Abstract
Background
Our aim was to identify genetic variants associated with blood pressure (BP) in childhood and adolescence.
Methods and Results
Genome-wide association study data from participating European ancestry cohorts of the Early Genetics and Lifecourse Epidemiology (EAGLE) Consortium was meta-analyzed across 3 epochs; prepuberty (4–7 years), puberty (8–12 years), and postpuberty (13–20 years). Two novel loci were identified as having genome-wide associations with systolic BP across specific age epochs: rs1563894 (ITGA11, located in active H3K27Ac mark and transcription factor chromatin immunoprecipitation and 5′-C-phosphate-G-3′ methylation site) during prepuberty (P=2.86×10–8) and rs872256 during puberty (P=8.67×10–9). Several single-nucleotide polymorphism clusters were also associated with childhood BP at P<5×10–3. Using a P value threshold of <5×10–3, we found some overlap in variants across the different age epochs within our study and between several single-nucleotide polymorphisms in any of the 3 epochs and adult BP-related single-nucleotide polymorphisms.
Conclusions
Our results suggest that genetic determinants of BP act from childhood, develop over the lifecourse, and show some evidence of age-specific effects.
Keywords: blood pressure, children, genetic epidemiology, Genome-Wide Association Study, hypertension, prehypertension
Systolic and diastolic blood pressure (SBP and DBP) are complex phenotypes, with known environmental genetic risk factors.1 Elevated SBP and DBP are associated with premature mortality2,3 and cardiovascular diseases.2,4–9
Recent genome-wide association studies (GWAS) have identified genetic variants associated with adult SBP and DBP and hypertension.10–24 Several of these loci are in biologically plausible candidate genes, for example, those that influence the renin–angiotensin system.25 There are established patterns of age-related changes in BP in industrialized populations, which support a potential interaction of genetic variants with age-related changes to environmental exposures in these populations.26,27 For genetic variants that directly modulate childhood BP, effects might change with age, might differ between developmental periods (early life, childhood, and adolescence), and might also differ to those variants that act in adulthood.
Children and adolescents are rarely treated with antihypertensives, whereas from middle age onwards, an increasing proportion of adults are using such medications.26 Consequently, it is easier to examine genetic variants that are associated with untreated SBP and DBP in children. Variation in SBP, and to a lesser extent DBP, in adolescence and early adulthood is associated with subsequent adult risk of coronary heart disease and stroke.28,29 Understanding the risk factors, including genetic variation, that are associated with SBP and DBP through childhood and into adolescence may therefore inform an improved understanding of the life course pathogenesis of adult hypertension and cardiovascular disease. GWAS of BP in children to date have been limited to investigations of cross-sectional BP in individual cohorts,30 often in non-European populations.31,32
The principal aim of the current study was to identify age-specific genetic associations with SBP and DBP across childhood and adolescence. The secondary aim was to compare results from this GWAS to published results from adult GWAS of BP.
Methods
EAGLE Consortium: BP Working Group
Seven pregnancy/birth or childhood cohorts of European ancestry from the Early Genetics and Lifecourse Epidemiology (EAGLE) Consortium BP working group with completed genome-wide genotyping and measures of SBP and DBP on participants between the ages of 4 and 20 years contributed to this study.
Each study was conducted with appropriate institutional ethics approval, and written informed consent was obtained from participants or their main caregivers. Each study conducted their own GWAS analyses, and results were then pooled using meta-analysis. The methodology for recruitment of participants and measurement of BP for each cohort are described in the Data Supplement.
Only those participants who had at least one BP measurement at, or before, 20 years of age were included in analyses. With the exception of the Brisbane Longitudinal Twin Study (BLTS), all analyses were restricted to children from singleton pregnancies, the first born (with relevant genotype and phenotype data) from cohorts that included siblings, or a randomly selected child from those that included twins. BLTS, which included all twins and siblings, took into account both zygosity and relatedness using the statistical package Merlin33 during their GWAS.
ICBP
We used GWAS data from studies contributing to the International Consortium of Blood Pressure (ICBP) on adults who were recruited for classical or genetic epidemiological purposes from general population samples17 to compare with associations identified in EAGLE.
Genotyping and Imputation
Single nucleotide polymorphisms (SNPs) were genotyped on one of the following platforms: Affymetrix, Illumina, or Perlegen. Predefined marker filters were applied before imputation (Hardy–Weinberg equilibrium >10–6, MAF >0.01, SNP call rate >95%). Each study imputed SNPs by combining their study’s genotyped SNPs with HapMap Phase II CEU SNPs samples, preferably using release 22 of HapMap, build 36. Imputation results are summarized as an allele dosage defined as the expected number of copies of the minor allele at that SNP (a fractional value between 0.0 and 2.0) for each genotype. Further details are provided in Data Supplement and Table 1.
Table 1. List of Participating Cohorts With GWAS and SBP and DBP Data Available in Children.
| Cohort | Time Frames, epochs | ||
|---|---|---|---|
| Prepubertal, (4–7 y) | Pubertal, (8–12 y) | Postpubertal, (13–20 y) | |
| Raine | 1276 | 1251 | 1009 |
| GenerationR | 1847 | ||
| ALSPAC | 5967 | 5750 | 4050 |
| Lisa Plus | 282 | ||
| YFS | 400 | 842 | |
| INMA | 600 | ||
| BLTS | 298 | 117 | |
| Total | 10 090 | 8423 | 5176 |
Number of subjects per cohort per time frame used in analyses. ALSPAC indicates The Avon Longitudinal Study of Parents and Children Bristol, UK; BLTS, Brisbane Longitudinal Twin Study, Brisbane, Queensland, Australia; GenerationR, The Generation R Study Group, Rotterdam, The Netherlands; INMA, Spanish INMA—Infancia y Medio Ambiente, Barcelona, Catalonia, Spain; Lisa Plus, Influence of lifestyle factors on the development of the immune system and allergies in East and West Germany Plus the influence of traffic emissions and genetics, Neuherberg, Germany; Raine, The Western Australian Pregnancy (Raine) Cohort, Perth, Western Australia; YFS; The Cardiovascular Risk in Young Finns Study, Turku, Finland.
Statistical Analysis
Cross-Sectional GWAS
Cross-sectional associations were performed for SBP and DBP in 3 age epochs over childhood/adolescence. These epochs were chosen before any analyses and were based on previous evidence of the ages in European populations that girls go through mencarche, boy’s voices break, and both sexes have a growth spurt and in part pragmatically, relating to available data in the participating studies. These epochs were the following: prepuberty (4 to ≤7 years), puberty (8 to ≤12 years), and postpuberty (13 to ≤20 years; Table 1 and Figure I in the Data Supplement).
Genetic analyses assumed an additive model for SNP-based main effects. All analyses were adjusted for age, height, and weight, consistent with analyses used in GWAS of BP in adults (adjusted for body mass index).14,18,20,24,34 The Z-score was calculated per individual as the difference between an individual’s BP measure and the average BP measure for that given age epoch and sex for each cohort. This was then divided by the standard deviation of the BP measure across the particular cohorts’ age, epoch, and sex. Those Z-scores were then regressed against each SNP (additive) adjusting for age, sex, and height of the individual. Participants could contribute to >1 epoch but only one measurement time point contributed to each epoch. The median measurement time was selected in the event that a cohort had repeatedly measured data within a single epoch.
Meta-Analyses
Separate genome-wide meta-analyses were run for SBP and DBP for each epoch using the inverse-variance weighting method in Meta-Analysis (software for GWAS) (www.sph.umich.edu/csg/abecasis/metal).38 Before meta-analysis, rare variants were excluded (MAF>0.01; Table I in the Data Supplement). Double genomic control correction was applied (once during the study-specific analyses [before the meta-analysis] and repeated on the statistics resulting from the meta-analysis). Heterogeneity between results from individual studies was assessed using I2 and a Q-statistic. Further filtering based on N-effectives >70% was also used. QQ-plots for the meta-analysis of each BP outcome across each epoch and data set are presented in Figure II in the Data Supplement. A threshold of P≤5×10−8 was used to define genome-wide levels of significance. An association of an SNP cluster was defined as ≥2 nearby variants each reaching a threshold of P≤5×10−3. This more liberal statistical significance level (P≤5×10−3) was used to ensure the inclusion of variants that could potentially have been identified in larger samples of children.
Exploring Functionality
We used the Encyclopedia of DNA Elements35 to investigate the functionality (transcriptional active region, H3K27Ac active regulatory elements, DNase hypersensitivity transcription factor–binding site information from chromatin immunoprecipitation (ChIP) analysis, and 5′-C-phosphate-G-3′ (CpG) methylation levels) of identified BP-related SNPs.
Comparing Findings Across Epochs and in Adults
Any variants identified as associated with SBP or DBP at P≤5×10−3 in any age epoch was examined in (1) the other 2 epochs in our study; (2) in adult participants from the ICBP, and (3) in SNPs identified as associated with adult CHD using the results from Coronary Artery Disease Genome-Wide Replication and Meta-Analysis (CARDIoGRAMplusC4D),36 a consortium designed to identify novel susceptibility loci for coronary artery disease and myocardial infarction.37
We also examined whether variants associated with adult BP in ICBP at P≤5 x 10−3 for association with BP in any of the 3 epochs in our study.
Meta-analyses were conducted in Meta-Analysis (software for GWAS). The statistical software package R version 2.12.139 was used to produce all Manhattan Plots. Regional association plots were produced using LocusZoom.40
Results
Seven cohorts contributed to meta-analyses, a total of 10 090 individuals for the prepubertal epoch, 8423 individuals for the pubertal epoch, and 5176 individuals for the postpubertal epoch (Table 1). Manhattan plots for SBP and DBP by epoch are presented in Figure III in the Data Supplement. Regional association plots for the most significant SNP (by outcome and epoch, as listed in Table 2) are shown in Figure IV in the Data Supplement. Regional association plots for SNP clusters are presented in Figure V in the Data Supplement (SBP) and Figure VI in the Data Supplement (DBP). Regional association plots and linkage disequilibrium plots for significant SNP clusters for SBP and DBP are presented by epoch in Figures VII to IX in the Data Supplement. Forest plots for all significant SNP clusters and most significant association by outcome and epoch are presented in Figures X to XV in the Data Supplement. Overview figures (forest plot, regional association plot, and Manhattan plot) are presented by epoch and outcome in Figures XVI to XVIII in the Data Supplement. Plots relating to the Encyclopedia of DNA Elements functionality are displayed in Figures XIX to XXII in the Data Supplement. Descriptive characteristics of each cohort are detailed in Table II in the Data Supplement. Summary of known BP (and BP-related effects) for significant genes and variants associated from EAGLE are shown in Tables III and IV in the Data Supplement and ICBP in Tables V and VI in the Data Supplement. Comparison of EAGLE meta-analysis results with CARDIOGRAM are in given in Table VII in the Data Supplement. Top SNP clusters are summarized in Tables VIII and IX in the Data Supplement. Comparisons between EAGLE and ICBP are given in Tables X and XI in the Data Supplement. Comparing significant SNPs from sex-stratificant meta-analyses to single cohort analyses with sex by SNP interactions are described in Table XII in the Data Supplement. Comparing meta-analyses across epochs for EAGLE are detailed in Tables XIII to XIV in the Data Supplement. Summary of known BP (and BP-related effects) for significant genes and variants associated with multiple epochs are listed in Tables XV and XVI in the Data Supplement.
Table 2. Most Significant Findings per Time Frame, Data Set, and BP Outcome Measure (Sex-Combined).
| Time Frame | SBP | DBP | ||
|---|---|---|---|---|
| SNPs <5×10−8 | Most Significant Finding | SNPs <5×10−8 | Most Significant Finding | |
| Prepuberty | 1 | rs1563894 | 0 | rs13040824 |
| Beta=−0.093 | Beta=−0.902 | |||
| 95% CI=[−0.126, −0.060] | 95% CI=[−0.127, −0.054] | |||
| P=2.86×10−8 | P=9.33×10−7 | |||
| Chromosome: 15 | Chromosome: 20 | |||
| Gene: ITGA11 | Gene: Unknown | |||
| Nearby genes: FEM1B, CORO2B, CALML4 | Nearby genes: None | |||
| MAF=0.19 | MAF=0.30 | |||
| R̂2 range =[0.67, 0.96] | R̂2 range =[0.49, 0.99] | |||
| Puberty | 1 | rs872256 | 0 | rs7897969 |
| Beta=0.096 | Beta=−0.749 | |||
| 95% CI=[0.063,0.129] | 95% CI=[−1.001, −0.496] | |||
| P=8.67×10−9 | P=4.82×10−7 | |||
| Chromosome: 9 | Chromosome: 10 | |||
| Gene: Unknown | Gene: Unknown | |||
| Nearby genes: SMARCA2, VLDLR | Nearby genes: None | |||
| MAF=0.41 | MAF=0.15 | |||
| R̂2 range =[0.94, 0.95] | R̂2 range =[0.09, 0.48] | |||
| Postpuberty | 0 | rs1010366 | 0 | rs12365302 |
| Beta=0.098 | Beta=0.139 | |||
| 95% CI=[0.057,0.139] | 95% CI=[0.086,0.192] | |||
| P=3.31×10−6 | P=3.96×10−7 | |||
| Chromosome: 7 | Chromosome:11 | |||
| Gene: C1GALT1 | Gene: Unknown | |||
| Nearby genes: None | Nearby genes: SLC35C1, CHST1 | |||
| MAF=0.39 | MAF=0.17 | |||
| R̂2 range =[0.97, 0.99] | R̂2 range =[0.97, 0.98] | |||
CI indicates confidence interval; DBP, diastolic blood pressure; and SBP, systolic blood pressure.
The sum of the number of SNPs reaching genome-wide levels of significance (P≤5×10−8) where all SNPs had an MAF>0.10 and all cohorts contributed to each SNP analyses (postpubertal) and at least 4 cohorts contributed to GWAS findings for the prepubertal and pubertal epochs. Beta values are in terms of Z-scores, the number of standard deviations away from the mean.
Systolic Blood Pressure
Genome-Wide Significance
Two SNPs reached genome-wide levels of significance (P<5×10−8) for childhood SBP (Table 2): in the prepubertal epoch, rs1563894 (chromosome position, 15.66422829; gene, ITGA11; per allele mean difference in SBP =−0.093SD [95% CI: −0.126, −0.060]; P=2.86×10−8; Tables 2 and 3 and Figures VI and XV in the Data Supplement); and during the pubertal epoch, rs872256 (chromosome position, 9.2496480; gene, unknown; β=0.096SD [95% CI: 0.063, 0.129]; P=8.67×10−9; Table 2). No SNPs reached genome-wide levels of significance for SBP in the postpubertal analyses.
Table 3. Top SNP Clusters From Systolic Blood Pressure and Diastolic Blood Pressure Meta-Analyses in EAGLE for Sex-Combined Data Set.
| Time Frame | Marker Name | Allele | CHR | POS | MAF | Gene | Beta (95% CI) | P Value | Direction | ICBP p | N Effective |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Systolic blood pressure | |||||||||||
| Prepubertal | rs1563894 | A/G | 15 | 66422829 | 0.19 | ITGA11 | −0.093 (−0.126, −0.060) | 2.86E-08* | −−−−− | 0.46 | 10 090 |
| Pubertal | rs3735398 | A/G | 7 | 36412646 | 0.12 | ANLN | 0.116 (0.073,0.159) | 1.28E-07 | +++++ | 0.75 | 8423 |
| rs3787159 | T/C | 20 | 56252573 | 0.46 | PPP4R1L | −0.064 (−0.091, −0.037) | 8.76E-06 | −−+−− | 0.46 | 8423 | |
| rs9667878 | T/C | 11 | 5180326 | 0.22 | OR51V1 | 0.135 (0.0740,0.196) | 9.74E-06 | +++++ | 1.00 | 8423 | |
| Postpubertal | rs1010366 | T/C | 7 | 7196351 | 0.39 | C1GALT1 | 0.098 (0.057,0.139) | 3.31E-06 | −++ | 0.90 | 5176 |
| rs4538187 | A/G | 2 | 63957245 | 0.16 | UGP2 | 0.102 (0.057,0.147) | 5.96E-06 | +++ | 0.70 | 5176 | |
| rs3901287 | A/T | 8 | 23240509 | 0.28 | LOXL2 | 0.108 (0.059,0.157) | 1.24E-05 | +++ | 0.68 | 5176 | |
| Diastolic blood pressure | |||||||||||
| Prepubertal | rs241264 | T/C | 1 | 4518898 | 0.31 | Between LOC284661 and AJAP1 | −0.082 (−0.0120, −0.043) | 2.95E-05 | −−−−− | 0.48 | 10 090 |
| rs1714524 | T/C | 3 | 159755790 | 0.44 | LOC100996447 | 0.154 (0.081,0.227) | 8.32E-05 | −−−−+ | 0.99 | 10 090 | |
| rs16875222 | A/T | 8 | 107955966 | 0.11 | Near ABRA, OXR1 | −0.119 (−0.179, −0.060) | 8.21E-05 | −−−−− | 0.72 | 8423 | |
| rs12237240 | T/G | 9 | 28329306 | 0.19 | LINGO2 | 0.068 (0.034,0.102) | 8.71E-05 | ++−+− | 0.89 | 8423 | |
| rs1387977 | T/G | 12 | 71307607 | 0.14 | TRHDE | 0.127 (0.064,0.190) | 6.92E-05 | ++−++ | 0.90 | 8423 | |
| Postpubertal | rs6949619 | T/C | 7 | 24396900 | 0.19 | Near NPY | −0.092 (−0.133, −0.051) | 1.13E-05 | −−− | 0.45 | 5176 |
| rs229038 | C/G | 21 | 27127300 | 0.22 | ADAMTS1 | 0.213 (0.115,0.311) | 2.26E-05 | +++ | 0.77 | 5176 | |
Bold text highlights SNPs (P<5×10−3) represented in regional association plots shown in Figures VII–IX in the Data Supplement. Beta values are in terms of Z-scores, the number of standard deviations away from the mean. CHR indicates chromosome; MAF, minor allele frequency; and POS, position.
SNPs which have reached genome-wide levels of significance (P<5×10−8).
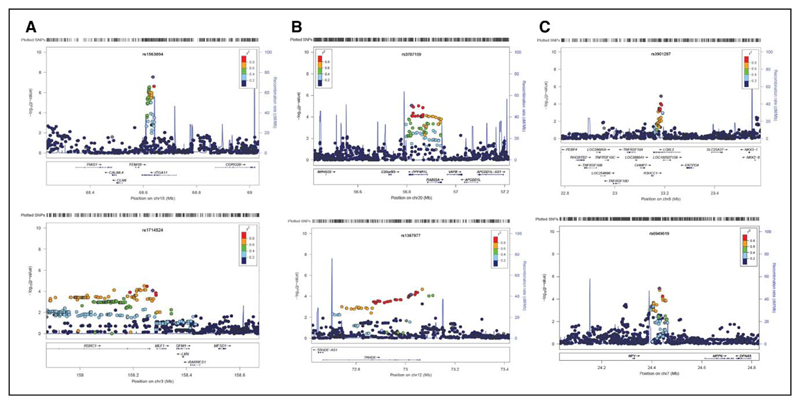
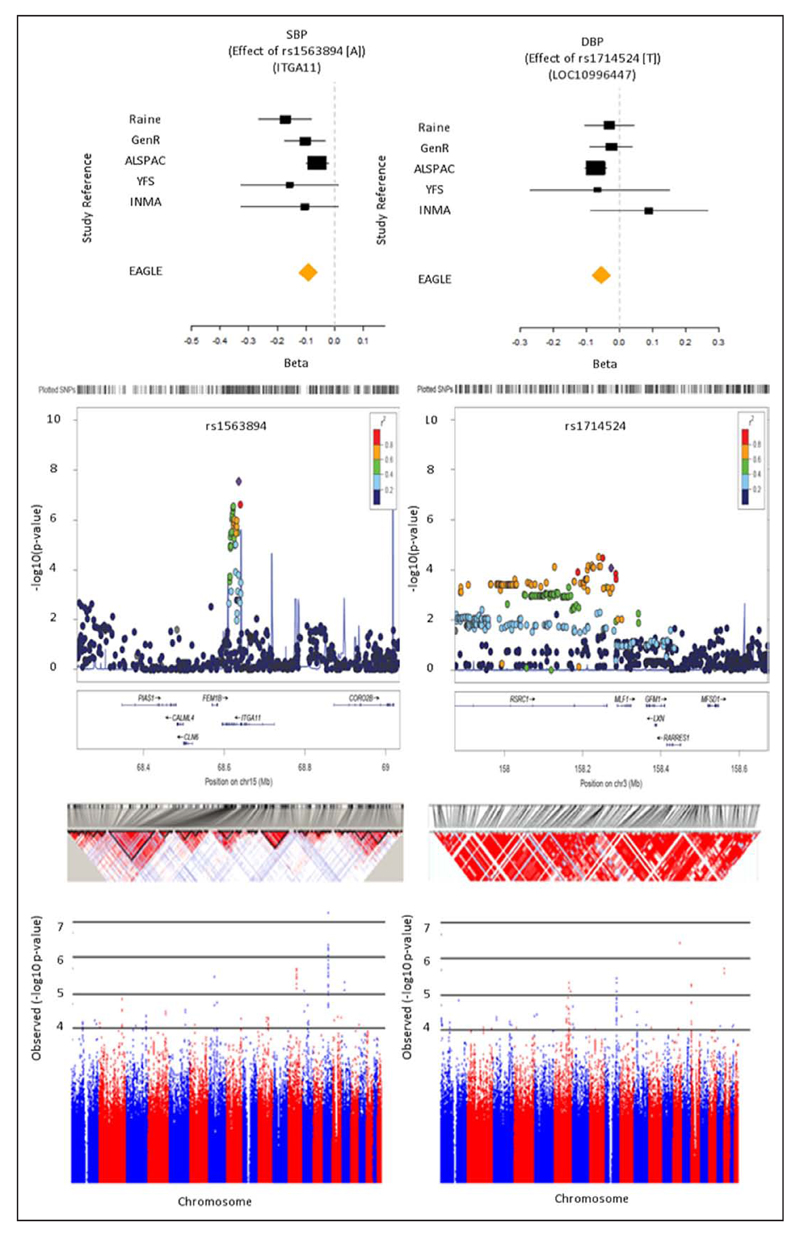
Clusters of variants in several genes were associated with SBP at P≤5×10−3 (Table 3, Figure 1, and Figure V in the Data Supplement): ITGA11 (prepuberty), ANLN and OR51V1 (puberty), and C1GALT1 and UGP2 (postpuberty). A cluster of SNPs was also identified surrounding SNP rs1563894 (ITGA11; prepuberty), highlighted above as showing genome-wide levels of significance with prepubertal SBP. We illustrate this significant SNP cluster in more depth (Figure 2).
Figure 1.
Regional association plots for significant SNP clusters –log10 (P values) are shown for all SNPs in the region, and color of circles indicates degree of LD with the most associated SNP in the region. SNPs correspond to those highlighted in bold text in Table 3. Plots shown on left represent systolic blood pressure, plots shown on right represent diastolic blood pressure. Rows represent prepubertal (A), pubertal (B), and postpubertal (C). For regional association, plots for all significant SNP clusters presented in Table 4 refer to Figures VII and VIII in the Data Supplement. LD indicates linkage disequilibrium.
Figure 2.
Most significant SNP clusters resulting from the sex-combined meta-analyses genome-wide association study (GWAS). From top to bottom: forest plot, regional association plot (–log10 (P values)), linkage disequilibrium plots (LOD scores), and Manhattan plots (–log10 (P values)). Green boxed areas on Manhattan plots highlight chromosome regions illustrated in regional association plots. Letters signify the following data subsets: SBP (A) and DBP (B) for the prepubertal epoch. ALSPAC indicates The Avon Longitudinal Study of Parents and Children Bristol, UK; EAGLE< Early Genetics and Lifecourse Epidemiology; GenR, The Generation R Study Group, Rotterdam, The Netherlands; ICBP, International Consortium of Blood Pressure; INMA, Spanish INMA—Infancia y Medio Ambiente, Barcelona, Catalonia, Spain; Raine, The Western Australian Pregnancy (Raine) Cohort, Perth, Western Australia; YFS; The Cardiovascular Risk in Young Finns Study, Turku, Finland.
Two SNPs were associated with the largest increases in SBP across these epochs: SNP rs3735398 (ANLN) during the pubertal epoch (β=0.116SD, 95% CI, 0.073, 0.159; P=1.28×10−7; Table 3 and Figure VIII in the Data Supplement) and a cluster surrounding SNP rs3901287 (LOXL2; β=0.108SD, 95% CI, 0.059, 0.157; P=1.24×10−5; Table 3 and Figure IX in the Data Supplement) was associated with postpubertal SBP.
Look-Up of Functionality of Childhood SBP Results
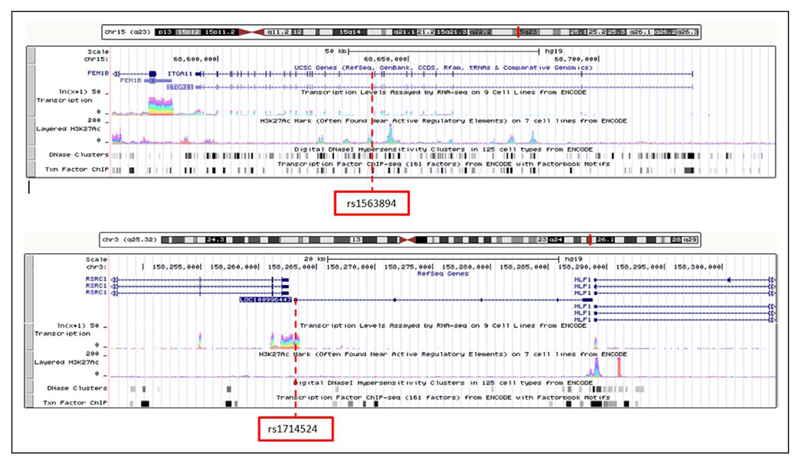
Several SNPs were found to be located either directly in or in close proximity to functionally active regions. For SNPs associated with SBP, rs1563894 was located on an active H3K27Ac, DNase hypersensitivity, transcription factor ChIP, and CpG methylation site (Figure 3 and Figure XIXa in the Data Supplement). Rs1010366 was located downstream and in close proximity to densely active H3K27Ac, DNase hypersensitivity, transcription factor ChIP, and CpG methylation site (Figure XIXc in the Data Supplement). Rs3735398 was located in a region of active transcription and DNase hypersensitivity. Rs3787159 was located directly upstream from areas with active transcription DNase hypersensitivity and transcription factor ChIP (Figure XXIb in the Data Supplement). Rs4538187 was located in a region of transcription factor ChIP and methylation and upstream from an active transcription site and downstream from an active H3K27Ac mark with dense methylation and transcription factor active H3K27Ac mark, DNase hypersensitivity, transcription factor ChIP, and CpG methylation site, whereas rs3901287 was located in close proximity to areas of active transcription, H3K27Ac mark, DNase hypersensitivity, transcription factor ChIP, and methylation (Figure XXIc in the Data Supplement).
Figure 3.
An overview of the Encyclopedia of DNA Elements functional activity (transcriptional active region, H3K27Ac active regulatory elements, DNase hypersensitivity transcription factor–binding site information from chromatin immunoprecipitation [ChIP] analysis, and CpG methylation levels) for rs1563894 (A; SBP) and rs1714524 (B; DBP) during the prepubertal epoch.
Comparisons Across Epochs and With Adult Outcomes
No SNPs reached associations of P≤5×10−3 in all 3 epochs. Variants in chromosome 2 (rs13025174, rs13032473 [TMEM247], and rs10186089 [FSHR]) were associated with elevated SBP during prepubertal and pubertal epochs (Table 4). Several SNPs in HORMAD2, HORMAD2-AS1, and MTMR3 were associated with SBP during the pubertal and postpubertal epochs (Table 4 and Table XIII in the Data Supplement).
Table 4. Comparing SBP and DBP Meta-Analyses Across Epochs in EAGLE.
| Marker Name | CHR | POS | Gene | MAF | Allele | Prepubertal | Pubertal | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta (95% CI) | P Value | Direction | Beta (95% CI) | P Value | Direction | ||||||
| Systolic blood pressure | |||||||||||
| rs13025174 | 2 | 46557999 | 0.56 | A/G | 0.075 (0.031,0.12) | 9.37E-04 | ++++ | 0.054 (0.026,0.082) | 1.38E-04 | ++−++ | |
| rs13032473 | 2 | 46559169 | TMEM247 | 0.56 | A/G | 0.076 (0.031,0.121) | 8.60E-04 | ++++ | 0.054 (0.026,0.082) | 1.38E-04 | ++−++ |
| rs10186089 | 2 | 49072628 | FSHR | 0.83 | A/G | 0.123 (0.056,0.190) | 3.07E-04 | ++++ | 0.073 (0.031,0.114) | 5.45E-04 | ++−−+ |
| Pubertal | Postpubertal | ||||||||||
| rs1383450 | 8 | 56167423 | 0.87 | T/C | 0.073 (0.033,0.112) | 2.85E-04 | +++++ | 0.115 (0.063,0.167) | 1.47E-05 | ?++ | |
| rs276978 | 16 | 84783954 | 0.30 | T/C | 0.073 (0.030,0.117) | 9.88E-04 | −++−+ | 0.098 (0.043,0.153) | 5.26E-04 | +++ | |
| rs2239382 | 20 | 9449526 | LAMP5 | 0.25 | C/G | −0.069 (−0.108, −0.030) | 5.62E-04 | −−+−− | −0.083 (−0.131, −0.035) | 7.33E-04 | −−−− |
| rs5752974 | 22 | 28601630 | 0.96 | A/G | −0.088 (−0.137, −0.040) | 3.74E-04 | −−++− | −0.115 (−0.175,-0.055) | 1.79E-04 | −−− | |
| rs16988143 | 22 | 28704677 | MTMR3 | 0.04 | T/C | 0.09 (0.041,0.138) | 3.05E-04 | ++−−+ | 0.118 (0.057,0.179) | 1.61E-04 | +++ |
| rs11552852 | 22 | 28754377 | HORMAD2-AS1, MTMR3 | 0.98 | T/C | −0.093 (−0.142, −0.043) | 2.68E-04 | −−++− | −0.123 (−0.185, −0.062) | 7.87E-05 | −−− |
| rs16988244 | 22 | 28763373 | HORMAD2-AS1 | 0.04 | T/G | 0.091 (0.042,0.140) | 2.46E-04 | ++−−+ | 0.115 (0.055,0.175) | 1.71E-04 | +++ |
| rs16988333 | 22 | 28882813 | HORMAD2 | 0.04 | A/G | 0.090 (0.041,0.139) | 2.84E-04 | ++−−+ | 0.110 (0.050, 0.170) | 3.12E-04 | +++ |
| rs5753042 | 22 | 28917972 | HORMAD2, LOC105372988 | 0.06 | A/G | 0.088 (0.039,0.137) | 3.92E-04 | ++−−+ | 0.105 (0.044,0.167) | 7.64E-04 | +++ |
| Diastolic blood pressure | |||||||||||
| rs13004438 | 2 | 179421913 | CCDC141, LOC105373766 | 0.17 | T/C | 0.082 (0.034,0.13) | 8.31E-04 | +++ | −0.053 (−0.084, −0.022) | 8.10E-04 | −−−−− |
| Prepubertal | Postpubertal | ||||||||||
| rs12542146 | 8 | 10485173 | 0.23 | A/G | 0.100 (0.041,0.159) | 9.29E-04 | ++? | −0.073 (−0.115, −0.030) | 8.42E-04 | −−− | |
| rs9373002 | 6 | 132279972 | 0.28 | A/G | 0.101 (0.042,0.159) | 7.36E-04 | ++? | 0.072 (0.029,0.114) | 8.95E-04 | +++ | |
| Pubertal | Postpubertal | ||||||||||
| rs2944782 | 2 | 47750478 | 0.21 | A/G | 0.075 (0.033,0.117) | 4.35E-04 | +++++ | 0.090 (0.041,0.140) | 3.31E-04 | +++ | |
| rs12532038 | 7 | 124281701 | POT1 | 0.39 | T/C | -0.059 (−0.090,-0.028) | 2.06E-04 | —-+ | −0.067 (−0.106, −0.028) | 8.25E-04 | −−+ |
| rs734335 | 14 | 100648711 | 0.40 | A/G | −0.092 (−0.136, −0.047) | 5.23E-05 | −−−−− | −0.110 (−0.165, −0.055) | 9.20E-05 | −−− | |
| rs8044400 | 16 | 74923996 | CNTNAP4 | 0.84 | T/C | −0.116 (−0.164, −0.068) | 2.10E-06 | −−−−+ | −0.099 (−0.157, −0.040) | 9.17E-04 | −−− |
Beta values are in terms of Z-scores, the number of standard deviations away from the mean. CI indicates confidence interval; DBP, diastolic blood pressure; and SBP, systolic blood pressure.
None of the SNPs found to be associated (P≤5×10−3) with childhood SBP were associated with adult SBP in the ICBP (Table 3). However, several loci previously found to be associated with adult SBP in ICBP were also associated with SBP in at least one epoch of childhood in EAGLE children (Table 5): (1) B3GNTL1 and KLHL1 (prepuberty); (2) DOK6, NKAIN2, ARGHGEF10, and MECOM (puberty) and (2) FGD5 and CSMD2 (postpuberty). One SNP that was associated with SBP in the pubertal epoch showed some possible association with CHD in adults (Table VII in the Data Supplement; rs3735398; P=0.039).
Table 5. Comparing Systolic and Diastolic Blood Pressure Meta-Analyses in Childhood (EAGLE) and Adulthood (ICBP) for Sex-Combined Data Sets.
| Time Frame | Marker Name | CHR | POS | Gene | MAF | ICBP | EAGLE | ||
|---|---|---|---|---|---|---|---|---|---|
| Beta | P Value | Beta (95%CI) | P Value | ||||||
| Systolic blood pressure | |||||||||
| Prepubertal | rs4986146 | 17 | 78534809 | B3GNTL1 | 0.3 | 0.422 (0.157,0.686) | 1.77E-03 | −0.055 (−0.086, −0.024) | 5.98E-04 |
| rs17810777 | 13 | 69385636 | KLHL1 | 0.11 | 0.377 (0.127,0.628) | 3.17E-03 | 0.066 (0.031,0.101) | 2.05E-04 | |
| Pubertal | rs8096788 | 18 | 65451861 | DOK6 | 0.31 | 0.65 (0.299,1.001) | 2.80E-04 | −0.095 (−0.146, −0.044) | 2.61E-04 |
| rs38627 | 7 | 76275543 | 0.27 | 0.495 (0.209,0.782) | 6.89E-04 | 0.077 (0.034,0.120) | 3.72E-04 | ||
| rs4943826 | 11 | 80730697 | 0.23 | 0.459 (0.163,0.755) | 2.36E-03 | 0.075 (0.032,0.118) | 5.65E-04 | ||
| rs332607 | 6 | 124765053 | NKAIN2 | 0.25 | 0.317 (0.106,0.528) | 3.19E-03 | 0.054 (0.025,0.083) | 3.55E-04 | |
| rs3824137 | 8 | 1808899 | ARHGEF10 | 0.23 | 0.358 (0.115,0.601) | 3.87E-03 | 0.058 (0.023,0.093) | 9.13E-04 | |
| rs11711274 | 3 | 170597773 | MECOM | 0.01 | 1.404 (0.437,2.371) | 4.42E-03 | −0.400 (−0.606, −0.194) | 1.40E-04 | |
| rs17033041 | 4 | 156610757 | 0.14 | 0.375 (0.116,0.633) | 4.47E-03 | −0.066 (−0.105, −0.027) | 9.66E-04 | ||
| Postpubertal | rs293927 | 3 | 14907160 | FGD5 | 0.13 | 0.422 (0.183,0.661) | 5.42E-04 | −0.082 (−0.125, −0.039) | 2.06E-04 |
| rs1687304 | 3 | 14929257 | FGD5 | 0.13 | 0.404 (0.164,0.645) | 9.75E-04 | 0.089 (0.046,0.132) | 6.21E-05 | |
| rs625757 | 1 | 33922472 | CSMD2 | 0.07 | 0.601 (0.261,0.942) | 5.43E-04 | −0.104 (−0.163, −0.045) | 4.93E-04 | |
| Diastolic blood pressure | |||||||||
| Prepubertal | rs6760458 | 2 | 42938973 | 0.23 | 0.195 (0.06,0.329) | 4.63E-03 | 0.055 (0.022,0.088) | 8.67E-04 | |
| Pubertal | rs7578149 | 2 | 20175919 | 0.46 | 0.213 (0.087,0.339) | 9.08E-04 | −0.050 (−0.079, −0.021) | 9.83E-04 | |
| rs11713251 | 3 | 49315011 | USP4 | 0.01 | 0.81 (0.292,1.327) | 2.19E-03 | 0.213 (0.093,0.333) | 5.03E-04 | |
| rs7914808 | 10 | 120991173 | GRK5 | 0.31 | 0.344 (0.108,0.579) | 4.20E-03 | 0.091 (0.038,0.144) | 9.07E-04 | |
| rs1951930 | 6 | 33890633 | 0.15 | 0.218 (0.059,0.376) | 7.15E-03 | 0.066 (0.027,0.105) | 7.49E-04 | ||
| Postpubertal | rs4650447 | 1 | 80256759 | 0.41 | 0.205 (0.081,0.328) | 1.18E-03 | −0.063 (−0.100, −0.026) | 9.63E-04 | |
| rs17064088 | 5 | 174293917 | 0.05 | 0.415 (0.142,0.689) | 2.90E-03 | 0.122 (0.051,0.193) | 6.71E-04 | ||
Please note that the International Consortium of Blood Pressure (ICBP) released their GWAS results here: http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000585.v1.p1; however, the effects reported (beta) are absolute values of the regression coefficient. Beta values are in terms of Z-scores, the number of standard deviations away from the mean. EAGLE, Early Genetics and Lifecourse Epidemiology; and ICBP, International Consortium of Blood Pressure.
Diastolic Blood Pressure
Genome-Wide Significance
No SNPs reached genome wide levels of significance (P<5×10−8) for childhood DBP (Table 2).
Clusters of variants in 7 loci were associated with childhood DBP (P≤5×10−3; Table 3) and were investigated further using LocusZoom.40 Regional associations for all of these associations are presented in Figure VI in the Data Supplement. SNPs associated with the largest positive increases in DBP per epoch included a prepubertal epoch cluster led by rs1714524 (LOC100996447; β=0.154SD, 95% CI, 0.081, 0.227; P=8.32×10−5; Figure 2 and Figure VIa in the Data Supplement). For the pubertal epoch, a cluster led by rs1387977 (TRHDE; β=0.127SD, 95% CI, 0.064, 0.190; P=6.92×10−5; Figure VIb in the Data Supplement). For the postpubertal epoch, a cluster led by rs6949619 (gene unknown, near NPY; β=−0.092SD, 95% CI, −0.133, −0.051; P=1.13×10−5; Figure VIc in the Data Supplement).
Look-Up of Functionality of Childhood DBP Results
For SNPs associated with DBP, rs13040824 was located downstream from an active H3K27Ac mark and located between 2 regions of DNase hypersensitivity and transcription factor ChIP (Figure XXa in the Data Supplement). Rs7897969 was located between 2 regions that had high levels of DNase hypersensitivity, transcription factor ChIP, and methylation (Figure XXb in the Data Supplement). Rs1236530 was located in a region of DNase hypersensitivity and transcription factor ChIP (Figure XXc in the Data Supplement). Rs241264 was located between 2 areas of high methylation, whereas rs1714524 was located in an area of active transcription (Figure 3 and Figure XXIIa in the Data Supplement). Rs1687522 was located in an area of active transcription and methylation (Figure XXIIb in the Data Supplement). Rs229038 was located within a cluster of DNase hypersensitivity and upstream for a highly active region of transcription and regulation (Figure XXIIc in the Data Supplement).
Look-Up of DBP Results Across Epochs in Childhood/Adolescence
No SNPs were significantly associated (P≤5×10−3) with DBP across all epochs. Opposing effects were observed for rs13004438 (CCDC141) for increasing DBP (β=0.082SD, 95% CI, 0.034, 0.13; P=0.8.31×10−4) during the prepubertal epoch but reducing it during the pubertal epoch (β=−0.053SD, 95% CI, −0.115, −0.030; P=8.10×10−4; Table 4). Variants in POT1 and CNTNAP4 were associated with consistently reducing DBP during the pubertal and postpubertal epochs (Table 4 and Table XIV in the Data Supplement).
None of the SNPs found to be at least associated (P≤5×10−3) with childhood DBP were associated with adult BP in the ICBP (Table 4). A number of loci previously found to be associated with adult DBP in ICBP were also associated with DBP in at least one epoch of childhood in EAGLE children (Table 5): USP4 and GRK5 (all pubertal).
We also investigated SNPs from Tables 2 and 3 in CARDIoGRAMplusC4D36 and found 2 significant hits for rs12365302 (P=0.008) and rs16875222 (P=0.037; Table VII in the Data Supplement).
Additional Analyses
As a sensitivity analyses, we repeated the GWAS analyses separately in females and males and found similar directions and magnitudes of associations, though given the smaller sample sizes within these 2 subgroups were not as well powered to report on the sex-stratified associations (Tables VIII– XII in the Data Supplement).
Discussion
To our knowledge, this is the first GWAS to examine genetic associations with BP cross-sectionally across childhood and adolescence. This builds on our previous work in which we found that an allelic score summing the established adult GWAS hits for SBP was associated with SBP at mean age 6 years but not with age-related change in SBP between age 6 and 17.41,42 In the current study, we have identified 2 novel SNPs: one associated with SBP measured prepubertally and one with SBP measured during puberty. We did not find any SNPs reaching genome-wide significance with DBP or with either SBP- or DBP-measured postpuberty, but when we reduced our statistical significance threshold to P≤5×10−3, there were several additional variants in gene clusters relating to SBP or DBP across the 3 epochs. At this more liberal P value threshold, we found some evidence of overlap in novel SNPs across childhood/adolescent age epochs within our study and some overlap with those that have been found by previous GWAS to be associated with adult BP.
ITGA11 was genome-wide associated with SBP during the prepubertal epoch and has been shown to be associated with hypertrophic cardiomyopathy43 and coronary artery disease.44 An SNP in close proximity to the SMARCA and VLDLR genes (rs872256) was genome-wide associated with SBP during the pubertal epoch. SMARCA is a member of the SWI/SNF family of proteins and is highly similar to the brahma protein, where it has been hypothesized that cardiac hypertrophy and the fetal gene expression program are associated with distinguishable binding of brahma and SMARCA4 on genes.45 From animal studies, brahma gene expression is found to be restricted to mesodermal tissues involved in early vasculogenesis and heart morphogenesis.46 VLDLR has been shown to be associated with obesity from both animal studies47–49 and human GWAS.50 Recently, a pathway analyses based on results from a GWAS has identified plausible biological links between VLDR with vascular endothelial growth factor, which is known to affect angiogenesis and atherosclerosis.51 Even with a reduced P value threshold of P≤5×10−3, these 2 genome-wide novel variants did not overlap with variants at this threshold in any other age group.
Some of the variants that we identified as associated with SBP or DBP in any of the different age epochs were in gene clusters that also had some evidence of potentially relevant functionality. FSHR which was associated with SBP in prepubertal and puberty has been shown to influence SBP vascular responses in hypertensive rats with hyperhomocysteinemia52 and is known to be involved in the regulation of systemic arterial BP (GeneCards). POT1 which was associated with lower DBP in pubertal and postpubertal children is an important molecular marker for biological aging.53 MTMR3 which was associated with lower SBP in puberty and postpuberty has been hypothesized to be a mediator for miR-4513, which is significantly associated with BP and related metabolic outcomes, such as cholesterol.54
The fact that we have found more variants associated with SBP than with DBP may reflect differences in changes in SBP and DBP across childhood and adolescence. SBP increases across childhood to a peak at around age 15/16 years and thereafter levels off, whereas DBP seems to increase monotonically across childhood into late adolescence/early twenties.26,27 This is the first GWAS of BP in children at different ages, and we have maximized the sample size by collaborating across several studies with relevant data. We limited analyses to Europeans only to minimize population stratification and followed-up hits by looking up for functionality and for overlap with SNPs from GWAS in adults.
We used age groups to define epochs during which BP was measured as prepubertal, pubertal, and postpubertal. We acknowledge that some participants will have been incorrectly categorized by this method. However, it was not possible to use Tanner scores for all participants, and to have used those data would have compromised our sample size. Furthermore, assessment of pubertal stages using self- (or parental) assessment of Tanner scales, which were the methods used in most cohorts in EAGLE, is also prone to misclassification.55 Misclassification because of using age is likely to be random, whereas Tanner scores could be systematic in relation to characteristics, such as body mass, that are related to BP.
Our sample size was too small to definitely test for sex differences, but consistent with findings from adult GWAS17,20,55–57; there did not seem to be notable differences between females and males. In many of our analyses, we used a P value threshold that was larger than conventional genome-wide thresholds. This decision was made a priori and was intended to ensure that we did not miss potentially important associations and overlaps (between epochs and with adult GWAS findings) given the relatively modest sample size. However, we acknowledge that these findings should be treated with caution until they are replicated.
Conclusions
To conclude, we have identified 2 novel loci related to SBP in childhood (one at prepuberty and one during puberty), but none related to SBP postpuberty or in any age epoch for DBP in childhood, at genome-wide significance. The 2 novel SBP SNPs were specific to those epochs and did not relate to SBP in other epochs even with a higher P value threshold of P≤5×10−3. With this more liberal P value, we identified more variants related to SBP and 2 variants related to DPB measured during puberty. Most of these were specific to the particular epoch in which they were found, though for a small number, we did find overlap with adjacent epochs and also some overlap with published adult variants. Thus, our results provide some support for age-specific associations, as well as for associations that might be present across most ages. The observed genetic associations with no previous history of association with adult BP may be true novel associations, but require further investigation and replication.
Supplementary Material
The Data Supplement is available at http://circgenetics.ahajournals.org/lookup/suppl/doi:10.1161/CIRCGENETICS.115.001190/-/DC1.
Clinical Perspective.
This study was designed to identify genetic variants associated with blood pressure in childhood and adolescence to improve our understanding of the lifecourse pathogenesis of hypertension and cardiovascular disease. Variation in systolic and diastolic blood pressure across the lifecourse is associated with subsequent adult risk of coronary heart disease and stroke. The age-related changes in blood pressure in industrialized countries suggest a varying gene environment interaction with age, but to date, relatively little is known about the genetic determinants of blood pressure in childhood and adolescence. Two novel loci were identified as having genome-wide associations with systolic blood pressure in specific age epochs. These loci were rs1563894 [ITGA11], associated with systolic blood pressure assessed prepuberty, and rs872256, which was associated with blood pressure during puberty. ITGA11 has been shown to be associated with hypertrophic cardiomyopathy and coronary artery disease. Rs872256 is in close proximity to the SMARCA and VLDLR genes. SMARCA is thought to influence cardiac hypertrophy and fetal gene expression. VLDLR has been shown to be associated with obesity and has plausible biological links with angiogenesis and atherosclerosis. We also found some evidence of gene clusters associated with blood pressure in childhood. Most of the effects we observed were specific to the particular epoch in which they were found, though a small number overlapped with adjacent epochs and with published adult variants. Our results suggest that genetic determinants of blood pressure act from childhood, develop over the life course, and show some evidence of age-specific effects.
Acknowledgments
Raine: We are grateful to the Raine Study participants and their families and to the Raine Study research staff, which includes data collectors, cohort managers, data managers, clerical staff, research scientists, and volunteers for cohort coordination and data collection. We gratefully acknowledge the assistance of the Western Australian DNA Bank (National Health and Medical Research Council of Australia National Enabling Facility). ALSPAC: We are extremely grateful to all the families who took part in this study, the midwives for their help in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists, and nurses. The Generation R Study is conducted by the Erasmus Medical Center in close collaboration with the School of Law and Faculty of Social Sciences of the Erasmus University, Rotterdam; the Municipal Health Service Rotterdam area, Rotterdam; the Rotterdam Homecare Foundation, Rotterdam; and the Stichting Trombosedienst & Artsenlaboratorium Rijnmond (STAR-MDC), Rotterdam. We gratefully acknowledge the contribution of children and parents, general practitioners, hospitals, midwives, and pharmacies in Rotterdam. The generation and management of GWAS genotype data for the Generation R Study were done at the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, The Netherlands. We thank Karol Estrada, Dr Tobias A. Knoch, Anis Abuseiris, Luc V. de Zeeuw, and Rob de Graaf for their help in creating GRIMP, BigGRID, MediGRID, and Services@MediGRID/D-Grid (funded by the German Bundesministerium fuer Forschung und Technology; grants 01 AK 803 A-H, 01 IG 07015 G) for access to their grid computing resources. We thank Mila Jhamai, Manoushka Ganesh, Pascal Arp, Marijn Verkerk, Lizbeth Herrera, and Marjolein Peters for their help in creating, managing, and QC of the GWAS database. Also, we thank Karol Estrada and Carolina Medina-Gomez for their support in creation and analysis of imputed data. We also acknowledge the contribution of Cornelia van Duijn for her intellectual input in this project. The Generation R Study is made possible by financial support from the Erasmus Medical Center, Rotterdam, the Erasmus University Rotterdam, and the Netherlands Organization for Health Research and Development. The study has been approved by the Medical Ethics Committee of the Erasmus Medical Center, Rotterdam. Written informed consent was obtained from all participants or their parent(s). INMA: We particularly thank all the participants for their generous collaboration. A full roster of the INMA Project Investigators can be found at http://www.proyectoinma.org/presentacion-inma/listado-investigadores/en_listado-investigadores.html. LISAplus: We are extremely grateful to all of the children and their parents for the contributions, to the obestric units for recruitment, and to the LISAplus study team. The Brisbane Longitudinal Twin Study (BLTS) was supported by grants from the Australian National Health and Medical Research Council (NHMRC; 241944, 389875 1031119, 1049894) and the Australian Research Council (DP0212016, DP0343921) for phenotyping and by the NHMRC Medical Bioinformatics Genomics Proteomics Program, 389891, for genotyping. We are grateful for the twins who participated in the study and all those who helped in carrying it out. We thank Ann Eldridge, Marlene Grace, and Kerrie McAloney (sample collection); David Smyth, Harry Beeby, and Daniel Park (IT support); Sarah Medland, Dale Nyholt, and Scott Gordon (imputation and genotyping QC), and Anjali Henders and Leanne McNeil (laboratory support). The Young Finns Study: The expert technical assistance in the statistical analyses by Irina Lisinen is gratefully acknowledged. Data on coronary artery disease/myocardial infarction have been contributed by CARDIoGRAMplusC4D investigators and have been downloaded from www.CARDIOGRAMPLUSC4D.ORG
Sources of Funding
Raine: We acknowledge the support of the Healthway Western Australia, the National Health and Medical Research Council of Australia (Grants 572613, 403981, 963209, 211912, 003209, and 353514), and the Canadian Institutes of Health Research (Grant MOP 82893). We gratefully acknowledge the assistance of the University of Western Australia (UWA), Raine Medical Research Foundation, Wind Over Water Foundation, the Telethon Institute for Child Health Research, UWA Faculty of Medicine, Dentistry and Health Sciences, Women and Infants Research Foundation, and Curtin University. ALSPAC: The UK Medical Research Council and the Wellcome Trust (Grant ref: 092731) and the University of Bristol provide core support for ALSPAC. We thank the Sanger Centre, Centre National de Genotypage, and 23andMe for generating the ALSPAC GWA data. The Generation R Study: The Generation R Study recieves financial support from the Erasmus Medical Center, Rotterdam, and The Netherlands Organization for Health Research and Development (ZonMw). Additional support was provided by a grant from the Dutch Kidney Foundation (C08.2251). Dr Jaddoe received an additional grant from the Netherlands Organization for Health Research and Development (ZonMW VIDI: 016.136.361). INMA: This study was funded by grants from Instituto de Salud Carlos III (FIS 11/01007, G03/176, FISFEDER 03/1615, 04/1509, 04/1112, 04/1931, 05/1052, 05/1079, 06/1213, 07/0314, and 09/02467, PS09/00432,and CB06/02/0041), European Commission (ENGAGE project and grant agreement HEALTH-F4-2007-201413), Fundació La Marató de TV3, and Fundación Roger Torné. Part of the DNA extractions and genotyping was performed at the Spanish National Genotyping Centre (CEGEN-Barcelona). The LISAplus study was supported by innovative research priority project Munich Center of Health Sciences (MC-Health, subproject I) of the Ludwig-Maximilians University Munich (LMU) and is funded by the German Bundesministerium fuer Forschung und Technology; grants 01 AK 803 A-H, 01 IG 07015 G. BLTS: This study was supported by the Australian Research Council (No A79600334, A79906588, A79801419, DP0212016, and DP0343921). The Young Finns Study has been financially supported by the Academy of Finland: grants 126925, 121584, 124282, 129378, 117797, and 41071, the Social Insurance Institution of Finland, Kuopio, Tampere, and Turku University Hospital Medical Funds, Juho Vainio Foundation, Paavo Nurmi Foundation, Finnish Foundation of Cardiovascular Research and Finnish Cultural Foundation, Sigrid Juselius Foundation, Tampere Tuberculosis Foundation, and Emil Aaltonen Foundation.
Appendix
From the Department of Biostatistics & Epidemiology, Auckland University of Technology, Auckland, New Zealand (P.G.P.); Departments of Epidemiology (H.R.T., G.V., A.G.U., F.R., A.H., V.W.V.J., C.M.v.D.), Pediatrics (H.R.T., A.G.U., V.W.V.J.), and Internal Medicine (A.G.U., F.R.), Erasmus Medical Center, Rotterdam, The Netherlands; The Medical Research Council Integrative Epidemiology Unit (N.J.T., L.D.H., D.M.E., B.S.P., D.A.L.), School of Social & Community Medicine (N.J.T., L.D.H., D.A.L.), School of Oral & Dental Sciences (B.S.P.), and School of Experimental Psychology (B.S.P.), University of Bristol, Bristol, UK; Helmholtz Zentrum Muenchen, German Research Centre for Environmental Health, Institute of Epidemiology, Neuherberg, Germany (E.T., J.H.); Department of Clinical Chemistry, Fimlab Laboratories (T.L., L.-P.L.), and Department of Clinical Physiology (M.K.), University of Tampere, Tampere, Finland; Municipal Institute of Medical Research (IMIM), Barcelona, Catalonia, Spain (M.M.); Quantitative Genetics (P.A.L., M.J.W.), Genetic Epidemiology (J.B.W., N.G.M.), and Molecular Epidemiology (G.W.M.), QIMR Berghofer Medical Research Institute, Brisbane, Queensland, Australia; Research Centre of Applied & Preventive Cardiovascular Medicine (V.A.), and Department of Medicine (J.V.), University of Turku, Turku, Finland; Lunenfeld-Tanenbaum Research Institute, University of Toronto, Ontario, Canada (L.B.); University of Queensland Diamantina Institute, Translational Research Institute, Brisbane, Queensland (D.M.E.); School of Women’s & Infants’ Health (J.P.N., C.E.P.), and The Western Australian Pregnancy Cohort (Raine) Genetic Epidemiology Team, School of Medicine & Pharmacology Royal Perth Hospital Unit (L.J.B.), The University of Western Australia, Perth, Western Australia, Australia; Department of Biological Psychology, Vrije Universiteit Amsterdam, Amsterdam, the Netherlands (D.I.B., J.-J.H.); Department of Population Medicine, Harvard Medical School & Harvard Pilgrim Health Care Institute, Boston, MA (M.W.G.); Departments of Health Sciences & Genetics, University of Leicester, Leicestershire, UK (M.D.T.); Department of Clinical Physiology & Nuclear Medicine, Turku University Hospital, University of Turku, Turku, Finland (O.R.); Department de Salud Pública, Universidad Miguel Hernández, San Juan de Alicante, Spain (J.V.); Department of Epidemiology & Biostatistics, School of Public Health, Imperial College London, London, UK (M.-R.J.); Institute of Health Sciences (M.-R.J.), Biocenter (M.-R.J.), University of Oulu, Oulu; Unit of Primary Care, Oulu University Hospital, Kajaanintie Oulu (M.-R.J.); Department of Children & Young People & Families, National Institute for Health & Welfare, Oulu, Finland (M.-R.J.); and Joanna Briggs Institute & School of Translational Health Science, University of Adelaide, Adelaide, Australia (L.J.P.).
Footnotes
Disclosures
Dr Lawlor, Timpson, Howe, Evans work in a unit that receives funding from the UK Medical Research Council (MC_UU_12013/3, MC_UU_12013/4, MC_UU_12013/5, and MC_UU_12013/9). Dr Howe is funded by a UK Medical Research Council fellowship (G1002375) and is supported by a fellowship from the UK Medical Research Council (MR/M020894/1). Dr Howe has also received grant income from the UK Economic and Social Research Council, the UK Biotechnology and Biological Sciences Research Council, and the US National Institute on Aging; these grants are not related to the content of this publication. Dr Gillman received royalties from Cambridge University Press for Maternal Obesity (2012), which they coedited, and from UpToDate for a chapter on dietary fat. Dr Pennell received the National Health and Medical Research Council of Australia (Grants 572613, 403981, 963209, 211912, 003209, and 353514) and the Canadian Institutes of Health Research (Grant MOP 82893). The other authors report no conflicts.
References
- 1.Ehret GB, Caulfield MJ. Genes for blood pressure: an opportunity to understand hypertension. Eur Heart J. 2013;34:951–961. doi: 10.1093/eurheartj/ehs455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stamler J, Stamler R, Neaton JD. Blood pressure, systolic and diastolic, and cardiovascular risks. US population data. Arch Intern Med. 1993;153:598–615. doi: 10.1001/archinte.153.5.598. [DOI] [PubMed] [Google Scholar]
- 3.Vasan RS, Larson MG, Leip EP, Evans JC, O’Donnell CJ, Kannel WB, et al. Impact of high-normal blood pressure on the risk of cardiovascular disease. N Engl J Med. 2001;345:1291–1297. doi: 10.1056/NEJMoa003417. [DOI] [PubMed] [Google Scholar]
- 4.Mahoney LT, Burns TL, Stanford W, Thompson BH, Witt JD, Rost CA, et al. Coronary risk factors measured in childhood and young adult life are associated with coronary artery calcification in young adults: the Muscatine Study. J Am Coll Cardiol. 1996;27:277–284. doi: 10.1016/0735-1097(95)00461-0. [DOI] [PubMed] [Google Scholar]
- 5.Muntner P, He J, Cutler JA, Wildman RP, Whelton PK. Trends in blood pressure among children and adolescents. JAMA. 2004;291:2107–2113. doi: 10.1001/jama.291.17.2107. [DOI] [PubMed] [Google Scholar]
- 6.WHO. World health statistics 2012. Global Health Observatory (GHO) 2012 [Google Scholar]
- 7.Levy D, DeStefano AL, Larson MG, O’Donnell CJ, Lifton RP, Gavras H, et al. Evidence for a gene influencing blood pressure on chromosome 17. Genome scan linkage results for longitudinal blood pressure phenotypes in subjects from the framingham heart study. Hypertension. 2000;36:477–483. doi: 10.1161/01.hyp.36.4.477. [DOI] [PubMed] [Google Scholar]
- 8.1999 World Health Organization-International Society of Hypertension Guidelines for the Management of Hypertension. Guidelines Subcommittee. J Hypertens. 1999;17:151–183. [PubMed] [Google Scholar]
- 9.Burt VL, Whelton P, Roccella EJ, Brown C, Cutler JA, Higgins M, et al. Prevalence of hypertension in the US adult population. Results from the Third National Health and Nutrition Examination Survey, 1988-1991. Hypertension. 1995;25:305–313. doi: 10.1161/01.hyp.25.3.305. [DOI] [PubMed] [Google Scholar]
- 10.Lieb W, Jansen H, Loley C, Pencina MJ, Nelson CP, Newton-Cheh C, et al. CARDIoGRAM Genetic predisposition to higher blood pressure increases coronary artery disease risk. Hypertension. 2013;61:995–1001. doi: 10.1161/HYPERTENSIONAHA.111.00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franceschini N, Fox E, Zhang Z, Edwards TL, Nalls MA, Sung YJ, et al. Asian Genetic Epidemiology Network Consortium Genome-wide association analysis of blood-pressure traits in African-ancestry individuals reveals common associated genes in African and non-African populations. Am J Hum Genet. 2013;93:545–554. doi: 10.1016/j.ajhg.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hong KW, Min H, Heo BM, Joo SE, Kim SS, Kim Y. Recapitulation of genome-wide association studies on pulse pressure and mean arterial pressure in the Korean population. J Hum Genet. 2012;57:391–393. doi: 10.1038/jhg.2012.31. [DOI] [PubMed] [Google Scholar]
- 13.Wain LV, Verwoert GC, O’Reilly PF, Shi G, Johnson T, Johnson AD, et al. LifeLines Cohort Study; EchoGen consortium; AortaGen Consortium; CHARGE Consortium Heart Failure Working Group; KidneyGen consortium; CKDGen consortium; Cardiogenics consortium; CardioGram Genome-wide association study identifies six new loci influencing pulse pressure and mean arterial pressure. Nat Genet. 2011;43:1005–1011. doi: 10.1038/ng.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu C, Li H, Qi Q, Lu L, Gan W, Loos RJ, et al. Common variants in or near FGF5, CYP17A1 and MTHFR genes are associated with blood pressure and hypertension in Chinese Hans. J Hypertens. 2011;29:70–75. doi: 10.1097/HJH.0b013e32833f60ab. [DOI] [PubMed] [Google Scholar]
- 15.Kraja AT, Vaidya D, Pankow JS, Goodarzi MO, Assimes TL, Kullo IJ, et al. A bivariate genome-wide approach to metabolic syndrome: STAMPEED consortium. Diabetes. 2011;60:1329–1339. doi: 10.2337/db10-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kato N, Takeuchi F, Tabara Y, Kelly TN, Go MJ, Sim X, et al. Meta-analysis of genome-wide association studies identifies common variants associated with blood pressure variation in east Asians. Nat Genet. 2011;43:531–538. doi: 10.1038/ng.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Padmanabhan S, Melander O, Johnson T, Di Blasio AM, Lee WK, Gentilini D, et al. Global BPgen Consortium Genome-wide association study of blood pressure extremes identifies variant near UMOD associated with hypertension. PLoS Genet. 2010;6:e1001177. doi: 10.1371/journal.pgen.1001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soranzo N, Spector TD, Mangino M, Kühnel B, Rendon A, Teumer A, et al. A genome-wide meta-analysis identifies 22 loci associated with eight hematological parameters in the HaemGen consortium. Nat Genet. 2009;41:1182–1190. doi: 10.1038/ng.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, Coin L, et al. Wellcome Trust Case Control Consortium Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009;41:666–676. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A, et al. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009;41:677–687. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ganesh SK, Tragante V, Guo W, Guo Y, Lanktree MB, Smith EN, et al. CARDIOGRAM, METASTROKE; LifeLines Cohort Study Loci influencing blood pressure identified using a cardiovascular gene-centric array. Hum Mol Genet. 2013;22:1663–1678. doi: 10.1093/hmg/dds555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson T, Gaunt TR, Newhouse SJ, Padmanabhan S, Tomaszewski M, Kumari M, et al. Cardiogenics Consortium; Global BPgen Consortium Blood pressure loci identified with a gene-centric array. Am J Hum Genet. 2011;89:688–700. doi: 10.1016/j.ajhg.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tragante V, Barnes MR, Ganesh SK, Lanktree MB, Guo W, Franceschini N, et al. Gene-centric meta-analysis in 87,736 individuals of European ancestry identifies multiple blood-pressure-related loci. Am J Hum Genet. 2014;94:349–360. doi: 10.1016/j.ajhg.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Milsted A, Underwood AC, Dunmire J, DelPuerto HL, Martins AS, Ely DL, et al. Regulation of multiple renin-angiotensin system genes by Sry. J Hypertens. 2010;28:59–64. doi: 10.1097/HJH.0b013e328332b88d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wills AK, Lawlor DA, Matthews FE, Sayer AA, Bakra E, Ben-Shlomo Y, et al. Life course trajectories of systolic blood pressure using longitudinal data from eight UK cohorts. PLoS Med. 2011;8:e1000440. doi: 10.1371/journal.pmed.1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Labarthe DR, Dai S, Fulton JE, Harrist RB, Shah SM, Eissa MA. Systolic and fourth- and fifth-phase diastolic blood pressure from ages 8 to 18 years: Project HeartBeat! Am J Prev Med. 2009;37(1 suppl):S86–S96. doi: 10.1016/j.amepre.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 28.Lawlor DA, Smith GD. Early life determinants of adult blood pressure. Curr Opin Nephrol Hypertens. 2005;14:259–264. doi: 10.1097/01.mnh.0000165893.13620.2b. [DOI] [PubMed] [Google Scholar]
- 29.McCarron DA. Sodium excretion and cardiovascular mortality. Lancet. 2001;358:665. doi: 10.1016/S0140-6736(01)05793-2. author reply 666. [DOI] [PubMed] [Google Scholar]
- 30.Melka MG, Bernard M, Mahboubi A, Abrahamowicz M, Paterson AD, Syme C, et al. Genome-wide scan for loci of adolescent obesity and their relationship with blood pressure. J Clin Endocrinol Metab. 2012;97:E145–E150. doi: 10.1210/jc.2011-1801. [DOI] [PubMed] [Google Scholar]
- 31.Xi B, Shen Y, Zhao X, Chandak GR, Cheng H, Hou D, et al. Association of common variants in/near six genes (ATP2B1, CSK, MTHFR, CYP17A1, STK39 and FGF5) with blood pressure/hypertension risk in Chinese children. J Hum Hypertens. 2014;28:32–36. doi: 10.1038/jhh.2013.50. [DOI] [PubMed] [Google Scholar]
- 32.Bhatnagar P, Barron-Casella E, Bean CJ, Milton JN, Baldwin CT, Steinberg MH, et al. Genome-wide meta-analysis of systolic blood pressure in children with sickle cell disease. PLoS One. 2013;8:e74193. doi: 10.1371/journal.pone.0074193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 34.Takeuchi F, Isono M, Katsuya T, Yamamoto K, Yokota M, Sugiyama T, et al. Blood pressure and hypertension are associated with 7 loci in the Japanese population. Circulation. 2010;121:2302–2309. doi: 10.1161/CIRCULATIONAHA.109.904664. [DOI] [PubMed] [Google Scholar]
- 35.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes TL, Thompson JR, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Preuss M, König IR, Thompson JR, Erdmann J, Absher D, Assimes TL, et al. CARDIoGRAM Consortium Design of the Coronary ARtery DIsease Genome-Wide Replication And Meta-Analysis (CARDIoGRAM) Study: A Genome-wide association meta-analysis involving more than 22 000 cases and 60 000 controls. Circ Cardiovasc Genet. 2010;3:475–483. doi: 10.1161/CIRCGENETICS.109.899443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2015. https://www.R-project.org/. [Google Scholar]
- 40.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Howe LD, Parmar PG, Paternoster L, Warrington NM, Kemp JP, Briollais L, et al. Genetic influences on trajectories of systolic blood pressure across childhood and adolescence. Circ Cardiovasc Genet. 2013;6:608–614. doi: 10.1161/CIRCGENETICS.113.000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Punwasi RV, Monnereau C, Hofman A, Jaddoe VW, Felix JF. The Influence of Known Genetic Variants on Subclinical Cardiovascular Outcomes in Childhood: Generation R Study. Circ Cardiovasc Genet. 2015;8:596–602. doi: 10.1161/CIRCGENETICS.114.000915. [DOI] [PubMed] [Google Scholar]
- 43.Talior-Volodarsky I, Connelly KA, Arora PD, Gullberg D, McCulloch CA. α11 integrin stimulates myofibroblast differentiation in diabetic cardiomyopathy. Cardiovasc Res. 2012;96:265–275. doi: 10.1093/cvr/cvs259. [DOI] [PubMed] [Google Scholar]
- 44.Duan S, Luo X, Dong C. Identification of susceptibility modules for coronary artery disease using a genome wide integrated network analysis. Gene. 2013;531:347–354. doi: 10.1016/j.gene.2013.08.059. [DOI] [PubMed] [Google Scholar]
- 45.Chang L, Kiriazis H, Gao XM, Du XJ, El-Osta A. Cardiac genes show contextual SWI/SNF interactions with distinguishable gene activities. Epigenetics. 2011;6:760–768. doi: 10.4161/epi.6.6.16007. [DOI] [PubMed] [Google Scholar]
- 46.Dauvillier S, Ott MO, Renard JP, Legouy E. BRM (SNF2alpha) expression is concomitant to the onset of vasculogenesis in early mouse postimplantation development. Mech Dev. 2001;101:221–225. doi: 10.1016/s0925-4773(00)00560-8. [DOI] [PubMed] [Google Scholar]
- 47.Kim OY, Lee SM, Chung JH, Do HJ, Moon J, Shin MJ. Arginase I and the very low-density lipoprotein receptor are associated with phenotypic biomarkers for obesity. Nutrition. 2012;28:635–639. doi: 10.1016/j.nut.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 48.Perman JC, Boström P, Lindbom M, Lidberg U, StÅhlman M, Hägg D, et al. The VLDL receptor promotes lipotoxicity and increases mortality in mice following an acute myocardial infarction. J Clin Invest. 2011;121:2625–2640. doi: 10.1172/JCI43068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Urban D, Pöss J, Böhm M, Laufs U. Targeting the proprotein convertase subtilisin/kexin type 9 for the treatment of dyslipidemia and atherosclerosis. J Am Coll Cardiol. 2013;62:1401–1408. doi: 10.1016/j.jacc.2013.07.056. [DOI] [PubMed] [Google Scholar]
- 50.Asselbergs FW, Guo Y, van Iperen EP, Sivapalaratnam S, Tragante V, Lanktree MB, et al. LifeLines Cohort Study Large-scale gene-centric meta-analysis across 32 studies identifies multiple lipid loci. Am J Hum Genet. 2012;91:823–838. doi: 10.1016/j.ajhg.2012.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Debette S, Visvikis-Siest S, Chen MH, Ndiaye NC, Song C, Destefano A, et al. Identification of cis- and trans-acting genetic variants explaining up to half the variation in circulating vascular endothelial growth factor levels. Circ Res. 2011;109:554–563. doi: 10.1161/CIRCRESAHA.111.243790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yen CH, Lau YT. Vascular responses in male and female hypertensive rats with hyperhomocysteinemia. Hypertension. 2002;40:322–328. doi: 10.1161/01.hyp.0000028489.29543.58. [DOI] [PubMed] [Google Scholar]
- 53.Zee RY, Ridker PM, Chasman DI. Genetic variants in eleven telomere-associated genes and the risk of incident cardio/cerebrovascular disease: The Women’s Genome Health Study. Clin Chim Acta. 2011;412:199–202. doi: 10.1016/j.cca.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ghanbari M, de Vries PS, de Looper H, Peters MJ, Schurmann C, Yaghootkar H, et al. A genetic variant in the seed region of miR-4513 shows pleiotropic effects on lipid and glucose homeostasis, blood pressure, and coronary artery disease. Hum Mutat. 2014;35:1524–1531. doi: 10.1002/humu.22706. [DOI] [PubMed] [Google Scholar]
- 55.Raman A, Lustig RH, Fitch M, Fleming SE. Accuracy of self-assessed Tanner staging against hormonal assessment of sexual maturation in overweight African-American children. J Pediatr Endocrinol Metab. 2009;22:609–622. doi: 10.1515/jpem.2009.22.7.609. [DOI] [PubMed] [Google Scholar]
- 56.Oikonen M, Tikkanen E, Juhola J, Tuovinen T, Seppälä I, Juonala M, et al. Genetic variants and blood pressure in a population-based cohort: the Cardiovascular Risk in Young Finns study. Hypertension. 2011;58:1079–1085. doi: 10.1161/HYPERTENSIONAHA.111.179291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang X, Ding X, Su S, Yan W, Harshfield G, Treiber F, et al. Genetic inflences on daytime and night-time blood pressure: similarities and differences. J Hypertens. 2009;27:2358–2364. doi: 10.1097/HJH.0b013e328330e84d. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.