Abstract
Large-scale whole genome sequence datasets offer novel opportunities to identify genetic variation underlying human traits. Here we apply genotype imputation based on whole genome sequence data from the UK10K and the 1000 Genomes Projects into 35,981 study participants of European ancestry, followed by association analysis with twenty quantitative cardiometabolic and hematologic traits. We describe 17 novel associations, including six rare (minor allele frequency [MAF]<1%) or low frequency variants (1%<MAF<5%) with platelet count (PLT), red cell indices (MCH, MCV) and high-density lipoprotein (HDL) cholesterol. Applying fine-mapping analysis to 233 known and novel loci associated with the twenty traits, we resolve associations of 59 loci to credible sets of 20 or less variants, and describe trait enrichments within regions of predicted regulatory function. These findings augment understanding of the allelic architecture of risk factors for cardiometabolic and hematologic diseases, and provide additional functional insights with the identification of potentially novel biological targets.
Introduction
Heritable influences to cardiometabolic and hematologic traits have been identified across the allele frequency spectrum. Rare (defined here as minor allele frequency [MAF] < 1%) and highly penetrant variants with large phenotypic effects have been identified, but account for a small proportion of phenotypic variance 1,2. At the other end of the allelic frequency spectrum, genome and exome wide association analyses based on sparse arrays have identified thousands of common (MAF ≥ 5%) and low frequency (MAF 1-5%), single nucleotide variants (SNVs), with modest effects 3–11. To investigate the influence of rare, less-frequent, and common variation on complex traits, we applied whole genome sequencing (WGS) in individuals from two British cohorts, the St Thomas’ Twin Registry (TwinsUK)12 and the Avon Longitudinal Study of Parents and Children (ALSPAC) 13 as part of the UK10K project. Sequencing was performed at an average depth of 7x across 3,781 individuals. The final dataset is described in 14 and consists of 42 million single nucleotide variants (SNVs), 3.5 million insertion/deletion polymorphisms (INDELs) and nearly 18,000 large deletions.
The initial phase of the UK10K Project applied a variety of statistical tests to identify rare alleles associated with a broad range of complex phenotypes. Besides yielding the first examples of novel trait associations identified through population-based WGS 15,16, the project provided a large-scale empirical evaluation of strategies for testing associations in the low and rare allele frequency range. First, the study demonstrated an overall paucity of low-frequency alleles with high-penetrance in the space where it was powered (defined by each variant’s effect >1.2 standard deviations and MAF ~0.5%), suggesting that in this frequency range novel discoveries required larger samples with greater statistical power. Further, it defined through simulations and empirical evidence the allelic space where genotype imputation was expected to be most beneficial for association studies. Finally, it developed a new genotype imputation panel based on WGS that significantly enhances imputation accuracy for low-frequency and rare variants in populations of European descent 17, substantially improving resolution and power in this frequency range.
Capitalising on these tools and discoveries, we sought to increase the representation of rare variation in association studies of cardiometabolic and hematologic traits through imputation using the UK10K and 1000 Genomes haplotype reference panels, studying up to 35,981 individuals of European descent from 18 different studies. After testing for association between 17 million sequence variants and 20 quantitative traits, we report on 17 novel variants associated with seven different traits. We applied fine-mapping approaches that exploit these more comprehensive imputation reference panels to identify sets of variants with high (>95%) joint probability of being causal at 59 different loci. By expanding the number of discovered loci for seven cardiometabolic traits and narrowing down known association signals to small sets of variants, our results demonstrate the utility of large imputation reference panels for the discovery and refinement of associations with complex quantitative traits.
Results
Common, low frequency and rare variant associations
We considered 20 different quantitative traits representing five biomedical trait groups: serum lipids (HDL, LDL, TC, TG), inflammatory biomarkers (CRP, IL6), renal function (uric acid, creatinine), fasting glycemic traits (glucose, insulin, HOMA-B, HOMA-IR) and haematological indices (HGB, RBC, MCH, MCHC, MCV, PCV, PLT, WBC) (Figure 1; see Figure legend for trait abbreviations). In the discovery stage, we tested associations of up to 15,188,514 autosomal and 468,312 X-linked SNVs and 1,311,244 biallelic indels (MAF≥0.1%) in up to 3,210 study participants with shallow WGS data available (depending on trait), and combined them with up to 32,904 participants from independent population based samples with SNPs imputed to the UK10K panel, or a combination of WGS reference panels 17,18 (Supplementary Note and Supplementary Table 1). We tested associations within each study using linear regression (Online Methods, Supplementary Table 1, Supplementary Table 2 and Supplementary Figure 1), and combined summary statistics from different studies with inverse-variance weighted meta-analyses.
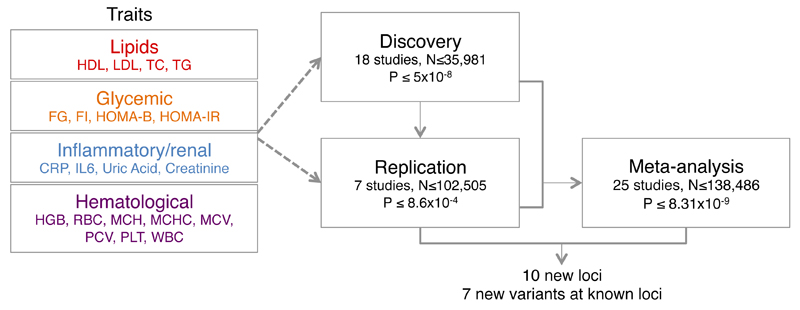
Figure 1. Study design.
Summary of traits and studies investigated in this study. Study-specific information is given in Supplementary Table 1. HDL = high-density lipoprotein cholesterol; LDL = low-density lipoprotein cholesterol; TC = total cholesterol; TG = triglycerides; FG = fasting glucose; FI = fasting insulin; HOMA-B = homeostatic model assessment of β-cell function; HOMA-IR = homeostatic model assessment of insulin resistance; CRP = C-reactive protein; IL6 = interleukin-6; HGB = haemoglobin; RBC = red blood cell count; MCH = mean cell haemoglobin; MCHC = mean cell haemoglobin concentration; MCV = mean cell volume; PCV = packed cell volume or hematocrit; PLT = platelet count; WBC = white blood cell count.
This effort yielded 171 independent associations (p-value ≤5x10-8) in the discovery meta-analysis, of which 110 represent previously reported GWAS signals, 48 mapped to statistically independent variants at known GWAS signals (secondary signals) and 13 to putative new associations. We obtained replication for 58/61 variants in as many as 102,505 independent samples from 5 different studies. We detected a total of 17 novel associations that were robustly replicated (defined as a replication p-value < 0.05/58 and meta-analysis p-value <8.31x10-9) in independent samples (Table 1 and Supplementary Table 3). Of these, ten were novel loci, or regions of the genome not previously associated with the trait of interest. Additionally, we identified seven variants defined as secondary signals, where the genetic variant mapped to within 1Mb of a locus already associated with the trait, but was statistically independent of any previously reported association (Online Methods). Of the 17 variants reported, three were coding and the rest were located in non-coding putative regulatory regions (see Box 1).
Table 1. List of novel variants and loci identified in this study.
| Associated Trait | Marker Name | Chr | Pos (hg19) | Locus/Nearest Gene | Coded allele | Non-Dcoded allele | MAF (WGS) | Beta (Joint) | SE(Joint) | P-value (Joint) | N (Joint) | Primary/Secondary Signal | Variant annotation |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PCV | rs10008637 | 4 | 77414144 | SHROOM3 | C | T | 0.463 | 0.032 | 0.004 | 1.08E-14 | 124,890 | Primary | intronic |
| PLT | rs2546979 | 5 | 159595612 | FABP6 | C | G | 0.291 | -0.049 | 0.004 | 1.81E-31 | 134,858 | Primary | intergenic |
| WBC | rs3130725 | 6 | 29118747 | ZNF311 | G | T | 0.131 | -0.008 | 0.001 | 2.70E-26 | 121,238 | Primary | intergenic |
| WBC | rs113164910 | 6 | 32427005 | HLA-DRA | AAC | A | 0.327 | 0.008 | 0.000 | 4.19E-54 | 122,412 | Primary | intergenic |
| PLT | rs61750929 | 9 | 91495135 | S1PR3 | T | C | 0.059 | -0.081 | 0.008 | 2.20E-21 | 134,858 | Primary | intergenic |
| PLT | rs150813342 | 9 | 135864513 | GFI1B | T | C | 0.004 | -0.408 | 0.026 | 4.73E-57 | 111,278 | Primary | synonymous |
| PLT | rs113373353 | 12 | 65007682 | RASSF3 | T | C | 0.111 | 0.055 | 0.006 | 1.76E-17 | 134,858 | Primary | intronic |
| PLT | rs575505283 | 15 | 43703277 | TP53BP1 | AT | A | 0.014 | -0.160 | 0.019 | 6.89E-17 | 121,073 | Primary | intronic |
| PLT | rs1801689 | 17 | 64210580 | APOH | C | A | 0.033 | 0.106 | 0.012 | 3.92E-19 | 134,858 | Primary | non-synonymous |
| PLT | rs75570992 | 22 | 50570755 | TRABD-MOV10L1 | C | G | 0.072 | 0.096 | 0.008 | 7.75E-32 | 134,377 | Primary | intronic |
| PLT | rs41315846 | 1 | 247712303 | GCSAML | C | T | 0.479 | 0.048 | 0.004 | 3.03E-34 | 134,858 | Secondary | intronic |
| PLT | rs78565404 | 3 | 184090242 | THPO | T | C | 0.057 | 0.136 | 0.009 | 1.65E-50 | 134,858 | Secondary | 3' UTR |
| UricAcid | rs56223908 | 4 | 9918492 | SLC2A9 | C | A | 0.080 | 0.137 | 0.018 | 9.21E-15 | 26,727 | Secondary | intronic |
| WBC | rs2442735 | 6 | 31346653 | HLA-B | G | A | 0.140 | -0.010 | 0.001 | 1.93E-46 | 121,528 | Secondary | intergenic |
| MCV | rs112233623 | 6 | 41924998 | CCND3 | T | C | 0.011 | 0.723 | 0.049 | 5.65E-49 | 107,036 | Secondary | intronic |
| HDL | rs3824477 | 9 | 107588328 | ABCA1 | A | G | 0.026 | 0.122 | 0.016 | 1.43E-13 | 56,306 | Secondary | intronic |
| MCH | rs117747069 | 16 | 170076 | NPRL3 | C | G | 0.037 | -0.172 | 0.024 | 4.20E-13 | 119,687 | Secondary | intronic |
Box 1. Biological and functional annotation of novel genetic variants and loci.
| Locus/Trait | Description of most likely functional SNP |
|---|---|
| GFI1B/PLT | Index SNP (iSNP) rs150813342 is a synonymous variant altering a predicted GFI1B exon 5 splice site. GFI1B is a transcription factor involved in the regulation of red cell and platelet production 34. Rare, heterozygous LoF mutations of GFI1B have been reported in hereditary thrombocytopenia [OMIM #187900]. rs150813342 has no LD proxies and it is predicted to be causative by CAVIARBF (PP=1). Furthermore, it lies within a region enriched for H3K4me1 and H3k36me3 in megakaryocytes 52. |
| NPRL3/MCH | iSNP rs117747069 is a low-frequency intronic variant of NPRL3 with no LD proxies, predicted as the most likely causal variant (CAVIARBF PP=0.84) and conditionally independent of the common NPRL3 variant rs11248850 previously associated with MCH 8. NPRL3 is known to contain nucleosome-depleted regions involved in the regulation of the alpha-globin genes on chr 16 8. rs117747069 is located in erythroid-specific super-enhancer 53 54,55,52, which is hypersensitive, enriched for H3K27ac marks in erythroblasts, and overlapping ChIP-Seq signal for erythroid transcription factors GATA-1, GATA-2 and TAL-1 in K562 cells 56. While the nearest gene NPRL3 is a potential target of the enhancer element, chromatin interactions in K562 cells 56 suggest that the super-enhancer element interacts with several downstream genes including HBA1 and HBA2. |
| CCND3/MCV | iSNP rs112233623 is a low-frequency intronic variant of CCND3, conditionally independent of the previously reported common association of rs9349204 with red cell traits8. Cyclin D3 plays a critical role in cell cycle regulation. The iSNP is located within an erythroid-specific enhancer 37,55,52 enriched for H3K27ac mark in erythroblasts and is bound by GATA-2 and TAL1 in K562 cells 56. Its association with hemoglobin A2 levels 36 also supports the role of this variant in the regulation of alpha-globin. |
| HLA-DRA/WBC | Index variant rs113164910 is a 2 bp indel lying in the class II MHC region, 14 kb 3’ of HLA-DRA. The most likely fSNP rs9268781 (8 kb 3’ of HLA-DR) is a strong eQTL for various HLA-DR and –DQ genes in blood 57 and overlaps a DNaseI hypersensitive (DHS) site in blood monocytes 58. Another LD proxy rs7763262 has been previously associated with IgA nephropathy 59. |
| HLA-B/WBC | iSNP rs2442735 is located ~20 kb 5’ of the HLA-B locus and is conditionally independent of another HLA-B intronic SNP in the class I MHC region rs2853946 associated with WBC 60. The most likely fSNP rs2853999, 1 kb 5’ of HLA-B, is a blood eQTL for HLA-C, C4A, and C4B and overlaps blood cell promoter and enhancer, DNase and histone marks. A proxy SNP has been associated with marginal zone lymphoma 61. |
| THPO/PLT | iSNP rs78565404 is a second THPO signal, conditionally independent of the previously reported platelet GWAS variant rs6141 62. Both SNPs fall in the 3’ UTR and have no LD proxies. THPO is a key regulator of platelet production. THPO gain-of-function mutations have been identified in hereditary thrombocythemia [OMIM #187950]. rs78565404 binds the transcription factor MAFK (ChIPSeq in HepG2 cells), a component of the NF-E2 complex involved in erythropoiesis and megakaryopoiesis 38,39. |
| GCSAML/PLT | iSNP rs41315846 is located in a hematopoietic cell-lineage specific promoter of GCSAML (C1orf150) 58. It is conditionally independent of previously reported GCSAML intronic iSNP rs7550918 and has no LD proxies. GCSAML encodes a protein thought to be a signaling molecule associated with germinal centers, the sites of proliferation and differentiation of mature B lymphocytes. rs41315846 lies within a putative enhancer overlapping DHS site, RUNX1, GATA1 and FLI1 ChIP-Seq peaks and H3k27ac enriched region in megakaryocytes 52. |
| FABP6/PLT | iSNP rs2546979 is a common intronic variant of FABP6, which encodes a fatty acid binding protein not known to play a role in platelet biology. It lies in a region of high LD spanning the region 5’ to the first intron of FABP6. The most likely fSNP (r2=0.7) rs2546372 (located ~22kb upstream of FABP6) overlaps regions enriched for H3k4me1 and H3k27ac signal in megakaryocytes, DNase, RUNX1, and FLI1 ChIP-Seq peaks 52. Another gene in this region, the transcription factor gene PTTG1 is highly expressed in bone marrow stem cells 63 and in megakaryocytes and erythroid precursors. Platelet promoter capture data from Blueprint shows that rs2546979 physically interacts with neighbouring gene CCNJL, which belongs to the family of cyclin genes involved in cell cycle regulation. The presence of H3K27ac (active promoter/enhancer) in the CCNJL promoter region and H3K36me3 (elongation) marks in the body of this gene indicates CCNJL is actively expressed in megakaryocytes 52. |
| TRABD-MOV10L1/ PLT | iSNP rs75570992 is intronic to MOV10L1, a predicted RNA helicase of unknown function. It is predicted to be causal (CAVIARBF PP=1) and associated with expression of the neighbouring gene TRABD in transformed fibroblasts, colon, and lymphoblastoid cells 57,33. However, another likely fSNP is proxy SNP rs75107793 (r2=0.5), which overlaps promoter and enhancer histone marks in many cell types 58, but more importantly, is located in a putative enhancer overlapping RUNX1 ChIP-Seq and DHS site and H3K4me1 enriched region in megakaryocytes 52. fSNP rs75107793 is also located within a DHS peak in erythroblasts, and lies upstream of TRABD promoter (GENCODE, FANTOM5). Based on RNA-Seq and epigenetic marks (H3K27ac, H3K4me3, H3H36me3), TRABD is expressed in megakaryocytes 52. |
| ZNF311/WBC | iSNP rs3130725 is located in an intergenic region on chromosome 6 containing extensive LD (>50 proxy SNPs [r2>0.8]), all of which (including rs3130725) are whole blood eQTLs for several genes in the region of class I HLA, including ZFP57, HLA-F, and HLA-H 57. The most likely fSNP is rs3129794, which is located in the promoter region of ZNF311 and overlaps an active promoter in K562 cells 58. |
| APOH/PLT | iSNP rs1801689 encodes a Cys325Arg amino acid substitution of APOH, also known as beta2-glycoprotein I, a platelet phospholipid-binding protein. It is the most likely fSNP (PP = 0.37 by CAVIARBF), though another proxy SNP rs8178824 (r2=1; PP = 0.22) is located in a liver-specific promoter (Roadmap). Platelet promoter capture data (BLUEPRINT) shows that rs1801689 physically interacts with neighbouring gene PRKCA (protein kinase C alpha), which also plays a role in platelet function and platelet production in mouse models of megakaryopoiesis 64,65. |
| S1PR3/PLT | iSNP rs61750929 is located ~100kb upstream of S1PR3, which encodes a receptor for sphingosine 1-phosphate (S1P) and likely contributes to the regulation of angiogenesis and vascular endothelial cell function.66,67. S1PR3 overlaps with C9orf47, a gene of unknown function. The iSNP has 33 strong LD proxies, in an inter-genic region between MIR4289 and S1PR3/C9orf47, several of which are cis-eQTLs for S1PR3 in whole blood 68 positioned within megakaryocytic DHS sites (rs62549698, rs9410336) or H3K4Me1-enriched enhancer regions (rs9410196, rs142550358, rs9410336) 52. Two lower LD (r2=0.5) proxies are synonymous (rs11795137) or 3’ UTR variants (rs62551536) of C9orf47. |
| RASSF3/PLT | iSNP rs113373353 and all 33 of its proxies are intronic to RASSF3, a tumor suppressor that also promotes apoptosis. The most likely fSNP rs77164989 (r2=0.8) lies within a putative enhancer that overlaps with DNase, H3K4me1, and RUNX1 ChIP-Seq peaks in megakaryocytes 52. |
| SHROOM3/HCT | iSNP rs10008637 is intronic to SHROOM3, which encodes a protein that binds and regulates the subcellular distribution of F-actin 69. An intronic LD proxy rs13146355 of SHROOM3 is associated with lower serum creatinine 20 and higher serum magnesium 21. Another LD proxy (r2=0.8), rs17319721, overlaps DHS sites in endothelial cells and is located in a TCF7L2-dependent enhancer increasing SHROOM3 transcription and influencing TGF-β1 signaling and renal function70. |
| ABCA1/HDL | The ABCA1 intronic variant rs3824477 (MAF=0.02) is in strong LD (r2 = 0.94) with ABCA1 missense variant (rs2066718 = p.Val771Leu), previously nominally associated with HDL (P=10-4)23. Both SNPs are independent of the common ABCA1 iSNP rs1883025 for HDL 71 and the secondary ABCA1 signal rs11789603 72. ABCA1 regulates cholesterol and phospholipid homeostasis. Rare loss-of-function variants of ABCA1 are associated with Tangier’s disease [OMIM #205400]. |
| TP53BP1/PLT | Index variant chr15:43703277 is a 1bp intronic indel of TP53BP1 located at a DHS site and binding site for several hematopoietic transcription factors including MAFK, GATA1, GATA2, and TAL1. A chromosomal aberration involving TP53BP1 is found in a form of myeloproliferative disorder with eosinophilia 73. The translocation t(5;15)(q33;q22) with PDGFRB creates a TP53BP1-PDGFRB fusion protein. |
| SLC2A9/Uric acid | iSNP rs56223908 (MAF=0.08) is intronic to the urate transporter SLC2A9 74. It has no LD proxies and it is conditionally independent of the more common, known SLC2A9 uric acid GWAS variant rs12498742 75. Rare mutations in SLC2A9 are a cause of autosomal recessive renal hypouricemia-2 [OMIM #612076]. The iSNP overlaps H3K4me1 enhancer histone marks in several Roadmap cells/tissues (blood, adrenal, muscle, heart, and lung) and is predicted as an active promoter in pancreas. |
The ten novel associations all involved hematologic traits and included seven variants associated with platelet count (PLT), two variants associated with white blood cell count (WBC), and one variant associated with haematocrit (PCV). Two of the ten loci were previously associated with other traits. The rs1801689 missense variant (p.Cys325Gly) in APOH associated with higher PLT was previously associated with higher low-density lipoprotein (LDL) cholesterol 19. SHROOM3 rs10008637 associated with higher PCV is an LD proxy (r2=0.98) for rs13146355, a common intronic variant associated with lower serum creatinine in East Asians 20 and higher serum magnesium in Europeans 21. One of the PLT-associated loci, synonymous variant rs150813342 of GFI1B, is reported concurrently in an independent exome sequencing data set 22.
Among the seven secondary signals within 1Mb of a known locus, one was associated with HDL (an intronic variant of ABCA1), one with uric acid (an intronic variant of SLC2A9) and five with hematological indices (PLT, WBC, MCV and MCH). Four loci harboured both common and independent, lower frequency variants (CCDN3 and MCV; THPO and PLT; GCSAML and PLT; ABCA1 and HDL). The low-frequency ABCA1 intronic variant rs3824477 (MAF=0.02) was in strong LD (r2=0.94) with an ABCA1 missense variant (rs2066718 = p.Val771Leu) nominally associated with HDL (p-value=10-4) in a targeted lipid gene re-sequencing study 23.
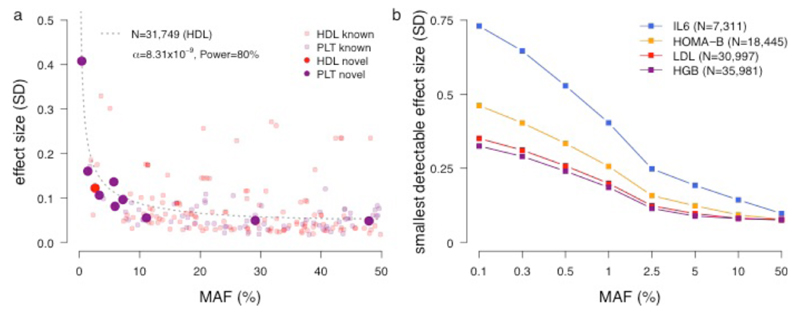
Three out of the ten novel loci, and three out of the seven secondary signal associations were observed for low frequency (MAF 1-5%) or rare (MAF <1%) variants, extending our understanding of the genetic architecture of cardiometabolic traits. To illustrate, we considered the effect sizes and allele frequencies of both known and novel variants for HDL and PLT (Figure 2a). Although we identified one rare variant with a large effect size (rs150813342 in GFI1B), the effect sizes of the other novel low-frequency variants were similar to those that have been previously reported in GWAS of common variants. Indeed, for variants with MAF ≥0.5%, we had 80% power to detect associations with effect sizes of 0.25, 0.25, 0.35 and 0.55 trait standard deviations for HGB, LDL, HOMA-B and IL6, respectively (Figure 2b). Although there may be rare variants of large effect that we were unable to identify in the current study, we likely did not miss large effect variants with MAF ≥0.5% and sufficient sequencing quality in European populations.
Figure 2. Allelic spectrum of cardiometabolic trait variants.
(a) For each variant surpassing the genome-wide threshold in this study, the effect size (measured in standard deviations) is plotted as a function of the minor allele frequency proportion (MAF%). Loci discovered in this study are plotted with larger symbols. Associations for HDL (black) and PLT (purple) for which novel variants were discovered are shown. The dotted line represents the curve for 80% power with a sample size of 31,749 (for HDL) and an alpha of 8.31x10–9. The power line for PLT (sample size 31,555) is similar and therefore not shown here. (b) Plot of smallest detectable effect size for a range of MAF%. Power calculations were performed for 4 traits of different trait groups and with different sample sizes: IL6, HOMA-B, LDL, and HGB.
Functional enrichment analysis of trait associated variants
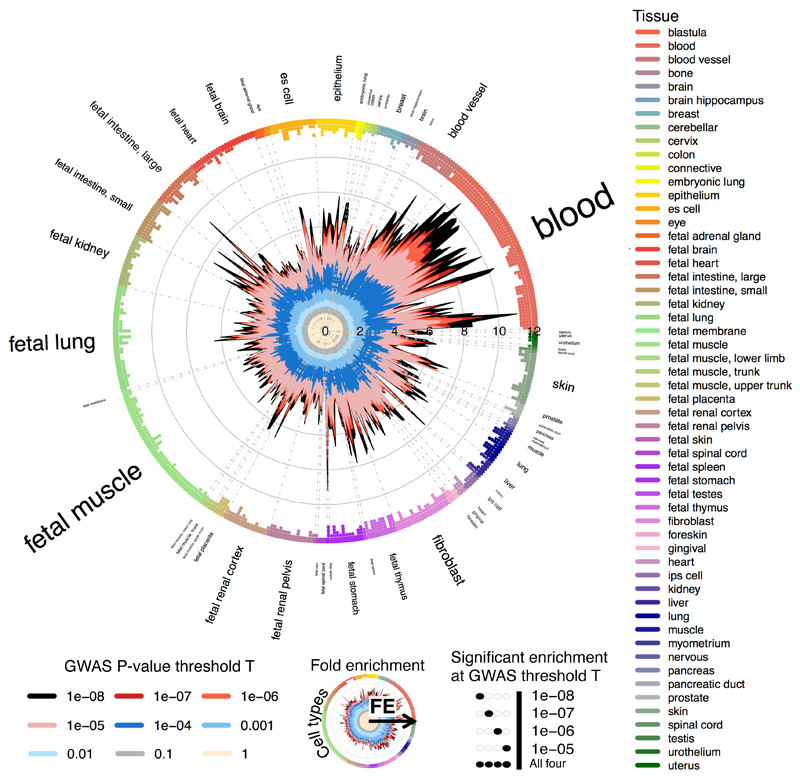
The majority of the associations we identified are found in non-coding regions, where the underlying cellular or molecular mechanisms are poorly defined. To evaluate the functional and regulatory properties of this set of variants, we estimated the extent to which associations for each of the 20 traits were non-randomly distributed across various coding, non-coding regulatory, and cell type-specific elements across the genome. We retrieved experimentally derived annotations from 1,005 genome-wide datasets from the GENCODE, ENCODE and Roadmap projects (Supplementary Table 4). We then used a novel nonparametric approach (GARFIELD) (Supplementary Note) to derive fold enrichment (FE) statistics for trait associated SNPs within each annotation, where SNPs were selected from genome-wide datasets based on their strength of association with each trait (Online Methods). An example of the results for one trait (PLT) and one annotation type (DHS hotspots) is shown in Figure 3, with all results summarised in Supplementary Table 5 and Supplementary Figure 2.
Figure 3. GARFIELD functional enrichment analyses.
Wheel plot displaying functional enrichment of associations with PLT within DHS hotspot regions in ENCODE and Roadmap studies. Radial axis shows the fold enrichment (FE) values calculated at each of eight GWAS P-value thresholds (T<10–1 to T<10–8) for each of 424 cell types. Cell types are sorted by tissue, represented on the outer circle with font size proportional to the number of cell types from that tissue. Boxes and circles next to the tissue labels are coloured with respect to tissue (right legend). FE values at the different thresholds T areplotted with different colours on the inner side of the circle (e.g. T<10–8 in black, bottom-left legend).Dots in the outer side of the circle denote significant enrichment (if present) for a given cell type at T<10–5 (outermost) to T<10–8 (innermost) (bottom-right legend). Results show overall well spread enrichment, with largest FE values obtained in blood, fetal spleen and fetal intestine tissues.
Lipid and hematological traits displayed ubiquitous and marked enrichment patterns, with 151 (p<10-8) and 906 (p<10-5) overall significant FE statistics for serum lipids, and 237 (p<10-8) and 749 (p<10-5) for hematological traits, respectively. As the most extreme cases, we found that associations with RBC were enriched in enhancers of the erythroid cell line K562 (FE=39.63, empirical p-value=2x10-5), while associations with WBC were enriched in footprints of CD20+ cells (FE=22.16, empirical p-value<10-5). The most significant association for LDL was within TSS chromatin states measured in the liver HepG2 cell line (FE=19.53, empirical p-value<10-5). Conversely, inflammatory and renal traits displayed weak patterns of enrichment. There was a significant enrichment of associations (FE=4.44-fold, empirical p-value=10-5) with creatinine within DHS hotspots of fetal kidney. Uric acid associations were weakly enriched in a small number of liver and fetal intestine annotations. Unexpectedly, we observed enrichment of triglycerides (TG) in HMVEC-LLy (lymphatic microvascular endothelial cells) footprints for SNPs with p<10-5 (FE=9.75, empirical p-value<10-5), which is much larger than that observed for the broader DHS hotspots (FE=4.30, empirical p-value<10-5). By contrast, there was no significant enrichment for footprints of the expected most relevant HepG2 (a well established hepatocyte cellular model for cholesterol metabolism) cell type.
Fine mapping of loci using dense imputation from WGS
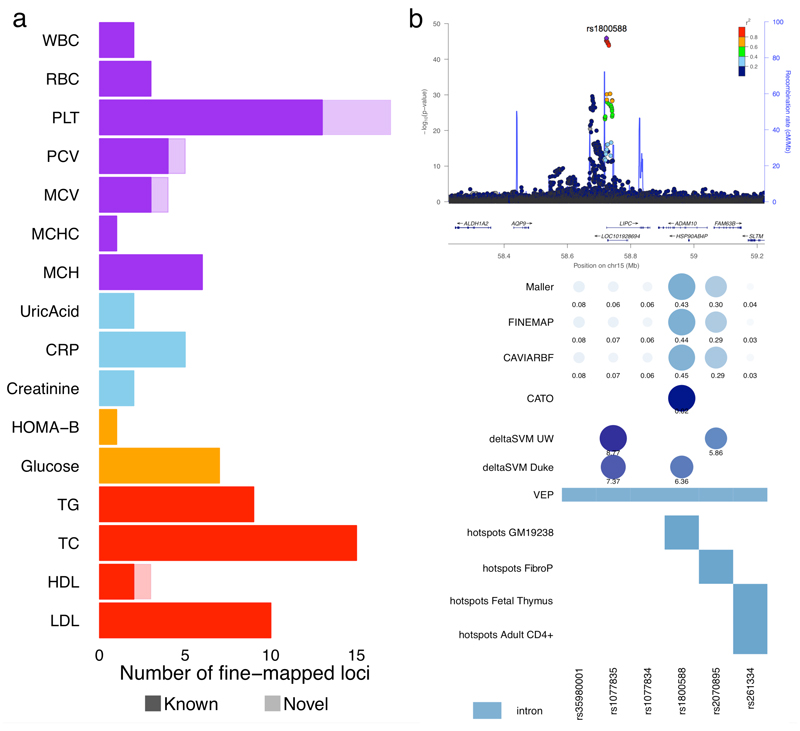
Linkage disequilibrium and incomplete ascertainment of variants in a given region of interest present significant challenges for pinpointing the causal variant[s] driving an association. To fine-map the causal variant[s] at associated loci, we exploited the high density of our whole-genome sequence reference panels to define the posterior probability of each variant being causal given all other variants in the region. We selected 417 regions with informative associations (p-value≤10-5, Online Methods) in the initial discovery meta-analysis and applied three distinct Bayesian approaches (namely ‘Maller’ 24, ‘FINEMAP’ 25 and ‘CAVIARBF’ 26) (Online Methods). For each of the three methods, we created 95% credible sets by ranking variants based on their decreasing posterior probability (PP) of association. These credible sets contain the minimum list of variants that jointly have at least 95% probability of containing the causal variant. We focused on 59 known or novel loci where the three methods identified a credible set of less than 20 variants, and where all variants were either directly genotyped or well imputed (Figure 4, Supplementary Table 6 and Supplementary Figure 3).
Figure 4. Fine mapping experiments.
Regional association plots for loci showing fine-mapped variants. (a) Numerical summary of 59 loci that were fine-mapped. (b) Example of fine-mapping and annotation at the LIPC locus for association with HDL. Panels show the regional association locuszoom plot, the PP statistics from the fine-mapping methods, the CATO and deltaSVM scores, VEP genic annotations and overlap of regulatory annotations found significant (coloured in blue) from GARFIELD enrichment analysis. Circles sizes and colours for all scores have been scaled with respect to score type (i.e. PP, CATO or deltaSVM) and numbers have been plotted below each circle.
Overall, 95% credible sets contained an average of 6.9 (standard deviation = 5.9) variants per locus when considering the union of all methods, or 5.5 (standard deviation = 4.7) when considering the intersection. In 45 cases the three methods yielded identical 95% credible sets, including 13 known and 5 novel loci where a single variant was predicted to be causative with posterior probability ~1 by all three methods. Of these 18 loci, five involved well-characterised missense variants (rs11591147 at PCSK9, rs1260326 at GCKR, rs855791 at TMPRSS6, rs7412 at APOE and rs429358 as a secondary signal at APOE). Missense variants were included in the 95% credible sets at several other loci (ABCG2, APOB, CD300LG, CILP2, HFE, PSORCS1, SH2B3, SLC30A8 and APOH). At four loci the credible interval included a variant predicted to alter an essential splicing donor/acceptor motif (GCSAML, MLXIPL, BET1L and CETP), and at the other three (DNAH11, IKZF1 and GFI1B) the 95% credible set included synonymous sites. For all other loci, the causative set included UTR, intergenic and intronic sites.
For each known locus we compared the variants in the fine-mapped set with published evidence from functional validation studies (Supplementary Table 6). Of the 59 discrete genomic regions, 40 were associated with one trait and 19 were associated with multiple traits. Further, 25 (42%) were known to have at least one causative variant previously experimentally or functionally validated. At 20 of the 25 loci, the previously validated functional variant was contained within the 95% credible interval identified using one or more fine-mapping methods. In 11 regions, the known causal variant was ranked with the highest posterior probability by at least one fine-mapping method. We also identified several other examples where the credible sets define high-priority variants for downstream follow-up. Among these are CRP rs1205 a 3’ UTR variant associated with C-reactive protein (CRP) that is located in a predicted liver enhancer region that alters a glucocorticoid receptor (NR3C1) transcription factor binding site; rs1822534 a regulatory region variant upstream of PPARG associated with PLT; ARHGEF3 rs1354034, an intronic variant associated with PLT located in a predicted enhancer region in hematopoietic and primary T cells (Roadmap epigenomics chromatin state) and predicted to alter a GATA motif; the total cholesterol (TC)-associated variant rs2169387 located in a predicted liver/muscle enhancer region several hundred kb upstream of PPP1R3B; the TC-associated ABCA1 rs2740488 variant located in a liver-specific promoter region; the PLT-associated variant rs12005199 located in a putative enhancer region upstream of AK3 bound by GATA1/2 and TAL1; PCV-associated HK1 intronic variant rs17476364 located in a hematopoietic cell enhancer region; and the TG-associated variant rs964184 located in a liver and fat enhancer within the ZNF259 3′ UTR.
Regulatory annotation of locus-specific findings
To inform our statistical fine-mapping approach, for every variant in a credible set we applied two scores for regulatory function based on cell type specific DNase I hypersensitivity sites (DHS): the deltaSVM score and the Contextual Analysis of Transcription Factor Occupancy (CATO) score (Online Methods) 27,28 (Supplementary Table 6). The functional activity of a variant’s effect allele is predicted by the magnitude of the deltaSVM score, with the sign indicating the increase or decrease of DNase I hypersensitivity, and therefore transcription factor (TF) binding potential, at the site. Similarly, the functional activity of a variant’s effect allele is predicted by the CATO score, where scores of 0.1 have a 51% true positive rate for perturbing known TF motifs, with the true positive rate increasing as the score increases to 1 28. To identify putatively causal variants, we considered deltaSVM scores greater than 10 in absolute value, CATO scores > 0.1, and high PP from the statistical fine-mapping methods.
This union-of-methods approach identified several strong cases for causal variants. At the TRIB1 locus associated with TG, TC, and LDL traits, rs112875651 has the strongest supporting evidence for causality from all three fine-mapping methods (0.517, 0.532 and 0.526) and also from extreme CATO and deltaSVM scores (0.315 and -12.31, respectively). Other functional variants have been suggested for the TRIB1 region, namely rs2001844 29 (r2=0.8) and rs6982502 30 (r2=0.7), but these SNPs were four orders of magnitude less significant than rs112875651 in our TG analysis, suggesting that rs112875651 may be a causal variant at TRIB1. At the CELSR2 locus associated with LDL and TC, all three fine-mapping methods provide evidence for causality (0.205,0.202 and 0.200) of rs12740374, though rs646776 (r2>0.8) is a stronger predicted causal variant from the PP estimates. However, additional supportive evidence for rs12740374 as the causal variant comes from a high CATO score (0.199) and an extreme deltaSVM score (14.37) for cell types with significant enrichment predicted by GARFIELD (liver and epithelial cells). The CATO and deltaSVM scores are also helpful when there are no obvious causal candidates from statistical fine-mapping. For example, at the CXCL2 locus associated with WBC, the PPs do not provide sufficiently strong evidence for a single causal variant. However index variant rs13128896 has strong functional evidence from its high CATO score (0.146) and its extreme deltaSVM score (-10.71) for blood and skin cell types, with the former cell type being enriched for WBC associations in the GARFIELD analysis.
Integration of methods to prioritize variants for follow-up
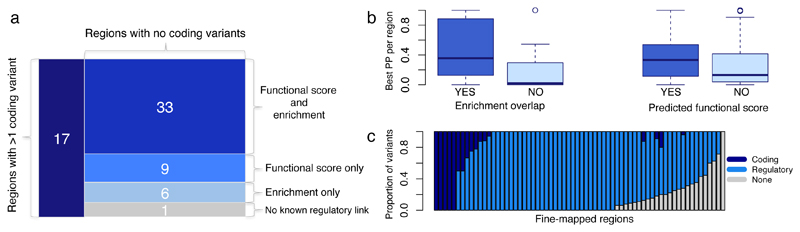
We next combined information from fine-mapping analysis, genome-wide functional enrichment results and regulatory scores to assess the overall evidence supporting functional and causal interpretation at 66 independent regions (in 59 loci). Overall, there were 17 regions with at least one coding variant, 33 regions with support from both functional enrichment and regulatory scores, 9 with functional scores only, and 6 with enrichment only (Figure 5a). Variants with functional enrichment overlap and those with regulatory scores had larger PPs of causality (average PP increase of 0.3 and 0.1, respectively) (Figure 5b), in contrast to variants with no such regulatory support, highlighting them as statistically more likely to be causal. For 24 of the 66 regions we found functional or regulatory support for only a fraction of the variants within credible sets (Figure 5c), ranging between 29% and 94% of variants with annotation from at least type of evidence (mean = 74%, standard deviation = 18%), resulting in up to a 71% reduction in the size of the credible set. Of note, there was only one fine-mapped region (G6PC2 locus associated to glucose) with statistical support alone and no regulatory support; however the credible set contained a single causal variant with PP>0.999 from all three fine-mapping approaches and the variant has previously been shown to enhance G6PC2 pre-mRNA splicing 31.
Figure 5. Fine-mapping summary of variant consequences.
(a) Number of fine-mapped trait-region pairs containing at least one variant in the 95% credible set with consequences (i) coding; (ii) functional score and overlapping annotation significantly enriched for the given trait; (iii) functional score only; (iv) significant enrichment overlap only; (v) none of the above. (b) Distribution of the top posterior probability (PP) per region for variants with significant enrichment overlap and predicted functional score (after removing regions containing a coding variant). Boxplots depict the median (thick horizontal line), 1st and 3rd quartiles (coloured box), maximum and minimum values (whiskers) and outlying values (circles). (c) Proportions of variants within credible sets with coding, regulatory or no annotations.
Discussion
Our analysis demonstrates the utility of low-pass WGS data combined with SNP array data deeply imputed to WGS reference panels for informing studies of quantitative cardio-metabolic and hematologic traits. By combining the UK10K and 1000 Genomes Project sequence data, we constructed a dense imputation reference panel that substantially improves upon the HapMap2 and 1000 Genomes panels. With this dense imputation reference panel, we investigated associations with variants as rare as 0.5% frequency.
Consistent with previous reports 17,32, our imputation accuracy declined with decreasing allele frequencies. Therefore, we did not consider very rare variants (MAF ≤ 0.001) or variants with poor imputation quality (INFO ≤ 0.4). This resulted in a substantial culling of the total number of variants that were identified in the UK10K project. Thus, our study may have missed rare-variant associations that would be identifiable in a larger study. Because genotype imputation provides model-based estimates of allelic probabilities in the study subjects, rather than hard-called empirically based genotypes, we could not reference cluster plots or intensity files in order to validate our findings. In this context, independent replication serves a critical function for validating associations from an imputation-based discovery effort.
Our dense imputation reference panels expanded the set of variants amenable to association analysis. Only one of the 17 novel loci we report was well tagged (r2 > 0.8) in HapMap2 or 1000 Genomes Phase 1. Markers assessed in previous GWAS of PLT, haemoglobin and WBC poorly tagged nine of the novel loci associated with hematologic traits. However, for platelet count (the trait for which we observed the most and strongest associations), the novel loci identified here increased the percentage of phenotypic variance explained from 7.71% to 8.23%. Though increasingly large imputation panels are useful for investigating low frequency and rare-variants, considerably larger sample sizes are needed identify rare-variants of modest-to-large effects.
For each novel locus identified we undertook epigenomic, tissue expression, and fine-mapping analyses to describe the potential mechanism of these associations (Box 1). Our results implicate several genes or loci not previously known to be involved in regulation of blood cell counts. For example, the chromosome 22 PLT index variant rs75570992 is located upstream of TRABD, a gene of unknown function. Based on RNA-Seq and epigenomic data from BLUEPRINT, TRABD is expressed in megakaryocytes. The index variant rs75570992 is associated with differential expression of TRABD in blood cells 33. Notably, the index variant is in partial LD with rs75107793 (r2=0.5), which lies upstream of the TRABD promoter in an H3K4me1-enriched putative megakaryocyte enhancer overlapping a ChIP-Seq site for the hematopoietic transcription factor RUNX1.
Another newly discovered locus leading to new mechanistic insights is GFI1B rs150813342, a synonymous variant predicted to alter an exonic splicing enhancer. GFI1B is a hematopoietic transcription factor required for normal red blood cell and platelet production 34. In a companion paper, we demonstrate that the rs150813342 variant influences the relative amounts of two GFI1B transcript isoforms, a full-length (long) and short isoform lacking the alternatively spliced exon 5 22. We further demonstrate the lineage-specific role of the long GFI1B isoform on megakaryocyte development. Prior studies have suggested that the short GFIB isoform is required for red cell production 35.
We identify several secondary, independent signals in genes previously implicated in regulation of blood cell counts (CCDN3, NLPR3, THPO). The new MCV-associated CCDN3 low-frequency variant rs112233623 was also associated with hemoglobin A2 levels 36. rs112233623 is located within an erythroid-specific enhancer 37 and is bound by the hematopoietic transcription factors GATA-2 and TAL1. Similarly, NLPR3 rs117747069 is located in an erythroid enhancer element involved in alpha-globin gene regulation and overlaps GATA-2 and TAL-1 ChIP-Seq sites. A 3’ UTR variant of the thrombopoietin gene (THPO rs6141) was previously associated with higher PLT. We identify a second, independent 3’ UTR THPO signal rs78565404. By ChIPSeq, rs78565404 is bound in liver HepG2 cells by musculoaponeurotic fibrosarcoma oncogene homolog K (MAFK), a component of the hematopoietic NF-E2 transcription factor complex involved in megakaryopoiesis 38,39.
Several of our newly identified variants are located within genes for congenital (GFI1B, THPO) or acquired (APOH) platelet disorders, underscoring that more subtle genetic variation within genes known to contain loss-of-function variants may reflect inter-individual differences in these complex traits. Rare loss-of-function GFI1B mutations have been identified in patients with congenital thrombocytopenia 40,41, while THPO mutations have more often been found in pedigrees with hereditary thrombocytosis. Most of the THPO mutations described in patients with familial thrombocytosis have involved non-coding sequences (splice site, 5’ UTR, intronic) gain-of-function mutations that lead to enhanced THPO mRNA translation efficiency 42–45. It remains to be determined whether the two common 3’ UTR variants of THPO associated with higher PLT similarly enhance mRNA translation and thrombopoietin synthesis. Recently, the first “loss-of-function” THPO missense mutation (p.Arg38Cys) was associated with aplastic anemia in the homozygous state and mild thrombocytopenia in the heterozygous state 46.
Apolipoprotein H (ApoH) is also known as β2–glycoprotein I (β2–GPI), a major autoantigen for the antiphospholipid antibody syndrome (APS), a clinical disorder characterized by arterial and venous thrombosis 47,48. Thrombocytopenia is also sometimes a feature of the APS. Interestingly, the p.Cys325Gly variant encoded by APOH rs1801689 disrupts the β2–GPI phospholipid binding site 49 ApoH/β2–GPI is also a component of LDL and binds to members of the LDL receptor family. The same APOH rs1801689 missense variant associated with higher platelet count was recently associated with higher LDL 19. β2–GPI/antiphospholipid antibody complexes bind to LRP8, an LDL receptor present on platelets and endothelial cells; this interaction has been postulated to play a role in β2–GPI-mediated thrombosis 50,51. However, even when we controlled for LDL levels, the rs1801689 association with platelet count remained intact, suggesting independent mechanisms driving the associations.
We undertook extensive fine-mapping of previously reported loci, identifying 59 loci where we could reduce associated signals to credible sets of 20 or less variants. We observed that the number of variants in the credible set was negatively correlated with the allele frequency of the index SNP, as expected since rare variants have fewer proxies on average. The newly identified loci had lower average minor allele frequencies and lower number of proxies, making the identification of causative variants more straightforward. Rare variants were also more likely to have severe consequences or lead to changes in the protein code, facilitating the identification of likely causative genes.
Our enrichment analyses showed that SNPs significantly associated with a phenotype of interest are over-represented within "functional" regions that were derived in a broad range of cell types and tissues. We evaluated the extent to which genetic associations for each of the 20 traits were enriched in different functional domains, and found that lipids and platelet counts were enriched in a large number of tissues and cell types compared to other traits displaying more localised (red cell traits) or null (renal, inflammatory traits) enrichment patterns. Combined with the fine-mapping experiments, we observed a positive correlation between the PP of causality and overlap with significantly enriched annotations. Overall this suggests that the process of sifting through putative causal variants can benefit from multi-pronged approaches incorporating fine mapping analysis to additional regulatory information obtained from epigenomes and deltaSVM and CATO scores. This information in turn empowers downstream functional experiments by guiding explorations of the functional consequences for sets of associated variants.
By performing detailed epigenomic and functional annotation, we were able to suggest several novel mechanisms for variants at known loci (e.g., differential splicing for GFI1B, experimentally demonstrated in a companion paper) or posit strong biologic candidates for further functional and cellular study on platelet production (e.g., TRABD), and highlight potential genetic connections between platelet count and traditional CVD risk factors such as cholesterol levels (APOH). Imputation using dense genotype maps affords a greater understanding of the relative contribution of rare and low frequency variants to complex traits, and allows the fine mapping of common variant association signals to manageable credible sets. In parallel, the development of robust functional enrichment methods and the overlap of fine-mapped associations with genome functional maps allowed us to pinpoint variants with high probability of being causal.
Online Methods
Imputation
Whole-genome sequence based haplotype reference panel
A joint reference panel was created as described in 17 by combining two large-scale, low read depth whole-genome sequencing datasets, TwinsUK and ALSPAC. The UK10K final release WGS data of 3,781 samples and 49,826,943 sites was used. From this dataset, multi-allelic sites, sites containing alleles inconsistent with that of the 1000 Genomes Project (1000GP) data, and singletons not existing in 1000GP were removed, leaving 28,615,640 sites. SHAPEIT v2 76 was used to re-phase the haplotypes in 3MB chunks with +/-250kb flanking regions. The phased chunks were then recombined with vcf-phased-join from the vcftools package 77. The 1000GP Phase I integrated variant set release (v3) for low-coverage whole-genomes in NCBI build 37 (hg19) coordinates was downloaded from 1000GP FTP site (23 Nov 2010 data freeze). This call-set includes phased haplotypes for 1,092 individuals and 39,293,751 variants (22 autosomes and chromosome X). For each chromosome, a summary file was generated and merged with that of the UK10K WGS data to identify multi-allelic sites and singletons not polymorphic in UK10K. These sites were excluded to create a new set of VCF files. The final reference panel included all 1,092 samples and 32,506,604 sites. The VCF-QUERY tool was used to convert the new VCF files into phased haplotypes and legend files for IMPUTE V2 78.
Pre-phasing and imputation of target GWAS
Genome-wide SNP data was obtained from each individual study, having undergone study-specific quality control (Supplementary Note). These samples were pre-phased using SHAPEIT v2, with the mean size of the windows in which conditioning haplotypes were defined set to 0.5MB. Due to the significantly higher number of variants in the WGS data, the re-phasing was conducted by 3MB chunks with 250kb buffering regions. Phased genotypes were then imputed to one of the three WGS reference panels (UK10K alone or UK10K+1000GP or 1000GP+Genomes of the Netherland (GoNL)) as detailed in Supplementary Table 1. Imputation was carried out using IMPUTE V2 using standard settings 78.
Association Testing
Phenotype preparation
All traits were available from previous studies. Information on trait measurements is summarised in Supplementary Table 1. Traits were transformed by inverse normalization (Creatinine, Glucose, HDL, HGB, HOMA-B, HOMA-IR, CRP, IL6, Insulin, LDL, PCV, PLT, TC, TG and Uric Acid), square root transform (MCH), log transform (WBC) or left untransformed (MCHC, MCV, RBC) in order to meet the normality assumption for linear model association testing. Traits were further residualised on associated covariables for each trait and each population sample, following detailed information given in the UK10K project manuscript 14 (summarised in Supplementary Table 4 therein). Finally, 10 principal components (PCs) were additionally regressed out from all traits for cohorts with unrelated individuals to further control for potential confounding. Information on individual study characteristics, including trait values and potential additional cohort-specific covariates applied are given in Supplementary Note and Supplementary Table 2. Histograms of trait residuals for which inverse normalization was not applied are shown in Supplementary Figure 4.
Study design for association testing
The study design is shown in Figure 1. Briefly, a total of 12,267 to 35,981 participants from 18 different studies were included in the discovery sample. Each cohort carried out single-marker association testing using linear additive models. Genotype dosages were used to account for the genotype uncertainty that might arise from sequencing, where each genotype was expressed on a quantitative scale between [0:2]. Variants that did not pass a low allele frequency threshold (MAF<0.1%), or imputed with low accuracy (defined by an imputation info score <0.4) were excluded from the analysis. Meta-analyses of cohort summary statistics were performed using GWAMA v 2.1 79 assuming a fixed effect model. Genomic control was used to adjust the summary statistics for both input and output data. We prioritised for replication all variants at the p-value ≤ 5x10-8 cutoff from the meta-analysis of 23 studies. During the course of the study, we updated our meta-analyses several times; variants were prioritised for replication if they met our cutoff (5x10-8) during any of these updates. These variants were taken forward into 2,141 - 102,505 additional independent samples from 7 cohorts (Supplementary Table 1), depending on the trait. Evidence for validation was based on a Bonferroni-corrected Stage 2 p-value of 8.6x10-4 (0.05/58) and joint meta-analysis p-value of 8.31x10-9 14.
Fine-Mapping of Associated Loci (Novel and Previously Identified Gwas Regions)
Annotation and selection of index variants for previously reported loci
For each trait we compiled a list of known loci by selecting all index SNPs associated with our traits of interest (lipids, fasting glucose, HOMA, uric acid, CRP, and blood cell counts and indices) from the NHGRI GWAS catalog (p-value ≤ 5x10-8, last updated in May 2014), supplemented by manual curation of all associations reported in the literature reaching the same genome-wide significance cutoff. Only those index variants with a marginal significance in the UK10K WGS cohort single-marker association statistics (p-value ≤ 0.05) were considered for conditional tests. Using TwinsUK and ALSPAC sequence data, we selected those variants with P-value less than 10-3 in the two-way meta-analysis. For each such variant we extracted regions for fine-mapping based on HapMap estimates of recombination rates. Where a region contained multiple correlated index variants associated with a given trait in the GWAS catalog, we clumped the set of index variants to remove highly correlated ones (using a LD metric r2>0.8 applied to within a 2 Mb sliding window from each known index SNP (+/-1 Mb)). This avoids collinearity errors when a variant is conditioned against multiple correlated index variants.
LD Pruning of UK10K index variants
We next applied an additional LD clumping procedure to thin the list of variants associated with each traits, assigning sets of variants to discrete LD bins if their pairwise metrics r2 was ≥ 0.2. For each LD bin, the variant most associated with the trait in question was retained for assessment in conditional analyses. Index variants for previously reported loci that mapped to within +/- 1Mb of an index variant for a known locus were also annotated.
Conditional analyses
Sequential conditional single-variant association analyses were carried out to confirm statistical independence between associations. In the initial round of conditional analysis, associations of SNVs with the respective quantitative trait were conditioned on the index variants for known loci clumped (r2>0.8) as described before (this step was carried out only for SNVs within +/- 1Mb of a known locus); in further rounds, associations were conditioned against all nearby known loci plus the best novel variant identified in the previous round of conditional analysis. The conditional analysis was tested independently for each cohort, and a meta-analysis was conducted at the end of each round until the conditional association p-value was no longer significant (p-value>10-5). A variant was considered independent if it had a conditional p-value ≤ 10-5 (corresponding to r2<0.2 in our data).
Finally, variants were classified as ‘known’ (denoting either a previously reported GWAS index variant, or a variant for which the association signal disappears after conditioning on the known locus) or ‘novel’ (denoted as variant which still is conditionally independent on known loci, and on eventual other novel independent signals in that region). For novel signals, the variant with the lowest conditional p-value between multiple associated variants was reported.
Bayesian Fine Mapping methods
For each previously reported (known) association and each novel index variant we extracted regions for fine-mapping based on HapMap estimates of recombination rates according to Maller et.al. 24. Specifically, the boundaries were chosen to be at a distance of at least 0.1 centimorgans on either side of the index or known SNP and if necessary extended further to include all its tagging variants (r2>0.1 within 1Mb windows). From the previously reported loci, only informative associations (p-value ≤10-5 the discovery stage analysis) were taken forward. Regions with multiple SNPs reported to be associated to the same trait were merged if overlapping. Analysis of each region was then performed separately using three different methods. We implemented the method of Maller et al 24, by converting our discovery stage meta-analysis p-values to Bayes’ factors (BF) of association using Wakefield’s approximation80. Additionally, we employed the fine-mapping methods CAVIARBF 81 and FINEMAP 25, both Bayesian approaches that utilize association summary statistics (rather than the original genotypic data) and SNP correlations to compute BFs. The BFs from each method were then used to calculate posterior probabilities, based on the assumption that there is a single causal SNP in each region. Conditional association analysis on the top fine-mapped variant was additionally carried out and (conditional) fine-mapping performed in order to fine-map secondary associations. For all regions, 95% credible sets were constructed in order to assess the uncertainty of the fine-mapping analyses. To assess the suitability of our two stage fine-mapping approach (conditional steps) in the presence of multiple causal variants, we further compared our results to those obtained from FINEMAP under a relaxed assumption of multiple causal variants (Supplementary Note, Supplementary Table 7).
Enrichment of GWAS SNPs in Functional and Regulatory Elements
In order to systematically characterize the functional, cellular and regulatory contribution of genetic variation implicated in each quantitative trait, we used GARFIELD, a non-parametric enrichment analysis approach taking genome-wide association summary statistics to calculate fold enrichment (FE) values at given significance thresholds, and then test them for significance via permutation testing while accounting for linkage disequilibrium, minor allele frequency, and local gene density. We used a range of functional annotations, including genic elements (GENCODE), DNaseI hypersensitive sites, transcription factor binding sites, histone modifications, and chromatin states (ENCODE and Roadmap Epigenomics) (Supplementary Table 4) and included different cell types and tissues in order to capture and characterize possible cell type specific patterns of enrichment. We calculated FE statistics at eight genome-wide significance thresholds T (in powers of 10) and tested their significance at the four most stringent ones (10-8 to 10-5) to analyse both stringent association findings as well as nominal ones. Multiple testing correction was further performed on the effective number of annotations used, resulting in enrichment p-value threshold of 1x10-4. Further information on the approach is provided in the Supplementary Note.
Scoring Credible Set Variants for Regulatory Function
DeltaSVM scores were generated as previously published by training the gapped k-mer support vector machine (gkmSVM) on cell type specific DHSs, computing weights for all possible 10-mers of the genome based on the SVM classifier, and calculating the difference in weights of 10-mers encompassing the reference and effect alleles for the variant of interest 27. Pre-computed weights were available from a total of 222 ENCODE DHS samples—99 from the Duke University (Duke) set and 123 from the University of Washington (UW) set 82. Genetic variants were scored for deltaSVM in all 222 cell lines and filtered for those with at least one deltaSVM score greater than absolute 5, allowing putative inference of relevant cell types or tissues. CATO scores were generated as described in 28. Briefly, logistic models were fit to imbalance in DNA accessibility in 443 DNase-seq datasets from the ENCODE and Roadmap Epigenomics projects. An independent model was fit for each of 44 TF families and included terms for both the effect of the variant on the TF position weight matrix as well as terms for genomic context. Genetic variants were then scored by taking the maximum prediction of all overlapping TF models. CATO scores greater than 0.1 were shown to have a 51% true positive rate on the initial training set and are therefore of interest 28.
Supplementary Material
Acknowledgements
This study makes use of data generated by the UK10K Consortium, derived from samples from the ALSPAC and TwinsUK datasets. A full list of the investigators who contributed to the generation of the data is available from www.UK10K.org. Funding for UK10K was provided by the Wellcome Trust under award WT091310. Nicole Soranzo's research is supported by the Wellcome Trust (Grant Codes WT098051 and WT091310), the EU FP7 (EPIGENESYS Grant Code 257082 and BLUEPRINT Grant Code HEALTH-F5-2011-282510) and the National Institute for Health Research Blood and Transplant Research Unit (NIHR BTRU) in Donor Health and Genomics at the University of Cambridge in partnership with NHS Blood and Transplant (NHSBT). The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR, the Department of Health or NHSBT. P.L. Auer was supported by NHLBI R21 HL121422-02.
Footnotes
URLs
The GARFIELD software is available in a standalone version at http://www.ebi.ac.uk/birney-srv/GARFIELD/ and as Bioconductor package at http://bioconductor.org/packages/release/bioc/html/garfield.htm. The deltaSVM scores were downloaded from http://www.beerlab.org/deltasvm/.
Author Contributions
Designed and or managed individual studies and contributed data: A.B., A.D., A.G.U., A.Ha., A.Ho., A.P.R., C.L., C.L.K., C.v.D., D.M., D.T., E.Z., G.G., H.W., J.C.C., J.S.K., L.F., M.A.S., M.Fra., M.Fro., N.J.T., N.S., P.G., P.L.A., R.A.S., R.P., W.M.; Generated and or quality controlled data: A.F., A.Ha., A.Ho., A.I., A.M., B.S., C.S.F., E.M.v.L., F.R., G.L., G.M., G.Z., H.E., I.N., J.H., J.L., J.L.M., J.R.B.P., K.P., K.W., L.C., L.S., M.C., M.E.K., M.S., M.T., N.A., O.H.F., S.S., T.J., T.R.G., W.A., Y.M.; Analysed the data and provided critical interpretation of results: A.F., A.Ha., A.Ho., C.B., C.S.F., D.J., F.v.D., H.E., J.A.M., J.H., J.L.M., J.R.B.P., K.P., K.W., L.C., M.C., M.T.M., P.D., P.L.A., S.S., T.J., T.R.G., V.I., W.A., W.Z., Y.M.; Provided tools or materials: A.P.R., E.Z., F.v.D., G.D., M.T.M., N.J.T., N.S., P.D.; Wrote the manuscript: A.P.R., C.B., D.J., J.A.M., J.H., J.L.M., K.W., L.C., L.F., M.A.S., N.J.T., N.S., P.L.A., V.I.; Evaluated the manuscript: A.B., A.D., A.F., A.G.U., A.Ha., A.Ho., A.I., A.M., A.P.R., B.S., C.B., C.L., C.L.K., C.S.F., C.v.D., D.J., D.M., D.T., E.M.v.L., E.Z., F.R., F.v.D., G.D., G.G., G.L., G.M., G.Z., H.E., H.W., I.N., J.A.M., J.C.C., J.H., J.L., J.L.M., J.R.B.P., J.S.K., K.P., K.W., L.C., L.F., L.S., M.A.S., M.C., M.E.K., M.Fra., M.Fro., M.S., M.T., M.T.M., N.A., N.J.T., N.S., O.H.F., P.D., P.G., P.L.A., R.A.S., R.P., S.S., T.J., T.R.G., V.I., W.A., W.M., W.Z., Y.M.; Designed and or managed the project: A.P.R., N.J.T., N.S., P.L.A.
Competing Financial Interests
The authors have no competing financial interests to declare.
References
- 1.Cohen J, et al. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science (New York, NY) 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 2.Johansen CT, et al. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat Genet. 2010;42:684–7. doi: 10.1038/ng.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Auer PL, et al. Rare and low-frequency coding variants in CXCR2 and other genes are associated with hematological traits. Nat Genet. 2014;46:629–34. doi: 10.1038/ng.2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Willer CJ, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–83. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huyghe JR, et al. Exome array analysis identifies new loci and low-frequency variants influencing insulin processing and secretion. Nat Genet. 2013;45:197–201. doi: 10.1038/ng.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morris AP, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–90. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peloso GM, et al. Association of low-frequency and rare coding-sequence variants with blood lipids and coronary heart disease in 56,000 whites and blacks. Am J Hum Genet. 2014;94:223–32. doi: 10.1016/j.ajhg.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van der Harst P, et al. Seventy-five genetic loci influencing the human red blood cell. Nature. 2012;492:369–375. doi: 10.1038/nature11677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Auer PL, et al. Imputation of exome sequence variants into population-based samples and blood-cell-trait-associated loci in African Americans: NHLBI GO Exome Sequencing Project. Am J Hum Genet. 2012;91:794–808. doi: 10.1016/j.ajhg.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steinthorsdottir V, et al. Identification of low-frequency and rare sequence variants associated with elevated or reduced risk of type 2 diabetes. Nat Genet. 2014;46:294–8. doi: 10.1038/ng.2882. [DOI] [PubMed] [Google Scholar]
- 11.Surakka I, et al. The impact of low-frequency and rare variants on lipid levels. Nat Genet. 2015;47:589–97. doi: 10.1038/ng.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moayyeri A, Hammond CJ, Hart DJ, Spector TD. Effects of age on genetic influence on bone loss over 17 years in women: the Healthy Ageing Twin Study (HATS) J Bone Miner Res. 2012;27:2170–8. doi: 10.1002/jbmr.1659. [DOI] [PubMed] [Google Scholar]
- 13.Boyd A, et al. Cohort Profile: the 'children of the 90s'--the index offspring of the Avon Longitudinal Study of Parents and Children. Int J Epidemiol. 2013;42:111–27. doi: 10.1093/ije/dys064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walter K, et al. The UK10K project identifies rare variants in health and disease. Nature. 2015;526:82–90. doi: 10.1038/nature14962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Timpson NJ, et al. A rare variant in APOC3 is associated with plasma triglyceride and VLDL levels in Europeans. Nat Commun. 2014;5:4871. doi: 10.1038/ncomms5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taylor PN, et al. Whole-genome sequence-based analysis of thyroid function. Nat Commun. 2015;6:5681. doi: 10.1038/ncomms6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang J, et al. Improved imputation of low-frequency and rare variants using the UK10K haplotype reference panel. Nat Commun. 2015;6:8111. doi: 10.1038/ncomms9111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Genome of the Netherlands Consortium. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat Genet. 2014;46:818–25. doi: 10.1038/ng.3021. [DOI] [PubMed] [Google Scholar]
- 19.Do R, et al. Common variants associated with plasma triglycerides and risk for coronary artery disease. Nature Genetics. 2013:1–9. doi: 10.1038/ng.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okada Y, et al. Meta-analysis identifies multiple loci associated with kidney function-related traits in east Asian populations. Nature Genetics. 2012;44:904–909. doi: 10.1038/ng.2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyer TE, et al. Genome-wide association studies of serum magnesium, potassium, and sodium concentrations identify six Loci influencing serum magnesium levels. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Polfus LM, et al. Whole-Exome Sequencing Identifies Loci Associated with Blood Cell Traits and Reveals a Role for Alternative GFI1B Splice Variants in Human Hematopoiesis. Am J Hum Genet. 2016;99:481–8. doi: 10.1016/j.ajhg.2016.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Service SK, et al. Re-sequencing expands our understanding of the phenotypic impact of variants at GWAS loci. PLoS Genet. 2014;10:e1004147. doi: 10.1371/journal.pgen.1004147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maller JB, et al. Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat Genet. 2012;44:1294–301. doi: 10.1038/ng.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benner C, et al. FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics. 2016;32:1493–501. doi: 10.1093/bioinformatics/btw018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hormozdiari F, Kostem E, Kang EY, Pasaniuc B, Eskin E. Identifying causal variants at loci with multiple signals of association. Genetics. 2014;198:497–508. doi: 10.1534/genetics.114.167908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee D, et al. A method to predict the impact of regulatory variants from DNA sequence. Nat Genet. 2015;47:955–61. doi: 10.1038/ng.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maurano MT, et al. Large-scale identification of sequence variants influencing human transcription factor occupancy in vivo. Nat Genet. 2015;47:1393–401. doi: 10.1038/ng.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Douvris A, et al. Functional analysis of the TRIB1 associated locus linked to plasma triglycerides and coronary artery disease. J Am Heart Assoc. 2014;3:e000884. doi: 10.1161/JAHA.114.000884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iwamoto S, et al. The role of TRIB1 in lipid metabolism; from genetics to pathways. Biochem Soc Trans. 2015;43:1063–8. doi: 10.1042/BST20150094. [DOI] [PubMed] [Google Scholar]
- 31.Baerenwald DA, et al. Multiple functional polymorphisms in the G6PC2 gene contribute to the association with higher fasting plasma glucose levels. Diabetologia. 2013;56:1306–16. doi: 10.1007/s00125-013-2875-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duan Q, Liu EY, Croteau-Chonka DC, Mohlke KL, Li Y. A comprehensive SNP and indel imputability database. Bioinformatics. 2013;29:528–31. doi: 10.1093/bioinformatics/bts724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lappalainen T, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–11. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moroy T, Vassen L, Wilkes B, Khandanpour C. From cytopenia to leukemia: the role of Gfi1 and Gfi1b in blood formation. Blood. 2015;126:2561–9. doi: 10.1182/blood-2015-06-655043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laurent B, et al. A short Gfi-1B isoform controls erythroid differentiation by recruiting the LSD1-CoREST complex through the dimethylation of its SNAG domain. J Cell Sci. 2012;125:993–1002. doi: 10.1242/jcs.095877. [DOI] [PubMed] [Google Scholar]
- 36.Danjou F, et al. Genome-wide association analyses based on whole-genome sequencing in Sardinia provide insights into regulation of hemoglobin levels. Nat Genet. 2015;47:1264–71. doi: 10.1038/ng.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sankaran VG, et al. Cyclin D3 coordinates the cell cycle during differentiation to regulate erythrocyte size and number. Genes Dev. 2012;26:2075–87. doi: 10.1101/gad.197020.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ono Y, et al. Induction of functional platelets from mouse and human fibroblasts by p45NF-E2/Maf. Blood. 2012;120:3812–21. doi: 10.1182/blood-2012-02-413617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shavit JA, et al. Impaired megakaryopoiesis and behavioral defects in mafG-null mutant mice. Genes Dev. 1998;12:2164–74. doi: 10.1101/gad.12.14.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stevenson WS, et al. GFI1B mutation causes a bleeding disorder with abnormal platelet function. J Thromb Haemost. 2013;11:2039–47. doi: 10.1111/jth.12368. [DOI] [PubMed] [Google Scholar]
- 41.Monteferrario D, et al. A dominant-negative GFI1B mutation in the gray platelet syndrome. N Engl J Med. 2014;370:245–53. doi: 10.1056/NEJMoa1308130. [DOI] [PubMed] [Google Scholar]
- 42.Wiestner A, Schlemper RJ, van der Maas AP, Skoda RC. An activating splice donor mutation in the thrombopoietin gene causes hereditary thrombocythaemia. Nat Genet. 1998;18:49–52. doi: 10.1038/ng0198-49. [DOI] [PubMed] [Google Scholar]
- 43.Ghilardi N, Wiestner A, Kikuch M, Ohsaka A, Skoda RC. Hereditary thrombocythaemia in a Japanese family is caused by a novel point mutation in the thrombopoietin gene. Br J Haematol. 1999;107:310–6. doi: 10.1046/j.1365-2141.1999.01710.x. [DOI] [PubMed] [Google Scholar]
- 44.Kondo T, et al. Familial essential thrombocythemia associated with one-base deletion in the 5'-untranslated region of the thrombopoietin gene. Blood. 1998;92:1091–6. [PubMed] [Google Scholar]
- 45.Liu K, et al. A de novo splice donor mutation in the thrombopoietin gene causes hereditary thrombocythemia in a Polish family. Haematologica. 2008;93:706–14. doi: 10.3324/haematol.11801. [DOI] [PubMed] [Google Scholar]
- 46.Dasouki MJ, et al. Exome sequencing reveals a thrombopoietin ligand mutation in a Micronesian family with autosomal recessive aplastic anemia. Blood. 2013;122:3440–9. doi: 10.1182/blood-2012-12-473538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Giannakopoulos B, Krilis SA. The pathogenesis of the antiphospholipid syndrome. N Engl J Med. 2013;368:1033–44. doi: 10.1056/NEJMra1112830. [DOI] [PubMed] [Google Scholar]
- 48.De Groot PG, Meijers JC, Urbanus RT. Recent developments in our understanding of the antiphospholipid syndrome. Int J Lab Hematol. 2012;34:223–31. doi: 10.1111/j.1751-553X.2012.01414.x. [DOI] [PubMed] [Google Scholar]
- 49.Sanghera DK, Wagenknecht DR, McIntyre JA, Kamboh MI. Identification of structural mutations in the fifth domain of apolipoprotein H (beta 2-glycoprotein I) which affect phospholipid binding. Hum Mol Genet. 1997;6:311–6. doi: 10.1093/hmg/6.2.311. [DOI] [PubMed] [Google Scholar]
- 50.Korporaal SJ, et al. Binding of low density lipoprotein to platelet apolipoprotein E receptor 2' results in phosphorylation of p38MAPK. J Biol Chem. 2004;279:52526–34. doi: 10.1074/jbc.M407407200. [DOI] [PubMed] [Google Scholar]
- 51.Lutters BC, et al. Dimers of beta 2-glycoprotein I increase platelet deposition to collagen via interaction with phospholipids and the apolipoprotein E receptor 2'. J Biol Chem. 2003;278:33831–8. doi: 10.1074/jbc.M212655200. [DOI] [PubMed] [Google Scholar]
- 52.Adams D, et al. BLUEPRINT to decode the epigenetic signature written in blood. Nat Biotechnol. 2012;30:224–6. doi: 10.1038/nbt.2153. [DOI] [PubMed] [Google Scholar]
- 53.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–47. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khan A, Zhang X. dbSUPER: a database of super-enhancers in mouse and human genome. Nucleic Acids Res. 2016;44:D164–71. doi: 10.1093/nar/gkv1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu J, et al. Combinatorial assembly of developmental stage-specific enhancers controls gene expression programs during human erythropoiesis. Dev Cell. 2012;23:796–811. doi: 10.1016/j.devcel.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Consortium, E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Consortium, G.T. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–60. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roadmap Epigenomics, C et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kiryluk K, et al. Discovery of new risk loci for IgA nephropathy implicates genes involved in immunity against intestinal pathogens. Nat Genet. 2014;46:1187–96. doi: 10.1038/ng.3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Keller MF, et al. Trans-ethnic meta-analysis of white blood cell phenotypes. Hum Mol Genet. 2014;23:6944–60. doi: 10.1093/hmg/ddu401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vijai J, et al. A genome-wide association study of marginal zone lymphoma shows association to the HLA region. Nat Commun. 2015;6:5751. doi: 10.1038/ncomms6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gieger C, et al. New gene functions in megakaryopoiesis and platelet formation. Nature. 2011;480:201–8. doi: 10.1038/nature10659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Menicanin D, Bartold PM, Zannettino AC, Gronthos S. Identification of a common gene expression signature associated with immature clonal mesenchymal cell populations derived from bone marrow and dental tissues. Stem Cells Dev. 2010;19:1501–10. doi: 10.1089/scd.2009.0492. [DOI] [PubMed] [Google Scholar]
- 64.Konopatskaya O, et al. PKCalpha regulates platelet granule secretion and thrombus formation in mice. J Clin Invest. 2009;119:399–407. doi: 10.1172/JCI34665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Williams CM, Harper MT, Poole AW. PKCalpha negatively regulates in vitro proplatelet formation and in vivo platelet production in mice. Platelets. 2014;25:62–8. doi: 10.3109/09537104.2012.761686. [DOI] [PubMed] [Google Scholar]
- 66.Kong Y, Wang H, Lin T, Wang S. Sphingosine-1-phosphate/S1P receptors signaling modulates cell migration in human bone marrow-derived mesenchymal stem cells. Mediators Inflamm. 2014;2014:565369. doi: 10.1155/2014/565369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang L, et al. Sphingosine 1-Phosphate Receptor 2 and 3 Mediate Bone Marrow-Derived Monocyte/Macrophage Motility in Cholestatic Liver Injury in Mice. Sci Rep. 2015;5:13423. doi: 10.1038/srep13423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Westra HJ, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45:1238–43. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hildebrand JD. Shroom regulates epithelial cell shape via the apical positioning of an actomyosin network. J Cell Sci. 2005;118:5191–203. doi: 10.1242/jcs.02626. [DOI] [PubMed] [Google Scholar]
- 70.Menon MC, et al. Intronic locus determines SHROOM3 expression and potentiates renal allograft fibrosis. J Clin Invest. 2015;125:208–21. doi: 10.1172/JCI76902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Do R, et al. Common variants associated with plasma triglycerides and risk for coronary artery disease. Nat Genet. 2013;45:1345–52. doi: 10.1038/ng.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Teslovich TM, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–13. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grand FH, et al. p53-Binding protein 1 is fused to the platelet-derived growth factor receptor beta in a patient with a t(5;15)(q33;q22) and an imatinib-responsive eosinophilic myeloproliferative disorder. Cancer Res. 2004;64:7216–9. doi: 10.1158/0008-5472.CAN-04-2005. [DOI] [PubMed] [Google Scholar]
- 74.Caulfield MJ, et al. SLC2A9 is a high-capacity urate transporter in humans. PLoS Med. 2008;5:e197. doi: 10.1371/journal.pmed.0050197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kottgen A, et al. Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat Genet. 2013;45:145–54. doi: 10.1038/ng.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods. 2012;9:179–81. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 77.Danecek P, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–8. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–9. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Magi R, Morris AP. GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics. 2010;11:288. doi: 10.1186/1471-2105-11-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wakefield J. Bayes factors for genome-wide association studies: comparison with P-values. Genet Epidemiol. 2009;33:79–86. doi: 10.1002/gepi.20359. [DOI] [PubMed] [Google Scholar]
- 81.Chen W, et al. Fine Mapping Causal Variants with an Approximate Bayesian Method Using Marginal Test Statistics. Genetics. 2015;200:719–36. doi: 10.1534/genetics.115.176107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.