Abstract
Diabetes is the leading cause of ESRD. Despite evidence for a substantial heritability of diabetic kidney disease, efforts to identify genetic susceptibility variants have had limited success. We extended previous efforts in three dimensions, examining a more comprehensive set of genetic variants in larger numbers of subjects with type 1 diabetes characterized for a wider range of cross-sectional diabetic kidney disease phenotypes. In 2843 subjects, we estimated that the heritability of diabetic kidney disease was 35% (P=6.4×10−3). Genome-wide association analysis and replication in 12,540 individuals identified no single variants reaching stringent levels of significance and, despite excellent power, provided little independent confirmation of previously published associated variants. Whole-exome sequencing in 997 subjects failed to identify any large-effect coding alleles of lower frequency influencing the risk of diabetic kidney disease. However, sets of alleles increasing body mass index (P=2.2×10−5) and the risk of type 2 diabetes (P=6.1×10−4) associated with the risk of diabetic kidney disease. We also found genome-wide genetic correlation between diabetic kidney disease and failure at smoking cessation (P=1.1×10−4). Pathway analysis implicated ascorbate and aldarate metabolism (P=9.0×10−6), and pentose and glucuronate interconversions (P=3.0×10−6) in pathogenesis of diabetic kidney disease. These data provide further evidence for the role of genetic factors influencing diabetic kidney disease in those with type 1 diabetes and highlight some key pathways that may be responsible. Altogether these results reveal important biology behind the major cause of kidney disease.
Keywords: diabetic kidney disease, genetics and development, genome-wide association study, whole exome sequencing
Diabetes is the leading cause of ESRD.1 Among the patients with type 1 diabetes (T1D), as many as one third develop serious renal complications,2 characterized by increasing urinary albumin excretion rates (AER) and decreasing kidney function, measured by eGFR, with 10%–20% of the subjects progressing to ESRD.3,4 In the vast majority of patients with T1D, renal complications reflect the condition of diabetic nephropathy,5 although we use the term diabetic kidney disease (DKD) here to reflect the fact that not all have histologic evidence of diabetic nephropathy. Whereas DKD is associated with high risk of cardiovascular disease6 and premature mortality,7 those who manage to avoid DKD have much better prognoses with survival rates comparable to subjects without diabetes.7 The treatment of DKD, primarily relying on the control of blood glucose levels and on the use of antihypertensive medication, can only retard disease progression rather than restore kidney function or efficiently prevent ESRD. This highlights the need for a better mechanistic understanding of DKD which would provide improved therapeutic targets and biomarkers of progression, both of which are expected to lead to improved personalization of care.
Family studies have shown familial clustering of DKD with a 2.1–2.3-fold increased risk of DKD in T1D siblings of probands with DKD.8–10 There is also evidence for shared genetic background between DKD and other diseases: the parents of the subjects with T1D and DKD have more type 2 diabetes (T2D) and cardiovascular disease.11–13 Despite evidence for genetic predisposition, few compelling signals have been identified for DKD. Positive reports of association with a range of candidate genes have generally failed to find support in larger analyses of independent samples,14 and only a few loci have been identified with genome-wide association studies (GWAS).15–17 In contrast, there have been a number of robust, replicated associations described for CKD in the general population.18–20 However, evidence that these variants influence predisposition to CKD in diabetes is mixed.15,20
The limited success of GWAS on DKD thus far may be due to multiple factors: relatively small sample sizes (in the thousands at the discovery stage); imprecise and variable diagnostics or phenotypic heterogeneity; and a focus on common variants (minor allele frequency [MAF] ≥5%), that neglects the possible contribution of lower frequency variants to DKD predisposition. In other complex traits, such as schizophrenia, initial challenges in identifying robust association signals have been overcome as sample sizes have increased.21,22
To overcome the limitations of previous studies in DKD, in this study we: increased sample size through GWAS meta-analysis; analyzed a range of phenotypic comparisons that encompass different stages and severity of DKD in subjects with T1D; and extended the genome-wide association screen from common variants to lower frequency coding variants through whole exome sequencing (WES).
Results
DKD Traits Are Heritable
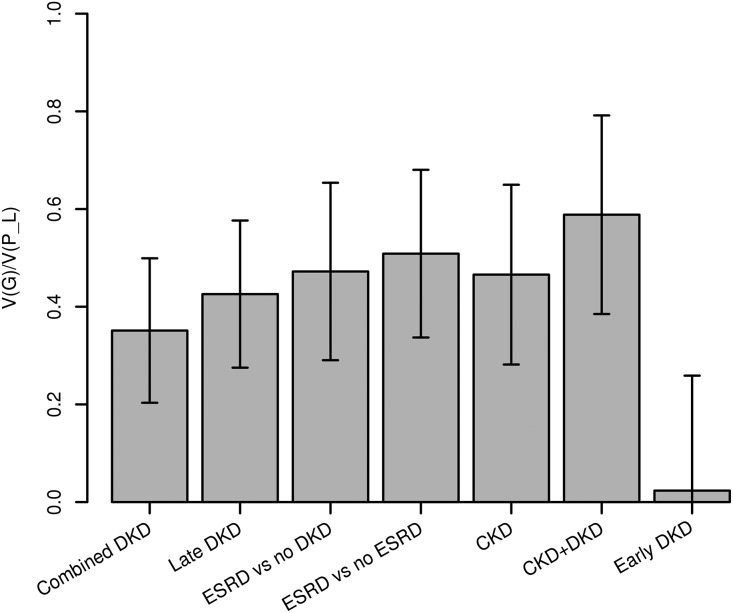
Because no comprehensive evaluation of heritability exists for the various stages of DKD, we estimated the narrow-sense, ‘chip’ heritability (i.e., the proportion of phenotypic variance explained by genome-wide single nucleotide polymorphisms (SNPs), h2) for the seven applied DKD phenotypic comparisons (Figure 1) in the context of T1D using data from the largest included GWAS study, the Finnish Diabetic Nephropathy (FinnDiane) Study.23 Heritability estimates varied greatly across the comparisons (Figure 2). For the primary, ‘combined DKD’ phenotypic comparison (micro- or macroalbuminuria or ESRD versus normal AER), h2 was 35% (P=6×10−3; Supplemental Table 1). The highest value, h2=59%, was obtained for ‘CKD+DKD’ (those with both CKD and DKD assigned as cases, and those with no CKD and no DKD as controls; P=1×10−3). Other late-stage phenotype definitions also yielded high estimates of heritability, e.g., 47% for ‘ESRD versus no DKD’ (P=3×10−3). When we included gender, diabetes duration, and age at diabetes diagnosis as covariates, the proportion of the remaining phenotypic variance explained by the genotyped SNPs increased for each phenotypic comparison, e.g., from 35% to 50% for ‘combined DKD’ (P=2.5×10−4). The estimates represent lower limits for the total heritability of DKD traits, as the method considers only those causal variants captured by the variants genotyped on the array.
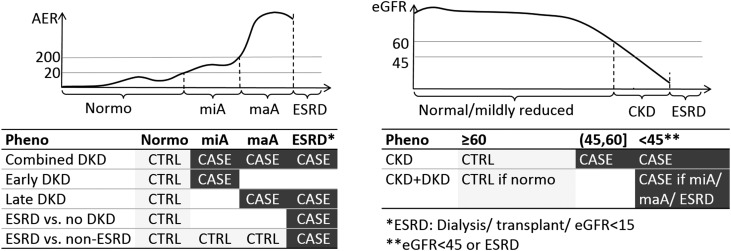
Figure 1.
Schematic picture of the DKD phenotypic comparisons on the basis of measured AER and eGFR, encompassing different stages and severities of DKD. ‘Combined DKD’ and ‘Late DKD’ (conventionally used in many previous genetic studies of DKD) are expected to capture genetic factors affecting DKD in general; ‘Early DKD’ comparison targets the genetic factors affecting the initiation of DKD, or with milder effect on the phenotype, whereas the two ESRD-based case definitions are expected to capture factors related to the late progression of DKD such as fibrotic processes, or genetic factors with particularly strong effect on the phenotype. Although the ‘CKD’ phenotype may reveal genetic factors for reduced renal function irrespective of the presence of albuminuria, the ‘CKD+DKD’ phenotype is an extreme phenotype that requires that controls have no signs of renal complications (neither AER or eGFR based). AER thresholds are given in µg/min, eGFR thresholds in ml/min per 1.73 m2. Number of samples per subphenotype: Normo: 2593; miA: 800; maA: 944; ESRD: 813; no CKD: 2909; CKD: 1255; no CKD, no DKD: 2018; CKD+DKD: 1117. Normo, normal AER; miA, microalbuminuria; maA, macroalbuminuria.
Figure 2.
The proportion of phenotypic variance explained by genotypes (V[G]/V[p_L]), i.e., the narrow-sense heritability, of the seven studied DKD phenotypic comparisons indicate high heritability especially for the more extreme phenotype definitions. Error bars represent the SEM.
GWAS
The GWAS discovery stage included 3135–5156 subjects with T1D, depending on phenotypic comparison (Supplemental Figure 1), from four studies: FinnDiane,23 the EURODIAB Family Study,24 the Scania Diabetes Registry (SDR),25 and the UK Nephropathy Family Study and Oxford Regional Prospective Study (NFS-ORPS/Cambridge)26,27 (Supplemental Tables 2 and 3, Table 1).
Table 1.
Clinical characteristics of the subjects, divided into cases and controls on the basis of the ‘combined DKD’ phenotype definition
| Characteristics | FinnDiane (N=3415) | EURODIAB (N=789) | SDR (N=558) | Cambridge (N=396) | ||||
|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | Cases | Controls | Cases | Controls | |
| N | 1802 | 1613 | 298 | 491 | 266 | 292 | 197 | 199 |
| Males, N (%) | 1067 (59) | 669 (41) | 170 (57.2) | 222 (45.3) | 150 (56) | 160 (55) | 109 (54) | 93 (47) |
| Age of onset of diabetes, yra | 13.4 (8.4) | 15.6 (8.8) | 16.7 (8.3) | 17.9 (8.1) | 15.9 (9.4) | 17.1 (9.5) | 9.1 (4.0) | 7.6 (6.0) |
| Age, yra | 45.1 (11.1) | 43.6 (11.7) | 40.3 (10.3) | 42.9 (10.2) | 48.2 (14.5) | 48.7 (13.2) | 17.0 (5.7) | 23.4 (10.2) |
| Duration of diabetes, yra | 31.7 (9.9) | 27.9 (9.6) | 23.7 (9.07) | 25.0 (7.7) | 32.3 (14.0) | 31.5 (12.3) | 7.9 (5.4) | 15.8 (6.8) |
| BMI, kg/m2a | 25.7 (4.1) | 25.3 (3.5) | 24.6 (3.7) | 25.1 (3.42) | 24.6 (3.3) | 24.4 (3.2) | NA | NA |
| HbA1C, %a | 8.7 (1.6) | 8.1 (1.2) | 8.3 (1.9) | 7.7 (1.6) | 7.9 (1.1) | 7.1 (0.9) | 10.5 (2.3) | 8.7 (1.6) |
| HbA1C, mmol/mola | 72 (17) | 65 (13) | 68 (21) | 61 (18) | 63 (12) | 54 (10) | 91 (25) | 72 (18) |
NA, not available.
Data displayed as mean (SD).
With 2563 T1D DKD patients and 2593 T1D non-DKD controls for the ‘combined DKD’ phenotype in the discovery cohorts, we estimated >80% statistical power to detect variants with MAF≥10% and allelic odds ratios (OR) ≥1.55 at genome-wide significance (P<5×10−8; Supplemental Table 4). Results from meta-analyses were well calibrated (genomic control [λGC], 1.01–1.05; Supplemental Figure 2) but showed little deviation from the null with no SNP reaching P<5×10−8 (no adjustment for multiple phenotypes). Across seven DKD case-control comparisons, a total of 101 regional lead SNPs reached a suggestive P value of <5×10−6 in at least one analysis (Supplemental Table 5). These 101 associations were tested for all DKD phenotypes available in the various stage 2 samples (Supplemental Tables 2 and 3), although meta-analysis results were only compiled across data for equivalent phenotypes. The two-stage design provided ≥80% power for the primary ‘combined DKD’ comparison to reach P<5×10−8 for variants with an OR≥1.47 and MAF≥10% (Supplemental Table 6).
In the joint meta-analysis of stage 1 and 2 samples, no SNP reached P<5×10−8, although three achieved P<1×10−6, all for the ESRD-based case definitions: rs1989248 near CNTNAP2, rs61277444 in PTPN13, and rs7562121 in AFF3 (Supplemental Figure 3, Supplemental Table 5). We also identified rs72809865 near NRG3 for further replication on the basis of nominal replication (P=0.05) for ‘combined DKD’ in stage 2. These four SNPs were examined in 1087 additional subjects using de novo genotyping (stage 3), although direct genotyping was unsuccessful for rs1989248 (near CNTNAP2). After the joint analysis of stage 1–3 results, the association at rs7562121 in AFF3 remained suggestive (P<1×10−6; Table 2).
Table 2.
Association analysis results for the top SNPs from the GWAS discovery + in silico replication analysis, selected for additional de novo replication
| Stages | rs61277444a CHR 4 | rs7562121 CHR 2 | rs1989248 CHR 7 | rs72809865 CHR 10 | ||
|---|---|---|---|---|---|---|
| Geneb: in PTPN13 | Geneb: in AFF3 | Geneb: 23kb to CNTNAP2 | Geneb: 350kb to NRG3 | |||
| Pheno:c,dESRD Versus Non-ESRD | Pheno:dESRD Versus No DKD | Pheno:c,dESRD Versus Non-ESRD | Pheno:dCKD+DKD | Pheno:cESRD Versus No DKD | Pheno:cCombined DKD | |
| Stage 1 | ||||||
| N | 813/3995 | 813/2394 | 813/3995 | 1117/2018 | 813/2394 | 2563/2593 |
| EAF | 0.09 | 0.09 | 0.23 | 0.28 | 0.28 | 0.16 |
| OR | 1.55 | 1.58 | 1.46 | 1.28 | 1.40 | 1.30 |
| 95% CI | 1.26 to 1.91 | 1.26 to 1.97 | 1.28 to 1.66 | 1.12 to 1.47 | 1.21 to 1.62 | 1.16 to 1.46 |
| P | 4.3×10−6 | 8.7×10−6 | 8.9×10−8 | 1.8×10−4 | 4.0×10−6 | 5.0×10−6 |
| Stage 2 | ||||||
| N | 964/4806 | 964/3187 | 964/4806 | 1104/1981 | 964/3187 | 2659/3842 |
| P | 2×10−3 | 3×10−3 | 0.04 | 1×10−3 | 0.02 | 0.05 |
| Stage 3 | ||||||
| N | 94/993 | 94/627 | 67/822 | 367/516 | ||
| P | 0.19 | 0.14 | 0.59 | 0.54 | ||
| All | ||||||
| N | 1871/9794 | 1871/6208 | 1844/9623 | 2221/3999 | 1777/5581 | 5589/6951 |
| N total | 11,665 | 8079 | 11,467 | 6220 | 7358 | 12,540 |
| OR | 1.41 | 1.42 | 1.27 | 1.26 | 1.29 | 1.17 |
| 95% CI | 1.21 to 1.65 | 1.20 to 1.67 | 1.17 to 1.39 | 1.15 to 1.38 | 1.17 to 1.43 | 1.09 to 1.26 |
| P | 1.9×10−6 | 6.0×10−6 | 3.5×10−7 | 6.0×10−7 | 1.8×10−6 | 7.4×10−6 |
The association in FinnDiane was moderately attenuated from OR=1.49 (95% CI, 1.19 to 1.86; P=4.5×10−4; ESRD versus non-ESRD) and OR=1.50 (95% CI, 1.18 to 1.91; P=1×10−3; ESRD versus no DKD) with the imputed data to OR=1.36 (95% CI, 1.11 to 1.66; P=3.0×10−3; ESRD versus non-ESRD) and OR=1.40 (95% CI, 1.11 to 1.75; P=3.9×10−3; ESRD versus no DKD) using directly genotyped data. CHR, chromosome; Pheno, phenotypic comparison; N, number of cases/controls; EAF, effect allele frequency; 95% CI, 95% confidence interval.
Because of moderate imputation quality at the discovery stage, rs61277444 was directly genotyped in 2913/3415 FinnDiane subjects.
The closest gene/genes.
Selection criteria for de novo replication: P value <10−6 for discovery + stage two meta-analysis.
Selection criteria for de novo replication: P value <0.05 in the in silico replication for the phenotype selected for replication.
Rs7562121 in AFF3 is in moderate linkage disequilibrium (LD) with rs7583877 (r2=0.67; D’= 0.94), the variant previously associated with ESRD in T1D.15 Conditional analysis showed that both SNPs represent the same signal (Supplemental Table 7). However, this study cannot be considered replication of previous reports because of substantial overlap of study samples. Analysis in nonoverlapping samples (222 ESRD cases, 1640 non-ESRD controls) provided no independent corroboration of the AFF3 association (OR=1.02; P=0.82).
The substantial heritability captured by SNPs on the genotyping array, and the failure to detect genome-wide significant signals, suggest that the causal variants are beyond detection in the present study, due to more modest effect size (e.g., OR<1.47 for MAF≥10%) and/or imperfect capture by common variant-focused GWAS analyses.
Re-Evaluation of Previous DKD Associations
The enlarged sample size provided an opportunity to investigate the previous, often contradictory reports of SNP associations.28 We selected signals from three sources: a literature-based meta-analysis of candidate gene associations28; GWASs on DKD in subjects with either T1D15,17,29–31 or T2D32,33; and GWASs on CKD irrespective of underlying pathology.20 Many of these previous studies included subjects overlapping with those in this analysis.
We estimate >80% statistical power to detect associations with allelic OR≥1.25 and MAF≥10% at nominal significance (α=0.05; Supplemental Table 8) for the ‘late DKD’ phenotype, which was most commonly used in previous publications. We observed only a modest deviation from the null P value distribution for the set of previously-associated variants, suggesting that many of these have been false positive findings (Supplemental Figure 4). Nevertheless, in addition to the AFF3 SNP associations described above, the associations between rs2838302 (in SIK1) and ‘ESRD versus non-ESRD’, and between rs17709344 (RGMA–MCTP2) and ‘CKD+DKD’ were directionally consistent and significant (P<1.1×10−3 to correct for 46 loci). The association between rs2838302 (SIK1) and ‘ESRD versus no DKD’ remained nominally significant (P=0.04) even after exclusion of the substantial overlap of the subjects (Supplemental Table 9).
We observed directionally consistent associations (less stringent P<0.05) for variants near WNT4-ZBTB40, SEMA6D-SLC24A5, ELMO1, ERBB4, 4p15.1, and 13q regions (Supplemental Figure 5). However, at four of these loci, the modest evidence of association was lost when overlapping samples were removed. Independent replication of previously reported signals (directionally consistent P<0.05) was therefore restricted to the signals at ELMO1, 13q, and SIK1 (Supplemental Table 9).
DKD Shows Shared Genetic Background with Obesity, T2D, and Smoking Cessation
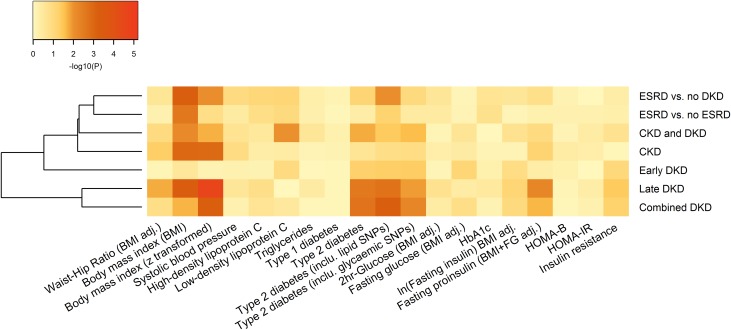
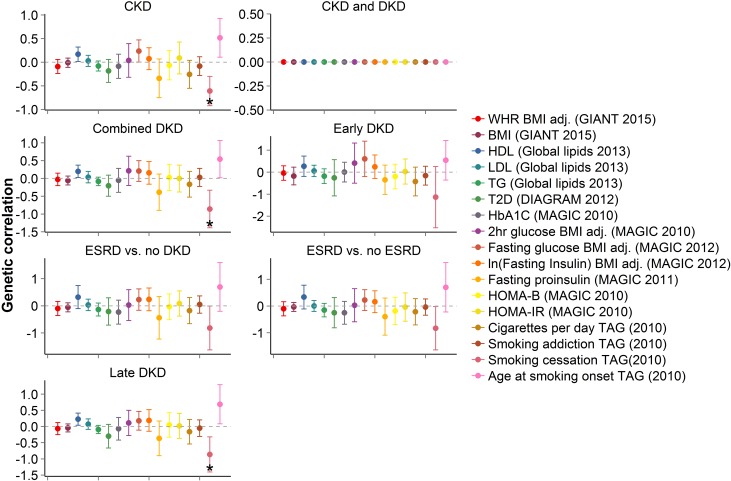
Body mass index (BMI), systolic BP, serum lipids, and smoking have been associated with DKD in epidemiologic studies24,34–36 whereas T2D, obesity, hypertension, and lipid disorders have been reported to cluster in families with DKD,11,12,37 suggesting a shared genetic background or a close correlation among these phenotypes. We constructed weighted genetic risk scores (GRS) for 19 diabetes,38–45 obesity,46–48 hypertension,49 or lipid-related50 intermediate phenotypes on the basis of 10–96 established SNPs (P<5×10−8) for each phenotype from previously published GWAS. The GRS for increasing BMI46 was associated (P<2.6×10−3 to correct for 19 traits) with multiple DKD traits (P=2.2×10−5 for ‘late DKD’; Figure 3, Supplemental Table 10), implicating a causal role of BMI for DKD. Although observational studies have shown contradictory results,51,52 these findings are consistent with those from a recent Mendelian Randomization study including FinnDiane subjects.53 The GRS predisposing to T2D was associated with increased risk of multiple DKD traits (e.g., P=1.9×10−3 for ‘combined DKD’ excluding pleiotropic lipid and glycemic SNPs; P=6.1×10−4 for ‘combined DKD’ including pleiotropic lipid SNPs) in line with previous reports of parental T2D being associated with DKD in subjects with T1D.12 These data support a causal, mechanistic link between metabolic syndrome and DKD in T1D.23
Figure 3.
The GRS for BMI and T2D were associated with DKD phenotypes in subjects with T1D. A GRS for MBI (z-transformed) was associated (P<2.6×10−3 to account for multiple testing; 19 GRS traits) with combined and late DKD, and CKD phenotypes; GRS for T2D was associated with late DKD.
We also applied the LD score regression method to estimate the genetic correlation between traits. In contrast to the GRS approach which uses only the most associated variants, this method examines the allelic effects of all SNPs in the meta-analysis.54,55 As expected, the seven DKD phenotypes were highly correlated with each other, at least in part due to overlapping phenotypic definitions, with the exception of ‘early DKD’ (Supplemental Figure 6). Smoking cessation was inversely correlated with ‘CKD’ (P=1.1×10−4) and other DKD traits (Figure 4). This is in line with a previous epidemiologic study showing higher risk of DKD for smokers, whereby exsmokers had similar risk of developing DKD as nonsmokers.36
Figure 4.
Genome-wide comparison of DKD traits and other phenotypes, evaluated with LD score regression, shows negative correlation between DKD traits and smoking cessation. Significant correlations (P<0.003 to account for multiple testing; 17 related phenotypes) are indicated with *. Bars represent 95% confidence intervals.
Gene Set Enrichment Analyses Suggest Glucuronidation and Other Pathways Affecting DKD
We performed gene set enrichment analyses (GSEA) of the GWAS results to identify biologic pathways and processes enriched among the most significant GWAS signals. The ascorbate and aldarate metabolism, and pentose and glucuronate interconversions pathways showed significant enrichment (P<1.6×10−5 to correct for multiple tested pathways). Literature linking ascorbate metabolism to DKD is sparse, but cell studies have suggested a reduced uptake rate of ascorbic acid (vitamin C) in DKD,56 and vitamin C plus E supplementation was reported to improve glomerular function in T2D.57 In the latter pathway, the flagged genes overlapped especially the glucuronate subpathway (Supplemental Figure 7). Negatively charged glucuronic acid units on heparan sulfate side chains participate in the maintenance of the negative charge selectivity of the glomerular basement membrane in the kidneys, and their cleavage by heparanase (endo-β-D-glucuronidase) has been suggested as an underlying cause for DKD.58 Although knock-out of the HSPE (heparanase) gene protects mice from DKD,59 no specific genetic variants in or near HSPE have been associated with DKD. Other gene sets with less marked enrichment (false discovery rate <0.05; Supplemental Table 11) included the cholesterol biosynthesis pathway (P=8.0×10−4) flagging among others the HMGCR gene (lowest P=0.02 for rs11726245 for ‘CKD’), which encodes the main target of statins, commonly used for prevention of diabetic macrovascular complications. Cholesterol levels are reported to be related to eGFR decline in T1D, whereas statin use improved eGFR in subjects with T2D in some60 but not all studies.61
Single-Marker Association Tests of WES Show Suggestive Associations for ESRD
To identify low frequency (1%≤MAF<5%) and rare (MAF<1%) coding variants contributing to DKD, we performed WES of 997 subjects with T1D from the FinnDiane, SDR, and Steno Diabetes Center. To maximize power, subjects were ascertained from the tails of the liability distribution: we selected patients with onset of macroalbuminuria or ESRD relatively soon after diagnosis of T1D, and controls with normal AER despite prolonged duration of T1D (Supplemental Table 12). This setting allowed us to study the ‘late DKD’ and ‘ESRD versus no DKD’ case-control contrasts (Figure 1).
No associations were observed at exome-wide significance (P<5×10−7; Supplemental Figure 8). Variants in five loci reached a P value of <1×10−5 for association with ‘ESRD versus no DKD’ (Supplemental Tables 13 and 14) with the strongest associations obtained for an intronic SNP rs188427269 within NVL (MAF 0.2%; P=3.3×10−7) and for rs13003941 in the 3′UTR of ERBB4 (MAF 35%; P=3.5×10−6). Other variants in ERBB4 (not in LD with rs13003941) were previously suggestively associated with DKD,15 and ErbB4 was shown important for the development of kidneys in mouse models.62
Gene Aggregate Tests of WES
To improve power to detect low frequency and rare variant associations, we performed gene-level aggregate tests using three complementary approaches: a variable threshold burden test,63 a dispersion test (SKAT),64 and a hybrid test (SKAT-O).65 No gene achieved exome-wide significance (P<2.5×10−6, adjusted for 20,000 genes; Supplemental Figures 9 and 10). For the ‘late DKD’ phenotype, the lowest P value was for GGA1 (P=3.1×10−5; Supplemental Figure 11), showing rare (MAF≤0.13%)66 missense alleles in seven cases but no controls across five variant sites (Supplemental Tables 15–17). For the ‘ESRD versus no DKD’ phenotype, the strongest associations were found at CCDC77 (P=2.1×10−5), THADA (P=8.3×10−5), and CHPT1 (P=1.9×10−5; Supplemental Figure 12, Supplemental Tables 18–20).
Although it has been hypothesized that common disease diagnoses are partially comprised of rare monogenic forms of the disease, we did not see enrichment in genes causing rare human glomerular diseases in our WES results (Supplemental Table 21). In addition, there was no enrichment between the WES signals and those arising from our GWAS studies. Thus, the variants and genes generating suggestive evidence for association in WES seem independent of rare variants identified for monogenic forms of kidney disease, and of the common variant contribution to DKD in subjects with T1D.
Bivariate Analysis of GWAS and WES Data
As some genetic factors may affect disease outcome only in the presence of interacting genetic factors, we complemented the GWAS and WES analyses using a multivariate variant selection method (Algorithm based on a BivAriate CUmulative Statistic, ABACUS)67 applied to both GWAS and WES data. Although no variant reached simulation P value (PS<5×10−8), suggestive evidence of association was obtained for variants in OSGIN1 in two different data sets (PS<5×10−6 for ‘ESRD versus no DKD’ in EURODIAB GWAS and in Steno WES; Supplemental Tables 22 and 23). Functional clustering of the results pointed at processes previously linked with DKD such as cholesterol and sucrose/glucose metabolism and inflammatory response pathway (Supplemental Tables 24 and 25).
Discussion
This study represents a systematic evaluation of common genetic risk factors and rare coding variants for a broad range of kidney complication phenotypes in T1D. To complement the association analyses, we performed various enrichment and correlation analyses considering the genome-wide content to obtain insight of the genetic architecture, biologic background, and correlation with other traits.
This study confirmed that renal complications in T1D are highly heritable, as a substantial proportion of the phenotypic variation can be captured by common variants on the GWAS chip, with the strongest heritability estimates observed for the most extreme phenotypic comparisons. This captured heritability could consist of common variants with modest effects, and/or variants of lower frequency, some of them imperfectly tagged by common SNPs. The limited evidence for linkage to DKD68 and general experience regarding the architecture of other common complex traits, suggest that it is unlikely that DKD risk is dominated by rare variants of large effect, although ultimately sequence based studies will be required for definitive evaluation.
We found suggestive common variant signals for DKD in or near AFF3, CNTNAP2, NRG3, and PTPN13 genes, but additional data sets will be needed to confirm (or refute) these signals and to increase power to detect others. For example, the effective sample size required to implicate a risk allele of MAF 20% and allelic OR 1.2 at genome wide significance is approximately 15,000. We demonstrated that many previously claimed DKD associations signals are likely to be false positives, although modest, independent evidence was found for associations near ELMO1, 13q, and SIK1 (P<0.05). Although the three-stage analysis of this study brings together the largest studies with the largest number of participants with T1D for GWAS on DKD, the total sample size remains modest. Future efforts to identify novel genetic risk factors for DKD will require investment in additional sample ascertainment and genotyping.
Different genetic factors may affect the various stages of DKD progression. To address the phenotypic heterogeneity within DKD, we applied selection of subphenotypes for the evaluation of renal complications. As various phenotypic comparisons yielded significant findings for different tests, these comparisons may address diverse aspects of the disease progression. Future studies would likely benefit from improved phenotyping and, in particular, from the detailed longitudinal follow-up of research participants.
Even though this study concentrated on patients with T1D, some of the results may be extended to DKD in T2D as well. However, as the pathology behind renal complications in T2D is more heterogenous,5 with ageing, atherosclerosis, and hypertension contributing appreciably to the renal function, the genetic background of DKD in T2D may have more features of the kidney disease in the general population.
Although no novel susceptibility loci were identified in this study, some signals started to appear when we aggregated genetic data as in pathway analysis, GRS, and LD score regression analysis. The pathway analyses, for example, highlighted involvement of the glucuronate interconversions pathway and supported the role of cholesterol metabolism in DKD. Analysis of GRS suggested that high BMI, as well as the genetic risk factors behind T2D, causally increase the risk of DKD in T1D. Finally, the LD score regression identified genetic correlation between smoking cessation and reduced risk of DKD in subjects with T1D, supporting the previous research finding of smoking as a risk factor for DKD.36 Altogether, these results may provide valuable clues to the biologic processes relevant to the pathogenesis of DKD.
Concise Methods
Subjects with T1D in GWAS and WES
The GWAS discovery stage included subjects with T1D from the FinnDiane Study,23 the EURODIAB Family Study,24 SDR,25 and NFS-ORPS.26,27 WES included subjects from FinnDiane, SDR, and Steno Diabetes Center (characterized in Supplemental Table 12). All studies were approved by the local ethics committees and conducted according to the principles of the Declaration of Helsinki. Written consent was obtained from the participants in FinnDiane, EURODIAB, SDR, and Steno Studies. In the NFS-ORPS study, written consent was obtained from parents, and verbal assent was obtained from children.
Phenotype Definitions
All subjects had T1D as diagnosed by their attending physician, with age at diabetes onset ≤40 years and insulin treatment initiated within 1 year of diagnosis. The kidney status was classified on the basis of AER and eGFR (please see the Supplemental Material and Supplemental Table 26). Patients receiving dialysis treatment, with a kidney transplant, or with an eGFR≤15 ml/min per 1.73m2 were defined to have ESRD. On the basis of these definitions, we analyzed seven different case-control phenotypes (Figure 1).
Patient Selection for WES
Patients were selected from the extreme ends of the liability distribution of DKD from each participating study. Patients had rapid onset of macroalbuminuria (within 20/25 years of diabetes onset in FinnDiane and Steno, respectively; no threshold in SDR) or ESRD (onset within 25 years of diabetes onset in FinnDiane and Steno). Controls were subjects with normal AER despite prolonged duration of T1D (≥32, 30, or 27 years in FinnDiane, Steno, and SDR, respectively). In addition, the FinnDiane controls were enriched for higher glycated hemoglobin (HbA1C) values (excluding subjects with HbA1C<6.5%).
Genotypes
Genome-Wide Genotyping and Imputation of the Discovery Cohorts
The genome-wide genotyping of the subjects in the SDR, NFS-ORPS, and EURODIAB was performed with the Illumina OmniExpress assays (Illumina, San Diego, CA). In quality control, samples with a call rate <98%, gender discrepancy, extremely high or low heterozygosity, or excess of estimated relatedness, were removed. Common SNPs (MAF≥5%) with genotyping rate <95% or not in Hardy–Weinberg equilibrium (P≤5.7×10−7) were removed. For low-frequency SNPs (MAF 1%–5%), the thresholds were 99% and P<10−4, respectively. In the FinnDiane Study, genotyping was performed with the Illumina 610Quad assay and the quality control was similar to the other studies, as described previously in detail.15 Principal component analysis was performed in all cohorts with Eigensoft.69 Imputation was performed with IMPUTE270 using the 1000 Genomes project (phase 1 v.3, released March 2012) as the imputation reference panel.71 Variants were filtered postimputation to those with MAF≥1%, minor allele count ≥10 in both patients and controls, and SNPtest INFO estimate of imputation quality ≥0.4.
WES and Variant Calling
Samples were sequenced at two centers. Sequencing was performed on an Illumina HiSeq2000. We required an average 20× target capture above 80% coverage, otherwise additional DNA was requested to ‘top up’ the sample. This resulted in mean sequencing depth of 54.97 (FinnDiane) and 42.23 (SDR and Steno) bases per position. After additional sequencing 497 samples were included from FinnDiane and 500 from SDR and Steno. Samples were mapped with Burrows–Wheeler aligner v7.4. Genome analysis toolkit v2.1 was used to refine and recalibrate the sequences and to call variants. Polymorphic variants (MAF>0) with a mapping quality <250, Hardy–Weinberg equilibrium P value >1×10−10, and call rate ≥75% were retained in the analysis. Samples with ≥10% missingness or extreme heterozygosity were excluded. Population outliers, duplicates, and related samples were removed. Variants were annotated using CHAos (http://www.well.ox.ac.uk/~kgaulton/chaos.shtml), snpEff,72 and VEP73 for functional class and transcript.
With 530,565 variants (491,553 SNPs and 39,012 indels) across 479 controls and 481 patients after the quality control, each individual carried a mean of 7566 synonymous, 6452 missense, and 103 protein truncating variants. The lower number of total variant sites compared with other, more outbred populations74 is in line with fewer variable sites seen in founder populations such as the Finns.75
Statistical Methods
Heritability Estimates
The narrow-sense heritability of the kidney phenotypes was estimated as the proportion of the phenotypic variance explained by the additive effects of the genotyped SNPs on the basis of the FinnDiane GWAS data using the GCTA v. 0.93.9, excluding samples with estimated relatedness ≥0.025.76 The observed variance explained was transformed to the underlying population scale on the basis of rough prevalence estimates as given in Supplemental Table 1. The heritability was estimated without covariates, and adjusting for sex, duration of T1D, and age at T1D onset.
GWAS Analysis
The genome-wide association analyses in stage 1 studies were performed with two methods in parallel. To obtain stable effect size estimates, we performed additive tests for association using SNPtest with the score method adjusted for sex, diabetes duration, age at diabetes onset, and the two first principal components. P values were obtained with a variance component based mixed model method, efficient mixed-model association expedited (EMMAX), which accounts for the sample structure, allowing to include relatives in the analysis.77 Models were adjusted for sex, diabetes duration, and age at diabetes onset, and the kinship matrix was calculated with EMMAX. EMMAX algorithm was implemented with the EPACTS software (www.sph.umich.edu/csg/kang/epacts/). Meta-analyses of the effect sizes were performed with the fixed-effect inverse variance method (GWAMA78 and METAL79), whereas P values were combined on the basis of the study-wise P values, sample sizes, and effect directions (METAL79). Meta-analysis results were further filtered to those with valid results from at least two studies. P values <5×10−8 were considered genome-wide significant, not correcting for multiple testing due to seven phenotypic comparisons, as the patient and control groups were overlapping and the traits were correlated with each other.
Power calculations were performed with the genetic power calculator (pngu.mgh.harvard.edu/∼purcell/gpc/) for simple case-control setting,80 and with Power Calculator for Two Stage Association Studies (CaTS; http://csg.sph.umich.edu//abecasis/cats/).81
Replication
Independent variants with P<5×10−6 were selected for in silico replication in six additional studies: the All Ireland–Warren 3–Genetics of Kidneys in Diabetes UK collection15 and the Genetics of Kidneys in Diabetes United States Study15 from the GENIE Consortium, the Diabetes Control and Complications Trial, and the Epidemiology of Diabetes Interventions and Complications (DCCT/EDIC) study,82,83 1073 subjects from the Joslin Diabetes Center T1D nephropathy collection, and the French, Belgian, and Danish subjects (the Steno Diabetes Center) from the French–Danish Effort84 (Supplemental Tables 2 and 3). After in silico replication, variants that replicated with a P<0.05 or had a combined P value <1×10−7 after meta-analysis were selected for de novo genotyping with TaqMan in 1095 additional FinnDiane patients, not part of the GWAS (Supplemental Table 2). rs1989248 was not successfully genotyped. Association testing was performed with logistic regression (implemented in PLINK or SNPtest, depending on the study), adjusted for sex, duration of diabetes, age at diabetes onset, and study specific covariates.
GRS Analysis
SNPs associated (P<5×10−8) in previous studies with waist/hip ratio (adjusted for BMI),47 BMI (untransformed48 and z-transformed46), systolic BP,49 low-density lipoprotein cholesterol, triglycerides, high-density lipoprotein cholesterol,50 T1D,39 T2D38 (including all SNPs, and without any other effects other than on T2D or lipids50 and T2D or glycemic traits41,85), 2-hour glucose (adjusted for BMI),40 fasting glucose (adjusted for BMI),41 HbA1C,42 fasting insulin (natural log transformed and adjusted for BMI),41 fasting proinsulin (adjusted for BMI and fasting glucose),43 HOMA-B, HOMA-IR,44 and insulin resistance45 were included in a GRS for each trait respectively. The GRS was weighted by the allelic effect of each variant on the DKD risk factor and associated with the DKD phenotypes using the stage 1 GWAS meta-analysis data.49 The lipid GRS were restricted to variants that predicted that specific trait and removed those that had effects on other lipid traits. We did not include a GRS for smoking behaviors as there were too few genome-wide significant associations to form a sufficient instrument.
LD Score Regression to Estimate Genetic Correlation
To estimate the genetic correlation between the GWAS stage 1 meta-analysis results for DKD phenotypes, and the related traits, we assembled the genome-wide summary statistics from all of the studies used to calculate GRS except for systolic BP and T1D as the full summary statistics were not available. We additionally computed genetic correlation with smoking behavior phenotypes (cigarettes per day, smoking addiction, smoking cessation, and age at smoking onset).86
Gene Set Enrichment Analyses
Gene set enrichment analysis was performed in the GWAS stage 1 meta-analysis results with the MAGENTA(vs.2)87 applied on 10,992 partially overlapping gene sets from GO, PANTHER, INGENUITY, KEGG, REACTOME, and BIOCARTA data bases; 3126 gene sets with ≥10 genes were analyzed. The 95 percentile cut-off for the gene scores was used to define the significant results.
Correction for Multiple Testing
The significance threshold for the results of the evaluation of previous loci, GRS, LD score regression, and pathway enrichment analyses were Bonferroni corrected for multiple testing with α=0.05 significance level, accounting for the number of tested loci, traits, or pathways. The results were not corrected for the seven phenotypic comparisons due to a considerable overlap of the patient and control groups.
WES Single Variant Analysis
Single variants were tested for association with DKD (patients, n=481; controls, n=479) and ESRD (patients, n=168; controls, n=479) using the logistic score test88 implemented in Epacts, with sex and two principal components as covariates. Related individuals, monomorphic SNPs, and those with SEM>10 were excluded from the analysis. Although the study setting provided low statistical power to detect rare variants (MAF<1%) with exome-wide significance (P<9×10−8 to correct for 530,776 tested variants) in line with previous reports on the statistical power to detect rare variants,89 we had sufficient power (80%) to detect a low frequency variant (MAF=5%) with a large OR of 5.65 (Supplemental Figure 13).
WES Gene-Based Analysis
We applied three series of gene based tests: a burden test (variable threshold)63 that assumes the direction of effect of grouped variants is the same, a dispersion test (SKAT)64 that performs well when the direction of variant effect differs, and a hybrid (SKAT-O)65 that uses multiple methods in a single test. Analyses were adjusted for sex and principal components. For all three tests we grouped variants into four categories: protein truncating variants (PTV; e.g., nonsense, frameshift, essential splice site), deleterious protein altering variants subdivided into ‘strict’ and ‘broad’ grouping if predicted deleterious by all five/at least one annotation algorithm, and any protein altering variants (e.g., missense, in-frame indel, nonessential splice site). These four groups are referred to as (1) PTV-only, (2) PTV+strict, (3) PTV+broad, and (4) PTV+missense, from the strictest to the most permissive class. A MAF threshold of 1% was applied to the more permissive masks PTV+missense and PTV+broad to exclude common variants from the WES analysis.
WES Gene Set Enrichment Analysis
A total of 43 gene sets were created, on the basis of top findings from the GWAS analyses, kidney-related functional terms in public databases, text mining approaches, and kidney gene expression (Supplemental Table 21). These gene sets were analyzed for enrichment in ‘Late DKD’ WES association results (SKAT-O, all four masks) using the GSEA method with SKAT-O’s P value as the ranking statistic.90 To guard against potential miscalibration of GSEA’s reported P values we conducted 100 permutations of case/control status and subsequently reran burden tests and GSEA analysis for each permutation. Significance of enrichment was validated with permutation. For validation of enrichment of GWAS result, the samples in WES were removed from the GWAS data and the analyses were repeated.
Bivariate Analysis of GWAS and WES Data
We applied ABACUS67 to the individual GWAS discovery cohorts on each of the seven different case-control phenotypes. In addition, ABACUS was applied to the WES cohorts (FinnDiane, Steno, and SDR) on the two phenotypic comparisons. SNP-sets were defined on the basis of REACTOME, KEGG, and GO Biologic Processes. To analyze nonannotated and intergenic SNPs, we also defined SNP-sets of continuous 3000 SNPs. Functional clustering of the ABACUS results was performed with DAVID software.91,92
Disclosures
P.-H.G. has received lecture honoraria from AstraZeneca (London, UK), Boehringer Ingelheim (Ingelheim am Rhein, Germany), Eli Lilly (Indianapolis, IN), Genzyme (Cambridge, MA), MSD (Kenilworth, NJ), Novartis (Basel, Switzerland), Novo Nordisk (Gladsaxe, Denmark), and Sanofi (Paris, France), and research grants from Eli Lilly and Roche (Basel, Switzerland). P.-H.G. is also an advisory board member for AbbVie (North Chicago, IL), Boehringer Ingelheim, Eli Lilly, Janssen (Beerse, Belgium), Medscape (New York, NY), MSD, Novartis (Basel, Switzerland), and Sanofi.
H.M.C. has acted as consultant/advisory panel member for Pfizer (New York, NY), Sanofi Aventis (Paris, France), Regeneron (Tarrytown, NY), and Eli Lilly, and on the data safety panel for Novartis; has received research support from Roche, Pfizer, Eli Lilly, Boehringer Ingelheim, AstraZeneca, and Sanofi; has participated in a lecturers’/speakers’ bureau and received honorarium from Pfizer; and is a shareholder in Roche.
J.T. has acted as consultant/ advisory panel member for Bayer Health Care (Leverkusen, Germany), Sanofi Aventis, Eli Lilly, Roche, Impeto Medical (Paris, France), and Novo Nordisk, and has received research support from Roche, Pfizer, Eli Lilly, Boehringer Ingelheim, AstraZeneca, and Sanofi; and is a shareholder in Orion Pharma (Espoo, Finland).
PR has acted as consultant/advisory panel member for Eli Lilly, Novo Nordisk, AbbVie, Boehringer Ingelheim, Astra Zeneca, Janssen, Astellas (Chuo-ku, Tokyo, Japan), BMS (New York, NY), and MSD (all honoraria to institution); has received research grants from Novo Nordisk, Astra Zeneca, and Novartis; has shares in Novo Nordisk AS.
J.C.F. has acted as consultant for Sanofi.
D.Z., M.J.B., and E.F. are employees and stockholders of Pfizer.
Supplementary Material
Acknowledgments
We thank A. Sandelin, A.-R. Salonen, T. Soppela, and J. Tuomikangas for skillful laboratory assistance. We also thank all of the subjects who participated in the FinnDiane study and gratefully acknowledge all of the physicians and nurses at each center involved in the recruitment of participants (Supplemental Table 28). We thank the participants in the EURODIAB Family Study and gratefully acknowledge the physicians and nurses at each center involved in subject recruitment. A complete list of participants in the DCCT/EDIC research group can be found in the New England Journal of Medicine, 2011, 365: 2366–2376.82
The research was supported by the European Union’s Seventh Framework Program (FP7/2007–2013) for the Innovative Medicine Initiative under grant agreement IMI/115006 (the SUMMIT consortium). The FinnDiane Study was supported by grants from the Folkhälsan Research Foundation, the Wilhelm and Else Stockmann Foundation, the Liv och Hälsa Foundation, Helsinki University Central Hospital Research Funds (EVO), the Signe and Ane Gyllenberg Foundation, Finska Läkaresällskapet, the Novo Nordisk Foundation (NNF14SA0003), and the Academy of Finland (134379 and 275614). The EURODIAB Family Study was funded by the European Union's Biomedicine and Health Programme. The NFS-ORPS Study was supported by NIHR/Wellcome Trust Cambridge Clinical Research Facility. The GENIE consortium was funded by the National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases (R01-DK081923) to J.N.H., J.C.F., and P.-H.G., the Northern Ireland Research and Development Office (A.P.M.) and Science Foundation Ireland. The DCCT/EDIC has been supported by cooperative agreement grants (1982–1993, 2012–2017), and contracts (1982–2012) with the Division of Diabetes Endocrinology and Metabolic Diseases of the National Institute of Diabetes and Digestive and Kidney Disease (current grant numbers U01 DK094176 and U01 DK094157), and through support by the National Eye Institute, the National Institute of Neurologic Disorders and Stroke, the General Clinical Research Centers Program (1993–2007), and Clinical Translational Science Center Program (2006 to present), Bethesda, MD. S.S.R. was supported by Juvenile Diabetes Research Foundation (grant 17-2012-542).
Parts of this study have been presented in abstract form at the 49th European Association for the Study of Diabetes (EASD) Annual Meeting September 23–27, 2013 in Barcelona, Spain; at the 74th American Diabetes Association (ADA) Scientific Session June 13–17, 2014 in San Francisco, CA; at the 75th ADA Scientific Session June 5–9, 2015 in Boston, MA; and at the 51st EASD Annual Meeting September 14–18, 2015 in Stockholm, Sweden.
Industry contributors have had no role in the DCCT/EDIC study but have provided free or discounted supplies or equipment to support participants’ adherence to the study: Abbott Diabetes Care (Alameda, CA), Animas (Westchester, PA), Bayer Diabetes Care (North America Headquarters, Tarrytown, NY), Becton Dickinson (Franklin Lakes, NJ), Eli Lilly (Indianapolis, IN), Extend Nutrition (St. Louis, MO), Insulet Corporation (Bedford, MA), Lifescan (Milpitas, CA), Medtronic Diabetes (Minneapolis, MN), Nipro Home Diagnostics (Ft. Lauderdale, FL), Nova Diabetes Care (Billerica, MA), Omron (Shelton, CT), Perrigo Diabetes Care (Allegan, MI), Roche Diabetes Care (Indianapolis, IN), and Sanofi-Aventis (Bridgewater NJ).
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
See related editorial, “Genetics of Diabetic Kidney Disease—From the Worst of Nightmares to the Light of Dawn?,” on pages 389–393.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2016020231/-/DCSupplemental.
Contributor Information
Collaborators: N. Sandholm, N. Van Zuydam, E. Ahlqvist, T. Juliusdottir, H.A. Deshmukh, N.W.Rayner, B. Di Camillo, C. Forsblom, J. Fadista, D. Ziemek, R.M. Salem, L.T. Hiraki, M. Pezzolesi, D. Trégouët, E. Dahlström, E. Valo, N. Oskolkov, C. Ladenvall, M.L. Marcovecchio, J. Cooper, F. Sambo, A. Malovini, M. Manfrini, A. J. McKnight, M. Lajer, V, Harjutsalo, D. Gordin, M. Parkkonen, J. Tuomilehto, V, Lyssenko, P.M. McKeigue, S.S. Rich, M.J. Brosnan, E. Fauman, R. Bellazzi, P. Rossing, S. Hadjadj, A. Krolewski, A.D. Paterson, J.C. Florez, J.N. Hirschhorn, A.P. Maxwell, D. Dunger, The DCCT, EDIC Study Group, GENIE Consortium, The FinnDiane Study Group, C. Cobelli, H.M. Colhoun, L. Groop, M.I. McCarthy, and P.-H. Groop
References
- 1.Collins AJ, Foley RN, Chavers B, Gilbertson D, Herzog C, Johansen K, Kasiske B, Kutner N, Liu J, St Peter W, Guo H, Gustafson S, Heubner B, Lamb K, Li S, Li S, Peng Y, Qiu Y, Roberts T, Skeans M, Snyder J, Solid C, Thompson B, Wang C, Weinhandl E, Zaun D, Arko C, Chen SC, Daniels F, Ebben J, Frazier E, Hanzlik C, Johnson R, Sheets D, Wang X, Forrest B, Constantini E, Everson S, Eggers P, Agodoa L: United States Renal Data System 2011 Annual Data Report: Atlas of chronic kidney disease & end-stage renal disease in the United States. Am J Kidney Dis 59(1 Suppl 1):e1–e420, 2012 [DOI] [PubMed] [Google Scholar]
- 2.Pambianco G, Costacou T, Ellis D, Becker DJ, Klein R, Orchard TJ: The 30-year natural history of type 1 diabetes complications: the Pittsburgh Epidemiology of Diabetes Complications Study experience. Diabetes 55: 1463–1469, 2006 [DOI] [PubMed] [Google Scholar]
- 3.Finne P, Reunanen A, Stenman S, Groop PH, Grönhagen-Riska C: Incidence of end-stage renal disease in patients with type 1 diabetes. JAMA 294: 1782–1787, 2005 [DOI] [PubMed] [Google Scholar]
- 4.Harjutsalo V, Maric C, Forsblom C, Thorn L, Wadén J, Groop PH; FinnDiane Study Group : Sex-related differences in the long-term risk of microvascular complications by age at onset of type 1 diabetes. Diabetologia 54: 1992–1999, 2011 [DOI] [PubMed] [Google Scholar]
- 5.Fioretto P, Mauer M: Histopathology of diabetic nephropathy. Semin Nephrol 27: 195–207, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jensen T, Borch-Johnsen K, Kofoed-Enevoldsen A, Deckert T: Coronary heart disease in young type 1 (insulin-dependent) diabetic patients with and without diabetic nephropathy: incidence and risk factors. Diabetologia 30: 144–148, 1987 [DOI] [PubMed] [Google Scholar]
- 7.Groop PH, Thomas MC, Moran JL, Wadèn J, Thorn LM, Mäkinen VP, Rosengård-Bärlund M, Saraheimo M, Hietala K, Heikkilä O, Forsblom C; FinnDiane Study Group : The presence and severity of chronic kidney disease predicts all-cause mortality in type 1 diabetes. Diabetes 58: 1651–1658, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seaquist ER, Goetz FC, Rich S, Barbosa J: Familial clustering of diabetic kidney disease. Evidence for genetic susceptibility to diabetic nephropathy. N Engl J Med 320: 1161–1165, 1989 [DOI] [PubMed] [Google Scholar]
- 9.Borch-Johnsen K, Nørgaard K, Hommel E, Mathiesen ER, Jensen JS, Deckert T, Parving HH: Is diabetic nephropathy an inherited complication? Kidney Int 41: 719–722, 1992 [DOI] [PubMed] [Google Scholar]
- 10.Harjutsalo V, Katoh S, Sarti C, Tajima N, Tuomilehto J: Population-based assessment of familial clustering of diabetic nephropathy in type 1 diabetes. Diabetes 53: 2449–2454, 2004 [DOI] [PubMed] [Google Scholar]
- 11.Thorn LM, Forsblom C, Fagerudd J, Pettersson-Fernholm K, Kilpikari R, Groop PH; FinnDiane Study Group : Clustering of risk factors in parents of patients with type 1 diabetes and nephropathy. Diabetes Care 30: 1162–1167, 2007 [DOI] [PubMed] [Google Scholar]
- 12.Fagerudd JA, Pettersson-Fernholm KJ, Grönhagen-Riska C, Groop PH: The impact of a family history of Type II (non-insulin-dependent) diabetes mellitus on the risk of diabetic nephropathy in patients with Type I (insulin-dependent) diabetes mellitus. Diabetologia 42: 519–526, 1999 [DOI] [PubMed] [Google Scholar]
- 13.Earle K, Walker J, Hill C, Viberti G: Familial clustering of cardiovascular disease in patients with insulin-dependent diabetes and nephropathy. N Engl J Med 326: 673–677, 1992 [DOI] [PubMed] [Google Scholar]
- 14.Williams WW, Salem RM, McKnight AJ, Sandholm N, Forsblom C, Taylor A, Guiducci C, McAteer JB, McKay GJ, Isakova T, Brennan EP, Sadlier DM, Palmer C, Söderlund J, Fagerholm E, Harjutsalo V, Lithovius R, Gordin D, Hietala K, Kytö J, Parkkonen M, Rosengård-Bärlund M, Thorn L, Syreeni A, Tolonen N, Saraheimo M, Wadén J, Pitkäniemi J, Sarti C, Tuomilehto J, Tryggvason K, Österholm AM, He B, Bain S, Martin F, Godson C, Hirschhorn JN, Maxwell AP, Groop PH, Florez JC; GENIE Consortium : Association testing of previously reported variants in a large case-control meta-analysis of diabetic nephropathy. Diabetes 61: 2187–2194, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sandholm N, Salem RM, McKnight AJ, Brennan EP, Forsblom C, Isakova T, McKay GJ, Williams WW, Sadlier DM, Mäkinen VP, Swan EJ, Palmer C, Boright AP, Ahlqvist E, Deshmukh HA, Keller BJ, Huang H, Ahola AJ, Fagerholm E, Gordin D, Harjutsalo V, He B, Heikkilä O, Hietala K, Kytö J, Lahermo P, Lehto M, Lithovius R, Osterholm AM, Parkkonen M, Pitkäniemi J, Rosengård-Bärlund M, Saraheimo M, Sarti C, Söderlund J, Soro-Paavonen A, Syreeni A, Thorn LM, Tikkanen H, Tolonen N, Tryggvason K, Tuomilehto J, Wadén J, Gill GV, Prior S, Guiducci C, Mirel DB, Taylor A, Hosseini SM, Parving HH, Rossing P, Tarnow L, Ladenvall C, Alhenc-Gelas F, Lefebvre P, Rigalleau V, Roussel R, Tregouet DA, Maestroni A, Maestroni S, Falhammar H, Gu T, Möllsten A, Cimponeriu D, Ioana M, Mota M, Mota E, Serafinceanu C, Stavarachi M, Hanson RL, Nelson RG, Kretzler M, Colhoun HM, Panduru NM, Gu HF, Brismar K, Zerbini G, Hadjadj S, Marre M, Groop L, Lajer M, Bull SB, Waggott D, Paterson AD, Savage DA, Bain SC, Martin F, Hirschhorn JN, Godson C, Florez JC, Groop PH, Maxwell AP; DCCT/EDIC Research Group : New susceptibility loci associated with kidney disease in type 1 diabetes. PLoS Genet 8: e1002921, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sandholm N, McKnight AJ, Salem RM, Brennan EP, Forsblom C, Harjutsalo V, Mäkinen VP, McKay GJ, Sadlier DM, Williams WW, Martin F, Panduru NM, Tarnow L, Tuomilehto J, Tryggvason K, Zerbini G, Comeau ME, Langefeld CD, Godson C, Hirschhorn JN, Maxwell AP, Florez JC, Groop PH; FIND Consortium; FinnDiane Study Group and the GENIE Consortium : Chromosome 2q31.1 associates with ESRD in women with type 1 diabetes. J Am Soc Nephrol 24: 1537–1543, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sandholm N, Forsblom C, Mäkinen VP, McKnight AJ, Osterholm AM, He B, Harjutsalo V, Lithovius R, Gordin D, Parkkonen M, Saraheimo M, Thorn LM, Tolonen N, Wadén J, Tuomilehto J, Lajer M, Ahlqvist E, Möllsten A, Marcovecchio ML, Cooper J, Dunger D, Paterson AD, Zerbini G, Groop L, Tarnow L, Maxwell AP, Tryggvason K, Groop PH; SUMMIT Consortium; FinnDiane Study Group : Genome-wide association study of urinary albumin excretion rate in patients with type 1 diabetes. Diabetologia 57: 1143–1153, 2014 [DOI] [PubMed] [Google Scholar]
- 18.Köttgen A, Glazer NL, Dehghan A, Hwang SJ, Katz R, Li M, Yang Q, Gudnason V, Launer LJ, Harris TB, Smith AV, Arking DE, Astor BC, Boerwinkle E, Ehret GB, Ruczinski I, Scharpf RB, Chen YD, de Boer IH, Haritunians T, Lumley T, Sarnak M, Siscovick D, Benjamin EJ, Levy D, Upadhyay A, Aulchenko YS, Hofman A, Rivadeneira F, Uitterlinden AG, van Duijn CM, Chasman DI, Paré G, Ridker PM, Kao WH, Witteman JC, Coresh J, Shlipak MG, Fox CS: Multiple loci associated with indices of renal function and chronic kidney disease. Nat Genet 41: 712–717, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pattaro C, Köttgen A, Teumer A, Garnaas M, Böger CA, Fuchsberger C, Olden M, Chen MH, Tin A, Taliun D, Li M, Gao X, Gorski M, Yang Q, Hundertmark C, Foster MC, O’Seaghdha CM, Glazer N, Isaacs A, Liu CT, Smith AV, O’Connell JR, Struchalin M, Tanaka T, Li G, Johnson AD, Gierman HJ, Feitosa M, Hwang SJ, Atkinson EJ, Lohman K, Cornelis MC, Johansson Å, Tönjes A, Dehghan A, Chouraki V, Holliday EG, Sorice R, Kutalik Z, Lehtimäki T, Esko T, Deshmukh H, Ulivi S, Chu AY, Murgia F, Trompet S, Imboden M, Kollerits B, Pistis G, Harris TB, Launer LJ, Aspelund T, Eiriksdottir G, Mitchell BD, Boerwinkle E, Schmidt H, Cavalieri M, Rao M, Hu FB, Demirkan A, Oostra BA, de Andrade M, Turner ST, Ding J, Andrews JS, Freedman BI, Koenig W, Illig T, Döring A, Wichmann HE, Kolcic I, Zemunik T, Boban M, Minelli C, Wheeler HE, Igl W, Zaboli G, Wild SH, Wright AF, Campbell H, Ellinghaus D, Nöthlings U, Jacobs G, Biffar R, Endlich K, Ernst F, Homuth G, Kroemer HK, Nauck M, Stracke S, Völker U, Völzke H, Kovacs P, Stumvoll M, Mägi R, Hofman A, Uitterlinden AG, Rivadeneira F, Aulchenko YS, Polasek O, Hastie N, Vitart V, Helmer C, Wang JJ, Ruggiero D, Bergmann S, Kähönen M, Viikari J, Nikopensius T, Province M, Ketkar S, Colhoun H, Doney A, Robino A, Giulianini F, Krämer BK, Portas L, Ford I, Buckley BM, Adam M, Thun GA, Paulweber B, Haun M, Sala C, Metzger M, Mitchell P, Ciullo M, Kim SK, Vollenweider P, Raitakari O, Metspalu A, Palmer C, Gasparini P, Pirastu M, Jukema JW, Probst-Hensch NM, Kronenberg F, Toniolo D, Gudnason V, Shuldiner AR, Coresh J, Schmidt R, Ferrucci L, Siscovick DS, van Duijn CM, Borecki I, Kardia SL, Liu Y, Curhan GC, Rudan I, Gyllensten U, Wilson JF, Franke A, Pramstaller PP, Rettig R, Prokopenko I, Witteman JC, Hayward C, Ridker P, Parsa A, Bochud M, Heid IM, Goessling W, Chasman DI, Kao WH, Fox CS; CARDIoGRAM Consortium; ICBP Consortium; CARe Consortium; Wellcome Trust Case Control Consortium 2 (WTCCC2) : Genome-wide association and functional follow-up reveals new loci for kidney function. PLoS Genet 8: e1002584, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Köttgen A, Pattaro C, Böger CA, Fuchsberger C, Olden M, Glazer NL, Parsa A, Gao X, Yang Q, Smith AV, O’Connell JR, Li M, Schmidt H, Tanaka T, Isaacs A, Ketkar S, Hwang SJ, Johnson AD, Dehghan A, Teumer A, Paré G, Atkinson EJ, Zeller T, Lohman K, Cornelis MC, Probst-Hensch NM, Kronenberg F, Tönjes A, Hayward C, Aspelund T, Eiriksdottir G, Launer LJ, Harris TB, Rampersaud E, Mitchell BD, Arking DE, Boerwinkle E, Struchalin M, Cavalieri M, Singleton A, Giallauria F, Metter J, de Boer IH, Haritunians T, Lumley T, Siscovick D, Psaty BM, Zillikens MC, Oostra BA, Feitosa M, Province M, de Andrade M, Turner ST, Schillert A, Ziegler A, Wild PS, Schnabel RB, Wilde S, Munzel TF, Leak TS, Illig T, Klopp N, Meisinger C, Wichmann HE, Koenig W, Zgaga L, Zemunik T, Kolcic I, Minelli C, Hu FB, Johansson A, Igl W, Zaboli G, Wild SH, Wright AF, Campbell H, Ellinghaus D, Schreiber S, Aulchenko YS, Felix JF, Rivadeneira F, Uitterlinden AG, Hofman A, Imboden M, Nitsch D, Brandstätter A, Kollerits B, Kedenko L, Mägi R, Stumvoll M, Kovacs P, Boban M, Campbell S, Endlich K, Völzke H, Kroemer HK, Nauck M, Völker U, Polasek O, Vitart V, Badola S, Parker AN, Ridker PM, Kardia SL, Blankenberg S, Liu Y, Curhan GC, Franke A, Rochat T, Paulweber B, Prokopenko I, Wang W, Gudnason V, Shuldiner AR, Coresh J, Schmidt R, Ferrucci L, Shlipak MG, van Duijn CM, Borecki I, Krämer BK, Rudan I, Gyllensten U, Wilson JF, Witteman JC, Pramstaller PP, Rettig R, Hastie N, Chasman DI, Kao WH, Heid IM, Fox CS: New loci associated with kidney function and chronic kidney disease. Nat Genet 42: 376–384, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, Sklar P; International Schizophrenia Consortium : Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460: 748–752, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schizophrenia Working Group of the Psychiatric Genomics Consortium : Biological insights from 108 schizophrenia-associated genetic loci. Nature 511: 421–427, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thorn LM, Forsblom C, Fagerudd J, Thomas MC, Pettersson-Fernholm K, Saraheimo M, Wadén J, Rönnback M, Rosengård-Bärlund M, Björkesten CG, Taskinen MR, Groop PH; FinnDiane Study Group : Metabolic syndrome in type 1 diabetes: association with diabetic nephropathy and glycemic control (the FinnDiane study). Diabetes Care 28: 2019–2024, 2005 [DOI] [PubMed] [Google Scholar]
- 24.Chaturvedi N, Bandinelli S, Mangili R, Penno G, Rottiers RE, Fuller JH: Microalbuminuria in type 1 diabetes: rates, risk factors and glycemic threshold. Kidney Int 60: 219–227, 2001 [DOI] [PubMed] [Google Scholar]
- 25.Lindholm E, Agardh E, Tuomi T, Groop L, Agardh CD: Classifying diabetes according to the new WHO clinical stages. Eur J Epidemiol 17: 983–989, 2001 [DOI] [PubMed] [Google Scholar]
- 26.Amin R, Widmer B, Prevost AT, Schwarze P, Cooper J, Edge J, Marcovecchio L, Neil A, Dalton RN, Dunger DB: Risk of microalbuminuria and progression to macroalbuminuria in a cohort with childhood onset type 1 diabetes: prospective observational study. BMJ 336: 697–701, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marcovecchio ML, Dalton RN, Schwarze CP, Prevost AT, Neil HA, Acerini CL, Barrett T, Cooper JD, Edge J, Shield J, Widmer B, Todd JA, Dunger DB: Ambulatory blood pressure measurements are related to albumin excretion and are predictive for risk of microalbuminuria in young people with type 1 diabetes. Diabetologia 52: 1173–1181, 2009 [DOI] [PubMed] [Google Scholar]
- 28.Mooyaart AL, Valk EJJ, van Es LA, Bruijn JA, de Heer E, Freedman BI, Dekkers OM, Baelde HJ: Genetic associations in diabetic nephropathy: a meta-analysis. Diabetologia 54: 544–553, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambo F, Malovini A, Sandholm N, Stavarachi M, Forsblom C, Mäkinen VP, Harjutsalo V, Lithovius R, Gordin D, Parkkonen M, Saraheimo M, Thorn LM, Tolonen N, Wadén J, He B, Osterholm AM, Tuomilehto J, Lajer M, Salem RM, McKnight AJ, Tarnow L, Panduru NM, Barbarini N, Di Camillo B, Toffolo GM, Tryggvason K, Bellazzi R, Cobelli C, Groop PH; GENIE Consortium; FinnDiane Study Group : Novel genetic susceptibility loci for diabetic end-stage renal disease identified through robust naive Bayes classification. Diabetologia 57: 1611–1622, 2014 [DOI] [PubMed] [Google Scholar]
- 30.Pezzolesi MG, Poznik GD, Mychaleckyj JC, Paterson AD, Barati MT, Klein JB, Ng DPK, Placha G, Canani LH, Bochenski J, Waggott D, Merchant ML, Krolewski B, Mirea L, Wanic K, Katavetin P, Kure M, Wolkow P, Dunn JS, Smiles A, Walker WH, Boright AP, Bull SB, Doria A, Rogus JJ, Rich SS, Warram JH, Krolewski AS; DCCT/EDIC Research Group : Genome-wide association scan for diabetic nephropathy susceptibility genes in type 1 diabetes. Diabetes 58: 1403–1410, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Craig DW, Millis MP, DiStefano JK: Genome-wide SNP genotyping study using pooled DNA to identify candidate markers mediating susceptibility to end-stage renal disease attributed to Type 1 diabetes. Diabet Med 26: 1090–1098, 2009 [DOI] [PubMed] [Google Scholar]
- 32.Shimazaki A, Kawamura Y, Kanazawa A, Sekine A, Saito S, Tsunoda T, Koya D, Babazono T, Tanaka Y, Matsuda M, Kawai K, Iiizumi T, Imanishi M, Shinosaki T, Yanagimoto T, Ikeda M, Omachi S, Kashiwagi A, Kaku K, Iwamoto Y, Kawamori R, Kikkawa R, Nakajima M, Nakamura Y, Maeda S: Genetic variations in the gene encoding ELMO1 are associated with susceptibility to diabetic nephropathy. Diabetes 54: 1171–1178, 2005 [DOI] [PubMed] [Google Scholar]
- 33.McDonough CW, Palmer ND, Hicks PJ, Roh BH, An SS, Cooke JN, Hester JM, Wing MR, Bostrom MA, Rudock ME, Lewis JP, Talbert ME, Blevins RA, Lu L, Ng MC, Sale MM, Divers J, Langefeld CD, Freedman BI, Bowden DW: A genome-wide association study for diabetic nephropathy genes in African Americans. Kidney Int 79: 563–572, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tolonen N, Forsblom C, Thorn L, Wadén J, Rosengård-Bärlund M, Saraheimo M, Feodoroff M, Mäkinen VP, Gordin D, Taskinen MR, Groop PH; FinnDiane Study Group : Lipid abnormalities predict progression of renal disease in patients with type 1 diabetes. Diabetologia 52: 2522–2530, 2009 [DOI] [PubMed] [Google Scholar]
- 35.Wiseman M, Viberti G, Mackintosh D, Jarrett RJ, Keen H: Glycaemia, arterial pressure and micro-albuminuria in type 1 (insulin-dependent) diabetes mellitus. Diabetologia 26: 401–405, 1984 [DOI] [PubMed] [Google Scholar]
- 36.Feodoroff M, Harjutsalo V, Forsblom C, Thorn L, Waden J, Tolonen N, Lithovius R, Groop PH: Smoking and progression of diabetic nephropathy in patients with type 1 diabetes. Acta Diabetol 53: 525–533, 2016 [DOI] [PubMed] [Google Scholar]
- 37.Hadjadj S, Péan F, Gallois Y, Passa P, Aubert R, Weekers L, Rigalleau V, Bauduceau B, Bekherraz A, Roussel R, Dussol B, Rodier M, Marechaud R, Lefebvre PJ, Marre M; Genesis France-Belgium Study : Different patterns of insulin resistance in relatives of type 1 diabetic patients with retinopathy or nephropathy: the Genesis France-Belgium Study. Diabetes Care 27: 2661–2668, 2004 [DOI] [PubMed] [Google Scholar]
- 38.DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium, Asian Genetic Epidemiology Network Type 2 Diabetes (AGEN-T2D) Consortium, South Asian Type 2 Diabetes (SAT2D) Consortium, Mexican American Type 2 Diabetes (MAT2D) Consortium, Type 2 Diabetes Genetic Exploration by Nex-generation sequencing in muylti-Ethnic Samples (T2D-GENES) Consortium, Mahajan A, Go MJ, Zhang W, Below JE, Gaulton KJ, Ferreira T, Horikoshi M, Johnson AD, Ng MC, Prokopenko I, Saleheen D, Wang X, Zeggini E, Abecasis GR, Adair LS, Almgren P, Atalay M, Aung T, Baldassarre D, Balkau B, Bao Y, Barnett AH, Barroso I, Basit A, Been LF, Beilby J, Bell GI, Benediktsson R, Bergman RN, Boehm BO, Boerwinkle E, Bonnycastle LL, Burtt N, Cai Q, Campbell H, Carey J, Cauchi S, Caulfield M, Chan JC, Chang LC, Chang TJ, Chang YC, Charpentier G, Chen CH, Chen H, Chen YT, Chia KS, Chidambaram M, Chines PS, Cho NH, Cho YM, Chuang LM, Collins FS, Cornelis MC, Couper DJ, Crenshaw AT, van Dam RM, Danesh J, Das D, de Faire U, Dedoussis G, Deloukas P, Dimas AS, Dina C, Doney AS, Donnelly PJ, Dorkhan M, van Duijn C, Dupuis J, Edkins S, Elliott P, Emilsson V, Erbel R, Eriksson JG, Escobedo J, Esko T, Eury E, Florez JC, Fontanillas P, Forouhi NG, Forsen T, Fox C, Fraser RM, Frayling TM, Froguel P, Frossard P, Gao Y, Gertow K, Gieger C, Gigante B, Grallert H, Grant GB, Grrop LC, Groves CJ, Grundberg E, Guiducci C, Hamsten A, Han BG, Hara K, Hassanali N, Hattersley AT, Hayward C, Hedman AK, Herder C, Hofman A, Holmen OL, Hovingh K, Hreidarsson AB, Hu C, Hu FB, Hui J, Humphries SE, Hunt SE, Hunter DJ, Hveem K, Hydrie ZI, Ikegami H, Illig T, Ingelsson E, Islam M, Isomaa B, Jackson AU, Jafar T, James A, Jia W, Jockel KH, Jonsson A, Jowett JB, Kadowaki T, Kang HM, Kanoni S, Kao WH, Kathiresan S, Kato N, Katulanda P, Keinanen-Kiukaanniemi KM, Kelly AM, Khan H, Khaw KT, Khor CC, Kim HL, Kim S, Kim YJ, Kinnunen L, Klopp N, Kong A, Korpi-Hyovalti E, Kowlessur S, Kraft P, Kravic J, Kristensen MM, Krithika S, Kumar A, Kumate J, Kuusisto J, Kwak SH, Laakso M, Lagou V, Lakka TA, Langenberg C, Langford C, Lawrence R, Leander K, Lee JM, Lee NR, Li M, Li X, Li Y, Liang J, Liju S, Lim WY, Lind L, Lindgren CM, Lindholm E, Liu CT, Liu JJ, Lobbens S, Long J, Loos RJ, Lu W, Luan J, Lyssenko V, Ma RC, Maeda S, Magi R, Mannisto S, Matthews DR, Meigs JB, Melander O, Metspalu A, Meyer J, Mirza G, Mihailov E, Moebus S, Mohan V, Mohlke KL, Morris AD, Muhleisen TW, Muller-Nurasyid M, Musk B, Nakamura J, Nakashima E, Navarro P, Ng PK, Nica AC, Nilsson PM, Njolstad I, Nothen MM, Ohnaka K, Ong TH, Owen KR, Palmer CN, Pankow JS, Park KS, Parkin M, Pechlivanis S, Pedersen NL, Peltonen L, Perry JR, Peters A, Pinidiyapathirage JM, Platou CG, Potter S, Price JF, Qi L, Radha V, Rallidis L, Rasheed A, Rathman W, Rauramaa R, Raychaudhuri S, Rayner NW, Rees SD, Rehnberg E, Ripatti S, Robertson N, Roden M, Rossin EJ, Rudan I, Rybin D, Saaristo TE, Salomaa V, Saltevo J, Samuel M, Sanghera DK, Saramies J, Scott J, Scott LJ, Scott RA, Segre AV, Sehmi J, Sennblad B, Shah N, Shah S, Shera AS, Shu XO, Shuldiner AR, Sigurdsson G, Sijbrands E, Silveira A, Sim X, Sivapalaratnam S, Small KS, So WY, Stancakova A, Stefansson K, Steinbach G, Steinthorsdottir V, Stirrups K, Strawbridge RJ, Stringham HM, Sun Q, Suo C, Syvanen AC, Takayanagi R, Takeuchi F, Tay WT, Teslovich TM, Thorand B, Thorleifsson G, Thorsteinsdottir U, Tikkanen E, Trakalo J, Tremoli E, Trip MD, Tsai FJ, Tuomi T, Tuomilehto J, Uitterlinden AG, Valladares-Salgado A, Vedantam S, Veglia F, Voight BF, Wang C, Wareham NJ, Wennauer R, Wickremasinghe AR, Wilsgaard T, Wilson JF, Wiltshire S, Winckler W, Wong TY, Wood AR, Wu JY, Wu Y, Yamamoto K, Yamauchi T, Yang M, Yengo L, Yokota M, Young R, Zabaneh D, Zhang F, Zhang R, Zheng W, Zimmet PZ, Altshuler D, Bowden DW, Cho YS, Cox NJ, Cruz M, Hanis CL, Kooner J, Lee JY, Seielstad M, Teo YY, Boehnke M, Parra EJ, Chambers JC, Tai ES, McCarthy MI, Morris AP: Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet 46: 234–244, 2014 [DOI] [PMC free article] [PubMed]
- 39.Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, Julier C, Morahan G, Nerup J, Nierras C, Plagnol V, Pociot F, Schuilenburg H, Smyth DJ, Stevens H, Todd JA, Walker NM, Rich SS; Type 1 Diabetes Genetics Consortium : Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet 41: 703–707, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saxena R, Hivert MF, Langenberg C, Tanaka T, Pankow JS, Vollenweider P, Lyssenko V, Bouatia-Naji N, Dupuis J, Jackson AU, Kao WH, Li M, Glazer NL, Manning AK, Luan J, Stringham HM, Prokopenko I, Johnson T, Grarup N, Boesgaard TW, Lecoeur C, Shrader P, O’Connell J, Ingelsson E, Couper DJ, Rice K, Song K, Andreasen CH, Dina C, Köttgen A, Le Bacquer O, Pattou F, Taneera J, Steinthorsdottir V, Rybin D, Ardlie K, Sampson M, Qi L, van Hoek M, Weedon MN, Aulchenko YS, Voight BF, Grallert H, Balkau B, Bergman RN, Bielinski SJ, Bonnefond A, Bonnycastle LL, Borch-Johnsen K, Böttcher Y, Brunner E, Buchanan TA, Bumpstead SJ, Cavalcanti-Proença C, Charpentier G, Chen YD, Chines PS, Collins FS, Cornelis M, J Crawford G, Delplanque J, Doney A, Egan JM, Erdos MR, Firmann M, Forouhi NG, Fox CS, Goodarzi MO, Graessler J, Hingorani A, Isomaa B, Jørgensen T, Kivimaki M, Kovacs P, Krohn K, Kumari M, Lauritzen T, Lévy-Marchal C, Mayor V, McAteer JB, Meyre D, Mitchell BD, Mohlke KL, Morken MA, Narisu N, Palmer CN, Pakyz R, Pascoe L, Payne F, Pearson D, Rathmann W, Sandbaek A, Sayer AA, Scott LJ, Sharp SJ, Sijbrands E, Singleton A, Siscovick DS, Smith NL, Sparsø T, Swift AJ, Syddall H, Thorleifsson G, Tönjes A, Tuomi T, Tuomilehto J, Valle TT, Waeber G, Walley A, Waterworth DM, Zeggini E, Zhao JH, Illig T, Wichmann HE, Wilson JF, van Duijn C, Hu FB, Morris AD, Frayling TM, Hattersley AT, Thorsteinsdottir U, Stefansson K, Nilsson P, Syvänen AC, Shuldiner AR, Walker M, Bornstein SR, Schwarz P, Williams GH, Nathan DM, Kuusisto J, Laakso M, Cooper C, Marmot M, Ferrucci L, Mooser V, Stumvoll M, Loos RJ, Altshuler D, Psaty BM, Rotter JI, Boerwinkle E, Hansen T, Pedersen O, Florez JC, McCarthy MI, Boehnke M, Barroso I, Sladek R, Froguel P, Meigs JB, Groop L, Wareham NJ, Watanabe RM; GIANT consortium; MAGIC investigators : Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nat Genet 42: 142–148, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manning AK, Hivert MF, Scott RA, Grimsby JL, Bouatia-Naji N, Chen H, Rybin D, Liu CT, Bielak LF, Prokopenko I, Amin N, Barnes D, Cadby G, Hottenga JJ, Ingelsson E, Jackson AU, Johnson T, Kanoni S, Ladenvall C, Lagou V, Lahti J, Lecoeur C, Liu Y, Martinez-Larrad MT, Montasser ME, Navarro P, Perry JR, Rasmussen-Torvik LJ, Salo P, Sattar N, Shungin D, Strawbridge RJ, Tanaka T, van Duijn CM, An P, de Andrade M, Andrews JS, Aspelund T, Atalay M, Aulchenko Y, Balkau B, Bandinelli S, Beckmann JS, Beilby JP, Bellis C, Bergman RN, Blangero J, Boban M, Boehnke M, Boerwinkle E, Bonnycastle LL, Boomsma DI, Borecki IB, Böttcher Y, Bouchard C, Brunner E, Budimir D, Campbell H, Carlson O, Chines PS, Clarke R, Collins FS, Corbatón-Anchuelo A, Couper D, de Faire U, Dedoussis GV, Deloukas P, Dimitriou M, Egan JM, Eiriksdottir G, Erdos MR, Eriksson JG, Eury E, Ferrucci L, Ford I, Forouhi NG, Fox CS, Franzosi MG, Franks PW, Frayling TM, Froguel P, Galan P, de Geus E, Gigante B, Glazer NL, Goel A, Groop L, Gudnason V, Hallmans G, Hamsten A, Hansson O, Harris TB, Hayward C, Heath S, Hercberg S, Hicks AA, Hingorani A, Hofman A, Hui J, Hung J, Jarvelin MR, Jhun MA, Johnson PC, Jukema JW, Jula A, Kao WH, Kaprio J, Kardia SL, Keinanen-Kiukaanniemi S, Kivimaki M, Kolcic I, Kovacs P, Kumari M, Kuusisto J, Kyvik KO, Laakso M, Lakka T, Lannfelt L, Lathrop GM, Launer LJ, Leander K, Li G, Lind L, Lindstrom J, Lobbens S, Loos RJ, Luan J, Lyssenko V, Mägi R, Magnusson PK, Marmot M, Meneton P, Mohlke KL, Mooser V, Morken MA, Miljkovic I, Narisu N, O’Connell J, Ong KK, Oostra BA, Palmer LJ, Palotie A, Pankow JS, Peden JF, Pedersen NL, Pehlic M, Peltonen L, Penninx B, Pericic M, Perola M, Perusse L, Peyser PA, Polasek O, Pramstaller PP, Province MA, Räikkönen K, Rauramaa R, Rehnberg E, Rice K, Rotter JI, Rudan I, Ruokonen A, Saaristo T, Sabater-Lleal M, Salomaa V, Savage DB, Saxena R, Schwarz P, Seedorf U, Sennblad B, Serrano-Rios M, Shuldiner AR, Sijbrands EJ, Siscovick DS, Smit JH, Small KS, Smith NL, Smith AV, Stančáková A, Stirrups K, Stumvoll M, Sun YV, Swift AJ, Tönjes A, Tuomilehto J, Trompet S, Uitterlinden AG, Uusitupa M, Vikström M, Vitart V, Vohl MC, Voight BF, Vollenweider P, Waeber G, Waterworth DM, Watkins H, Wheeler E, Widen E, Wild SH, Willems SM, Willemsen G, Wilson JF, Witteman JC, Wright AF, Yaghootkar H, Zelenika D, Zemunik T, Zgaga L, Wareham NJ, McCarthy MI, Barroso I, Watanabe RM, Florez JC, Dupuis J, Meigs JB, Langenberg C; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium; Multiple Tissue Human Expression Resource (MUTHER) Consortium : A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat Genet 44: 659–669, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soranzo N, Sanna S, Wheeler E, Gieger C, Radke D, Dupuis J, Bouatia-Naji N, Langenberg C, Prokopenko I, Stolerman E, Sandhu MS, Heeney MM, Devaney JM, Reilly MP, Ricketts SL, Stewart AF, Voight BF, Willenborg C, Wright B, Altshuler D, Arking D, Balkau B, Barnes D, Boerwinkle E, Bohm B, Bonnefond A, Bonnycastle LL, Boomsma DI, Bornstein SR, Bottcher Y, Bumpstead S, Burnett-Miller MS, Campbell H, Cao A, Chambers J, Clark R, Collins FS, Coresh J, de Geus EJ, Dei M, Deloukas P, Doring A, Egan JM, Elosua R, Ferrucci L, Forouhi N, Fox CS, Franklin C, Franzosi MG, Gallina S, Goel A, Graessler J, Grallert H, Greinacher A, Hadley D, Hall A, Hamsten A, Hayward C, Heath S, Herder C, Homuth G, Hottenga JJ, Hunter-Merrill R, Illig T, Jackson AU, Jula A, Kleber M, Knouff CW, Kong A, Kooner J, Kottgen A, Kovacs P, Krohn K, Kuhnel B, Kuusisto J, Laakso M, Lathrop M, Lecoeur C, Li M, Li M, Loos RJ, Luan J, Lyssenko V, Magi R, Magnusson PK, Malarstig A, Mangino M, Martinez-Larrad MT, Marz W, McArdle WL, McPherson R, Meisinger C, Meitinger T, Melander O, Mohlke KL, Mooser VE, Morken MA, Narisu N, Nathan DM, Nauck M, O’Donnell C, Oexle K, Olla N, Pankow JS, Payne F, Peden JF, Pedersen NL, Peltonen L, Perola M, Polasek O, Porcu E, Rader DJ, Rathmann W, Ripatti S, Rocheleau G, Roden M, Rudan I, Salomaa V, Saxena R, Schlessinger D, Schunkert H, Schwarz P, Seedorf U, Selvin E, Serrano-Rios M, Shrader P, Silveira A, Siscovick D, Song K, Spector TD, Stefansson K, Steinthorsdottir V, Strachan DP, Strawbridge R, Stumvoll M, Surakka I, Swift AJ, Tanaka T, Teumer A, Thorleifsson G, Thorsteinsdottir U, Tonjes A, Usala G, Vitart V, Volzke H, Wallaschofski H, Waterworth DM, Watkins H, Wichmann HE, Wild SH, Willemsen G, Williams GH, Wilson JF, Winkelmann J, Wright AF: WTCCC, Zabena C, Zhao JH, Epstein SE, Erdmann J, Hakonarson HH, Kathiresan S, Khaw KT, Roberts R, Samani NJ, Fleming MD, Sladek R, Abecasis G, Boehnke M, Froguel P, Groop L, McCarthy MI, Kao WH, Florez JC, Uda M, Wareham NJ, Barroso I, Meigs JB: Common variants at 10 genomic loci influence hemoglobin A1C levels via glycemic and nonglycemic pathways. Diabetes 59: 3229–3239, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strawbridge RJ, Dupuis J, Prokopenko I, Barker A, Ahlqvist E, Rybin D, Petrie JR, Travers ME, Bouatia-Naji N, Dimas AS, Nica A, Wheeler E, Chen H, Voight BF, Taneera J, Kanoni S, Peden JF, Turrini F, Gustafsson S, Zabena C, Almgren P, Barker DJ, Barnes D, Dennison EM, Eriksson JG, Eriksson P, Eury E, Folkersen L, Fox CS, Frayling TM, Goel A, Gu HF, Horikoshi M, Isomaa B, Jackson AU, Jameson KA, Kajantie E, Kerr-Conte J, Kuulasmaa T, Kuusisto J, Loos RJ, Luan J, Makrilakis K, Manning AK, Martínez-Larrad MT, Narisu N, Nastase Mannila M, Ohrvik J, Osmond C, Pascoe L, Payne F, Sayer AA, Sennblad B, Silveira A, Stancáková A, Stirrups K, Swift AJ, Syvänen AC, Tuomi T, van ’t Hooft FM, Walker M, Weedon MN, Xie W, Zethelius B, Ongen H, Mälarstig A, Hopewell JC, Saleheen D, Chambers J, Parish S, Danesh J, Kooner J, Ostenson CG, Lind L, Cooper CC, Serrano-Ríos M, Ferrannini E, Forsen TJ, Clarke R, Franzosi MG, Seedorf U, Watkins H, Froguel P, Johnson P, Deloukas P, Collins FS, Laakso M, Dermitzakis ET, Boehnke M, McCarthy MI, Wareham NJ, Groop L, Pattou F, Gloyn AL, Dedoussis GV, Lyssenko V, Meigs JB, Barroso I, Watanabe RM, Ingelsson E, Langenberg C, Hamsten A, Florez JC; DIAGRAM Consortium; GIANT Consortium; MuTHER Consortium; CARDIoGRAM Consortium; C4D Consortium : Genome-wide association identifies nine common variants associated with fasting proinsulin levels and provides new insights into the pathophysiology of type 2 diabetes. Diabetes 60: 2624–2634, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, Wheeler E, Glazer NL, Bouatia-Naji N, Gloyn AL, Lindgren CM, Mägi R, Morris AP, Randall J, Johnson T, Elliott P, Rybin D, Thorleifsson G, Steinthorsdottir V, Henneman P, Grallert H, Dehghan A, Hottenga JJ, Franklin CS, Navarro P, Song K, Goel A, Perry JR, Egan JM, Lajunen T, Grarup N, Sparsø T, Doney A, Voight BF, Stringham HM, Li M, Kanoni S, Shrader P, Cavalcanti-Proença C, Kumari M, Qi L, Timpson NJ, Gieger C, Zabena C, Rocheleau G, Ingelsson E, An P, O’Connell J, Luan J, Elliott A, McCarroll SA, Payne F, Roccasecca RM, Pattou F, Sethupathy P, Ardlie K, Ariyurek Y, Balkau B, Barter P, Beilby JP, Ben-Shlomo Y, Benediktsson R, Bennett AJ, Bergmann S, Bochud M, Boerwinkle E, Bonnefond A, Bonnycastle LL, Borch-Johnsen K, Böttcher Y, Brunner E, Bumpstead SJ, Charpentier G, Chen YD, Chines P, Clarke R, Coin LJ, Cooper MN, Cornelis M, Crawford G, Crisponi L, Day IN, de Geus EJ, Delplanque J, Dina C, Erdos MR, Fedson AC, Fischer-Rosinsky A, Forouhi NG, Fox CS, Frants R, Franzosi MG, Galan P, Goodarzi MO, Graessler J, Groves CJ, Grundy S, Gwilliam R, Gyllensten U, Hadjadj S, Hallmans G, Hammond N, Han X, Hartikainen AL, Hassanali N, Hayward C, Heath SC, Hercberg S, Herder C, Hicks AA, Hillman DR, Hingorani AD, Hofman A, Hui J, Hung J, Isomaa B, Johnson PR, Jørgensen T, Jula A, Kaakinen M, Kaprio J, Kesaniemi YA, Kivimaki M, Knight B, Koskinen S, Kovacs P, Kyvik KO, Lathrop GM, Lawlor DA, Le Bacquer O, Lecoeur C, Li Y, Lyssenko V, Mahley R, Mangino M, Manning AK, Martínez-Larrad MT, McAteer JB, McCulloch LJ, McPherson R, Meisinger C, Melzer D, Meyre D, Mitchell BD, Morken MA, Mukherjee S, Naitza S, Narisu N, Neville MJ, Oostra BA, Orrù M, Pakyz R, Palmer CN, Paolisso G, Pattaro C, Pearson D, Peden JF, Pedersen NL, Perola M, Pfeiffer AF, Pichler I, Polasek O, Posthuma D, Potter SC, Pouta A, Province MA, Psaty BM, Rathmann W, Rayner NW, Rice K, Ripatti S, Rivadeneira F, Roden M, Rolandsson O, Sandbaek A, Sandhu M, Sanna S, Sayer AA, Scheet P, Scott LJ, Seedorf U, Sharp SJ, Shields B, Sigurethsson G, Sijbrands EJ, Silveira A, Simpson L, Singleton A, Smith NL, Sovio U, Swift A, Syddall H, Syvänen AC, Tanaka T, Thorand B, Tichet J, Tönjes A, Tuomi T, Uitterlinden AG, van Dijk KW, van Hoek M, Varma D, Visvikis-Siest S, Vitart V, Vogelzangs N, Waeber G, Wagner PJ, Walley A, Walters GB, Ward KL, Watkins H, Weedon MN, Wild SH, Willemsen G, Witteman JC, Yarnell JW, Zeggini E, Zelenika D, Zethelius B, Zhai G, Zhao JH, Zillikens MC, Borecki IB, Loos RJ, Meneton P, Magnusson PK, Nathan DM, Williams GH, Hattersley AT, Silander K, Salomaa V, Smith GD, Bornstein SR, Schwarz P, Spranger J, Karpe F, Shuldiner AR, Cooper C, Dedoussis GV, Serrano-Ríos M, Morris AD, Lind L, Palmer LJ, Hu FB, Franks PW, Ebrahim S, Marmot M, Kao WH, Pankow JS, Sampson MJ, Kuusisto J, Laakso M, Hansen T, Pedersen O, Pramstaller PP, Wichmann HE, Illig T, Rudan I, Wright AF, Stumvoll M, Campbell H, Wilson JF, Bergman RN, Buchanan TA, Collins FS, Mohlke KL, Tuomilehto J, Valle TT, Altshuler D, Rotter JI, Siscovick DS, Penninx BW, Boomsma DI, Deloukas P, Spector TD, Frayling TM, Ferrucci L, Kong A, Thorsteinsdottir U, Stefansson K, van Duijn CM, Aulchenko YS, Cao A, Scuteri A, Schlessinger D, Uda M, Ruokonen A, Jarvelin MR, Waterworth DM, Vollenweider P, Peltonen L, Mooser V, Abecasis GR, Wareham NJ, Sladek R, Froguel P, Watanabe RM, Meigs JB, Groop L, Boehnke M, McCarthy MI, Florez JC, Barroso I; DIAGRAM Consortium; GIANT Consortium; Global BPgen Consortium; Anders Hamsten on behalf of Procardis Consortium; MAGIC investigators : New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet 42: 105–116, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scott RA, Fall T, Pasko D, Barker A, Sharp SJ, Arriola L, Balkau B, Barricarte A, Barroso I, Boeing H, Clavel-Chapelon F, Crowe FL, Dekker JM, Fagherazzi G, Ferrannini E, Forouhi NG, Franks PW, Gavrila D, Giedraitis V, Grioni S, Groop LC, Kaaks R, Key TJ, Kühn T, Lotta LA, Nilsson PM, Overvad K, Palli D, Panico S, Quirós JR, Rolandsson O, Roswall N, Sacerdote C, Sala N, Sánchez MJ, Schulze MB, Siddiq A, Slimani N, Sluijs I, Spijkerman AM, Tjonneland A, Tumino R, van der A DL, Yaghootkar H, McCarthy MI, Semple RK, Riboli E, Walker M, Ingelsson E, Frayling TM, Savage DB, Langenberg C, Wareham NJ; RISC Study Group; EPIC-InterAct Consortium : Common genetic variants highlight the role of insulin resistance and body fat distribution in type 2 diabetes, independent of obesity. Diabetes 63: 4378–4387, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, Helgadottir A, Styrkarsdottir U, Gretarsdottir S, Thorlacius S, Jonsdottir I, Jonsdottir T, Olafsdottir EJ, Olafsdottir GH, Jonsson T, Jonsson F, Borch-Johnsen K, Hansen T, Andersen G, Jorgensen T, Lauritzen T, Aben KK, Verbeek AL, Roeleveld N, Kampman E, Yanek LR, Becker LC, Tryggvadottir L, Rafnar T, Becker DM, Gulcher J, Kiemeney LA, Pedersen O, Kong A, Thorsteinsdottir U, Stefansson K: Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet 41: 18–24, 2009 [DOI] [PubMed] [Google Scholar]
- 47.Shungin D, Winkler TW, Croteau-Chonka DC, Ferreira T, Locke AE, Mägi R, Strawbridge RJ, Pers TH, Fischer K, Justice AE, Workalemahu T, Wu JM, Buchkovich ML, Heard-Costa NL, Roman TS, Drong AW, Song C, Gustafsson S, Day FR, Esko T, Fall T, Kutalik Z, Luan J, Randall JC, Scherag A, Vedantam S, Wood AR, Chen J, Fehrmann R, Karjalainen J, Kahali B, Liu CT, Schmidt EM, Absher D, Amin N, Anderson D, Beekman M, Bragg-Gresham JL, Buyske S, Demirkan A, Ehret GB, Feitosa MF, Goel A, Jackson AU, Johnson T, Kleber ME, Kristiansson K, Mangino M, Mateo Leach I, Medina-Gomez C, Palmer CD, Pasko D, Pechlivanis S, Peters MJ, Prokopenko I, Stančáková A, Ju Sung Y, Tanaka T, Teumer A, Van Vliet-Ostaptchouk JV, Yengo L, Zhang W, Albrecht E, Ärnlöv J, Arscott GM, Bandinelli S, Barrett A, Bellis C, Bennett AJ, Berne C, Blüher M, Böhringer S, Bonnet F, Böttcher Y, Bruinenberg M, Carba DB, Caspersen IH, Clarke R, Daw EW, Deelen J, Deelman E, Delgado G, Doney AS, Eklund N, Erdos MR, Estrada K, Eury E, Friedrich N, Garcia ME, Giedraitis V, Gigante B, Go AS, Golay A, Grallert H, Grammer TB, Gräßler J, Grewal J, Groves CJ, Haller T, Hallmans G, Hartman CA, Hassinen M, Hayward C, Heikkilä K, Herzig KH, Helmer Q, Hillege HL, Holmen O, Hunt SC, Isaacs A, Ittermann T, James AL, Johansson I, Juliusdottir T, Kalafati IP, Kinnunen L, Koenig W, Kooner IK, Kratzer W, Lamina C, Leander K, Lee NR, Lichtner P, Lind L, Lindström J, Lobbens S, Lorentzon M, Mach F, Magnusson PK, Mahajan A, McArdle WL, Menni C, Merger S, Mihailov E, Milani L, Mills R, Moayyeri A, Monda KL, Mooijaart SP, Mühleisen TW, Mulas A, Müller G, Müller-Nurasyid M, Nagaraja R, Nalls MA, Narisu N, Glorioso N, Nolte IM, Olden M, Rayner NW, Renstrom F, Ried JS, Robertson NR, Rose LM, Sanna S, Scharnagl H, Scholtens S, Sennblad B, Seufferlein T, Sitlani CM, Vernon Smith A, Stirrups K, Stringham HM, Sundström J, Swertz MA, Swift AJ, Syvänen AC, Tayo BO, Thorand B, Thorleifsson G, Tomaschitz A, Troffa C, van Oort FV, Verweij N, Vonk JM, Waite LL, Wennauer R, Wilsgaard T, Wojczynski MK, Wong A, Zhang Q, Hua Zhao J, Brennan EP, Choi M, Eriksson P, Folkersen L, Franco-Cereceda A, Gharavi AG, Hedman AK, Hivert MF, Huang J, Kanoni S, Karpe F, Keildson S, Kiryluk K, Liang L, Lifton RP, Ma B, McKnight AJ, McPherson R, Metspalu A, Min JL, Moffatt MF, Montgomery GW, Murabito JM, Nicholson G, Nyholt DR, Olsson C, Perry JR, Reinmaa E, Salem RM, Sandholm N, Schadt EE, Scott RA, Stolk L, Vallejo EE, Westra HJ, Zondervan KT, Amouyel P, Arveiler D, Bakker SJ, Beilby J, Bergman RN, Blangero J, Brown MJ, Burnier M, Campbell H, Chakravarti A, Chines PS, Claudi-Boehm S, Collins FS, Crawford DC, Danesh J, de Faire U, de Geus EJ, Dörr M, Erbel R, Eriksson JG, Farrall M, Ferrannini E, Ferrières J, Forouhi NG, Forrester T, Franco OH, Gansevoort RT, Gieger C, Gudnason V, Haiman CA, Harris TB, Hattersley AT, Heliövaara M, Hicks AA, Hingorani AD, Hoffmann W, Hofman A, Homuth G, Humphries SE, Hyppönen E, Illig T, Jarvelin MR, Johansen B, Jousilahti P, Jula AM, Kaprio J, Kee F, Keinanen-Kiukaanniemi SM, Kooner JS, Kooperberg C, Kovacs P, Kraja AT, Kumari M, Kuulasmaa K, Kuusisto J, Lakka TA, Langenberg C, Le Marchand L, Lehtimäki T, Lyssenko V, Männistö S, Marette A, Matise TC, McKenzie CA, McKnight B, Musk AW, Möhlenkamp S, Morris AD, Nelis M, Ohlsson C, Oldehinkel AJ, Ong KK, Palmer LJ, Penninx BW, Peters A, Pramstaller PP, Raitakari OT, Rankinen T, Rao DC, Rice TK, Ridker PM, Ritchie MD, Rudan I, Salomaa V, Samani NJ, Saramies J, Sarzynski MA, Schwarz PE, Shuldiner AR, Staessen JA, Steinthorsdottir V, Stolk RP, Strauch K, Tönjes A, Tremblay A, Tremoli E, Vohl MC, Völker U, Vollenweider P, Wilson JF, Witteman JC, Adair LS, Bochud M, Boehm BO, Bornstein SR, Bouchard C, Cauchi S, Caulfield MJ, Chambers JC, Chasman DI, Cooper RS, Dedoussis G, Ferrucci L, Froguel P, Grabe HJ, Hamsten A, Hui J, Hveem K, Jöckel KH, Kivimaki M, Kuh D, Laakso M, Liu Y, März W, Munroe PB, Njølstad I, Oostra BA, Palmer CN, Pedersen NL, Perola M, Pérusse L, Peters U, Power C, Quertermous T, Rauramaa R, Rivadeneira F, Saaristo TE, Saleheen D, Sinisalo J, Slagboom PE, Snieder H, Spector TD, Thorsteinsdottir U, Stumvoll M, Tuomilehto J, Uitterlinden AG, Uusitupa M, van der Harst P, Veronesi G, Walker M, Wareham NJ, Watkins H, Wichmann HE, Abecasis GR, Assimes TL, Berndt SI, Boehnke M, Borecki IB, Deloukas P, Franke L, Frayling TM, Groop LC, Hunter DJ, Kaplan RC, O’Connell JR, Qi L, Schlessinger D, Strachan DP, Stefansson K, van Duijn CM, Willer CJ, Visscher PM, Yang J, Hirschhorn JN, Zillikens MC, McCarthy MI, Speliotes EK, North KE, Fox CS, Barroso I, Franks PW, Ingelsson E, Heid IM, Loos RJ, Cupples LA, Morris AP, Lindgren CM, Mohlke KL; ADIPOGen Consortium; CARDIOGRAMplusC4D Consortium; CKDGen Consortium; GEFOS Consortium; GENIE Consortium; GLGC; ICBP; International Endogene Consortium; LifeLines Cohort Study; MAGIC Investigators; MuTHER Consortium; PAGE Consortium; ReproGen Consortium : New genetic loci link adipose and insulin biology to body fat distribution. Nature 518: 187–196, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, Powell C, Vedantam S, Buchkovich ML, Yang J, Croteau-Chonka DC, Esko T, Fall T, Ferreira T, Gustafsson S, Kutalik Z, Luan J, Mägi R, Randall JC, Winkler TW, Wood AR, Workalemahu T, Faul JD, Smith JA, Hua Zhao J, Zhao W, Chen J, Fehrmann R, Hedman AK, Karjalainen J, Schmidt EM, Absher D, Amin N, Anderson D, Beekman M, Bolton JL, Bragg-Gresham JL, Buyske S, Demirkan A, Deng G, Ehret GB, Feenstra B, Feitosa MF, Fischer K, Goel A, Gong J, Jackson AU, Kanoni S, Kleber ME, Kristiansson K, Lim U, Lotay V, Mangino M, Mateo Leach I, Medina-Gomez C, Medland SE, Nalls MA, Palmer CD, Pasko D, Pechlivanis S, Peters MJ, Prokopenko I, Shungin D, Stančáková A, Strawbridge RJ, Ju Sung Y, Tanaka T, Teumer A, Trompet S, van der Laan SW, van Setten J, Van Vliet-Ostaptchouk JV, Wang Z, Yengo L, Zhang W, Isaacs A, Albrecht E, Ärnlöv J, Arscott GM, Attwood AP, Bandinelli S, Barrett A, Bas IN, Bellis C, Bennett AJ, Berne C, Blagieva R, Blüher M, Böhringer S, Bonnycastle LL, Böttcher Y, Boyd HA, Bruinenberg M, Caspersen IH, Ida Chen YD, Clarke R, Daw EW, de Craen AJ, Delgado G, Dimitriou M, Doney AS, Eklund N, Estrada K, Eury E, Folkersen L, Fraser RM, Garcia ME, Geller F, Giedraitis V, Gigante B, Go AS, Golay A, Goodall AH, Gordon SD, Gorski M, Grabe HJ, Grallert H, Grammer TB, Gräßler J, Grönberg H, Groves CJ, Gusto G, Haessler J, Hall P, Haller T, Hallmans G, Hartman CA, Hassinen M, Hayward C, Heard-Costa NL, Helmer Q, Hengstenberg C, Holmen O, Hottenga JJ, James AL, Jeff JM, Johansson Å, Jolley J, Juliusdottir T, Kinnunen L, Koenig W, Koskenvuo M, Kratzer W, Laitinen J, Lamina C, Leander K, Lee NR, Lichtner P, Lind L, Lindström J, Sin Lo K, Lobbens S, Lorbeer R, Lu Y, Mach F, Magnusson PK, Mahajan A, McArdle WL, McLachlan S, Menni C, Merger S, Mihailov E, Milani L, Moayyeri A, Monda KL, Morken MA, Mulas A, Müller G, Müller-Nurasyid M, Musk AW, Nagaraja R, Nöthen MM, Nolte IM, Pilz S, Rayner NW, Renstrom F, Rettig R, Ried JS, Ripke S, Robertson NR, Rose LM, Sanna S, Scharnagl H, Scholtens S, Schumacher FR, Scott WR, Seufferlein T, Shi J, Vernon Smith A, Smolonska J, Stanton AV, Steinthorsdottir V, Stirrups K, Stringham HM, Sundström J, Swertz MA, Swift AJ, Syvänen AC, Tan ST, Tayo BO, Thorand B, Thorleifsson G, Tyrer JP, Uh HW, Vandenput L, Verhulst FC, Vermeulen SH, Verweij N, Vonk JM, Waite LL, Warren HR, Waterworth D, Weedon MN, Wilkens LR, Willenborg C, Wilsgaard T, Wojczynski MK, Wong A, Wright AF, Zhang Q, Brennan EP, Choi M, Dastani Z, Drong AW, Eriksson P, Franco-Cereceda A, Gådin JR, Gharavi AG, Goddard ME, Handsaker RE, Huang J, Karpe F, Kathiresan S, Keildson S, Kiryluk K, Kubo M, Lee JY, Liang L, Lifton RP, Ma B, McCarroll SA, McKnight AJ, Min JL, Moffatt MF, Montgomery GW, Murabito JM, Nicholson G, Nyholt DR, Okada Y, Perry JR, Dorajoo R, Reinmaa E, Salem RM, Sandholm N, Scott RA, Stolk L, Takahashi A, Tanaka T, Van’t Hooft FM, Vinkhuyzen AA, Westra HJ, Zheng W, Zondervan KT, Heath AC, Arveiler D, Bakker SJ, Beilby J, Bergman RN, Blangero J, Bovet P, Campbell H, Caulfield MJ, Cesana G, Chakravarti A, Chasman DI, Chines PS, Collins FS, Crawford DC, Cupples LA, Cusi D, Danesh J, de Faire U, den Ruijter HM, Dominiczak AF, Erbel R, Erdmann J, Eriksson JG, Farrall M, Felix SB, Ferrannini E, Ferrières J, Ford I, Forouhi NG, Forrester T, Franco OH, Gansevoort RT, Gejman PV, Gieger C, Gottesman O, Gudnason V, Gyllensten U, Hall AS, Harris TB, Hattersley AT, Hicks AA, Hindorff LA, Hingorani AD, Hofman A, Homuth G, Hovingh GK, Humphries SE, Hunt SC, Hyppönen E, Illig T, Jacobs KB, Jarvelin MR, Jöckel KH, Johansen B, Jousilahti P, Jukema JW, Jula AM, Kaprio J, Kastelein JJ, Keinanen-Kiukaanniemi SM, Kiemeney LA, Knekt P, Kooner JS, Kooperberg C, Kovacs P, Kraja AT, Kumari M, Kuusisto J, Lakka TA, Langenberg C, Le Marchand L, Lehtimäki T, Lyssenko V, Männistö S, Marette A, Matise TC, McKenzie CA, McKnight B, Moll FL, Morris AD, Morris AP, Murray JC, Nelis M, Ohlsson C, Oldehinkel AJ, Ong KK, Madden PA, Pasterkamp G, Peden JF, Peters A, Postma DS, Pramstaller PP, Price JF, Qi L, Raitakari OT, Rankinen T, Rao DC, Rice TK, Ridker PM, Rioux JD, Ritchie MD, Rudan I, Salomaa V, Samani NJ, Saramies J, Sarzynski MA, Schunkert H, Schwarz PE, Sever P, Shuldiner AR, Sinisalo J, Stolk RP, Strauch K, Tönjes A, Trégouët DA, Tremblay A, Tremoli E, Virtamo J, Vohl MC, Völker U, Waeber G, Willemsen G, Witteman JC, Zillikens MC, Adair LS, Amouyel P, Asselbergs FW, Assimes TL, Bochud M, Boehm BO, Boerwinkle E, Bornstein SR, Bottinger EP, Bouchard C, Cauchi S, Chambers JC, Chanock SJ, Cooper RS, de Bakker PI, Dedoussis G, Ferrucci L, Franks PW, Froguel P, Groop LC, Haiman CA, Hamsten A, Hui J, Hunter DJ, Hveem K, Kaplan RC, Kivimaki M, Kuh D, Laakso M, Liu Y, Martin NG, März W, Melbye M, Metspalu A, Moebus S, Munroe PB, Njølstad I, Oostra BA, Palmer CN, Pedersen NL, Perola M, Pérusse L, Peters U, Power C, Quertermous T, Rauramaa R, Rivadeneira F, Saaristo TE, Saleheen D, Sattar N, Schadt EE, Schlessinger D, Slagboom PE, Snieder H, Spector TD, Thorsteinsdottir U, Stumvoll M, Tuomilehto J, Uitterlinden AG, Uusitupa M, van der Harst P, Walker M, Wallaschofski H, Wareham NJ, Watkins H, Weir DR, Wichmann HE, Wilson JF, Zanen P, Borecki IB, Deloukas P, Fox CS, Heid IM, O’Connell JR, Strachan DP, Stefansson K, van Duijn CM, Abecasis GR, Franke L, Frayling TM, McCarthy MI, Visscher PM, Scherag A, Willer CJ, Boehnke M, Mohlke KL, Lindgren CM, Beckmann JS, Barroso I, North KE, Ingelsson E, Hirschhorn JN, Loos RJ, Speliotes EK; LifeLines Cohort Study; ADIPOGen Consortium; AGEN-BMI Working Group; CARDIOGRAMplusC4D Consortium; CKDGen Consortium; GLGC; ICBP; MAGIC Investigators; MuTHER Consortium; MIGen Consortium; PAGE Consortium; ReproGen Consortium; GENIE Consortium; International Endogene Consortium : Genetic studies of body mass index yield new insights for obesity biology. Nature 518: 197–206, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.International Consortium for Blood Pressure Genome-Wide Association Studies, Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, Smith AV, Tobin MD, Verwoert GC, Hwang SJ, Pihur V, Vollenweider P, O'Reilly PF, Amin N, Bragg-Gresham JL, Teumer A, Glazer NL, Launer L, Zhao JH, Aulchenko Y, Heath S, Sober S, Parsa A, Luan J, Arora P, Dehghan A, Zhang F, Lucas G, Hicks AA, Jackson AU, Peden JF, Tanaka T, Wild SH, Rudan I, Igl W, Milaneschi Y, Parker AN, Fava C, Chambers JC, Fox ER, Kumari M, Go MJ, van der Harst P, Kao WH, Sjogren M, Vinay DG, Alexander M, Tabara Y, Shaw-Hawkins S, Whincup PH, Liu Y, Shi G, Kuusisto J, Tayo B, Seielstad M, Sim X, Nguyen KD, Lehtimaki T, Matullo G, Wu Y, Gaunt TR, Onland-Moret NC, Cooper MN, Platou CG, Org E, Hardy R, Dahgam S, Palmen J, Vitart V, Braund PS, Kuznetsova T, Uiterwaal CS, Adeyemo A, Palmas W, Campbell H, Ludwig B, Tomaszewski M, Tzoulaki I, Palmer ND, CARDIoGRAM consortium, CKDGen Consortium, KidneyGen Consortium, EchoGen consortium, CHARGE-HF consortium, Aspelund T, Garcia M, Chang YP, O'Connell JR, Steinle NI, Grobbee DE, Arking DE, Kardia SL, Morrison AC, Hernandez D, Najjar S, McArdle WL, Hadley D, Brown MJ, Connell JM, Hingorani AD, Day IN, Lawlor DA, Beilby JP, Lawrence RW, Clarke R, Hopewell JC, Ongen H, Dreisbach AW, Li Y, Young JH, Bis JC, Kahonen M, Viikari J, Adair LS, Lee NR, Chen MH, Olden M, Pattaro C, Bolton JA, Kottgen A, Bergmann S, Mooser V, Chaturvedi N, Frayling TM, Islam M, Jafar TH, Erdmann J, Kulkarni SR, Bornstein SR, Grassler J, Groop L, Voight BF, Kettunen J, Howard P, Taylor A, Guarrera S, Ricceri F, Emilsson V, Plump A, Barroso I, Khaw KT, Weder AB, Hunt SC, Sun YV, Bergman RN, Collins FS, Bonnycastle LL, Scott LJ, Stringham HM, Peltonen L, Perola M, Vartiainen E, Brand SM, Staessen JA, Wang TJ, Burton PR, Soler Artigas M, Dong Y, Snieder H, Wang X, Zhu H, Lohman KK, Rudock ME, Heckbert SR, Smith NL, Wiggins KL, Doumatey A, Shriner D, Veldre G, Viigimaa M, Kinra S, Prabhakaran D, Tripathy V, Langefeld CD, Rosengren A, Thelle DS, Corsi AM, Singleton A, Forrester T, Hilton G, McKenzie CA, Salako T, Iwai N, Kita Y, Ogihara T, Ohkubo T, Okamura T, Ueshima H, Umemura S, Eyheramendy S, Meitinger T, Wichmann HE, Cho YS, Kim HL, Lee JY, Scott J, Sehmi JS, Zhang W, Hedblad B, Nilsson P, Smith GD, Wong A, Narisu N, Stancakova A, Raffel LJ, Yao J, Kathiresan S, O'Donnell CJ, Schwartz SM, Ikram MA, Longstreth WT,Jr, Mosley TH, Seshadri S, Shrine NR, Wain LV, Morken MA, Swift AJ, Laitinen J, Prokopenko I, Zitting P, Cooper JA, Humphries SE, Danesh J, Rasheed A, Goel A, Hamsten A, Watkins H, Bakker SJ, van Gilst WH, Janipalli CS, Mani KR, Yajnik CS, Hofman A, Mattace-Raso FU, Oostra BA, Demirkan A, Isaacs A, Rivadeneira F, Lakatta EG, Orru M, Scuteri A, Ala-Korpela M, Kangas AJ, Lyytikainen LP, Soininen P, Tukiainen T, Wurtz P, Ong RT, Dorr M, Kroemer HK, Volker U, Volzke H, Galan P, Hercberg S, Lathrop M, Zelenika D, Deloukas P, Mangino M, Spector TD, Zhai G, Meschia JF, Nalls MA, Sharma P, Terzic J, Kumar MV, Denniff M, Zukowska-Szczechowska E, Wagenknecht LE, Fowkes FG, Charchar FJ, Schwarz PE, Hayward C, Guo X, Rotimi C, Bots ML, Brand E, Samani NJ, Polasek O, Talmud PJ, Nyberg F, Kuh D, Laan M, Hveem K, Palmer LJ, van der Schouw YT, Casas JP, Mohlke KL, Vineis P, Raitakari O, Ganesh SK, Wong TY, Tai ES, Cooper RS, Laakso M, Rao DC, Harris TB, Morris RW, Dominiczak AF, Kivimaki M, Marmot MG, Miki T, Saleheen D, Chandak GR, Coresh J, Navis G, Salomaa V, Han BG, Zhu X, Kooner JS, Melander O, Ridker PM, Bandinelli S, Gyllensten UB, Wright AF, Wilson JF, Ferrucci L, Farrall M, Tuomilehto J, Pramstaller PP, Elosua R, Soranzo N, Sijbrands EJ, Altshuler D, Loos RJ, Shuldiner AR, Gieger C, Meneton P, Uitterlinden AG, Wareham NJ, Gudnason V, Rotter JI, Rettig R, Uda M, Strachan DP, Witteman JC, Hartikainen AL, Beckmann JS, Boerwinkle E, Vasan RS, Boehnke M, Larson MG, Jarvelin MR, Psaty BM, Abecasis GR, Chakravarti A, Elliott P, van Duijn CM, Newton-Cheh C, Levy D, Caulfield MJ, Johnson T: Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature 478: 103–109, 2011 [DOI] [PMC free article] [PubMed]
- 50.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, Strait J, Duren WL, Maschio A, Busonero F, Mulas A, Albai G, Swift AJ, Morken MA, Narisu N, Bennett D, Parish S, Shen H, Galan P, Meneton P, Hercberg S, Zelenika D, Chen WM, Li Y, Scott LJ, Scheet PA, Sundvall J, Watanabe RM, Nagaraja R, Ebrahim S, Lawlor DA, Ben-Shlomo Y, Davey-Smith G, Shuldiner AR, Collins R, Bergman RN, Uda M, Tuomilehto J, Cao A, Collins FS, Lakatta E, Lathrop GM, Boehnke M, Schlessinger D, Mohlke KL, Abecasis GR: Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet 40: 161–169, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klein R, Klein BE, Moss SE: Is obesity related to microvascular and macrovascular complications in diabetes? The Wisconsin Epidemiologic Study of Diabetic Retinopathy. Arch Intern Med 157: 650–656, 1997 [PubMed] [Google Scholar]
- 52.Hill CJ, Cardwell CR, Maxwell AP, Young RJ, Matthews B, O’Donoghue DJ, Fogarty DG: Obesity and kidney disease in type 1 and 2 diabetes: an analysis of the National Diabetes Audit. QJM 106: 933–942, 2013 [DOI] [PubMed] [Google Scholar]
- 53.Todd JN, Dahlström EH, Salem RM, Sandholm N, Forsblom C, McKnight AJ, Maxwell AP, Brennan E, Sadlier D, Godson C, Groop PH, Hirschhorn JN, Florez JC; FinnDiane Study Group : Genetic Evidence for a Causal Role of Obesity in Diabetic Kidney Disease. Diabetes 64: 4238–4246, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Patterson N, Daly MJ, Price AL, Neale BM; Schizophrenia Working Group of the Psychiatric Genomics Consortium : LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 47: 291–295, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR: ReproGen Consortium, Psychiatric Genomics Consortium, Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium 3, Duncan L, Perry JR, Patterson N, Robinson EB, Daly MJ, Price AL, Neale BM: An atlas of genetic correlations across human diseases and traits. Nat Genet 47: 1236–1241, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ng LL, Ngkeekwong FC, Quinn PA, Davies JE: Uptake mechanisms for ascorbate and dehydroascorbate in lymphoblasts from diabetic nephropathy and hypertensive patients. Diabetologia 41: 435–442, 1998 [DOI] [PubMed] [Google Scholar]
- 57.Farvid MS, Jalali M, Siassi F, Hosseini M: Comparison of the effects of vitamins and/or mineral supplementation on glomerular and tubular dysfunction in type 2 diabetes. Diabetes Care 28: 2458–2464, 2005 [DOI] [PubMed] [Google Scholar]
- 58.Deckert T, Feldt-Rasmussen B, Borch-Johnsen K, Jensen T, Kofoed-Enevoldsen A: Albuminuria reflects widespread vascular damage. The Steno hypothesis. Diabetologia 32: 219–226, 1989 [DOI] [PubMed] [Google Scholar]
- 59.Gil N, Goldberg R, Neuman T, Garsen M, Zcharia E, Rubinstein AM, van Kuppevelt T, Meirovitz A, Pisano C, Li JP, van der Vlag J, Vlodavsky I, Elkin M: Heparanase is essential for the development of diabetic nephropathy in mice. Diabetes 61: 208–216, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shepherd J, Kastelein JP, Bittner VA, Carmena R, Deedwania PC, Breazna A, Dobson S, Wilson DJ, Zuckerman AL, Wenger NK; Treating to New Targets Steering Committee and Investigators : Intensive lipid lowering with atorvastatin in patients with coronary artery disease, diabetes, and chronic kidney disease. Mayo Clin Proc 83: 870–879, 2008 [PubMed] [Google Scholar]
- 61.Sandhu S, Wiebe N, Fried LF, Tonelli M: Statins for improving renal outcomes: a meta-analysis. J Am Soc Nephrol 17: 2006–2016, 2006 [DOI] [PubMed] [Google Scholar]
- 62.Veikkolainen V, Naillat F, Railo A, Chi L, Manninen A, Hohenstein P, Hastie N, Vainio S, Elenius K: ErbB4 modulates tubular cell polarity and lumen diameter during kidney development. J Am Soc Nephrol 23: 112–122, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Price AL, Kryukov GV, de Bakker PI, Purcell SM, Staples J, Wei LJ, Sunyaev SR: Pooled association tests for rare variants in exon-resequencing studies. Am J Hum Genet 86: 832–838, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X: Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet 89: 82–93, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee S, Emond MJ, Bamshad MJ, Barnes KC, Rieder MJ, Nickerson DA, Christiani DC, Wurfel MM, Lin X; NHLBI GO Exome Sequencing Project—ESP Lung Project Team : Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am J Hum Genet 91: 224–237, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lek M, Karczewski K, Minikel E, Samocha K, Banks E, Fennell T, O’Donnell-Luria A, Ware J, Hill A, Cummings B, Tukiainen T, Birnbaum D, Kosmicki J, Duncan L, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Cooper D, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki M, Levy Moonshine A, Natarajan P, Orozco L, Peloso G, Poplin R, Rivas M, Ruano-Rubio V, Ruderfer D, Shakir K, Stenson P, Stevens C, Thomas B, Tiao G, Tusie-Luna M, Weisburd B, Won H, Yu D, Altshuler D, Ardissino D, Boehnke M, Danesh J, Roberto E, Florez J, Gabriel S, Getz G, Hultman C, Kathiresan S, Laakso M, McCarroll S, McCarthy M, McGovern D, McPherson R, Neale B, Palotie A, Purcell S, Saleheen D, Scharf J, Sklar P, Patrick S, Tuomilehto J, Watkins H, Wilson J, Daly M, MacArthur D: Analysis of protein-coding genetic variation in 60,706 humans. bioRxiv , DOI: 10.1101/030338, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Di Camillo B, Sambo F, Toffolo G, Cobelli C: ABACUS: an entropy-based cumulative bivariate statistic robust to rare variants and different direction of genotype effect. Bioinformatics 30: 384–391, 2014 [DOI] [PubMed] [Google Scholar]
- 68.Wessman M, Forsblom C, Kaunisto MA, Söderlund J, Ilonen J, Sallinen R, Hiekkalinna T, Parkkonen M, Maxwell AP, Tarnow L, Parving HH, Hadjadj S, Marre M, Peltonen L, Groop PH; FinnDiane Study Group : Novel susceptibility locus at 22q11 for diabetic nephropathy in type 1 diabetes. PLoS One 6: e24053, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D: Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38: 904–909, 2006 [DOI] [PubMed] [Google Scholar]
- 70.Howie BN, Donnelly P, Marchini J: A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 5: e1000529, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA; 1000 Genomes Project Consortium : An integrated map of genetic variation from 1,092 human genomes. Nature 491: 56–65, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cingolani P, Platts A, Wang L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM: A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6: 80–92, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F: Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics 26: 2069–2070, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ruark E, Münz M, Renwick A, Clarke M, Ramsay E, Hanks S, Mahamdallie S, Elliott A, Seal S, Strydom A, Gerton L, Rahman N: The ICR1000 UK exome series: A resource of gene variation in an outbred population. F1000Research 4: 883, 2015 [DOI] [PMC free article] [PubMed]
- 75.Lim ET, Würtz P, Havulinna AS, Palta P, Tukiainen T, Rehnström K, Esko T, Mägi R, Inouye M, Lappalainen T, Chan Y, Salem RM, Lek M, Flannick J, Sim X, Manning A, Ladenvall C, Bumpstead S, Hämäläinen E, Aalto K, Maksimow M, Salmi M, Blankenberg S, Ardissino D, Shah S, Horne B, McPherson R, Hovingh GK, Reilly MP, Watkins H, Goel A, Farrall M, Girelli D, Reiner AP, Stitziel NO, Kathiresan S, Gabriel S, Barrett JC, Lehtimäki T, Laakso M, Groop L, Kaprio J, Perola M, McCarthy MI, Boehnke M, Altshuler DM, Lindgren CM, Hirschhorn JN, Metspalu A, Freimer NB, Zeller T, Jalkanen S, Koskinen S, Raitakari O, Durbin R, MacArthur DG, Salomaa V, Ripatti S, Daly MJ, Palotie A; Sequencing Initiative Suomi (SISu) Project : Distribution and medical impact of loss-of-function variants in the Finnish founder population. PLoS Genet 10: e1004494, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang J, Lee SH, Goddard ME, Visscher PM: GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 88: 76–82, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kang HM, Sul JH, Service SK, Zaitlen NA, Kong SY, Freimer NB, Sabatti C, Eskin E: Variance component model to account for sample structure in genome-wide association studies. Nat Genet 42: 348–354, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Magi R, & Morris AP: GWAMA: Software for genome-wide association meta-analysis. BMC Bioinformatics 11: 288, 2010 [DOI] [PMC free article] [PubMed]
- 79.Willer CJ, Li Y, Abecasis GR: METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26: 2190–2191, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Purcell S, Cherny SS, Sham PC: Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics 19: 149–150, 2003 [DOI] [PubMed] [Google Scholar]
- 81.Skol AD, Scott LJ, Abecasis GR, Boehnke M: Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet 38: 209–213, 2006 [DOI] [PubMed] [Google Scholar]
- 82.de Boer IH, Sun W, Cleary PA, Lachin JM, Molitch ME, Steffes MW, Zinman B; DCCT/EDIC Research Group : Intensive diabetes therapy and glomerular filtration rate in type 1 diabetes. N Engl J Med 365: 2366–2376, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.DCCT/EDIC research group : Effect of intensive diabetes treatment on albuminuria in type 1 diabetes: long-term follow-up of the Diabetes Control and Complications Trial and Epidemiology of Diabetes Interventions and Complications study. Lancet Diabetes Endocrinol 2: 793–800, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Germain M, Pezzolesi MG, Sandholm N, McKnight AJ, Susztak K, Lajer M, Forsblom C, Marre M, Parving HH, Rossing P, Toppila I, Skupien J, Roussel R, Ko YA, Ledo N, Folkersen L, Civelek M, Maxwell AP, Tregouet DA, Groop PH, Tarnow L, Hadjadj S: SORBS1 gene, a new candidate for diabetic nephropathy: results from a multi-stage genome-wide association study in patients with type 1 diabetes. Diabetologia 58: 543–548, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Scott RA, Lagou V, Welch RP, Wheeler E, Montasser ME, Luan J, Mägi R, Strawbridge RJ, Rehnberg E, Gustafsson S, Kanoni S, Rasmussen-Torvik LJ, Yengo L, Lecoeur C, Shungin D, Sanna S, Sidore C, Johnson PC, Jukema JW, Johnson T, Mahajan A, Verweij N, Thorleifsson G, Hottenga JJ, Shah S, Smith AV, Sennblad B, Gieger C, Salo P, Perola M, Timpson NJ, Evans DM, Pourcain BS, Wu Y, Andrews JS, Hui J, Bielak LF, Zhao W, Horikoshi M, Navarro P, Isaacs A, O’Connell JR, Stirrups K, Vitart V, Hayward C, Esko T, Mihailov E, Fraser RM, Fall T, Voight BF, Raychaudhuri S, Chen H, Lindgren CM, Morris AP, Rayner NW, Robertson N, Rybin D, Liu CT, Beckmann JS, Willems SM, Chines PS, Jackson AU, Kang HM, Stringham HM, Song K, Tanaka T, Peden JF, Goel A, Hicks AA, An P, Müller-Nurasyid M, Franco-Cereceda A, Folkersen L, Marullo L, Jansen H, Oldehinkel AJ, Bruinenberg M, Pankow JS, North KE, Forouhi NG, Loos RJ, Edkins S, Varga TV, Hallmans G, Oksa H, Antonella M, Nagaraja R, Trompet S, Ford I, Bakker SJ, Kong A, Kumari M, Gigante B, Herder C, Munroe PB, Caulfield M, Antti J, Mangino M, Small K, Miljkovic I, Liu Y, Atalay M, Kiess W, James AL, Rivadeneira F, Uitterlinden AG, Palmer CN, Doney AS, Willemsen G, Smit JH, Campbell S, Polasek O, Bonnycastle LL, Hercberg S, Dimitriou M, Bolton JL, Fowkes GR, Kovacs P, Lindström J, Zemunik T, Bandinelli S, Wild SH, Basart HV, Rathmann W, Grallert H, Maerz W, Kleber ME, Boehm BO, Peters A, Pramstaller PP, Province MA, Borecki IB, Hastie ND, Rudan I, Campbell H, Watkins H, Farrall M, Stumvoll M, Ferrucci L, Waterworth DM, Bergman RN, Collins FS, Tuomilehto J, Watanabe RM, de Geus EJ, Penninx BW, Hofman A, Oostra BA, Psaty BM, Vollenweider P, Wilson JF, Wright AF, Hovingh GK, Metspalu A, Uusitupa M, Magnusson PK, Kyvik KO, Kaprio J, Price JF, Dedoussis GV, Deloukas P, Meneton P, Lind L, Boehnke M, Shuldiner AR, van Duijn CM, Morris AD, Toenjes A, Peyser PA, Beilby JP, Körner A, Kuusisto J, Laakso M, Bornstein SR, Schwarz PE, Lakka TA, Rauramaa R, Adair LS, Smith GD, Spector TD, Illig T, de Faire U, Hamsten A, Gudnason V, Kivimaki M, Hingorani A, Keinanen-Kiukaanniemi SM, Saaristo TE, Boomsma DI, Stefansson K, van der Harst P, Dupuis J, Pedersen NL, Sattar N, Harris TB, Cucca F, Ripatti S, Salomaa V, Mohlke KL, Balkau B, Froguel P, Pouta A, Jarvelin MR, Wareham NJ, Bouatia-Naji N, McCarthy MI, Franks PW, Meigs JB, Teslovich TM, Florez JC, Langenberg C, Ingelsson E, Prokopenko I, Barroso I; DIAbetes Genetics Replication and Meta-analysis (DIAGRAM) Consortium : Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat Genet 44: 991–1005, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tobacco and Genetics Consortium : Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet 42: 441–447, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Segrè AV, Groop L, Mootha VK, Daly MJ, Altshuler D; DIAGRAM Consortium; MAGIC investigators : Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet 6: e1001058, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lin DY, Tang ZZ: A general framework for detecting disease associations with rare variants in sequencing studies. Am J Hum Genet 89: 354–367, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Moutsianas L, Agarwala V, Fuchsberger C, Flannick J, Rivas MA, Gaulton KJ, Albers PK, McVean G, Boehnke M, Altshuler D, McCarthy MI; GoT2D Consortium : The power of gene-based rare variant methods to detect disease-associated variation and test hypotheses about complex disease. PLoS Genet 11: e1005165, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP: Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102: 15545–15550, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huang W, Sherman BT, Lempicki RA: Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1–13, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huang W, Sherman BT, Lempicki RA: Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57, 2009 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.