Abstract
Microbial burden of chronic wounds is believed to play an important role in impaired healing and development of infection-related complications. However, clinical cultures have little predictive value of wound outcomes, and culture-independent studies have been limited by cross-sectional design and small cohort size. We systematically evaluated the temporal dynamics of the microbiota colonizing diabetic foot ulcers (DFU), a common and costly complication of diabetes, and its association with healing and clinical complications. Dirichlet multinomial mixture modeling, Markov chain analysis, and mixed-effect models were used to investigate shifts in the microbiota over time and its associations with healing. Here we show to our knowledge previously unreported temporal dynamics of the chronic wound microbiome. Microbiota community instability was associated with faster healing and improved outcomes. DFU microbiota were found to exist in one of four community types that experienced frequent and non-random transitions. Transition patterns and frequencies associated with healing time. Exposure to systemic antibiotics destabilized the wound microbiota, rather than altering overall diversity or relative abundance of specific taxa. This study provides to our knowledge previously unreported evidence that the dynamic wound microbiome is indicative of clinical outcomes and may be a valuable guide for personalized management and treatment of chronic wounds.
INTRODUCTION
Chronic, non-healing wounds affect 6.5 million patients annually in the US and are an increasing public health and economic threat, exceeding estimated annual treatment costs of $9.7 billion (Bickers et al. 2006). Chronic wounds almost always affect individuals with an underlying predisposition (e.g. obesity, advanced age, diabetes) and are often disguised as a comorbid condition. A major type of chronic wound is the diabetic foot ulcer (DFU), a common complication of diabetes that results from neuropathy coupled with mechanical stress and tissue breakdown. Those with diabetes have a 15–25% lifetime incidence of DFU (Valensi et al. 2005) and result in amputation in 15.6% of cases (Ramsey et al. 1999). Projections estimate that diabetes will continue to increase in prevalence (Guariguata et al. 2014); thus addressing management and treatment strategies for this complication is critical.
Microbial bioburden is believed to contribute to impaired healing of chronic wounds and it is estimated that over 50% of DFUs are infected upon presentation (Prompers et al. 2007); however, infections are difficult to diagnose due to the diminished or absent clinical signs in DFUs resulting from peripheral neuropathy and/or vascular disease (Glaudemans et al. 2015). Without clinical suspicion, wound cultures provide little diagnostic value, as bacteria colonize all open wounds. Our previous work demonstrated that clinical cultures underestimate bacterial diversity and load when compared to culture-independent techniques, based on the prokaryote-specific 16S ribosomal RNA (rRNA) gene. Multiple dimensions of the microbiota may be important, including microbial diversity, microbial load, and abundance of potential pathogens (Gardner and Frantz 2008). Although other studies have used culture-independent methods to examine DFUs and other chronic wound microbiomes, these studies employed cross-sectional designs (Dowd et al. 2008; Price et al. 2009; Gontcharova et al. 2010; Gardner et al. 2013; Wittebole et al. 2014; Wolcott et al. 2015) and the relationship between the wound microbiome and outcomes has not been rigorously examined.
Microbial communities exhibit a wide range of stabilities across the human body (Ding and Schloss 2014; Flores et al. 2014); however, what these differing stabilities mean for the health of the community or the host remain poorly understood. Very little is known about the dynamics of the wound microbiota during healing, deterioration, or exposure to antibiotics. To date, no study has investigated the microbial dynamics of chronic wounds. These dynamics may contain information about the vulnerability of the wound to opportunistic infections or provide insight as to the origin of stalled wound healing. It is critical to study these dynamics to enhance our understanding of chronic wounds and improve our ability to effectively treat them.
We address several important limitations of previous studies by performing a study designed to capture the longitudinal dynamics of microbiota colonizing DFUs and examining the association between the DFU microbiome and clinical outcomes. Microbiota were sampled from DFUs every two weeks for 26 weeks or until healed. We employed high throughput sequencing of the 16S rRNA gene to define multiple metrics of the microbiome, including diversity, stability, and relative abundance of potential pathogens and identified microbiomic features associated with DFU clinical outcomes. Though our study was focused on the microbiota in DFU, many of these findings may be true of other chronic wounds and should be considered in future studies and treatments of chronic wounds.
RESULTS
We enrolled 100 subjects into a prospective, longitudinal cohort study to analyze temporal dynamics of DFU microbiota and association with outcomes using culture-independent approaches. DFU microbiota was collected at initial presentation (baseline) and resampled every two weeks until: 1) DFU healed; 2) lower extremity amputation; or 3) the conclusion of 26 weeks of follow up. All subject received standardized treatment of surgical debridement and offloading. Of the 100 enrolled subjects, 31 experienced an infection-related complication, defined as: 1) amputation; 2) wound deterioration, or 3) development of osteomyelitis. Table S1 summarizes clinical factors by complication status.
Characterization of the DFU microbiota at baseline
DFU microbiomes were determined by sequencing of hypervariable regions V1 through V3 of the 16S ribosomal RNA (rRNA) gene. The most abundant genus identified was Staphylococcus, present in 345 of the 349 samples, with an average relative abundance of 22.77%. The second, third, and fourth most abundant genera were Streptococcus (11.98%; 318 of 349 samples), Corynebacterium (11.46%; 346 of 349 samples), and Anaerococcus (7%; 300 of 349 samples), respectively. All other genera represented <5% of bacterial relative abundance in this dataset. A more detailed characterization can be found in Table S2. We further classified Staphylococcus operational taxonomic units (OTUs) to species level for 79.5% of the OTUs. Of the 22.77% attributed to Staphylococcus, 13.3% was classified as S. aureus, 5.3% was S. pettenkoferi, and 4% was not further classified. While S. aureus is a common DFU isolate, the high abundance of S. pettenkoferi was surprising as this species was only recently characterized in 2007 (Trülzsch et al. 2007), though it was identified as the cause of osteomyelitis in patient with a chronic DFU in France (Loïez et al. 2007).
DFU microbiota can be partitioned into four community types
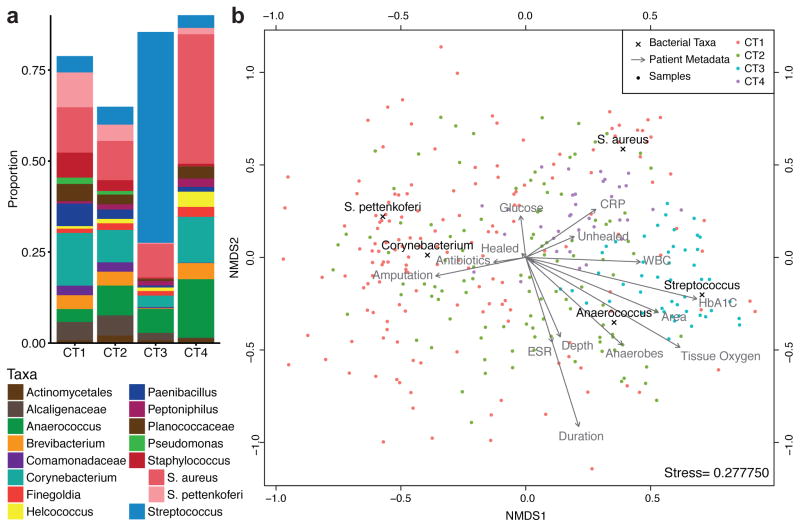
We assigned DFUs to community types with the Dirichlet multinomial mixture (DMM) model-based approach (Holmes et al. 2012). The DMM model supposes a more biologically relevant distribution of data, which overcomes limitations of alternative methods such as k-means (Holmes et al. 2012) and PAM clustering (Ding and Schloss 2014). The DFU microbiomes were clustered into 4 groups, or Community Types (CT), by minimizing the Laplace approximation (Fig. S1). The top five differentiating taxa contributed 48.9% of the total difference between a one and four component model, though the major distinguishing taxa were Streptococcus (25.6%) and S. aureus (11.8%) (Fig. 1A). CT3 DFUs were characterized by high relative abundances of Streptococcus (median = 64.0%). CT4 DFUs were comprised of relatively high levels of S. aureus (median=23.8%). CT1 and CT2 were highly heterogeneous with no dominant taxa contributing more than a median of 5% of total relative abundance. This was also reflected by Theta values, a measure of cluster variability with smaller values corresponding to highly variable communities, which were 3.7 and 6.9, for the CT1 and CT2 compared to 16.4 and 10.5 for CT3 and CT4, respectively. Community type summaries are described in greater detail in Table S3.
Figure 1. The DFU microbiome clusters into four Community Types.
(A) DFU samples partitioned into four clusters by Dirichlet multinomial mixture model. Mean relative abundances of bacterial taxa in DFU samples assigned to each Community Type. Relative abundance is shown on the Y-axis. Taxa are filtered to those with a mean abundance greater than 1%. (B) Sample similarity between DFU microbial communities were calculated using the Bray-Curtis distance and these distances were ordinated and visualized via non-metric multidimensional scaling (NMDS). Each taxonomic contribution to community differentiation is overlaid with black text and “x” indicating the exact location. The impacts of various metadata are depicted as vectors labeled with gray text. Success of NMDS ordination is represented by the stress score, which measures the agreement between the 2-D and multidimensional representations. Stress scores range from 0 to 1 and scores below 0.3 are considered good approximations. Samples, taxa, and metadata that are closer together are more related. Samples are color-coded based on community type.
To better visualize how CTs were associated with microbiota composition and clinical features, we generated a biplot depicting these relationships (Fig. 1B). As would be expected, the taxa vectors for Streptococcus and S. aureus are closely associated with the CT3 and CT4, respectively. Interestingly, the samples with the highest proportion of S. aureus are not included in CT4, demonstrating the importance of the whole community in distinguishing clusters. Streptococcus was closely associated with HbA1C levels and anaerobe levels with ulcer depth. Serum C-reactive protein levels (CRP) and white blood cell counts (WBC), both measures of inflammation used to inform the diagnosis of infections, localized separately with CT4 and CT3, respectively. Subject outcomes also contributed to data separation, with amputation localizing with CT1 and CT2, and unhealed subjects localizing with CT4. Further quantification of the correlation between clinical factors and DFU microbiota are provided in Table S4.
The frequency of Community Type transitions in DFU are associated with clinical outcomes
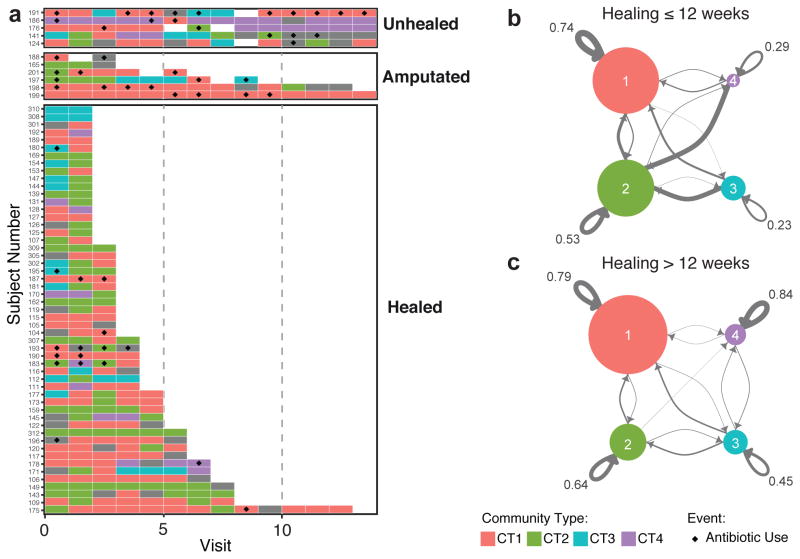
We next investigated the stability of the CTs by exploring the frequency and type of CT transitions. The DFU microbiota was highly dynamic with CT transitions occurring every 1.76 study visits (approximately 3.52 weeks) on average (Fig. 2A). Transition frequencies were significantly associated with subject outcomes (healed = 1.60, unhealed = 2.04, amputation = 3.08 study visits/CT-transition). We further subdivided healed subjects into those whose ulcers closed in <12 weeks and those closed in >12 weeks. Consistent with our analysis, the faster healing subjects experienced greater transition frequencies (<12 weeks = 1.45, >12 weeks 2.11 study visits/CT-transition, Wilcoxon p-value = 0.011).
Figure 2. DFU Community Types are dynamic.
(A) Per patient illustration of Community Type switching grouped by outcome. Depicted on the X-axis is visit number. Each row on the Y-axis represents a subject with a DFU. Colored boxes illustrate which Community Type was colonizing the DFU at the indicated visit number. Empty tiles represent a missed visit, whereas gray tiles indicate that a sample was not collected or available for analysis at that time point. The black diamonds indicate that the patient received antibiotics since the last visit. Only subjects that participated in >1 study visit are shown. (B) Markov chain visualization depicting the differential transition probabilities between community types of DFUs that healed in 12 weeks or did not. Each node represents a Community Type, arrows indicate the transition direction and probability (thickness), node size represents number of samples. Annotated are the self-transition probabilities.
We then questioned whether transition patterns between CTs were related to ulcer outcomes. By quantifying transitions between CTs we could represent the data as a Markov chain, with nodes representing CTs and edges representing transition frequencies by their weight (Fig. 2B). The transition patterns between those that healed in <12 weeks and those that healed in >12 weeks were significantly different (p-value < 0.0001). In those who healed in <12 weeks, CT1 and CT2 dominated the transitions and were noted to have high self-transition rates of 0.74 and 0.53, respectively. In contrast CT3 and CT4 experienced lower self-transition rates of 0.23 and 0.29, and had a predilection for transitioning to CT2. For subjects that took >12 weeks to heal, there is a marked increase in self-transitions, with ulcers stalling in CT3 and CT4 at rates of 0.45 and 0.84, respectively, indicating that the stability of these CTs may be detrimental to wound healing. Analysis of the stationary distribution and expected recurrence time revealed similar trends (Table S5). The presence or absence of transitions between CT3 and CT4 also differentiated the two groups, with no recorded instances in wounds healing in <12 weeks. Together these findings suggest that community stability reflects a delayed healing phenotype.
DFU with more dynamic microbiota heal faster than those with less dynamic microbiota
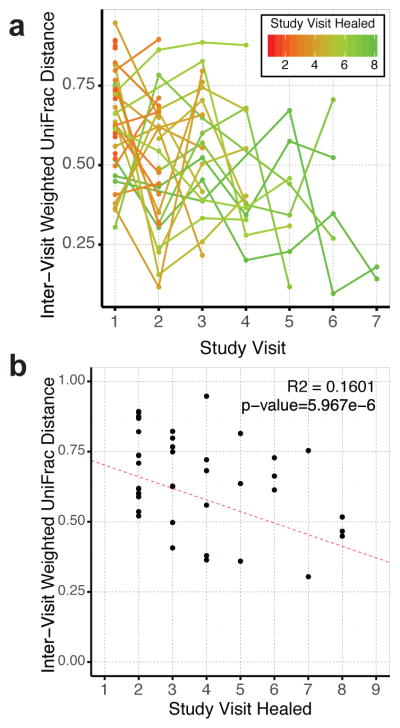
To address more subtle patterns of variation, which may not be apparent when examining broad community types, we used the inter-visit weighted UniFrac (WUF) distance as a proxy of stability. The weighted UniFrac metric measures the proportion of shared OTUs, their phylogenetic relationships, and their relative distributions on a scale of 0 to 1, with higher values indicating greater instability. We generated mixed-effect linear regressions to model the relationship between microbiota instability and time required to heal in those that healed within 24 weeks. This model suggests that all ulcers are slowly stabilizing at a rate of −0.024/visit; however, slow healing ulcers begin in a more stable state (−0.036 per visit required to heal) (Fig. 3A). Because mixed-effect models do not allow generation of a traditional R2 value, we calculated marginal and conditional pseudo-R2 values, which reveals an estimate of the variance due to the fixed effects alone and the combined model of fixed and random effects respectively. The marginal R2 was estimated to be 0.201 and the conditional to be 0.280, indicating that our model explains a moderate amount of the variation.
Figure 3.
Inter-visit Weighted UniFrac distances associations with healing time for subjects that healed during the study. X axes represent the study visit; study visits were 2 weeks apart. (A) Inter-visit distances are shown for each subject and depict a negative trend over time. Line and point colors represent the number of study visits that the ulcer persisted (red = 1, green = 8). Ulcers stabilize at a rate of −0.024/visit, but start at a lower rate in those ulcers that require more time to heal (−0.036 per visit required to heal). (B) Inter-visit distances between baseline and first study visit as a function of number of visits until healing. A negative correlation is found even within this initial comparison (R2 = 0.1601, p<0.0001).
The first inter-visit distance, between the baseline study visit and following visit, includes the effect of the initial surgical debridement. Thus it was possible that the high instability in faster healing wounds was an artifact of the first study visit being weighted more. To address this concern, we investigated the relationship between healing time and the amount of change between baseline and the following visit (2 weeks’ time) using a traditional linear model. We found the same negative association between healing time and the inter-visit distance (R2 = 0.16, p<0.0001) (Fig. 3B), suggesting the effect is independent of debridement.
Effect of antibiotics on temporal stability in DFU microbiota
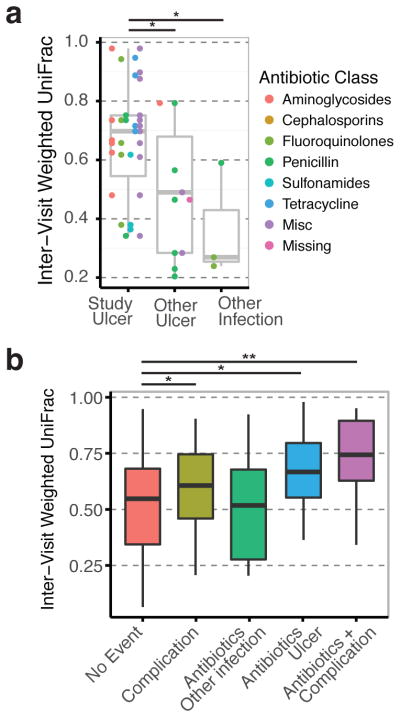
During the course of the study, 32 subjects required the administration of antibiotics, which afforded us the opportunity to glean the effects of antibiotics on ulcer microbiomes. Antibiotic exposure did not drive microbiota variation in our samples (Fig 1B). Furthermore, we did not detect any significant changes in community diversity as measured by the Shannon index or OTU richness, perhaps due to unique interactions between specific antibiotic classes and personal microbial communities. We binned antibiotics into categories based on their class and mechanism of action, and assessed their potential to disrupt microbial communities using the inter-visit WUF distances as before. We did not detect significant differences in microbial stability due to antibiotic class. However, in half of the cases, the antibiotics were prescribed to treat infections not involving the studied ulcer (e.g. other ulcers, urinary tract infection, upper respiratory infection, sinus infection). When we examined the subjects treated specifically for the study ulcer, we found that antibiotics administered produced significantly higher community disruption than if the antibiotic was given for a different indication (Fig. 4A).
Figure 4.
Effects of antibiotics on microbial communities in DFUs. (A) Boxplot showing the inter-visit Weighted UniFrac distances of subjects during exposure to antibiotics split by indication. Antibiotics given for the ulcer being studied produces greater community disruption than antibiotics given for other ulcers or other infections. Antibiotic class did not yield more information. (B) Boxplot showing the inter-visit distances of all samples binned by event type (complication, antibiotics, both, or none). Antibiotics and ulcer complications both disrupt the microbiota, and their combined effect is additive.
In some cases, during the same time period that antibiotics were administered, the ulcer was designated as having a complication (wound deterioration or osteomyelitis). We modeled how these complications interacted with the antibiotics using mixed-effect linear regressions as before (Fig. 4B). We found that both complications and antibiotics contributed to community disruption, though the larger effect was noted for antibiotics (WUF = 0.084 and 0.140 respectively). Furthermore, targeted antibiotics and complications had an additive effect on the amount of community disruption (WUF = 0.201).
DISCUSSION
Here, we explore the temporal dynamics of the human chronic wound microbiota. Microbiome studies in other body sites have shown that disease states are associated with less stability (Martinez et al. 2008; Jenq et al. 2012; DiGiulio et al. 2015). Surprisingly, DFUs that experienced delayed healing or resulted in amputation were associated with increased stability, while the inverse was true for faster healing wounds. One way of interpreting these findings is to conclude that there is no “normal” DFU community. A wound is by definition an abnormal and transient state in physiology. As such, colonizing bacteria should be considered opportunistic and unlikely to have evolved harmonious methods of existing with the host. From this perspective, instability in the microbiome is a reflection of effective control of wound bacteria, which prevents any community structure from stabilizing. In contrast, a DFU with a stable outgrowth of certain bacteria reflects a stalled healing state where the colonizing bacteria have overridden the host’s defenses.
We found that the DFU microbiome can be partitioned into 4 community types. Increased community type transitions were associated with improved healing rates; however, these community type transitions were not random. In quickly healing ulcers, CT1 and CT2 were substantially more likely to remain unchanged, whereas CT3 and CT4 were more likely to transition to CT2. In slow or unhealing wounds, we found that CT3 and CT4 became much more resilient. These findings suggest that the prognostic capacity of transition frequencies would be augmented by information of community structure. Further studies are needed to delineate cause and effect relationships of the microbiota with the wound environment.
Despite the regular use of antibiotics to treat infections, little is known about their impact on microbial communities in chronic wounds. We did not detect any differences in community diversity or composition due to antibiotic exposure, unlike the gut where exposure to certain antibiotics is known to decrease diversity levels, predisposing to infection by Clostridium difficile (Dethlefsen and Relman 2011; Stein et al. 2013). Instead, as in other body sites (Keeney et al. 2014; Modi et al. 2014; Zhang et al. 2014; Mayer et al. 2015), antibiotics disrupted the microbiota. The extent of community disruption was not dependent on the class of antibiotic; rather it was whether the antibiotic was targeted towards the ulcer being studied. However, our analysis is limited by the biweekly sampling frequency, limiting the detection of short-lived changes.
Another limitation of this study is that relatively few subjects required amputations or did not heal during the study, perhaps a reflection of the regular care the subjects received for their DFUs at 2 week intervals. Therefore, we could not robustly analyze these specific outcomes with respect to the microbiota. To circumvent this obstacle, we relied upon alternative endpoints, including rate of healing and aggregate infection related complications (i.e. wound deterioration, osteomyelitis, amputation). The cohort was also disproportionately white and male, a reflection of the demographic composition at the study site. While a homogeneous cohort is advantageous from a study design standpoint, limiting potential variability due to race and sex, the findings should be interpreted with caution. Studies in more diverse cohorts should be conducted to determine if the findings presented here are broadly applicable across race and sex.
In some reports, over half of DFUs are infected at the time of presentation (Prompers et al. 2007); however, identifying reliable criteria to diagnose an infection is complicated by the attenuated response to infections in diabetic persons (Brem and Tomic-Canic 2007). While our results would benefit from validation in larger cohorts, and their applicability to other types of chronic wounds needs to be tested, we provide evidence that the temporal dynamics of the wound microbiome may be useful for identifying stalled wounds requiring antibiotic treatment. We envision that these findings will ultimately guide clinicians in the management of chronic wounds in a personalized manner.
MATERIALS AND METHODS
Study Design
A prospective, longitudinal cohort design was used to examine DFU microbiota and outcomes in 100 subjects. DFU microbiota was collected at initial presentation (baseline) and resampled every two weeks until: 1) DFU healed; 2) lower extremity amputation; or 3) the conclusion of 26 weeks of follow up. The Institutional Review Boards at the University of Iowa and the University of Pennsylvania approved all study procedures.
Setting and Sample
Subjects were enrolled from September 2008 through October 2012 at the University of Iowa Hospitals and Clinics (UIHC) and the Iowa City Veteran’s Affairs Medical Center (VA). Subjects were recruited through local media advertisements and from outpatient clinics at UIHC and the VA. The target population was diabetic adults (i.e., 18 years of age or older) with a DFU on the plantar surface of the foot and ankle/brachial or toe/brachial indexes > 0.5 to ensure the sample was a homogenous group of neuropathic DFUs. Individuals meeting these criteria were enrolled after providing informed written consent.
We standardized the management of the study DFUs after enrollment, including ulcer dressings (i.e., Lyofoam®, Molnlycke Health Care), devices used for offloading (i.e., total contact casts were used for 87 subjects; DH boots for 13 subjects), and ulcer debridement (i.e., aggressive sharp debridement of necrotic tissue in the wound bed was completed at baseline and callus on the wound edge was removed every two weeks), in order to minimize the number of factors unrelated to ulcer bioburden that could impact DFU outcomes. DFU management did not include antimicrobial dressings, topical antimicrobials, and/or systemic antibiotics, unless an infection-related complication was present at enrollment or occurred during follow-up. Baseline data were collected immediately after enrollment. Study data were collected every two weeks until one of the study endpoints was reached.
Study Variables
Clinical factors
The research team measured a set of clinical factors in order to identify pertinent co-variates for the analyses and to comprehensively describe the study sample. At baseline, demographic data, diabetes type and duration, and duration of study ulcer were collected using subject self-report and medical records. Standard laboratory tests were used to measure baseline glycemic control (haemoglobin A1c levels), as well as immune (White blood cell count) and inflammatory markers (C reactive protein). The research team assessed each subject for ischemia using toe-brachial index and for neuropathy using 5.07 Semmes-Weinstein monofilament. Transcutaneous oxygen pressure was measured at baseline and at each follow-up visit, using a transcutaneous oxygen monitor (Novametrix 840®, Novametrix Medical Systems Inc.). Ulcer location was categorized as forefoot, midfoot, or heel.
Microbiome
Ulcer specimens were collected using the Levine technique. After cleansing with non-bacteriostatic saline, an Amies swab (Copan, Italy) was rotated over a 1-cm2 area of viable wound tissue in the center of the wound bed for five seconds, using sufficient pressure to extract wound-tissue fluid, DNA was isolated from swab specimens as previously described (Gardner et al. 2013). Levine’s swab technique was used because it samples the viable, deep wound tissue in a non-invasive manner, allowing for serial sampling of the wound over time. Levine’s swab produces comparable results to tissue specimens for microbial load and diversity (Gardner et al. 2006). Amplification of the 16S rRNA gene V1–V3 region was performed as described previously (Meisel et al. 2016), using the Illumina MiSeq platform with 300 bp paired-end ‘V3’ chemistry. This resulted in a dataset of 7,702,607 high quality, classifiable sequences used in the final analysis, with a mean of 22,070 (range 1,206–69,167) sequences per sample. Sequence pre-processing followed methods described previously (Meisel et al. 2016), modified by performing denovo OTU clustering via UCLUST, assigning taxonomy with BLAST, and subsampling at 1200 sequences per sample. Sequences corresponding to the taxa “Geobacillus”, “Bacillus”, and “Lactococcus” were removed as these were identified as contaminants in the negative controls. QIIME 1.9.0 (Caporaso et al. 2010) was used for initial stages of sequence analysis. Sequences were clustered into OTUs (operational taxonomic units, a proxy for ‘species’) using UCLUST (Edgar 2010) at 97% sequence similarity. Microbial diversity was calculated using the following alpha diversity indices: 1) Shannon diversity index; 2) Faith’s phylogenetic distance (PD); and 3) number of observed OTUs. Taxonomic classification of sequences were made using BLAST, as implemented in QIIME.
Outcomes
Members of the research team, who were blinded to the microbiota status, assessed healing and infection-related complications every two weeks. Ulcer closure was assessed using the Wound Healing Society’s definition of “an acceptably healed wound,” a valid and reliable definition (Margolis et al. 1996). The outcome “healed by 12 weeks” was defined as wound closure before or at 12 weeks of follow-up. “Development of infection-related complications” was defined as wound deterioration, new osteomyelitis, and/or amputations due to DFU infections.
Wound deterioration was defined as the new development of frank erythema and heat, and an increase in size > 50% over baseline. Two members of the research team independently assessed each DFU for erythema and heat. Two members of the research team independently assessed size using the VeVMD® digital software system (Vista Medical, Winnipeg, Manitoba, Canada), which was loaded on a Dell Latitude D630 laptop computer (Dell, Round Rock, Texas). Digital images were taken that contained the ulcer, a 3×3 square-centimeter image orientation card, and a single-point wound-depth indicator (i.e. A cotton-tipped swab that had been placed in the deepest aspect of the DFU and marked where the swab intersected with the plane of the peri-wound skin) and uploaded into the VeVMD program. VevMD tools were used to trace the ulcer outline and a line along the wound depth indicator to generate measures of depth and surface area.
Osteomyelitis was assessed using radiographs and MRI at baseline and during follow-up visits when subjects presented with new tracts to bone, wound deterioration, elevated temperature, elevated white count, elevated erythrocyte sedimentation rate, or elevated C-reactive protein. If these indicators were absent at follow-up, radiographs were not retaken. Subjects experiencing lower limb amputations had their medical records reviewed by the research team to ensure amputations were due to DFU infection, and not some other reason.
Data Analyses
The R Statistical Package (R Core Team 2016) was used for all computations. Non-parametric Wilcoxon rank-sum tests were used to compare differences between groups. Spearman correlations were used to correlate continuous variables. Kruskal-Wallis tests, followed by Wilcoxon rank sum post-hoc tests, were used for categorical variables. Linear models were calculated in base R; mixed-effect regressions were generated using the NLME package (Pinheiro et al. 2007). Partial and conditional pseudo-R2 values were calculated using the piecewiseSEM package (Lefcheck 2016). Sample biplot was generated using the Breadcrumbs package as done in (Morgan et al. 2015). Differences in Markov chain transition frequencies were tested with a Fisher’s test and simulated p-value. Dirichlet multinomial mixture modeling was performed using the R package Dirichlet Multinomial (v1.10.0). Counts were calculated at the highest level of taxonomic classification. The number of community types was determined by selecting the number of Dirichlet components that minimized the Laplace approximation of the model evidence (Holmes et al. 2012). Each sample was assigned to the community type that had the largest posterior probability. Inter-visit distances were calculated using the weighted UniFrac distance between consecutive visits. If visits were discontinuous (i.e. missing sample) no distances were reported.
Supplementary Material
Acknowledgments
We thank John E. Femino, MD and Phinit Phisitkul, MD (University of Iowa) for managing the patient population and assisting with recruitment; Kris Heilmann and the University of Iowa Microbiology Lab for DFU cultures; and the patients who participated in this study. This work was supported by grants from the National Institutes of Health [NINR R01NR009448 to SEG, NIAMS R00AR060873, NIAMS R01AR066663 and NINR R01NR015639 to EAG] and a grant from the Pennsylvania Department of Health to EAG. MAL and BPH were supported by an NIH grant to the Department of Dermatology at University of Pennsylvania (NIAMS T32AR007465). The content is solely the responsibility of the authors and does not necessarily represent the official view of the funding bodies. The PA Department of Health specifically disclaims responsibility for any analyses, interpretations, or conclusions. The views expressed in this article are those of the authors and do not necessarily reflect the position or policy of the Department of Veterans Affairs or the United States government.
Footnotes
AUTHOR CONTRIBUTIONS
Conceptualization, Supervision, and Funding Acquisition: SEG and EAG; Methodology: MAL, SEG, DJM, EAG; Formal Analysis: MAL, LK, SLH, DJM; Investigation: MAL, LK, JH, AST, BPH; Data Curation: QZ, CLF; Resources: SEG, SM, DJM, EAG; Visualization: MAL; Writing-Original Draft: MAL, SEG, EAG; Writing-Review and Editing: All authors
References
- Bickers DR, Lim HW, Margolis D, Weinstock MA, Goodman C, Faulkner E, et al. The burden of skin diseases: 2004. J Am Acad Dermatol. 2006 Sep;55(3):490–500. doi: 10.1016/j.jaad.2006.05.048. [DOI] [PubMed] [Google Scholar]
- Brem H, Tomic-Canic M. Cellular and molecular basis of wound healing in diabetes. J Clin Invest. 2007 May;117(5):1219–22. doi: 10.1172/JCI32169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Meth. 2010 May;7(5):335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. PNAS. 2011 Mar 15;108( Supplement 1):4554–61. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGiulio DB, Callahan BJ, McMurdie PJ, Costello EK, Lyell DJ, Robaczewska A, et al. Temporal and spatial variation of the human microbiota during pregnancy. PNAS National Acad Sciences. 2015 Aug 17;112(35):201502875–11065. doi: 10.1073/pnas.1502875112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding T, Schloss PD. Dynamics and associations of microbial community types across the human body. Nature. 2014 May 15;509(7500):357–60. doi: 10.1038/nature13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd SE, Wolcott RD, Sun Y, McKeehan T, Smith E, Rhoads D. Polymicrobial Nature of Chronic Diabetic Foot Ulcer Biofilm Infections Determined Using Bacterial Tag Encoded FLX Amplicon Pyrosequencing (bTEFAP) In: Egles C, editor. PLoS ONE. 10. Vol. 3. Public Library of Science; 2008. Oct 3, p. e3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Bioinformatics. 19. Vol. 26. Oxford University Press; 2010. Oct 1, Search and clustering orders of magnitude faster than BLAST; pp. 2460–1. [DOI] [PubMed] [Google Scholar]
- Flores GE, Caporaso J, Henley JB, Rideout J, Domogala D, Chase J, et al. Temporal variability is a personalized feature of the human microbiome. Genome Biol. 2014;15(12):531. doi: 10.1186/s13059-014-0531-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner SE, Frantz RA. Biol Res Nurs. 1. Vol. 10. SAGE Publications; 2008. Jul, Wound bioburden and infection-related complications in diabetic foot ulcers; pp. 44–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner SE, Frantz RA, Saltzman CL, Hillis SL, Park H, Scherubel M. Diagnostic validity of three swab techniques for identifying chronic wound infection. Wound Repair Regen. 2006 Sep;14(5):548–57. doi: 10.1111/j.1743-6109.2006.00162.x. [DOI] [PubMed] [Google Scholar]
- Gardner SE, Hillis SL, Heilmann K, Segre JA, Grice EA. The neuropathic diabetic foot ulcer microbiome is associated with clinical factors. Diabetes Am Diabetes Assoc. 2013;62(3):923–30. doi: 10.2337/db12-0771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaudemans AWJM, Uçkay I, Lipsky BA. Challenges in diagnosing infection in the diabetic foot. Diabet Med. 2015 Jun;32(6):748–59. doi: 10.1111/dme.12750. [DOI] [PubMed] [Google Scholar]
- Gontcharova V, Youn E, Sun Y, Wolcott RD, Dowd SE. A comparison of bacterial composition in diabetic ulcers and contralateral intact skin. Open Microbiol J. 2010;4:8–19. doi: 10.2174/1874285801004010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guariguata L, Whiting DR, Hambleton I, Beagley J, Linnenkamp U, Shaw JE. Global estimates of diabetes prevalence for 2013 and projections for 2035. Diabetes Res Clin Pract. 2014 Feb;103(2):137–49. doi: 10.1016/j.diabres.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Holmes I, Harris K, Quince C. Dirichlet Multinomial Mixtures: Generative Models for Microbial Metagenomics. In: Gilbert JA, editor. PLoS ONE. 2. Vol. 7. 2012. Feb 3, p. e30126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenq RR, Ubeda C, Taur Y, Menezes CC, Khanin R, Dudakov JA, et al. J Exp Med. 5. Vol. 209. Rockefeller Univ Press; 2012. May 7, Regulation of intestinal inflammation by microbiota following allogeneic bone marrow transplantation; pp. 903–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney KM, Yurist-Doutsch S, Arrieta M-C, Finlay BB. Effects of antibiotics on human microbiota and subsequent disease. Annu Rev Microbiol. 2014;68(1):217–35. doi: 10.1146/annurev-micro-091313-103456. [DOI] [PubMed] [Google Scholar]
- Lefcheck JS. piecewiseSEM: Piecewise structural equation modelling in r for ecology, evolution, and systematics. Methods Ecol Evol. 2016;7(5):573–9. [Google Scholar]
- Loïez C, Wallet F, Pischedda P, Renaux E, Senneville E, Mehdi N, et al. First case of osteomyelitis caused by “Staphylococcus pettenkoferi”. J Clin Microbiol American Society for Microbiology. 2007 Mar;45(3):1069–71. doi: 10.1128/JCM.02328-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis DJ, Berlin JA, Strom BL. Wound Repair Regen. 3. Vol. 4. Blackwell Science; 1996. Jul, Interobserver agreement, sensitivity, and specificity of a “healed” chronic wound; pp. 335–8. [DOI] [PubMed] [Google Scholar]
- Martinez C, Antolin M, Santos J, Torrejon A, Casellas F, Borruel N, et al. Unstable composition of the fecal microbiota in ulcerative colitis during clinical remission. Am J Gastroenterol. 2008 Mar;103(3):643–8. doi: 10.1111/j.1572-0241.2007.01592.x. [DOI] [PubMed] [Google Scholar]
- Mayer BT, Srinivasan S, Fiedler TL, Marrazzo JM, Fredricks DN, Schiffer JT. Rapid and Profound Shifts in the Vaginal Microbiota Following Antibiotic Treatment for Bacterial Vaginosis. J Infect Dis. 2015 Aug 6;212(5):793–802. doi: 10.1093/infdis/jiv079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel JS, Hannigan GD, Tyldsley AS, SanMiguel AJ, Hodkinson BP, Zheng Q, et al. Skin Microbiome Surveys Are Strongly Influenced by Experimental Design. J Invest Dermatol. 2016 May;136(5):947–56. doi: 10.1016/j.jid.2016.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modi SR, Collins JJ, Relman DA. J Clin Invest. 10. Vol. 124. American Society for Clinical Investigation; 2014. Oct 1, Antibiotics and the gut microbiota; pp. 4212–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan XC, Kabakchiev B, Waldron L, Tyler AD, Tickle TL, Milgrom R, et al. Associations between host gene expression, the mucosal microbiome, and clinical outcome in the pelvic pouch of patients with inflammatory bowel disease. Genome Biol BioMed Central. 2015;16(1):67. doi: 10.1186/s13059-015-0637-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro J, Bates D, DebRoy S, Sarkar D. R package version. 2007. Linear and nonlinear mixed effects models. [Google Scholar]
- Price LB, Liu CM, Melendez JH, Frankel YM, Engelthaler D, Aziz M, et al. Community analysis of chronic wound bacteria using 16S rRNA gene-based pyrosequencing: impact of diabetes and antibiotics on chronic wound microbiota. In: Ratner AJ, editor. PLoS ONE. 7. Vol. 4. 2009. p. e6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prompers L, Huijberts M, Apelqvist J, Jude E, Piaggesi A, Bakker K, et al. High prevalence of ischaemia, infection and serious comorbidity in patients with diabetic foot disease in Europe. Baseline results from the Eurodiale study. Diabetologia. 2007 Jan;50(1):18–25. doi: 10.1007/s00125-006-0491-1. [DOI] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: 2016. Available from: https://www.R-project.org. [Google Scholar]
- Ramsey SD, Newton K, Blough D, McCulloch DK, Sandhu N, Reiber GE, et al. Incidence, outcomes, and cost of foot ulcers in patients with diabetes. Diabetes Care. 1999 Mar;22(3):382–7. doi: 10.2337/diacare.22.3.382. [DOI] [PubMed] [Google Scholar]
- Stein RR, Bucci V, Toussaint NC, Buffie CG, Rätsch G, Pamer EG, et al. Ecological Modeling from Time-Series Inference: Insight into Dynamics and Stability of Intestinal Microbiota. In: Mering von C, editor. PLoS Comput Biol. 12. Vol. 9. 2013. Dec 12, p. e1003388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trülzsch K, Grabein B, Schumann P, Mellmann A, Antonenka U, Heesemann J, et al. Staphylococcus pettenkoferi sp. nov., a novel coagulase-negative staphylococcal species isolated from human clinical specimens. Int J Syst Evol Microbiol. 2007 Jul;57(Pt 7):1543–8. doi: 10.1099/ijs.0.64381-0. [DOI] [PubMed] [Google Scholar]
- Valensi P, Girod I, Baron F, Moreau-Defarges T, Guillon P. Quality of life and clinical correlates in patients with diabetic foot ulcers. Diabetes & Metabolism. 2005 Jun;31(3):263–71. doi: 10.1016/s1262-3636(07)70193-3. [DOI] [PubMed] [Google Scholar]
- Wittebole X, De Roock S, Opal SM. A historical overview of bacteriophage therapy as an alternative to antibiotics for the treatment of bacterial pathogens. Virulence Landes Bioscience. 2014 Jan 1;5(1):226–35. doi: 10.4161/viru.25991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolcott RD, Hanson JD, Rees EJ, Koenig LD, Phillips CD, Wolcott RA, et al. Analysis of the chronic wound microbiota of 2,963 patients by 16S rDNA pyrosequencing. Wound Repair Regen. 2015 Oct 14; doi: 10.1111/wrr.12370. n/a–n/a. [DOI] [PubMed] [Google Scholar]
- Zhang M, Jiang Z, Li D, Jiang D, Wu Y, Ren H, et al. Oral Antibiotic Treatment Induces Skin Microbiota Dysbiosis and Influences Wound Healing. Microb Ecol. 2014 Oct 10;69(2):415–21. doi: 10.1007/s00248-014-0504-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.