approach is of course limited in the inferences that can be made, although access to serial samples from the same individuals with prospectively collected exposure data can help to improve this.
Modelling methylation change over time
Longitudinal data sets are valuable in many epidemiological contexts. Repeated measures of epigenetic signatures allows the modelling of change in methylation over time in tandem with single or repeated measures of an exposure that occurs prior to the methylation measurement 47. One can assess the persistence of differential methylation observed at birth across the life course, whether any changes are reversible and what factors may explain reversibility. For example, an epigenome-wide association study identified that methylation levels in seven gene regions were associated with maternal smoking during pregnancy. Four of these remained associated with maternal smoking throughout childhood and adolescence 48. Furthermore, models can be developed and tested to evaluate the intensity, duration and timing of an exposure on DNA methylation 49. The extent to which methylation data can indicate the particular timing of exposures – for example, during the intrauterine period or during puberty – is currently poorly understood.
A DNA methylation score derived from smoking responsive DNA methylation sites has previously been used as an indicator of smoking status i.e. to categorise current, former and never smokers49 and the widely used ‘epigenetic clock’ has been used to predict age from methylation patterns 23,50. The use of the ‘epigenetic clock’ as an indicator of biological rather than chronological age is increasingly being mooted, based on the assumption that DNA methylation signatures provide an index of cellular ageing (as in the case of telomere length).
The forensic epigenome
The potential utility of DNA methylation in forensics is beginning to generate interest, to the extent that by 2009 a popular prime-time TV programme, Law and Order: Special Victims Unit ran an episode, entitled “Perverted” (Figure 2), in which DNA methylation analysis was utilised to demonstrate that DNA at a crime scene had been generated in vitro (and was unmethylated rather than the in vivo methylated copy) and thus appeared to have been planted 51. Whilst a far-fetched storyline, the methods alluded to in the programme had been reported in Forensic Science International: Genetics 52. The body fluid or tissue source of DNA obtained at crime scenes can also be of forensic importance, and DNA methylation can help in this identification 53,54.
Figure 2.
Law and Order: Special Victims Unit: Season 10, Episode 8 Perverted. First Aired: November 18 2009 DNA links a suspect to the murder of a biker in Central Park whose gang specialized in hits and prostitution. Genetic analysis matched the murder weapon with the suspect but DNA methylation analysis showed the two samples to differ and was used to prove that the suspect was innocent.
https://en.wikipedia.org/wiki/Law_%26_Order:_Special_Victims_Unit_(season_10)
Other potential forensic uses include distinguishing between a monozygotic twin pair to establish which twin left a DNA sample at a crime scene. Methylation patterns, unlike the germ-line genome, can be identifying due to the phenotypic information that they reflect 55,56 (although potentially, somatic mutations detected in complete high coverage genome sequencing could also identify a particular MZ twin). This is not entirely of purely theoretical interest, as identical twins have indeed gone unprosecuted in such situations 57. Between-twin methylation differences appear stable enough to be useful even when there is a substantial time interval between when a sample from one twin was recovered from a crime scene and the twin pair had samples collected and examined 58. Indeed, if it transpires that there is any non-germ-line genetic variation based paternal-to-offspring transmission of methylation, as some epigenetic enthusiasts claim, this could even be used in paternity tests involving an offspring of one of a pair of male MZ twins. Transgenerational epigenetic inheritance has been reviewed in detail 59 but remains a contentious area of epigenetic research, in particular with respect to the public health importance of epigenetic variations transmitted across generations 60.
Despite over-blown claims regarding potential forensic uses for DNA-based face shape prediction 61, common genetic variation cannot provide much useful information about individual characteristics beyond sex and (probabilistically and problematically) ethnicity and related characteristics including eye, hair and skin colour 62,63,64, known collectively as “externally visible characteristics”, or EVCs 65.
DNA methylation offers the possibility of moving beyond conventional EVCs and adding identification of other aspects of the bodily habitus of the (generally unwitting) source of forensic blood (or other tissue) samples. .The epigenetic clock, mentioned above, is producing mean absolute differences of chronological and estimated age of only 3-4 years in adults 51which would certainly be useful for narrowing the range of those who could have been the source of recovered samples. Approaches using fewer markers have been formally tested within a forensics framework66,67,68, with demonstration of reasonable robustness to long-term room temperature blood sample storage 68, and application to non-blood DNA sources 69.
As outlined above, smoking behaviour is a characteristic that can be reasonably reliably predicted using DNA methylation data, with separation of current from ex-smokers being possible 50. Alcohol consumption has also been investigated, and useful indicators based on DNA methylation data may be developed, but evidence is too limited to generate these at present70,71. For other aspects of habitus, such as body mass index, the combination of genetic and epigenetic data can improve prediction over the use of genetic data only 72, but this does not approach the level of being useful for identification.
Knowledge of smoking and alcohol drinking behaviours could certainly help in the identification of recovered samples. In other situations DNA methylation data could in principle point to both identifying characteristics and to illegal activity. For example, there has been considerable interest in potential epigenetic effects of cocaine, methamphetamine and other substance use, although this is mainly limited to animal studies at present73,74,75,76.
Speculating further, maternal behaviours during pregnancy could leave marks on the offspring epigenome that have forensic implications as well as the epidemiological implications discussed earlier. As alluded to above, maternal smoking during pregnancy leaves such identifying and in some cases persistent offspring DNA methylation changes 49, providing retrospective indicators of this behaviour. There is also mounting evidence that maternal obesity, underweight or other dietary perturbation during pregnancy is associated with changes to the offspring’s epigenome 20,77,78. This has more than theoretical interest, since in the US various states have introduced legislation (in one case referred to as “the cocaine mom act”) through which mothers who have participated in behaviours that could damage their foetuses during pregnancy can be subject to criminal prosecution 79. The role of epigenetic understanding in formulating evidence as to whether in general maternal behaviours during pregnancy can influence foetal development and outcomes has been discussed in the legal context 80, but beyond this, DNA methylation data could be used retrospectively to establish that such behaviours have been practised during a particular pregnancy. In addition to smoking, maternal alcohol use during pregnancy has also been suggested to produce identifiable methylation changes in offspring 81, although this is not well-established as yet. Offspring methylation indicators of other maternal behaviours, largely in the context of animal model studies, are currently subject to intense research activity 82,83,84. Indeed, continuing the speculation beyond the realms that it should probably go, paternal behaviours that lead to detrimental effects on the offspring through transmitted epigenetic changes could also become a topic for investigation, taking the exclusive focus away from mothers (with its potential of compromising the constitutional rights of one societal group) 85.
The biosocial archive
Clearly the very same persistent epigenetic markers that allow for a potential forensic epigenome can also usefully serve in the context of epidemiological studies, contributing to the delineation of the full range of exposures to which individuals are subject (sometimes referred to as the exposome)86. The potential synthesis and integration of exposure history across the life course, during which social and biological processes are inscribed on the epigenome, suggests the possibility of a biosocial archive. The ability to better characterize exposures, including prenatal exposures using samples collected postnatally, offers a tool of considerable value to epidemiologists.
Just as researching the health effects of smoking acted as an impetus for the birth of chronic disease epidemiology 87, smoking has probably been the most widely investigated exposure in epigenetic epidemiology, as discussed above. Smoking-related methylation of the F2RL3 locus predicts mortality from cancer, cardiovascular disease and other causes, even after statistical adjustment for reported smoking behaviour 88 This probably indicates that the methylation measure provides a better indicator of long-term exposure to smoking than do reports of the behaviour, a conclusion strengthened by the finding that the associations of reported smoking behaviour with mortality were virtually abolished following adjustment for F2RL3 methylation. It is obvious that smoking is an underlying cause of these categories of mortality, but adjustment for a better indicator of the exposure attenuates the apparent effect of reported smoking behaviour. One important implication of this is that use of such methylation markers could reduce the residual confounding that remains after adjustment for reported smoking in observational epidemiological studies, and given that such residual confounding can 89- and does 90- generate misleading evidence from such studies this is potentially of considerable value.
Investigating the effects of maternal smoking on offspring health is an area of considerable research activity, and some studies rely on retrospective reports of whether mothers smoked during pregnancy. Persistent methylation markers assayed on offspring blood samples could clearly add to exposure classification in such situations 49.
There are many studies of environmental exposures and DNA methylation, but differences in technologies used for methylation assessment and statistical analysis makes drawing firm conclusions, of the sort available for smoking and age, difficult 91,92. Potentially exciting findings, for example, an apparent methylation marker of prenatal lead exposure on umbilical cord blood samples 93, require robust replication. The very considerable value of such exposure indicators makes this an exciting area of research, which is likely to yield much that is worthwhile and can be implemented in ongoing epidemiological studies with banked blood samples.
In some cases markers of particular exposures that have been searched for have not been found. For example, it would be very valuable to have a persistent indicator of having been born pre-term, given the association of pre-term birth with many later-life health outcomes. Whilst many gestational age related methylation differences were observed in umbilical cord blood samples, these were not found to persist at later ages48.
A final issue relating to the epidemiological context is that the potential to identify previously unrevealed characteristics of individuals from methylation array data has led to suggestions that this produces particular ethical concerns with respect to the sharing of such data 94, and has led to debate about this 95. Epigenetic data can and are considered in the same ethical and governance framework as other personal data in epidemiological studies, although their potential to reflect a more detailed exposure history than is revealed by questionnaire or other data sources may warrant further consideration when consenting study participants.
Mechanisms to explain the stability of environmentally-induced variation in DNA methylation
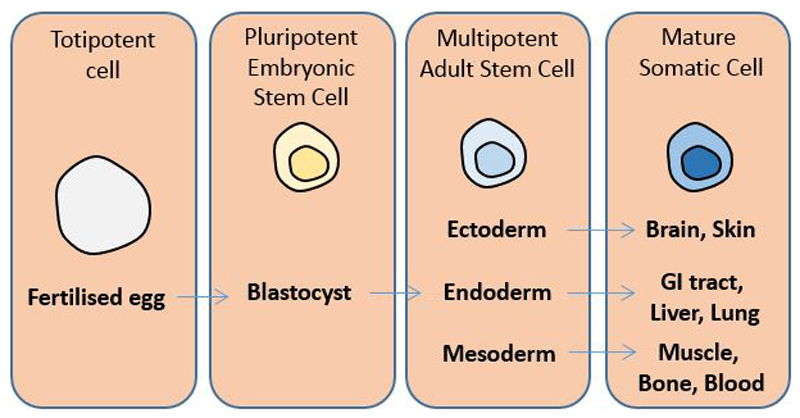
The concepts of a biosocial archive and forensic epigenome both rely upon the perpetuation (or measurable attenuation over time) of environmentally-induced epigenetic changes however, the mechanisms explaining such persistence remain unclear. The majority of evidence available from epigenetic epidemiology studies to date is based on blood cells or cells from the oral mucosa5,8). Both blood and oral epithelium are tissues with a rapid turnover of cells and therefore are dependent on adult stem cells (ASCs) for their maintenance, or tissue homeostasis as this has been termed96, involving both tissue repair following injury and physiological tissue renewal 97. Briefly, ASCs are a cell sub-type that promotes tissue renewal by replenishing more differentiated cells (Figure 3) 98, 99. There are many differences between ASCs and more differentiated cells within the same tissue, including in gene expression 98, 100 and cellular environment 98, 101.
Figure 3.
Lineage potential and adult stem cells. ASCs can give rise to cells of certain lineages.
A biological model where ASCs can act as reservoirs of environmental stresses, by continued tissue replenishment with cells harbouring a somatic event that is mitotically transmissible (e.g. a genetic mutation or an epigenetic modification), has been proposed in the context of cancer102. The general idea is that, if an exposure causes an oncogenic somatic event in non-ASCs, any increased cancer risk associated with such an exposure would be short-lived, especially in high turnover tissues. However, this is not consistent with the notion that cancer (and many other non-communicable diseases) result from long-term exposure to different risk factors 103including past exposures – such as a history of tobacco smoking in lung cancer 42. Such increased disease risk that persists after cessation of exposure to a particular environmental factor must involve modifications occurring during the exposure period that are maintained across cell division.
Evidence to date on epigenetic responsiveness to environmental, life style, behavioural and other factors is largely limited to peripheral tissues, due to ease of sampling. The question arises as to whether other tissues (with lower turnover) provide an equal or better archive of exposure. Brain tissue from the prefrontal cortex has been linked to methylation changes induced by chronic pain 104and other such examples can be found. However, the presence of persistent epigenetic changes in high turnover tissues, such as blood, may represent a more robust and responsive measure of dynamic change than a tissue with low turnover.
Recent literature, reviewed above, has provided evidence of sustained methylation differences according to past exposures 43,49. However, relatively little attention has been devoted to the cellular mechanisms underlying such maintenance. An obvious interpretation of sustained differential methylation is that there are still cells remaining in which the methylation modification arose. Given the high turnover of hematopoietic and epithelial tissues, this hypothesis is unlikely to apply in such cases. It is possible that some methylation modifications favour cell survival, thus increasing the lifespan beyond that of other cells. However, this explanation is ad hoc and is unlikely to apply in many cases – although this could occur if such modifications affect the function of genes associated with cell senescence or apoptosis. Furthermore, the increase in lifespan would have to be substantial to account for long-term effects after several years of exposure cessation. The counter argument has also been posited, that is, that DNA methylation changes promote genomic instability 105 and may therefore promote cell death rather than survival.
A second possibility is that the signal maintenance arises from the daughter cells of the parent cells where the methylation modification took place and was transmitted during mitosis (mitotic stability). Although this is a more plausible hypothesis, it does not explain why some exposure-associated methylation patterns are maintained over time, while others are not. One possibility is that a methylation change will only be maintained across (long) time periods – especially in high turnover tissues – if it arises in one or more ASCs 102. Although exposure continuation would likely increase the methylation signals, the epigenetic consequences of an exposure in the past (such as past smoking) could be maintained in the tissue due to cell replenishment by epigenetically-modified ASCs. This model would be a plausible biological mechanism for the maintenance of the absolute risk over time among individuals no longer exposed to the risk factor – as in the elevated lung cancer in former smokers 42. Importantly in the epidemiological context, whilst it is understandable that the development of such models has focused on epigenetic changes which influence disease risk, methylation and other epigenetic responses to exposures which do not have any consequences for disease, but can serve as exposure indicators, may be maintained in the same way.
This model also provides a plausible explanation for short-term associations between an exposure and methylation levels. According to this model, such associations can result from methylation modifications not having occurred in ASCs (although other processes could lead to the same outcome). This could happen either by chance (which types of cells happen to get most exposure, or which respond with a methylation change, a process that is likely to be probabilistic) or through biological mechanisms. For example, it is possible that modifiability of some CpG regions depends on the cell sub-type within a tissue hierarchy 106. It is also well-known that ASCs have protective mechanisms, including protection against external stresses in their microenvironment 101. Therefore, it would be expected that the proportion of cells epigenetically affected by a given exposure in the ASC compartment would be influenced by the proportion of cell sub-types in a given tissue.
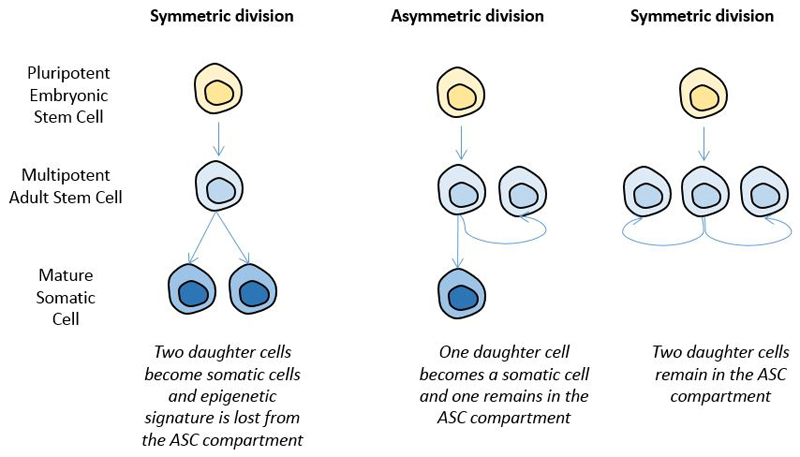
An additional implication of this model is that even a methylation modification that occurred in an ASC might not persist over time. Methylation modifications in these cells could be reversed before cell division or after a few cell divisions 106, and additional mechanisms might also be involved. When an ASC divides, there are three potential outcomes: one committed cell (i.e. a cell that will generate differentiated cells) and one ASC (which is referred to as an asymmetric division), two committed cells or two ASCs (symmetric divisions). Such flexibility is important for prioritizing between ASC niche maintenance and replenishment of differentiated cells in response to specific stimuli 107. If the ASC in which the epigenetic event occurred undergoes a symmetric division producing two committed cells, the epigenetic event will not be maintained in the ASC compartment 102 and, therefore, will be detectable for a shorter period of time after the end of the exposure. A summary of the models of proliferation and maintenance of ASCs are provided in Figure 4.
Figure 4.
Symmetric and asymmetric stem cell division and maintenance of environmentally induced epigenetic change
Mitotic heritability of cellular phenotypes is one of the two historical origins of the notion of “epigenetics” (attributed to David Nanney, and contrasting to the developmental notion of how genes result in phenotypes, associated with Conrad Waddington) 108,109 and methylation is, of course, only one of several mechanisms that could lead to such stability across mitosis 110,111. The stage of development (from embryonic stem cells through subsequent somatic stem cells and the niches they enter) at which a mitotically stable change occurs could influence the range of tissues in which this change is observed. The understanding of these processes of cellular differentiation and the evolution and dynamic nature of methylation signatures, both in general 112,113 and in particular lineages (such as B cells) 114, is rapidly developing. The influence of particular exposures (e.g. diet, smoking, lead) at particular stages and levels of chronicity on DNA methylation within cellular lineages is being explored 115 and greater knowledge of these processes will help to inform theories of epigenetic persistence in different tissue types.
The ASC model, Mendelian randomization, ageing and interventions
Implications of adopting an ASC model to explain methylation stability over time extend to conceptual issues regarding Mendelian randomization116. This technique has been applied in different fields of epidemiology, including epigenetic epidemiology. In the latter, Mendelian randomization can be used to obtain more robust evidence regarding (a) the effects of an exposure on methylation levels, (b) the effect of a methylation levels on a disease outcome, and (c) combining both in order to evaluate whether methylation levels are mediators of the association between the exposure and the outcome (a strategy referred to as two-step Mendelian randomization)121.
Because a genetic instrument for methylation levels has the same genotype regardless of the cell type and life course stage, it approximates the situation of long-term exposure-associated methylation modifications better than the case of short-term associations. This is particularly important if two-step Mendelian randomization is used to assess the mediating role of methylation modifications121 in relation to the effects of a time-restricted exposure, such as maternal smoking in pregnancy. If maternal smoking in pregnancy (in a hypothetical situation) only has short-term effects on methylation, then genetic instruments for such methylation differences would not necessarily provide valid evidence regarding the consequences of maternal smoking. This implies that using Mendelian randomization to investigate methylation modifications as mediators would be more reasonable in cases where there are long-term associations between the exposure and methylation. Understanding the (possible) role of ASCs in long-term epigenetic modifications may help in producing a broader understanding of conceptual considerations necessary for optimal study design and causal inference.
Another implication of considering the importance of cell lineages relates to physiological processes, such as ageing. As discussed above there are considerable age-related methylation modifications, useful in age-prediction at the population level 23, 51. If such modifications occurred in committed cells, they would not accumulate – in a single cell – over time. Therefore, it would be more plausible to think that they occur in ASCs, thus re-emphasizing (assuming that the proposed model is true) the importance of this cell sub-type to the ageing process117.
In the context of epigenetic markers as potential targets for intervention, the proposed role of ASCs in long-term maintenance of methylation modification would also be important. Referring back to the example of the association of past smoking with methylation 43 and lung cancer risk 42; if long-term methylation modifications mediate sustained increase in risk of lung cancer risk in former smokers, then intervening on such modifications towards a “never smoker pattern” could contribute to reduce cancer risk. If such long-term modifications result from tissue renewal by epigenetically-modified ASCs, then a drug (or any other intervention) would have to be able to reach the ASC compartment of the relevant tissue and reverse the methylation modifications in these cells. In this regard, the protection mechanisms of ASCs 101,118,119 would likely influence the efficacy of methylation-targeted interventions.
Relevance to the epidemiologist
In considering the concepts of a biosocial archive, a forensic epigenome and an ASC model for epigenetic persistence, one can speculate upon their relevance to the epidemiologist. The epigenome, or more specifically DNA methylation patterns, could enhance (and in some instances replace) exposure measurement or be considered as a biomarker in a diagnostic, predictive or prognostic context, adding a molecular dimension to a conventional observational study.
Epigenetic measures could also be considered as a surrogate end point in a clinical trial setting. Given the current state of knowledge, setting up randomized controlled trials to evaluate long-term effects of an exposure on methylation levels might be unjustifiable in most cases. However, such an investigation could be incorporated in trials with other primary goals – for example, in a randomized controlled trial evaluating the efficacy of an intervention to quit smoking. If the intervention is successful (e.g. increases smoking quitting rates) and the time from the first to the last measurement of the trial is sufficient to evaluate long-term effects on methylation, then an intention-to-treat analysis with methylation levels as the outcome could be performed. Moreover, strategies to improve causal inference in observational studies – such as Mendelian randomization120,121 – can be applied to observational data to investigate whether an exposure produces (at an aggregate level) a tendency towards a particular methylation change.
It is unlikely that epidemiological approaches will be able to resolve the outstanding questions regarding the hypothetical role of ASCs in epigenetic persistence of environmentally responsive DNA methylation changes; molecular biology will perhaps play a more important role here. Nevertheless, the ASC hypothesis may serve to help in the interpretation of epidemiological observations where data are generated from the longitudinal analysis of exposure-induced epigenetic changes.
Conclusions
The field of epigenetic epidemiology continues to provide evidence that the epigenome may serve as a very useful exposure indicator. There are broad applications in epidemiological studies; as a refined exposure indicator and biosocial archive but also in more specialised instances where prediction of phenotype can be gleaned from molecular signatures captured in the epigenome. Initial evidence that some epigenetic marks persist over decades whereas others are short-lived raises the question as to the possible mechanisms underlying the stability of environmentally induced epigenetic changes. ASCs provide a plausible biological mechanism (unlikely to be the only one) underlying epigenetic persistence, although considerably more empirical data are required on this. ASCs can be considered as potential biological archives of events that have occurred throughout the life course (which also contributes to the broader notion of the forensic epigenome). Such considerations also imply that in addition to the well-recognised between-tissue specificity complication in epigenetic epidemiological studies within-tissue (in this case, cell differentiation stage) specificity is of importance.
Acknowledgements
G.D.S. and C.L.R. are partially supported by the ESRC (RES-060-23-0011) ‘The biosocial archive: transforming lifecourse social research through the incorporation of epigenetic measures’. This work was supported by the UK Medical Research Council Integrative Epidemiology Unit, Cancer Research UK [C18281/A19169] and the University of Bristol (MC_UU_12013_1, MC_UU_12013_2).
Footnotes
Conflict of interest statement
GDS’ mother smoked and drank alcohol during her pregnancy with him, and (jokingly) suggested he sue her for the damage done.
References
- 1.Kaelin WG, Jr, McKnight SL. Influence of metabolism on epigenetics and disease. Cell. 2013;153:56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Relton CL, Davey Smith G. Epigenetic epidemiology of common complex disease: prospects for prediction, prevention, and treatment. PLoS Med. 2010;7:e1000356. doi: 10.1371/journal.pmed.1000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han L, Su B, Li WH, Zhao Z. CpG island density and its correlations with genomic features in mammalian genomes. Genome Biol. 2008;9:R79. doi: 10.1186/gb-2008-9-5-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rakyan VK, Down TA, Balding DJ, Beck S. Epigenome-wide association studies for common human diseases. Nat Rev Genet. 2011;12:529–541. doi: 10.1038/nrg3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bakulski KM, Fallin MD. Epigenetic epidemiology: promises for public health research. Environ Mol Mutagen. 2014;55:171–183. doi: 10.1002/em.21850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michels KB, Binder AM, Dedeurwaerder S, et al. Recommendations for the design and analysis of epigenome-wide association studies. Nat Methods. 2013;10:949–955. doi: 10.1038/nmeth.2632. [DOI] [PubMed] [Google Scholar]
- 7.Paul DS, Beck S. Advances in epigenome-wide association studies for common diseases. Trends Mol Med. 2014;20:541–543. doi: 10.1016/j.molmed.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Relton CL, Davey Smith G. Is epidemiology ready for epigenetics? Int J Epidemiol. 2012;41:5–9. doi: 10.1093/ije/dys006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chambers JC, Loh M, Lehne B, et al. Epigenome-wide association of DNA methylation markers in peripheral blood from Indian Asians and Europeans with incident type 2 diabetes: a nested case-control study. Lancet Diabetes Endocrinol. 2015;3:526–34. doi: 10.1016/S2213-8587(15)00127-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang L, Willis-Owen SA, Laprise C, et al. An epigenome-wide association study of total serum immunoglobulin E concentration. Nature. 2015;520:670–4. doi: 10.1038/nature14125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ng JW, Barrett LM, Wong A, Kuh D, Davey Smith G, Relton CL. The role of longitudinal cohort studies in epigenetic epidemiology: challenges and opportunities. Genome Biol. 2012;13:246. doi: 10.1186/gb-2012-13-6-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Relton CL, Gaunt T, McArdle W, et al. Data Resource Profile: Accessible Resource for Integrated Epigenomic Studies (ARIES) Int J Epidemiol. 2015 doi: 10.1093/ije/dyv072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feinberg JI, Bakulski KM, Jaffe AE, et al. Paternal sperm DNA methylation associated with early signs of autism risk in an autism-enriched cohort. Int J Epidemiol. 2015 doi: 10.1093/ije/dyv028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tobi EW, Slieker RC, Stein AD, et al. Early gestation as the critical time-window for changes in the prenatal environment to affect the adult human blood methylome. Int J Epidemiol. 2015 doi: 10.1093/ije/dyv043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kupers LK, Xu X, Jankipersadsing SA, et al. DNA methylation mediates the effect of maternal smoking during pregnancy on birthweight of the offspring. Int J Epidemiol. 2015 doi: 10.1093/ije/dyv048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hernandez-Vargas H, Castelino J, Silver MJ, et al. Exposure to aflatoxin B1 in utero is associated with DNA methylation in white blood Cells of infants in The Gambia. Int J Epidemiol. 2015 doi: 10.1093/ije/dyv027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bakulski KM, Lee H, Feinberg JI, et al. Prenatal mercury concentration is associated with changes in DNA methylation at TCEANC2 in newborns. Int J Epidemiol. 2015 doi: 10.1093/ije/dyv032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lillycrop KA, Costello PM, Teh AL, et al. Association between perinatal methylation of the neuronal differentiation regulator HES1 and later childhood neurocognitive function and behaviour. Int J Epidemiol. 2015 doi: 10.1093/ije/dyv052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agha G, Houseman EA, Kelsey KT, Eaton CB, Buka SL, Loucks EB. Adiposity is associated with DNA methylation profile in adipose tissue. Int J Epidemiol. 2014 doi: 10.1093/ije/dyu236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharp GC, Lawlor DA, Richmond RC, et al. Maternal pre-pregnancy BMI and gestational weight gain, offspring DNA methylation and later offspring adiposity: findings from the Avon Longitudinal Study of Parents and Children. Int J Epidemiol. 2015 doi: 10.1093/ije/dyv042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stringhini S, Polidoro S, Sacerdote C, et al. Life-course socioeconomic status and DNA methylation of genes regulating inflammation. Int J Epidemiol. 2015 doi: 10.1093/ije/dyv060. [DOI] [PubMed] [Google Scholar]
- 22.Sung H, Yang HH, Zhang H, et al. Common genetic variants in epigenetic machinery genes and risk of upper gastrointestinal cancers. Int J Epidemiol. 2015 doi: 10.1093/ije/dyv050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marioni RE, Shah S, McRae AF, et al. The epigenetic clock is correlated with physical and cognitive fitness in the Lothian Birth Cohort 1936. Int J Epidemiol. 2015 doi: 10.1093/ije/dyu277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Campanella G, Polidoro S, Di Gaetano C, et al. Epigenetic signatures of internal migration in Italy. Int J Epidemiol. 2014 doi: 10.1093/ije/dyu198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang K, Fan G. DNA methylation in Cell differentiation and reprogramming: an emerging systematic view. Regen Med. 2010;5:531–544. doi: 10.2217/rme.10.35. [DOI] [PubMed] [Google Scholar]
- 26.Bock C, Beerman I, Lien WH, et al. DNA methylation dynamics during in vivo differentiation of blood and skin stem Cells. Mol Cell. 2012;47:633–647. doi: 10.1016/j.molcel.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheaffer KL, Kim R, Aoki R, et al. DNA methylation is required for the control of stem Cell differentiation in the small intestine. Genes Dev. 2014;28:652–664. doi: 10.1101/gad.230318.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spivakov M, Fisher AG. Epigenetic signatures of stem-Cell identity. Nat Rev Genet. 2007;8:263–271. doi: 10.1038/nrg2046. [DOI] [PubMed] [Google Scholar]
- 29.Johannes F, Colot V, Jansen RC. Epigenome dynamics: a quantitative genetics perspective. Nat Rev Genet. 2008;9:883–890. doi: 10.1038/nrg2467. [DOI] [PubMed] [Google Scholar]
- 30.Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014;156:45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramchandani S, Bhattacharya SK, Cervoni N, Szyf M. DNA methylation is a reversible biological signal. PNAS. 1999;96:6107–6112. doi: 10.1073/pnas.96.11.6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carmona JJ, Sofer T, Hutchinson J, et al. Short-term airborne particulate matter exposure alters the epigenetic landscape of human genes associated with the mitogen-activated protein kinase network: a cross-sectional study. Environ Health. 2014;13:94. doi: 10.1186/1476-069X-13-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yi S. Birds do it, bees do it, worms and ciliates do it too: DNA methylation from unexpected corners of the tree of life. Genome Biology. 2012;13:174. doi: 10.1186/gb-2012-13-10-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karathia H, Vilaprinyo E, Sorribas A, Alves R. Saccharomyces cerevisiae as a Model Organism: A Comparative Study. PLoS ONE. 2011;6:e16015. doi: 10.1371/journal.pone.0016015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Capuano F, Mulleder M, Kok R, Blom HJ, Ralser M. Cytosine DNA Methylation Is Found in Drosophila melanogaster but Absent in Saccharomyces cerevisiae, Schizosaccharomyces pombe, and Other Yeast Species. Anal Chem. 2014;86:3697–3702. doi: 10.1021/ac500447w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng S, Cokus SJ, Zhang X, et al. Conservation and divergence of methylation patterning in plants and animals. PNAS. 2010;107:8689–8694. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mill J, Heijmans BT. From promises to practical strategies in epigenetic epidemiology. Nat Rev Genet. 2013;14:585–594. doi: 10.1038/nrg3405. [DOI] [PubMed] [Google Scholar]
- 38.Godfrey KM, Lillycrop KA, Burdge GC, Gluckman PD, Hanson MA. Epigenetic mechanisms and the mismatch concept of the developmental origins of health and disease. Pediatr Res. 2007;61:5R–10R. doi: 10.1203/pdr.0b013e318045bedb. [DOI] [PubMed] [Google Scholar]
- 39.Gluckman PD, Hanson MA, Mitchell MD. Developmental origins of health and disease: reducing the burden of chronic disease in the next generation. Genome Med. 2010;2:14. doi: 10.1186/gm135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waterland RA, Michels KB. Epigenetic epidemiology of the developmental origins hypothesis. Annu Rev Nutr. 2007;27:363–388. doi: 10.1146/annurev.nutr.27.061406.093705. [DOI] [PubMed] [Google Scholar]
- 41.Peto J. That lung cancer incidence falls in ex-smokers: misconceptions 2. Br J Cancer. 2011;104:389. doi: 10.1038/sj.bjc.6606080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guida F, Sandanger TM, Castagne R, et al. Dynamics of smoking-induced genome-wide methylation changes with time since smoking cessation. Hum Mol Genet. 2015;24:2349–2359. doi: 10.1093/hmg/ddu751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cole DA, Preacher KJ. Manifest variable path analysis: potentially serious and misleading consequences due to uncorrected measurement error. Psychol Methods. 2014;19:300–315. doi: 10.1037/a0033805. [DOI] [PubMed] [Google Scholar]
- 44.Vanderweele TJ, Valeri L, Ogburn EL. The role of measurement error and misclassification in mediation analysis. Epidemiology. 2012;23:561–564. doi: 10.1097/EDE.0b013e318258f5e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blakely T, McKenzie S, Carter K. Misclassification of the mediator matters when estimating indirect effects. J Epidemiol Community Health. 2013;67:458–66. doi: 10.1136/jech-2012-201813. [DOI] [PubMed] [Google Scholar]
- 46.Le Cessie S, Debeij J, Rosendaal FR, Cannegieter SC, Vandenbroucke JP. Quantification of bias in direct effects estimates due to different types of measurement error in the mediator. Epidemiology. 2012;23:551–560. doi: 10.1097/EDE.0b013e318254f5de. [DOI] [PubMed] [Google Scholar]
- 47.Simpkin AJ, Suderman M, Gaunt TR, et al. Longitudinal analysis of DNA methylation associated with birth weight and gestational age. Hum Mol Genet. 2015;24:3752–3763. doi: 10.1093/hmg/ddv119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Richmond RC, Simpkin AJ, Woodward G, et al. Prenatal exposure to maternal smoking and offspring DNA methylation across the lifecourse: findings from the Avon Longitudinal Study of Parents and Children (ALSPAC) Hum Mol Genet. 2015;24:2201–2217. doi: 10.1093/hmg/ddu739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Elliott HR, Tillin T, McArdle WL, et al. Differences in smoking associated DNA methylation patterns in South Asians and Europeans. Clin Epigenetics. 2014;6:4. doi: 10.1186/1868-7083-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DNA methylation solves crimes. 2009 Law and Order: Special Victims Unit, Perverted (TV series) http://www.whatisepigenetics.com/dna-methylation-solves-crimes/
- 52.Frumkin D, Wasserstrom A, Davidson A, Grafit A. Authentication of forensic DNA samples. Forensic Science. 2010;4:95–103. doi: 10.1016/j.fsigen.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 53.Madi T, Balamurugan K, Bombardi R, Duncan G, McCord B. The determination of tissue-specific DNA methylation patterns in forensic biofluids using bisulfite modification and pyrosequencing. Electrophoresis. 2012;33:1736–1745. doi: 10.1002/elps.201100711. [DOI] [PubMed] [Google Scholar]
- 54.Lee HY, An JH, Jung SE, et al. Genome-wide methylation profiling and a multiplex construction for the identification of body fluids using epigenetic markers. Forensic Sci Int Genet. 2015;17:17–24. doi: 10.1016/j.fsigen.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 55.Levesque ML, Casey KF, Szyf M, et al. Genome-wide DNA methylation variability in adolescent monozygotic twins followed since birth. Epigenetics. 2014;9:1410–1421. doi: 10.4161/15592294.2014.970060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stewart L, Evans N, Bexon KJ, van der Meer DJ, Williams GA. Differentiating between monozygotic twins through DNA methylation-specific high-resolution melt curve analysis. Analytical Biochem. 2015;476:36–39. doi: 10.1016/j.ab.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 57.Himmelreich C. Despite DNA Evidence, Twins Charged in Heist Go Free. Time. 2009 Mar 23; [Google Scholar]
- 58.Zhang N, Zhao S, Zhang S-H, et al. Intra-monozygotic twin pair discordance and Longitudinal Variation of Whole-Genome Scale DNA Methylation in Adults. PLoS One. 2015;10:e0135022. doi: 10.1371/journal.pone.0135022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heard E, Martienssen RA. Transgenerational epigenetic inheritance: myths and mechanisms. Cell. 2014;157:95–109. doi: 10.1016/j.cell.2014.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Davey Smith G. Epigenesis for epidemiologists: does evo-devo have implications for population health research and practice? Int J Epidemiol. 2012;41:236–47. doi: 10.1093/ije/dys016. [DOI] [PubMed] [Google Scholar]
- 61.Pollack A. Building a Face, and a Case, on DNA. Thew New York Times. 2015 [Google Scholar]
- 62.Fullwiley D. Can DNA "Witness" Race?: Forensic Uses of an Imperfect Ancestry Testing Technology. Genewatch. 2008;21:12–14. [Google Scholar]
- 63.Krimsky S, Sloan K. Race and the Genetic Revolution: Science, Myth and Culture. Columbia University Press; 2011. [Google Scholar]
- 64.Duster T. The molecular reinscription of race: unanticipated issues in biotechnology and forensic science. Patterns of Prejudice. 2006;40:427–441. [Google Scholar]
- 65.Gunn P, Walsh S, Roux C. The nucleic acid revolution continues – will forensic biology become forensic molecular biology? Frontiers in Genetics. 2014;5:44. doi: 10.3389/fgene.2014.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang Y, Yan J, Hou J, Fu X, Li L, Hou Y. Developing a DNA methylation assay for human age prediction in blood and bloodstain. Forensic Sci Int Genet. 2015;17:129–136. doi: 10.1016/j.fsigen.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 67.Zbieć-Piekarska R, Spólnicka M, Kupiec T, et al. Examination of DNA methylation status of the ELOVL2 marker may be useful for human age prediction in forensic science. Forensic Sci Int Genet. 2015;14:161–167. doi: 10.1016/j.fsigen.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 68.Yi SH, Xu LC, Mei K, Yang RZ, Huang DX. Isolation and identification of age-related DNA methylation markers for forensic age-prediction. Forensic Sci Int Genet. 2014;11:117–125. doi: 10.1016/j.fsigen.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 69.Lee HY, Jung SE, Oh YN, Choi A, Yang WI, Shin KJ. Epigenetic age signatures in the forensically relevant body fluid of semen: a preliminary study. Forensic Sci Int Genet. 2015;19:28–34. doi: 10.1016/j.fsigen.2015.05.014. [DOI] [PubMed] [Google Scholar]
- 70.Weng JT, Wu LS, Lee CS, Hsu PW, Cheng AT. Integrative epigenetic profiling analysis identifies DNA methylation changes associated with chronic alcohol consumption. Comput Biol Med. 2015;64:299–306. doi: 10.1016/j.compbiomed.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 71.Philibert RA, Penaluna B, White T, et al. A pilot examination of the genome-wide DNA methylation signatures of subjects entering and exiting short-term alcohol dependence treatment programs. Epigenetics. 2014;9:1212–1219. doi: 10.4161/epi.32252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shah S, Bonder MJ, Marioni RE, et al. Improving Phenotypic Prediction by Combining Genetic and Epigenetic Associations. Am J Hum Genet. 2015;97:75–85. doi: 10.1016/j.ajhg.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Godino A, Jayanthi S, Cadet JL. Epigenetic landscape of amphetamine and methamphetamine addiction in rodents. Epigenetics. 2015;10:574–580. doi: 10.1080/15592294.2015.1055441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zumbrun EE, Sido JM, Nagarkatti PS, Nagarkatti M. Epigenetic Regulation of Immunological Alterations Following Prenatal Exposure to Marijuana Cannabinoids and its Long Term Consequences in Offspring. J Neuroimmune Pharmacol. 2015;10:245–254. doi: 10.1007/s11481-015-9586-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ploense K, Carr A, Baker-Andresen D, et al. Prolonged Access to Cocaine Results in Distinct Epigenetic Changes in the Prefrontal Cortex. Faseb Journal. 2014;28:supplement LB622. [Google Scholar]
- 76.Jayanthi S, McCoy MT, Chen B, et al. Methamphetamine downregulates striatal glutamate receptors via diverse epigenetic mechanisms. Biol Psychiatry. 2014;76:47–56. doi: 10.1016/j.biopsych.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Green BB, Marsit CJ. Select Prenatal Environmental Exposures and Subsequent Alterations of Gene-Specific and Repetitive Element DNA Methylation in Fetal Tissues. Curr Environ Health Rep. 2015;2:126–36. doi: 10.1007/s40572-015-0045-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Allard C, Desgagné V, Patenaude J, et al. Mendelian randomization supports causality between maternal hyperglycemia and epigenetic regulation of leptin gene in newborns. Epigenetics. 2015;10:342–51. doi: 10.1080/15592294.2015.1029700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Calhoun A. The criminalisation of bad mothers. New York Times Magazine. 2012 Apr 25; [Google Scholar]
- 80.Smith TF, Maccani MA, Knopik VS. Symposium Article: Maternal Smoking During Pregnancy and Offspring Health Outcomes: The Role of Epigenetic Research in Informing Legal Policy and Practice. Hastings Law Journal. 2013;64:1619–1648. [Google Scholar]
- 81.Masemola ML, van der Merwe L, Lombard Z, Viljoen D, Ramsay M. Reduced DNA methylation at the PEG3 DMR and KvDMR1 loci in children exposed to alcohol in utero: a South African Fetal Alcohol Syndrome cohort study. Front Genet. 2015;6:85. doi: 10.3389/fgene.2015.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yohn NL, Bartolomei MS, Blendy JA. Multigenerational and transgenerational inheritance of drug exposure: The effects of alcohol, opiates, cocaine, marijuana, and nicotine. Progress in Biophysics and Molecular Biology. 2015;118:21–33. doi: 10.1016/j.pbiomolbio.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Itzhak Y, Ergui I, Young JI. Long-term parental methamphetamine exposure of mice influences behavior and hippocampal DNA methylation of the offspring. Mol Psychiatry. 2015;20:232–9. doi: 10.1038/mp.2014.7. [DOI] [PubMed] [Google Scholar]
- 84.Zhao Q, Hou J, Chen B, et al. Prenatal cocaine exposure impairs cognitive function of progeny via insulin growth factor II epigenetic regulation. Neurobiol Dis. 2015;82:54–65. doi: 10.1016/j.nbd.2015.05.014. [DOI] [PubMed] [Google Scholar]
- 85.Henricks J. What to Expect When You’re Expecting: Fetal Protection Laws that Strip Away the Constitutional Rights of Pregnant Women. Boston College Journal of Law and Social Justice. 2015;35:117–152. [Google Scholar]
- 86.Wild CP. The exposome: from concept to utility. IJE. 2012;41:24–32. doi: 10.1093/ije/dyr236. [DOI] [PubMed] [Google Scholar]
- 87.Vandenbroucke JP. Commentary: 'Smoking and lung cancer'--the embryogenesis of modern epidemiology. Int J Epidemiol. 2009;38:1193–6. doi: 10.1093/ije/dyp292. [DOI] [PubMed] [Google Scholar]
- 88.Zhang Y, Schöttker B, Ordóñez-Mena J, et al. F2RL3 methylation, lung cancer incidence and mortality. Int J Cancer. 2015;137:1739–48. doi: 10.1002/ijc.29537. [DOI] [PubMed] [Google Scholar]
- 89.Philips AN, Davey Smith G. How independent are “independent” effects? Relative risk estimation when correlated exposures are measured imprecisely. J Clin Epidemiol. 1991;44:1223–31. doi: 10.1016/0895-4356(91)90155-3. [DOI] [PubMed] [Google Scholar]
- 90.Davey Smith G, Ebrahim S. Data dredging, bias, or confounding. BMJ. 2002;325:1437–1438. doi: 10.1136/bmj.325.7378.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ruiz-Hernandez A, Kuo CC, Rentero-Garrido P, et al. Environmental chemicals and DNA methylation in adults: a systematic review of the epidemiologic evidence. Clinical Epigenetics. 2015;7:55. doi: 10.1186/s13148-015-0055-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Green BB, Marsit CJ. Select Prenatal Environmental Exposures and Subsequent Alterations of Gene-Specific and Repetitive Element DNA Methylation in Fetal Tissues. Current Environmental Health Reports. 2015;2:126–136. doi: 10.1007/s40572-015-0045-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sen A, Cingolani P, Senut MC, et al. Lead exposure induces changes in 5-hydroxymethylcytosine clusters in CpG islands in human embryonic stem cells and umbilical cord blood. Epigenetics. 2015;10:607–621. doi: 10.1080/15592294.2015.1050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Philbert RA, Terry N, Erwin C, Philibert WJ, Beach SRH, Brody GH. Methylation array data can simultaneously identify individuals and convey protected health information: an unrecognized ethical concern. Clinical Epigenetics. 2014;6:28. doi: 10.1186/1868-7083-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Joly Y, Dyke SOM, Cheung WA, Rothstein MA, Pastinen T. Risk of re-identification of epigenetic methylation data: a more nuanced response is needed. Clinical pigenetics. 2015;7:45. doi: 10.1186/s13148-015-0079-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Simson BD, Clevers H. Strategies for Homeostatic Stem Cell Self-Renewal in Adult Tissues. Cell. 2011;145:851–862. doi: 10.1016/j.cell.2011.05.033. [DOI] [PubMed] [Google Scholar]
- 97.Chen Y, Xie N, Jin P, Wang T. DNA methylation and hydroxymethylation in stem cells. Cell Biochem Funct. 2015;33:161–73. doi: 10.1002/cbf.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fuchs E, Chen T. A matter of life and death: self-renewal in stem cells. EMBO Rep. 2013;14:39–48. doi: 10.1038/embor.2012.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gonzalez MA, Bernad A. Characteristics of adult stem cells. Adv Exp Med Biol. 2012;741:103–120. doi: 10.1007/978-1-4614-2098-9_8. [DOI] [PubMed] [Google Scholar]
- 100.Tarnok A, Ulrich H, Bocsi J. Phenotypes of stem cells from diverse origin. Cytometry A. 2010;77:6–10. doi: 10.1002/cyto.a.20844. [DOI] [PubMed] [Google Scholar]
- 101.Morrison SJ, Spradling AC. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132:598–611. doi: 10.1016/j.cell.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hartwig FP, Nedel F, Collares T, Tarquinio SB, Nor JE, Demarco FF. Oncogenic somatic events in tissue-specific stem cells: a role in cancer recurrence? Ageing Res Rev. 2014;13:100–106. doi: 10.1016/j.arr.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 103.Ezzati M, Riboli E. Behavioral and dietary risk factors for noncommunicable diseases. N Engl J Med. 2013;369:954–964. doi: 10.1056/NEJMra1203528. [DOI] [PubMed] [Google Scholar]
- 104.Alvarado S, Tajerian M, Suderman M, et al. An epigenetic hypothesis for the genomic memory of pain. Front Cell Neurosci. 2015;9:88. doi: 10.3389/fncel.2015.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Skinner MK, Guerrero-Bosagna C, Haque MMM. Environmentally induced epigenetic transgenerational inheritance of sperm epimutations promote genetic mutations. Epigenetics. 2015;10:762–71. doi: 10.1080/15592294.2015.1062207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hochberg Z, Feil R, Constancia M, Fraga M, Junien C, Carel JC, et al. Child health, developmental plasticity, and epigenetic programming. Endocr Rev. 2011;32:159–224. doi: 10.1210/er.2009-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Morrison SJ, Kimble J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 2006;441:1068–1074. doi: 10.1038/nature04956. [DOI] [PubMed] [Google Scholar]
- 108.Haig D. Commentary: The epidemiology of epigenetics. Int J Epidemiology. 2012;41:13–16. doi: 10.1093/ije/dyr183. [DOI] [PubMed] [Google Scholar]
- 109.C, Maggert KA. What Do You Mean, “Epigenetic”? Genetics. 2015;199:887–896. doi: 10.1534/genetics.114.173492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rinalidi L, Benitah SA. Epigenetic regulation of adult stem cell function. FEBS J. 2015;282:1589–604. doi: 10.1111/febs.12946. [DOI] [PubMed] [Google Scholar]
- 111.Nashun B, Hill PW, Hajkova P. Reprogramming of cell fate: epigenetic memory and the erasure of memories past. The EMBO Journal. 2015;34:1296–308. doi: 10.15252/embj.201490649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Beerman I, Derrick JR. Epigenetic Control of Stem Cell Potential during Homeostasis, Aging, and Disease. Cell. 2015:613–625. doi: 10.1016/j.stem.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Xie W, Schultz MD, Lister R, et al. Epigenomic Analysis of Multilineage Differentiation of Human Embryonic Stem Cells. Cell. 2013;153:1134–1148. doi: 10.1016/j.cell.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kulis M, Merkel A, Health S, et al. Whole-genome fingerprint of the DNA methylome during human B cell differentiation. Nat Genet. 2015;47:746–56. doi: 10.1038/ng.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Senut MC, Sen A, Cingolani P, Shaik A, Land SJ, Ruden DM. Lead exposure disrupts global DNA methylation in human embryonic stem cells and alters their neuronal differentiation. Toxicological Sciences. 2014;139:142–161. doi: 10.1093/toxsci/kfu028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Human Molecular Genetics. 2014;23:R89–R98. doi: 10.1093/hmg/ddu328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chaudhary PM, Roninson IB. Expression and activity of P-glycoprotein, a multidrug efflux pump, in human hematopoietic stem cells. Cell. 1991;66:85–94. doi: 10.1016/0092-8674(91)90141-k. [DOI] [PubMed] [Google Scholar]
- 119.Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5:275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 120.Smith GD, Ebrahim S. 'Mendelian randomization': can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 121.Relton CL, Davey Smith G. Two-step epigenetic Mendelian randomization: a strategy for establishing the causal role of epigenetic processes in pathways to disease. Int J Epidemiol. 2012;41:161–176. doi: 10.1093/ije/dyr233. [DOI] [PMC free article] [PubMed] [Google Scholar]