Abstract
Therapeutic nucleic acids hold great promise for the treatment of disease but require vectors for safe and effective delivery. Synthetic nanoparticle vectors composed of poly(β-amino esters) (PBAEs) and nucleic acids have previously demonstrated potential utility for local delivery applications. To expand potential utility to include systemic delivery of mRNA, here we develop hybrid polymer-lipid nanoformulations for systemic delivery to the lung. Through co-formulation of PBAEs with lipid-polyethylene glycol (PEG), mRNA formulations were developed with increased serum stability and increased in vitro potency. The formulations were capable of functional delivery of mRNA to the lungs following intravenous administration in mice. To our knowledge, this is the first description of systemic administration of mRNA for delivery to the lungs using a degradable polymer-lipid nanoparticle.
Keywords: Nanoparticles, mRNA, Drug delivery, PBAE, Stability
Nucleic acid therapeutics have great potential to treat disease at the genetic level, especially in cases where the aberrant phenotype arises from missing or mutated protein(s).[1,2] Recent advances with mRNA, including improvements with in vitro transcription, has increased interest in the therapeutic potential of this biomolecule.[3] Unlike DNA, mRNA need only reach the cytoplasm to induce protein expression, and furthermore bears no apparent risk of insertional mutagenesis[4]. However, like DNA delivery, the application of mRNA therapy is limited by the need for safe and effective delivery systems.[5] Nanoparticles prepared from cationic polymers have been shown to deliver nucleic acids.[6] Polyethyleneimine (PEI) has been broadly investigated as a nucleic acid delivery vehicle, as its high density of amine groups facilitates efficient complexation with negatively charged nucleic acids and buffers the acidic endosomal environment following nanoparticle endocytosis.[7,8] However, PEI is not degradable and there are concerns regarding its toxicity, features that could limit its use for clinical applications.[9] Poly(β-amino esters) (PBAEs) are degradable polycations with demonstrated potential as delivery systems for nucleic acid delivery[10] both for in vitro[11,12] and in vivo[13,14] applications.
Previous studies have suggested that serum stability may limit the efficacy of PBAEs for systemic administration.[15] To this end, PBAE terpolymer copolymers containing internal alkyl tails were developed.[16] The alkyl amine was incorporated to enable formulation of terpolymers with other hydrophobic components in order to increase particle stability in physiological conditions. Here, we demonstrate that PBAE terpolymer nanoparticles can be formulated with a polyethylene glycol-modified lipid (PEG-lipid) and result in serum stabilized nanoparticles and improved efficacy. PBAEs that were found to be serum stabilized in vitro upon incorporation of PEG-lipid at 7 mol% were evaluated in vivo and shown to be capable of mRNA delivery to the lungs in mice following intravenous administration.
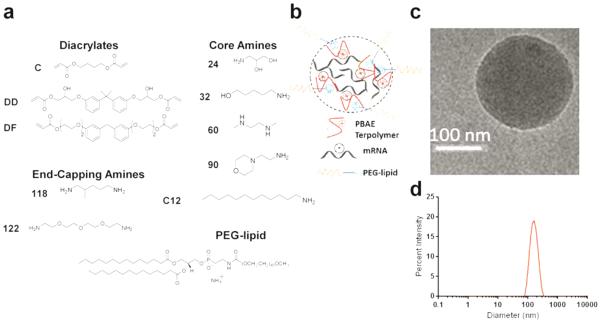
The PBAEs selected for synthesis in this study were based on analysis of literature results from this class of materials for DNA delivery.[16] We synthesized 7 PBAEs that included 6 lead terpolymers and 1 lead non-terpolymer (i.e. not including the alkyl amine) from the monomers shown in Figure 1a. Synthetic methodology was adapted for synthesis in dimethylformamide (DMF) and purification in diethylether in order to reduce the possibility of toxicity arising from excess monomer (Scheme S1).[16] Polymer characterization was performed via GPC, 1H-NMR, and FTIR (Figures S1-3). Nanoparticles (Figure 1b) were prepared by mixing each synthesized polymer with mRNA coding for firefly luciferase diluted in acidic (pH 5.2) buffer. Nanoparticles were formulated such that the ratio of amine groups in the polymer to phosphate groups in the nucleic acid (N/P) was 57; this is consistent with a weight ratio of approximately 50:1, similar to what has been used for successful PBAE transfections previously.[17]Stable nanoparticles were formed both with and without addition of 7 mol% PEG-lipid (C14-PEG2000), based on an in vitro optimization study (Figure S4), and particle formation was confirmed by CryoTEM imaging (Figure 1c and Figure S5) and dynamic light scattering (Figure 1d, Table S1).
Figure 1.
PBAE polymer synthesis and nanoparticle formation. (a) Monomers from top-performing materials identified previously were chosen. Briefly, step-growth polymerization was performed via Michael addition of diacrylates and amines, with dodecyl amine added in the case of terpolymers to form a random copolymer. Chains were subsequently reacted with an end-capping amine. For nomenclature, “DD90-C12-122” refers to a terpolymer synthesized from the DD diacrylate reacted with amine 90 along with the C12 alkylamine, and then end-capped with amine 122 (see Representative Synthesis section in the Supplementary Information). Nanoparticles were then formulated by incorporation of PEG-lipid via hydrophobic interactions. (b) Cartoon depiction of PEGylated, mRNA-loaded terpolymer nanoparticles. (c) Representative cryoTEM image of DD90-C12-122 formulated with PEG-lipid. (d) Representative sizing data from dynamic light scattering of DD90-C12-122 formulated with PEG-lipid.
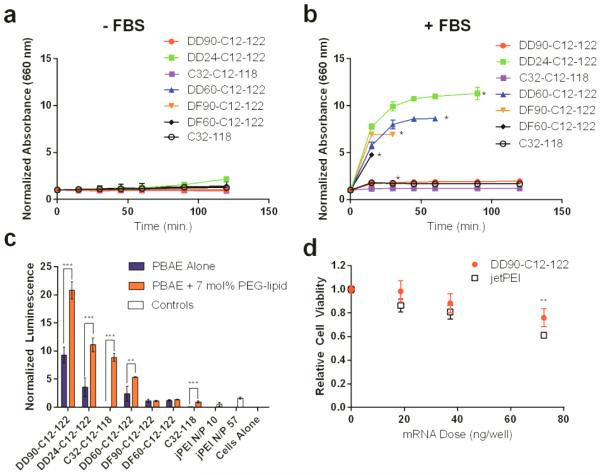
Co-formulation with PEG-lipid was intended to promote PBAE nanoparticle stability under physiological conditions, especially in the presence of serum proteins.[16] Polymers formulated without PEG-lipid exhibited a dramatic increase in size during dialysis against phosphate buffered saline (PBS) at pH 7.4, while nanoparticles formulated with 7 mol% PEG-lipid remained stable both throughout dialysis (Figure S6) and for at least an additional two hours of incubation in PBS at 37°C (Figure 2a), as indicated by turbidity measurements.[18] Nanoparticles prepared from the non-terpolymer, C32-118, co-formulated with PEG-lipid were also stable under these conditions, suggestive of some PEG-lipid incorporation despite the lack of dodecyl amine in C32-118. When nanoparticle solutions were spiked with fetal bovine serum (Figure 2b), only two PEGylated terpolymer formulations resisted aggregation: DD90-C12-122 and C32-C12-118. All other terpolymer nanoparticles agglomerated and eventually precipitated from solution, as confirmed by RNA quantification in the supernatant at the experiment endpoint (Figure S7). The rescue of serum stability for representative agglomerating nanoparticles (DD24-C12-122 and DD60-C12-122) was possible through addition of higher amounts of PEG-lipid (Figure S8a-b). Turbidity assays were carried out at a nanoparticle concentration relevant for in vivo injections (50 ng mRNA μL−1), suggesting that PEG-lipid can play a significant role in aggregation resistance in serum in vivo, independent of polymer properties.
Figure 2.
PBAE serum stability and in vitro efficacy. (a) PEGylated PBAE nanoparticles were stable for approximately 2 hours in PBS at 37°C, as indicated by turbidity measurements. These particles were previously dialyzed in PBS for 2 hours at 4°C, a process which results in non-PEGylated particles aggregating (mean ± SD, n = 3). (b) PEGylated DD90-C12-122 and C32-C12-118, as well as PEGylated C32-118 remained stable following addition of 10% v/v fetal bovine serum, while others aggregated (mean ± SD, n = 3). (c) PBAE terpolymer nanoparticles formulated with luciferase-encoding mRNA promote luciferase protein expression in HeLa cells (50 ng/well mRNA dose), as quantified by cellular luminescence. Luminescence was normalized to live cell signal using a fluorescent live/dead cell assay. In this case, transfection is compared to a commercial polymeric transfection reagent jetPEI. (d) Relative viability of cells treated with PEGylated DD90-C12-122 and jetPEI (N/P=57) **indicates p<0.01, *** indicates p < 0.001 (mean ± SD, n = 4).
To determine whether serum stability of PBAE nanoparticles correlated with in vitro efficacy, HeLa cells were transfected with PBAE nanoparticles (50 ng mRNA per well), with and without PEG-lipid, formulated with luciferase-encoding mRNA (Figure 2c). Cell luminescence was then measured as an indicator of transfection efficacy and normalized to cell viability as determined by fluorescent assay. Commercially acquired jetPEI, a polymeric transfection reagent, was used for comparison and formulated according to the protocol-recommended N/P ratio as well as N/P of 57. The only terpolymer nanoparticles that resisted aggregation in serum stability studies, PEGylated DD90-C12-122 and C32-C12-118, were also among the most potent transfection materials. Furthermore, these materials showed a significant increase in potency when compared with their non-PEGylated counterparts. Cytotoxicity data demonstrated that, at an N/P of 57, cells treated with PEGylated DD90-C12-122 at a dose of 0 – 70 ng mRNA per well retained >75% viability relative to untreated cells, despite the lack of a wash step following transfection, and at higher dose DD90-C12-122 was significantly less toxic than jetPEI (Figure 2d). Nanoparticles which had rapidly precipitated from solution in stability studies (i.e. DF90-C12-122 and DF60-C12-122) were the least effective, further supporting a correlation between serum stability and in vitro efficacy. Though the non-terpolymer control (C32-118) had improved efficacy following PEGylation, the improvement in C32-C12-118 potency was far greater after inclusion of PEG-lipid, potentially as a result of increased PEG-lipid incorporation in the nanoparticle mediated by the C12 group as observed in Eltoukhy et al.[16] Interestingly, nanoparticles with rescued serum stability by increased PEG-lipid had a decrease in in vitro potency (Figure S8c-d). This is suggestive of a limit above which increasing PEG-lipid content is detrimental to effective transfection, perhaps as a result of reduced cellular uptake or inefficient RNA unpackaging within endosomes.[19] In sum, the stability and in vitro efficacy data suggest that the effect of PEG-lipid inclusion on nanoparticle performance is dependent on PBAE chemical structure.
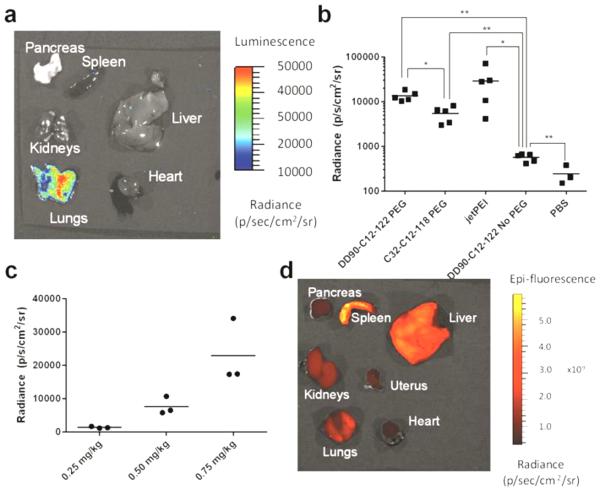
The three terpolymers which had the highest in vitro potencies, PEGylated DD90-C12-122, DD24-C12-122, and C32-C12-118, were further considered for in vivo testing. C57BL/6 mice were injected intravenously with all three PEGylated nanoparticles (7 mol% PEG-lipid) and a sample non-PEGylated nanoparticle (DD90-C12-122) at an mRNA dose of 0.5 mg kg−1 mouse. A positive control of jetPEI was included as well. It is noted that for jetPEI N/P = 8 was used (the maximum recommended N/P due to toxicity), whereas all PBAE nanoparticles were maintained at N/P = 57. In preliminary studies, nanoparticles prepared from DD24-C12-122 led to immediate mortality in most mice, an observation attributed to aggregation in circulation resulting in lung occlusions,[20] consistent with the serum-induced agglomeration demonstrated in Figure 2b. When DD24-C12-122 nanoparticles were formulated with a higher mol% PEG-lipid to resist aggregation (Figure S8a) no mortality was observed, but these nanoparticles also showed no efficacy (Figure S9), corroborating in vitro findings (Figure S8c). For mRNA delivered with PEGylated DD90-C12-122 and C32-C12-118, protein is expressed primarily in the lungs (Figure 3a-b). The impact on in vivo efficacy from improved serum stability imparted by PEG-lipid is most clearly seen in elevated luminescence following delivery of PEGylated C32-C12-118 compared to non-PEGylated DD90-C12-122, in spite of their similar potency in vitro. Additionally, the luminescence achieved using PEGylated DD90-C12-122 and C32-C12-118 is comparable to that obtained via in vivo jetPEI (Figure 3b), and PEGylated DD90-C12-122 mRNA delivery is dose dependent (Figure 3c). This stands in stark contrast to previous studies on intravenous delivery of PBAE-DNA complexes, in which jetPEI outperformed PBAEs by two orders of magnitude in the lungs.[15] A recent study demonstrated in vivo mRNA expression in the lungs following pulmonary administration of cationic polymeric nanoparticles [21], however, to our knowledge, ours is the first example of biodegradable polymeric nanoparticles to successfully delivering mRNA to the lungs following intravenous injection in mice.
Figure 3.
In vivo efficacy of PBAE nanoparticles. DD90-C12-122. DD90-C12-122 (formulated with and without 7 mol% PEG-lipid), PEGylated C32-C12-118, and in vivo jetPEI nanoparticles were formulated with luciferase-coding mRNA and delivered via tail vein injection to mice at a dose of 0.5 mg mRNA/kg mouse. (a) Representative luminescent image of excised organs following PEGylated DD90-C12-122 delivery demonstrate protein expression mainly in the lungs. (b) At a dose of 0.5 mg mRNA kg−1 mouse injected intravenously, PEGylated DD90-C12-122, as well as PEGylated C32-C12-118, yielded significantly greater luminescence than a non-PEGylated counterpart and had protein expression comparable to that of jetPEI in the lungs; * indicates p < 0.05, ** indicates p < 0.01. (c) Luminescent radiance is dependent on the dose of PEGylated DD90-C12-122 nanoparticles. (d) Imaging of mouse organs following injection of PEGylated DD90-C12-122 nanoparticles loaded with fluorescent mRNA revealed that nanoparticle biodistribution is similar across major organs (representative fluorescent image shown).
As PBAE nanoparticles were primarily effective in promoting mRNA translation in the lungs, we next studied biodistribution of these delivered mRNA. PEGylated DD90-C12-122 nanoparticles were formulated with fluorescently-labeled mRNA and injected intravenously in mice (0.5 mg kg-1 mRNA). As seen in Figure 3d, fluorescence is detected broadly in all organs excised at a similar level at 6 hours post-injection (Figure S10), despite the fact that mRNA is primarily expressed in the lungs. Interestingly, many lipid-mRNA nanoparticles in the literature also distribute throughout the organs, but deliver mRNA most effectively to the liver following systemic administration.[22,23] The disconnect between RNA biodistribution and the site of RNA efficacy has also been observed using lipopolymeric nanoparticles for siRNA delivery to the lungs[24]. Specific expression in the lungs could be related to selective tagging/trafficking by endogenous proteins: For some lipid-RNA nanoparticle formulations, liver specificity is due to adsorption of Apolipoprotein E (ApoE) onto the nanoparticles, which thereby traffics the formulation to hepatocytes which express the ApoE receptor[25]. While the mechanism by which formulations developed here result in preferential expression of mRNA in the lung remains unclear, it is possible that, like lipid nanoparticles, interaction with serum proteins or surface receptors on lung endothelium may account for the lung specificity of our particles. Regardless, the safe and effective delivery of nucleic acid to the lungs may have utility in the treatment of a variety of diseases at the genetic level, including pulmonary hypertension[26] and cancer.[27]
In conclusion, we have developed PBAE-lipid mRNA nanoparticles with improved serum stability, which are capable of delivering mRNA to the lung. To our knowledge, this is the first demonstration of systemic mRNA delivery to the lung using degradable polymeric nanoparticles. We hypothesize that degradability of the materials used in the delivery of RNA therapeutics may be particularly important for applications requiring repeat dosing of mRNA such as protein replacement therapies.[10] The function of PEGylated DD90-C12-122 and C32-C12-118 nanoparticles demonstrated here warrants continued development and optimization to further improve potency. One approach to potentially access even more potent formulations would utilize Design of Experiment methodology to vary the formulation components, which has been shown to lead to improved mRNA delivery efficacy in a separate liver-targeting system.[21] Importantly, these data demonstrate a correlation between serum stability and in vivo efficacy: the terpolymers that form serum-stable nanoparticles when formulated with a low amount (7 mol%) of PEG-lipid also led to the most potent in vivo mRNA delivery. The efficacy of these top-performing PEGylated PBAE candidates also compared favorably in vivo to commercially-available jetPEI. However, given the low in vivo potency of highly PEGylated serum-stable nanoparticles, serum stability alone is not a predictor of in vivo function. Still, in vitro assessment of serum stability, when used as a secondary metric following an efficacy screen, may prove useful in accelerating the discovery of PBAE formulations for intravenous nucleic acid delivery.
Experimental Section
Polymers were synthesized by dissolving diacrylate, amine, and alkyl amine monomers (concentration 1M) in anhydrous dimethyl formamide at a molar ratio of 1.2:0.7:0.3 and reacted for 48 hours at 90°C. End capping monomers were then added at room temperature and reacted for an additional 24 hours, followed by 2 washes with diethyl ether. Polymers were re-dissolved in anhydrous dimethyl sulfoxide and stored at −80°C. Nanoparticles were synthesized by dissolving mRNA in sodium acetate buffer (pH 5.2) and polymer/PEG-lipid (when applicable) in either sodium acetate buffer (for non-PEGylated nanoparticles) or ethanol (for PEGylated nanoparticles) as a separate phase. The two phases were mixed vigorously in order to form the nanocomplexes. PEGylated particles were subjected to dialysis in PBS in order to remove ethanol before experiments.
Supplementary Material
Acknowledgements
The authors would like to acknowledge project funding by Shire Pharmaceuticals (Lexington, MA) and the MIT Skoltech Initiative, in addition to partial funding from the Cancer Center Support (core) Grant P30-CA14051 from the NCI. The authors acknowledge the contribution of the Koch Institute Nanotechnology Materials Core for their assistance in the high-throughput particle sizing and CryoTEM imaging.
Footnotes
Supporting information for this article is given via a link at the end of the document.
More detailed experimental procedures can be found in the supplementary information provided.
References
- [1].Friedmann T. Science. 1989;244:1275–1281. doi: 10.1126/science.2660259. [DOI] [PubMed] [Google Scholar]
- [2].Draghici B, Ilies MA. J. Med. Chem. 2015;58:4091–4130. doi: 10.1021/jm500330k. [DOI] [PubMed] [Google Scholar]
- [3].Kauffman KJ, Webber MJ, Anderson DG. J. Control. Release. 2015 doi: 10.1016/j.jconrel.2015.12.032. DOI 10.1016/j.jconrel.2015.12.032. [DOI] [PubMed] [Google Scholar]
- [4].Yamamoto A, Kormann M, Rosenecker J, Rudolph C. Eur. J. Pharm. Biopharm. 2009;71:484–489. doi: 10.1016/j.ejpb.2008.09.016. [DOI] [PubMed] [Google Scholar]
- [5].Kanasty R, Dorkin JR, Vegas A, Anderson D. Nat. Mater. 2013;12:967–77. doi: 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]
- [6].Ibraheem D, Elaissari a, Fessi H. Int. J. Pharm. 2014;459:70–83. doi: 10.1016/j.ijpharm.2013.11.041. [DOI] [PubMed] [Google Scholar]
- [7].Boussif O, Lezoualc’h F, Zanta M. a, Mergny MD, Scherman D, Demeneix B, Behr JP. Proc. Natl. Acad. Sci. U. S. A. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bennett CF, Chiang MY, Chan H, Shoemaker JE, Mirabelli CK. Mol. Pharmacol. 1992;41:1023–1033. [PubMed] [Google Scholar]
- [9].Moghimi SM, Symonds P, Murray JC, Hunter a. C., Debska G, Szewczyk A. Mol. Ther. 2005;11:990–995. doi: 10.1016/j.ymthe.2005.02.010. [DOI] [PubMed] [Google Scholar]
- [10].Lynn DM, Langer R. J. Am. Chem. Soc. 2000;122:10761–10768. [Google Scholar]
- [11].Green JJ, Shi J, Chiu E, Leshchiner ES, Langer R, Anderson DG. Bioconjug. Chem. 2006;17:1162–1169. doi: 10.1021/bc0600968. [DOI] [PubMed] [Google Scholar]
- [12].Cutlar L, Zhou D, Gao Y, Zhao T, Greiser U, Wang W, Wang W. Biomacromolecules. 2015;16:2609–2617. doi: 10.1021/acs.biomac.5b00966. [DOI] [PubMed] [Google Scholar]
- [13].Guerrero-Cázares H, Tzeng SY, Young NP, Abutaleb AO, Quiñones-Hinojosa A, Green JJ. ACS Nano. 2014;8:5141–5153. doi: 10.1021/nn501197v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Mastorakos P, da Silva AL, Chisholm J, Song E, Choi WK, Boyle MP, Morales MM, Hanes J, Suk JS. Proc. Natl. Acad. Sci. 2015;112:201502281. doi: 10.1073/pnas.1502281112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zugates GT, Peng W, Zumbuehl A, Jhunjhunwala S, Huang Y-H, Langer R, a Sawicki J, Anderson DG. Mol. Ther. 2007;15:1306–1312. doi: 10.1038/sj.mt.6300132. [DOI] [PubMed] [Google Scholar]
- [16].Eltoukhy A. a., Chen D, Alabi C. a., Langer R, Anderson DG. Adv. Mater. 2013;25:1487–1493. doi: 10.1002/adma.201204346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Eltoukhy A. a., Siegwart DJ, Alabi C. a., Rajan JS, Langer R, Anderson DG. Biomaterials. 2012;33:3594–3603. doi: 10.1016/j.biomaterials.2012.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lauraeus S, Holopainen JM, Taskinen MR, Kinnunen PKJ. Biochim. Biophys. Acta-Biomembranes. 1998;1373:147–162. doi: 10.1016/s0005-2736(98)00102-3. [DOI] [PubMed] [Google Scholar]
- [19].Mishra S, Webster P, Davis ME. Eur. J. Cell Biol. 2004;83:97–111. doi: 10.1078/0171-9335-00363. [DOI] [PubMed] [Google Scholar]
- [20].Jung CW, Jacobs P. Magn. Reson. Imaging. 1995;13:661–674. doi: 10.1016/0730-725x(95)00024-b. [DOI] [PubMed] [Google Scholar]
- [21].Jarzębińska A, Pasewald T, Lambrecht J, Mykhaylyk O, Kümmerling L, Beck P, Hasenpusch G, Rudolph C, Plank C, Dohman C. Angew. Chem. Int. Ed. 2016;55:9591–95. doi: 10.1002/anie.201603648. [DOI] [PubMed] [Google Scholar]
- [22].Kauffman KJ, Dorkin JR, Yang JH, Heartlein MW, DeRosa F, Mir FF, Fenton OS, Anderson DG. Nano Lett. 2015;15:7300–7306. doi: 10.1021/acs.nanolett.5b02497. [DOI] [PubMed] [Google Scholar]
- [23].Fenton OS, Kauffman KJ, McClellan RL, Appel EA, Dorkin JR, Tibbitt MW, Heartlein MW, DeRosa F, Langer R, Anderson DG. Adv. Mater. 2016;28:2939–2943. doi: 10.1002/adma.201505822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Dahlman JE, Barnes C, Khan OF, Thiriot A, Jhunjunwala S, Shaw TE, Xing Y, Sager HB, Sahay G, Speciner L, et al. Nat. Nanotechnol. 2014;19 doi: 10.1038/nnano.2014.84. DOI 10.1038/nnano.2014.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Akinc A, Querbes W, De S, Qin J, Frank-Kamenetsky M, Jayaprakash KN, Jayaraman M, Rajeev KG, Cantley WL, Dorkin JR, et al. Mol. Ther. 2010;18:1357–64. doi: 10.1038/mt.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Budhiraja R, Tuder RM, Hassoun PM. Circulation. 2004;109:159–165. doi: 10.1161/01.CIR.0000102381.57477.50. [DOI] [PubMed] [Google Scholar]
- [27].Kumar V, Abbas AK, Aster JC. Robbins & Cotran Pathologic Basis of Disease. Elsevier; Philadelphia, PA: 2015. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.