Abstract
The total number of glomeruli is a fundamental parameter of kidney function but very difficult to determine using standard methodology. Here, we counted all individual glomeruli in murine kidneys and sized the capillary tufts by combining in vivo fluorescence labeling of endothelial cells, a novel tissue–clearing technique, lightsheet microscopy, and automated registration by image analysis. Total hands–on time per organ was <1 hour, and automated counting/sizing was finished in <3 hours. We also investigated the novel use of ethyl-3-phenylprop-2-enoate (ethyl cinnamate) as a nontoxic solvent–based clearing reagent that can be handled without specific safety measures. Ethyl cinnamate rapidly cleared all tested organs, including calcified bone, but the fluorescence of proteins and immunohistochemical labels was maintained over weeks. Using ethyl cinnamate–cleared kidneys, we also quantified the average creatinine clearance rate per glomerulus. This parameter decreased in the first week of experimental nephrotoxic nephritis, whereas reduction in glomerular numbers occurred much later. Our approach delivers fundamental parameters of renal function, and because of its ease of use and speed, it is suitable for high-throughput analysis and could greatly facilitate studies of the effect of kidney diseases on whole-organ physiology.
Keywords: glomerular endothelial cells, glomerular filtration rate, glomerulonephritis, immune complexes, Immunology and pathology, kidney anatomy
A highly organized vascular system is essential for kidney function, and impaired circulatory performance in nephrons results in organ failure. Every nephron contains a tuft of capillaries within the glomerulus that performs blood ultrafiltration.1 During chronic renal diseases, the numbers and function of nephrons are considered to decline. Until now, enumerating nephrons/glomeruli and validating the integrity of their tuft structures were only possible either by laborious tissue embedding/sectioning and manual stereologic counting2,3 or after kidney maceration.4 Therefore, many studies extrapolate glomerular numbers on sample counts, which is prone to errors. Furthermore, precise morphologic information relating to the structure and position of individual glomeruli within the whole organ is lost. The latter is amenable by high–field magnetic resonance imaging, which however, requires advanced laboratory equipment and suffers from relatively poor resolution that precludes distinction between Bowman’s capsule and tuft.5,6
High–resolution optical three–dimensional reconstructions of kidneys on the basis of physical sectioning and serial imaging can, in principle, distinguish between glomerular walls and tufts. This procedure, however, is highly labor intensive, slow, and prone to errors, and it requires extensive experience for satisfactory results.7 Lightsheet fluorescence microscopy (LSFM) has revolutionized the field by enabling the reconstruction of intact organs or whole animals with cellular resolution in three dimensions within minutes.8,9 This has primarily led to many studies focusing on the three-dimensional architecture of rodent brains,10–12 whereas a thorough quantification of glomeruli from LSFM data has so far not been performed, despite promising initial observations.13
Critical for the successful acquisition of lightsheet images and subsequent reconstruction are effective techniques for tissue clearing that can render opaque organs highly transparent.7 Although clearing can be performed using water-based agents, these methods generally result in samples that are less transparent and highly fragile7 compared with clearing procedures on the basis of the use of organic solvents.
Solvent-based clearing requires the dehydration of samples with, for example, tetrahydrofuran (THF) or tert-butanol before treatment with organic solvents that match the refractive index (RI) of dehydrated proteins. Although such techniques can result in highly transparent samples that are structurally robust, a number of significant drawbacks exist, which can include the dramatic loss of signal from fluorescent proteins (FPs) and the relatively long length of time required for full clearing. Perhaps more importantly, all clearing protocols in use until now rely on severely toxic substances. For example, clearing reagents, like benzylalcohol/benzylbenzoate (BABB), dibenzylether (DBE), dichlormethane, or methyl salicylate, are potent carcinogens.13 In addition, THF can form explosive peroxides.14 This requires comprehensive measures to protect researchers from exposure to aerosols or uncontrolled reactions,13,14 which has impaired the more widespread use of LSFM.
We have developed a new protocol for solvent-based clearing that overcomes these limitations. Because the stability of FPs is very pH sensitive, we made use of pH–adjusted ethanol series for sample dehydration,15 and for RI matching, we searched for nontoxic compounds with an RI>1.5. We discovered that ethyl-3-phenylprop-2-enoate (ethyl cinnamate [ECi]) (Supplemental Table 1) fulfills these criteria. ECi is a Food and Drug Administration–approved food flavor and additive for cosmetic products since 2007.16 It is also considered harmless according to the European directive 67/548/EWG (material safety data sheet; Sigma-Aldrich, St. Louis, MO). We discovered that ECi is also an excellent clearing reagent for mammalian tissues.
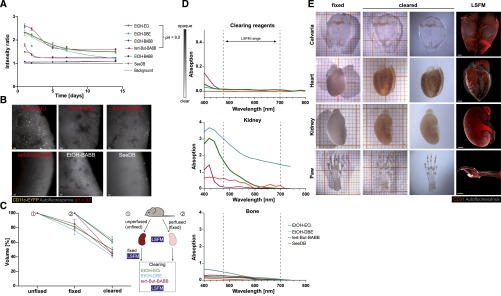
Maintaining the signal from FPs expressed in cells is of key importance for studying the cellular composition of cleared organs. However, the general current practice of organic clearing protocols using THF-DBE/dichlormethane or propanol/BABB,17 in addition to being toxic,7,17 results in FP fluorescence that lasts only days.7 We, therefore, tested and compared the stability of EYFP fluorescence in kidneys from CD11c-EYFP mice18 cleared using established protocols14,17,19,20 or ethanol-ECi. In general, we found a slow decay of the signal-to-background ratio of EYFP over time (Figure 1, A and B). However, fluorescence was still detectable in individual cells within organs after 14 days of constant exposure to the clearing reagent. Ethanol-ECi preservation of fluorescence was equivalent to the best available alternative protocol14 (Figure 1A), maintaining FP fluorescence for prolonged periods. Similar results were obtained for other FPs (Supplemental Figure 1), and the penetration depth in solvent-cleared kidneys was superior to that of water-based clearing (Figure 1B). Next, we measured kidney shrinkage induced by fixation and clearing. Here, ethanol-ECi was equivalent to the best alternative method (Figure 1C). Importantly, a 50% volume reduction only reflects approximately 20% changes in organ diameters, and completely avoiding shrinkage after fixation is not possible.21,22 We then estimated the clearing efficiency by measuring the absorption of bones and kidneys over the visible light spectrum. Here, ethanol-ECi was again equivalent to the best alternative method (Figure 1D).
Figure 1.
Optical clearing via Ethanol-ECi allows clearing of soft and hard tissues with prolonged fluorescence maintenance. (A) Long–term EYFP fluorescence over background in kidneys of CD11c-EYFP mice after clearing with different protocols. pH values of dehydration reagents were adjusted where indicated. Data are means±SEM of 100 measured cell-to-background ratios from one kidney per condition measured repetitively at all time points. (B) Representative images from kidneys at day 14 after clearing with the indicated protocols. Red text indicates pH adjustment of dehydration reagent. Scale bars, 50 μm. (C) Volume shrinkage of kidneys induced by perfusion/fixation and different clearing protocols measured from LSFM data using automated image analysis. The schematic of the experiment is shown on the right. Data are means±SEM of three kidneys each analyzed repetitively per condition. From each animal, one kidney was unperfused, and one was perfused. In total, nine animals (18 kidneys) were analyzed. (D) Transparency of clearing reagents, kidney, or bone after clearing with the indicated protocols measured spectrophotometrically. The wavelength range used for excitation/emission in LSFM is indicated with dashed lines. Data are means from five technical replicates of one sample measured per condition. SEMs are not shown because of extremely small variations of data. (E) Optical clearing via ECi is working for soft tissues (kidney and heart) and hard tissues (calvarial or long bones). In addition to EYFP preservation, fluorescence labeling by antibodies resists the clearing procedure as shown via endothelial specific staining (CD31; red). Paw =100 μm. Grid size (in left two columns) is 1 mm (small squares). Data are representative of 10 organs imaged similarly. EtOH, ethanol. Scale bars, 1000 μm.
In addition to endogenous fluorescence from FPs, the maintenance of chemical fluorescent tracers, such as fluorescence-labeled antibodies, is also of central importance, especially in relation to imaging of the blood vessel system in kidneys. We, therefore, performed ubiquitous endothelial labeling in whole animals by intravenous injection of anti-CD31 antibodies23 labeled with Alexa Fluor-647. Endothelial cells confined to blood vessels could then be imaged in the entire animal, enabling the three–dimensional blood vessel architecture to be visualized in different soft organs (Figure 1E), including brain, liver, and lung (not shown).
Our method allows clearing hard bones (Figure 1E, Supplemental Figures 2–4, Supplemental Movie 1), which may facilitate the analysis of renal osteodystrophy24 in future studies. In contrast to a recently published study,25 ECi clearing also preserved FP fluorescence in bone marrow, thus allowing direct imaging of individual cells in their native environment without the need for secondary antibody labeling (Supplemental Figure 1). Image rendering of CD31-labeled structures allowed the visualization of three–dimensional vessel trees in entire organs (Supplemental Figures 2–4, Supplemental Movies 1 and 2). Furthermore, the imaging of nonspecific autofluorescence was useful for visualizing the general tissue morphology of organs. In the heart, we were able to see the intracardial papillary muscles and the chordae tendinae in great anatomic detail using three–dimensional volume rendering of autofluorescence structures (Supplemental Figure 3, Supplemental Movie 2). Significantly, cardiac complications are closely associated with chronic renal failure.26 Consequently, LSFM has the potential to greatly improve the analysis of heart defects in chronic renal failure models.
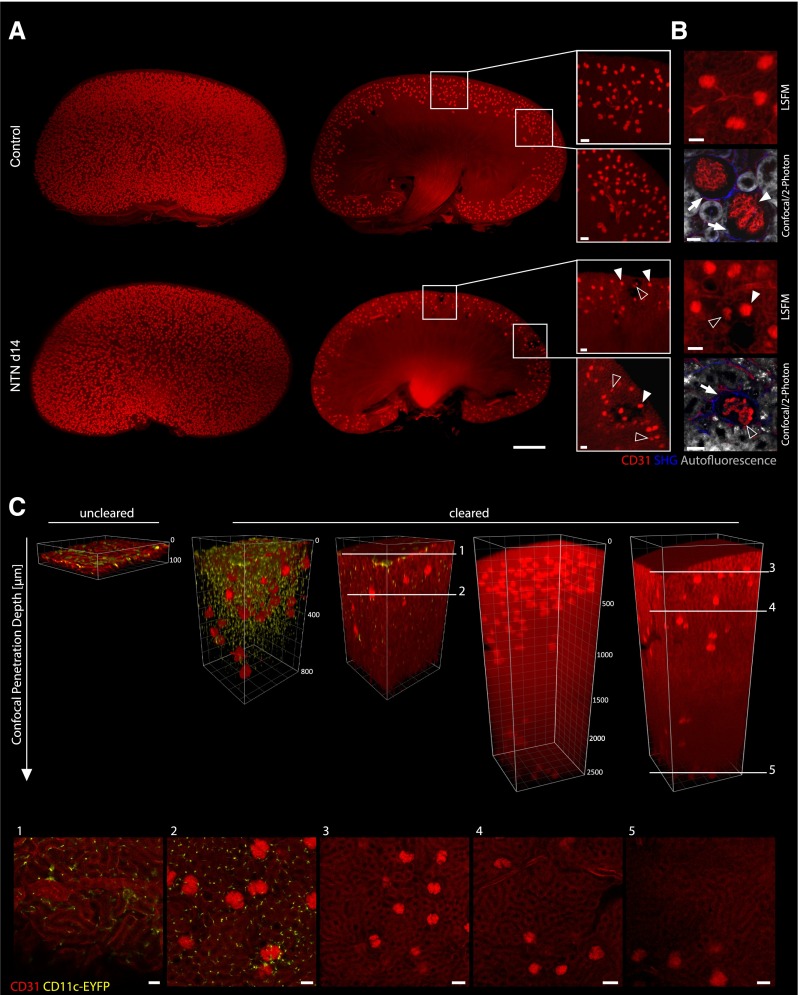
Having established and evaluated the ethanol-ECi clearing protocol, we further performed an in-depth analysis of the vascular system of murine kidneys. Ethanol-ECi clearing yielded transparent whole organs (Figures 1E and 2A) that could be completely scanned by LSFM. Image stacks revealed the endothelial network within all glomeruli as bright fluorescent areas in one three-dimensional stack (Figure 2A, Supplemental Movies 3 and 4). Thereby, antibody-marked capillaries enabled the selective visualization of glomerular tufts (Figure 2B). Because we used CD11c-EYFP animals, we could also simultaneously image and count dendritic cells within the kidneys in high resolution by confocal microscopy (Figure 2C, Supplemental Figure 5). Here, the use of cleared samples for confocal microscopy enabled extremely deep imaging, which was limited only by the working distance of the objective (Figure 2C).
Figure 2.
Visualization of 3D vessel trees and dendritic cells in entire ECi-cleared kidneys via LSFM and confocal/two-photon imaging. LSFM of specifically stained endothelial structures (CD31; red) allows three-dimensional reconstruction of whole kidneys. (A) Three-dimensional reconstructions of kidneys from a healthy control or an organ suffering from NTN (day 14). Note that the NTN kidney shows lower glomerular density, whereas two–dimensional optical sections reveal CD31-negative areas of corrupted glomeruli and the surrounding vasculature compared with a homogeneous field of equally sized glomeruli in healthy controls (white boxes). Furthermore, a higher heterogeneity of glomerular tuft sizes was observable in NTN kidneys at day 14. Many shrunken glomerular tufts (open arrowheads) were visible next to normally sized elements (white arrowheads). Scale bars, 50 μm. (B) Enhanced views of kidney structure (autofluorescence [gray] and fibrous tissue [second harmonic generation (SHG); blue]) and glomerular tuft (CD31; red) via combined confocal and two–photon laser scanning microscopy (LSM) compared with enhanced LSFM magnification. Compared with controls, NTN kidneys showed decreased tuft size (open arrowheads; as opposed to normal tuft size [white arrowheads]), decrease of endothelial CD31 label in their capillaries, and increased tissue fibrosis (indicated by higher SHG signal; white arrows) around damaged glomeruli. Scale bars, 20 μm in confocal/two-photon microscopy; 50 μm in LSFM. (C) Penetration depth of confocal LSM into ECi-cleared kidneys from CD11c-EYFP animals is enhanced compared with in uncleared samples. The maximum penetration depth of short wavelengths was increased from approximately 50 to 800 μm as shown via detection of CD11c-eYFP+ cells. The detected CD11c-eYFP+ cells still show the characteristic dendritic morphology after ECi clearing in the entire kidney tissue as emphasized at two exemplary focal planes of the shown Z stack (1 and 2). Longer wavelengths of the used endothelial marker CD31-Alexa Fluor-647 (red) can be detected even deeper in the tissue. The objective of the used confocal microscope allows a maximum penetration depth of 2500 μm because of the working distance. This penetration depth was reached in cleared kidneys and still allowed the discrimination of glomeruli from the surrounding tubular structures as indicated with the white lines at different focal planes (3–5).
The high contrast visibility of the kidney surface and all glomerular tufts facilitated the development of a fully automated image–processing routine for quantification. This routine measured the kidney volume and counted all glomeruli contained within the organ in <3 hours. (Figure 3, A and B, Supplemental Figures 6 and 7, Supplemental Movies 3 and 4). The total glomerular count in a healthy kidney was around 13,000–18,000, which agreed well with published data obtained by traditional techniques.3–5,27 When interpreting the automatically computed glomerular counts, it should be considered that our approach may slightly underestimate the number of tufts because of low detection sensitivity for dim glomeruli. Thus, experimental conditions should be optimized to yield maximum staining intensity of the investigated structures.
Figure 3.
Fully automated quantification of functional elements in healthy and nephritic kidneys. (A) On the basis of whole-organ images, the total kidney volume was calculated by automated image analysis and shows the typical swelling of NTN kidneys at day 7 and declining kidney size down to control levels at day 14. **P<0.05 (two–tailed, unpaired Kruskal–Wallis H test); ***P<0.001 (two–tailed, unpaired Kruskal–Wallis H test). (B) The total numbers of glomeruli per kidney were quantified with a fully automated image processing algorithm showing a highly significant loss of glomeruli at day 14 of NTN compared with day 7 and controls. **P<0.05 (two–tailed, unpaired Kruskal–Wallis H test). (C) Distribution of tuft volumes of NTN mice at day 7 and day 14 relative to the distribution in healthy controls. The glomerular tuft volumes at day 14 of NTN are increasing compared with control and day 7 of NTN as indicated via their median values (dashed lines). For the generation of B and C, a total number of 302,023 glomeruli from 23 kidneys was counted, and their tufts were individually sized by voxel counting. ****P<0.001 (chi-squared test). Daily urine analysis shows loss of glomerular filtration functionality indicated by (D) increasing albumin-to-creatinine (Alb/Crea) ratio and (E) decreasing creatinine (Crea) clearance (nonsignificant two–tailed, unpaired Kruskal–Wallis H test). (F) On the basis of automated quantification of glomeruli, the creatinine clearance efficiency per glomerulus was calculated, showing the decreasing clearance efficiency at day 7 and day 14 compared with in control mice (nonsignificant two–tailed, unpaired Kruskal–Wallis H test). For D–F, data of n=2 control animals and n=3 NTN-treated animals at day 7 and day 14 were analyzed.
The endothelial label also allowed the quantification of the volume of all glomerular tufts by voxel rendering (Figure 3C). Thereby, the voxel size was 5 μm isotropic, and each tuft was represented by >1000 voxels; this count is at least 40× more than voxel counts obtainable with magnetic resonance imaging.5,28 This allowed precise separation of tufts from the nonlabeled structures of, for example, the Bowman’s capsule. Tufts in control animals showed a relatively narrow distribution (median volume of 80×103 μm3), which corresponds to approximately 33% of the volume of entire glomeruli measured by stereology.5
Next, we investigated nephrotoxic nephritis (NTN), an antiserum-induced model of human crescentic GN that frequently leads to ESRD.1 Macroscopic inspection of kidney lightsheet image stacks 2 weeks after NTN induction revealed reduced numbers of glomeruli (Figure 2A) associated with patchy tissue defects (Figure 2A, insets, Supplemental Movie 3). High–resolution fluorescence imaging by three–dimensional confocal or two-photon microscopy several 100 μm into the tissue (Figure 2, B and C) showed that glomeruli from nephritic mice showed typical alterations of late NTN (that is, endothelia with strongly reduced CD31 expression and more periglomerular fibrotic tissue) as evidenced by a higher second harmonic generation signal (Figure 2B, Supplemental Movie 3). Also, at late-stage NTN, more irregularly shaped glomeruli were evident (Figure 2B) compared with the more spherical form observed in healthy mice.5
On day 7 after NTN induction, automated image analysis showed swelling of kidneys (Figure 3A) but no reduction of glomerular numbers (Figure 3B). Interestingly, distribution of tuft volumes in NTN showed increasing heterogeneity and swelling at day 7. By day 14, an approximately 32% glomerular loss was evident, especially of glomeruli with small- and medium-sized tufts (Figure 3C). An increasing albumin-to-creatinine ratio indicated deterioration of the glomerular filtration barrier from day 7 to day 14 (Figure 3D). Accordingly, creatinine clearance was reduced approximately 28% by day 7 and approximately 61% by day 14 (Figure 3E). The computed glomerular numbers at these time points enabled the average GFR to be calculated, a parameter unobtainable by standard analysis techniques. Average GFR was reduced approximately 33% by day 7 and approximately 42% by day 14 (Figure 3F), indicating that the decrease of total glomerular function at day 14 resulted from a combination of defects in individual tufts and gradual glomerular loss. These findings showed that LSFM enables unprecedented insight into the functionality of a whole organ and the effect of disease. This approach will be of great value in nephrology, where total organ function depends on the combined contributions of all individual nephrons. The usefulness of the generated data will depend on the functional structures being labeled. In NTN, defects in glomerular basement membrane or podocyte function will occur earlier than damage to capillary loops.29 Thus, future studies should apply double or triple labeling of these elements followed by time-resolved LSFM. Our demonstration of rapid automatic quantification of functional elements in kidneys has enormous applicative potential and serves as a blueprint for generating quantitative analyses from whole–organ lightsheet data.
Concise Methods
Mice
All animal experiments were in accordance with German guidelines and approved by the relevant local authorities in Essen and Bonn. For the experiments, female mice of age 7–14 weeks old were used unless stated otherwise.
NTN Induction
Nephrotoxic sheep serum was generated by immunizing a sheep with homogenized murine renal cortex in CFA followed by monthly boosting doses in IFA. NTN in mice was induced by intraperitoneal injection of 0.35 ml nephrotoxic serum per mouse as previously described.30,31
Antibody Staining and Sample Preparation
Eight- to 12-week-old female mice (CD11c-EYFP,18 CX3CR-EGFP,32 and CatchupIVM-red33) were injected intravenously with 10 μg CD31-AF647 (catalog no. 102516; BioLegend, San Diego, CA) per mouse in PBS (total volume of 150 μl) and euthanized by CO2 10 minutes after injection. Immediately after killing, mice were transcardially perfused with 15 ml cold PBS and 5 mM EDTA and perfusion fixed with 15 ml cold 4% PFA/PBS (pH 7.4). After perfusion, organs were removed and postfixed in cold 4% PFA/PBS (pH 7.4) at 4°C to 8°C considering organ–specific incubation times (Supplemental Table 2). To reconstruct the physiologic shape of the collapsed lung, the organ was filled with 800 μl low-melting/low-gelling agarose (catalog no. 50302; SeaPrep Agarose; Lonza) after PFA/PBS perfusion. For agarose hardening, the lung was transferred into ice cold water for 10 minutes. After this procedure, the organ was postfixed as described above. Small organs, like lymph nodes, were embedded in 1% low-melting/low-gelling agarose and afterward, postfixed with 4% PFA/PBS as described above.
Sample Dehydration and Clearing
After perfusion and postfixation with 4% PFA/PBS, samples were dehydrated with ethanol (pH 9.0), thereby considering organ-specific concentrations and incubation times (Supplemental Tables 2 and 3). Incubation of all samples was performed at 4°C to 8°C in gently shaking 5-ml tubes. For optimal tissue dehydration, the incubation with 100% ethanol had to be done twice to remove all leftover water molecules. After dehydration, the samples were transferred to ECi (catalog no. 112372; Sigma-Aldrich) and incubated while gently shaking at room temperature (the freezing/melting point of ECi is 6°C to 8°C) until they became transparent. For comparison of our new clearing reagent with three other established solvent–based clearing protocols, tissue clearing was performed as described14,17,19 with slight modifications. According to Schwarz et al.,17 Becker et al.,14 and Ertürk et al.,19 organs were dehydrated in graded ethanol or a tert-butanol series of 30% (vol/vol), 50% (vol/vol), 70% (vol/vol), 80% (vol/vol), 96% (vol/vol), and 100% (vol/vol) twice for 24 hours each with pH adjusted to 9.0 at RT or 30°C in gently shaking 5-ml tubes. Then, tissue clearing was performed via either BABB (benzyl alcohol [catalog no. 402834] and benzyl benzoate [catalog no. B6630]; Sigma-Aldrich) or DBE (catalog no. 108014; Sigma-Aldrich). For comparison, we also used the original ethanol-BABB protocol without pH adjustment.34 As an example of a water–based simple immersion tissue clearing, the protocol of SeeDB clearing was performed.20 Here, the samples were dehydrated via an increasing fructose solution series of 20% (wt/vol), 40% (wt/vol), and 60% (wt/vol) for 8 hours at RT and 80% (wt/vol) and 100% (wt/vol) for 12 hours at RT. The final clearing step was on the basis of 24 hours of incubation at RT with SeeDB (80.2% [wt/wt] fructose) in gently shaking 5-ml tubes. SeeDB does not recommend pH adjustment.
LSFM
To image whole organs, a LaVision BioTec Ultramicroscope (LaVision BioTec, Bielefeld, Germany) with an Olympus MVX10 Zoom Microscope Body (Olympus, Tokyo, Japan), a LaVision BioTec Laser Module, an Andor Neo sCMOS Camera with a pixel size of 6.5×6.5 μm2, and detection optics with an optical magnification range from 1.26× to 12.6× and an NA of 0.5 were used. For EYFP and EGFP excitation, a 488-nm optically pumped semiconductor laser was used. For tdTomato excitation, a 561-nm optically pumped semiconductor laser was used, and for CD31-AF647 excitation, a 647-nm diode laser was used. Emitted wavelengths were detected with specific detection filters: 525/50 nm for EGFP, 545/30 nm for EYFP, 595/40 nm for tdTomato, and 680/30 nm for CD31-AF647. Because the excitation optics of the microscope provided a lightsheet thickness of 5–40 μm, the Z-step size was set to 5 μm for all measurements. The optical zoom factor varied from 1.6× to 10×.
Single– and Two–Photon Laser Scanning Microscopy
For high-magnification imaging of the samples, a Leica TCS SP8 Fully Automated Epifluorescence Confocal Microscope (Leica Microsystems, Buffalo Grove, IL) with AOTF and AOBS scanoptics, HyD detection, and two-photon (MP) and compact OPO on a DM6000 CFS frame was used. Imaging of glomeruli in cleared kidneys was performed via a 25× HCX IRAPO L objective with an NA of 0.95 in water. Fluorescence signals were generated via sequential scans, exciting CD31-AF647 via single-photon excitation using a HeNE laser at 633 nm, and detecting in confocal mode with an internal HyD at 660–720 nm. The second sequence was performed with a two-photon laser tuned to 960 nm for SHG detection at 460/50 nm and autofluorescence detection at 520/50 nm, both detected with external PMTs NDD1 and NDD2.
Image Reconstruction
For image reconstruction, ImageJ (Image Processing and Analysis in Java; http://imagej.nih.gov/ij/) and Imaris 8.1.2 (Bitplane, Switzerland) were used. Three-dimensional rendering of LSFM data as well as laser scanning microscopy data were performed via Imaris software (final movies were put together and labeled using Microsoft Movie Maker; Microsoft, Redmond, WA). Supplemental Movies 1–4 were strongly downsampled to accommodate the extremely large original datasets (typically multigigabytes). The loss of optical image quality in the movies was kept at a minimum; however, it was impossible to avoid completely.
Statistical Analyses
Statistical analyses of kidney data were performed with GraphPad Prism 6 software (GraphPad Software Inc., San Diego, CA). Depending on the type of data, two–tailed, unpaired t tests or chi-squared test were used as indicated in the figures.35
Image Analyses
To evaluate the kidney volume as well as the number, diameter, and volume of glomeruli, we developed an automated analysis procedure of LSFM data. The image analysis algorithm was implemented in the programming language python (https://www.python.org) and carried out fully automatically. The procedure includes (1) preprocessing of image data, (2) segmentation of the kidney region, (3) segmentation of the glomeruli, and (4) quantification of the segmented regions (more detail is in Supplemental Material). To compare kidney volumes before and after clearing, the algorithm was adapted accordingly to handle low-contrast images before clearing.
Computing Time
The computations were carried out on a GNU/Linux Server (SUSE) and an Intel Xeon CPU E7–4890 v2 (2.80 GHz ×60; 757 GB RAM). The average computing time for one Z stack was 2.8±0.7 hours. The computations were carried out fully automatically, and the algorithm could be easily adjusted to run on computers with lower RAM and CPU resources. The code for the detection algorithm is available for use on request.
Disclosures
A patent application has been filed by A.K. for the use of ethyl cinnamate as a novel nontoxic clearing reagent.
Supplementary Material
Acknowledgments
We thank Marc Schuster (Institute for Experimental Immunology and Imaging) for helpful discussions, the Imaging Center Essen (http://imces.uk-essen.de) for help with imaging, and Anthony Squire for proofreading.
This work was supported by funds from German Research Foundation “Immunobone” grants GU769/4-1 (to M.G.) and GU769/4-2 (to M.G.) and European Union grants EU HEALTH-2013-INNOVATION-1, MATHIAS (to M.G.) and EU H2020 Research and Innovation Action MULTIMOT (to M.G.).
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2016020232/-/DCSupplemental.
References
- 1.Kurts C, Panzer U, Anders HJ, Rees AJ: The immune system and kidney disease: Basic concepts and clinical implications. Nat Rev Immunol 13: 738–753, 2013 [DOI] [PubMed] [Google Scholar]
- 2.Bertram JF, Douglas-Denton RN, Diouf B, Hughson MD, Hoy WE: Human nephron number: Implications for health and disease. Pediatr Nephrol 26: 1529–1533, 2011 [DOI] [PubMed] [Google Scholar]
- 3.Murawski IJ, Maina RW, Gupta IR: The relationship between nephron number, kidney size and body weight in two inbred mouse strains. Organogenesis 6: 189–194, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stelloh C, Allen KP, Mattson DL, Lerch-Gaggl A, Reddy S, El-Meanawy A: Prematurity in mice leads to reduction in nephron number, hypertension, and proteinuria. Transl Res 159: 80–89, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baldelomar EJ, Charlton JR, Beeman SC, Hann BD, Cullen-McEwen L, Pearl VM, Bertram JF, Wu T, Zhang M, Bennett KM: Phenotyping by magnetic resonance imaging nondestructively measures glomerular number and volume distribution in mice with and without nephron reduction. Kidney Int 89: 498–505, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chacon-Caldera J, Geraci S, Krämer P, Cullen-McEwen L, Bertram JF, Gretz N, Schad LR: Fast glomerular quantification of whole ex vivo mouse kidneys using Magnetic Resonance Imaging at 9.4 Tesla. Z Med Phys 26: 54–62, 2016 [DOI] [PubMed] [Google Scholar]
- 7.Richardson DS, Lichtman JW: Clarifying tissue clearing. Cell 162: 246–257, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stelzer EH: Light-sheet fluorescence microscopy for quantitative biology. Nat Methods 12: 23–26, 2015 [DOI] [PubMed] [Google Scholar]
- 9.Keller PJ, Schmidt AD, Wittbrodt J, Stelzer EH: Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science 322: 1065–1069, 2008 [DOI] [PubMed] [Google Scholar]
- 10.Chung K, Wallace J, Kim SY, Kalyanasundaram S, Andalman AS, Davidson TJ, Mirzabekov JJ, Zalocusky KA, Mattis J, Denisin AK, Pak S, Bernstein H, Ramakrishnan C, Grosenick L, Gradinaru V, Deisseroth K: Structural and molecular interrogation of intact biological systems. Nature 497: 332–337, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang B, Treweek JB, Kulkarni RP, Deverman BE, Chen CK, Lubeck E, Shah S, Cai L, Gradinaru V: Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell 158: 945–958, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Susaki EA, Tainaka K, Perrin D, Kishino F, Tawara T, Watanabe TM, Yokoyama C, Onoe H, Eguchi M, Yamaguchi S, Abe T, Kiyonari H, Shimizu Y, Miyawaki A, Yokota H, Ueda HR: Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell 157: 726–739, 2014 [DOI] [PubMed] [Google Scholar]
- 13.Renier N, Wu Z, Simon DJ, Yang J, Ariel P, Tessier-Lavigne M: iDISCO: A simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 159: 896–910, 2014 [DOI] [PubMed] [Google Scholar]
- 14.Becker K, Jährling N, Saghafi S, Weiler R, Dodt HU: Chemical clearing and dehydration of GFP expressing mouse brains. PLoS One 7: e33916, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campbell TN, Choy FYM: The effect of pH on green fluorescent protein: A brief review. Mol Biol Today 2: 1–4, 2001 [Google Scholar]
- 16.Wang Y, Zhang DH, Zhang JY, Chen N, Zhi GY: High-yield synthesis of bioactive ethyl cinnamate by enzymatic esterification of cinnamic acid. Food Chem 190: 629–633, 2016 [DOI] [PubMed] [Google Scholar]
- 17.Schwarz MK, Scherbarth A, Sprengel R, Engelhardt J, Theer P, Giese G: Fluorescent-protein stabilization and high-resolution imaging of cleared, intact mouse brains. PLoS One 10: e0124650, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindquist RL, Shakhar G, Dudziak D, Wardemann H, Eisenreich T, Dustin ML, Nussenzweig MC: Visualizing dendritic cell networks in vivo. Nat Immunol 5: 1243–1250, 2004 [DOI] [PubMed] [Google Scholar]
- 19.Ertürk A, Becker K, Jährling N, Mauch CP, Hojer CD, Egen JG, Hellal F, Bradke F, Sheng M, Dodt HU: Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nat Protoc 7: 1983–1995, 2012 [DOI] [PubMed] [Google Scholar]
- 20.Ke MT, Fujimoto S, Imai T: SeeDB: A simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat Neurosci 16: 1154–1161, 2013 [DOI] [PubMed] [Google Scholar]
- 21.Wehrl HF, Bezrukov I, Wiehr S, Lehnhoff M, Fuchs K, Mannheim JG, Quintanilla-Martinez L, Kohlhofer U, Kneilling M, Pichler BJ, Sauter AW: Assessment of murine brain tissue shrinkage caused by different histological fixatives using magnetic resonance and computed tomography imaging. Histol Histopathol 30: 601–613, 2015 [DOI] [PubMed] [Google Scholar]
- 22.Loqman MY, Bush PG, Farquharson C, Hall AC: A cell shrinkage artefact in growth plate chondrocytes with common fixative solutions: Importance of fixative osmolarity for maintaining morphology. Eur Cell Mater 19: 214–227, 2010 [DOI] [PubMed] [Google Scholar]
- 23.Duncan GS, Andrew DP, Takimoto H, Kaufman SA, Yoshida H, Spellberg J, de la Pompa JL, Elia A, Wakeham A, Karan-Tamir B, Muller WA, Senaldi G, Zukowski MM, Mak TW: Genetic evidence for functional redundancy of Platelet/Endothelial cell adhesion molecule-1 (PECAM-1): CD31-deficient mice reveal PECAM-1-dependent and PECAM-1-independent functions. J Immunol 162: 3022–3030, 1999 [PubMed] [Google Scholar]
- 24.Vanholder R, Fouque D, Glorieux G, Heine GH, Kanbay M, Mallamaci F, Massy ZA, Ortiz A, Rossignol P, Wiecek A, Zoccali C, London GM; European Renal Association European Dialysis; Transplant Association (ERA-EDTA) European Renal; Cardiovascular Medicine (EURECA-m) working group : Clinical management of the uraemic syndrome in chronic kidney disease. Lancet Diabetes Endocrinol 4: 360–373, 2016 [DOI] [PubMed] [Google Scholar]
- 25.Acar M, Kocherlakota KS, Murphy MM, Peyer JG, Oguro H, Inra CN, Jaiyeola C, Zhao Z, Luby-Phelps K, Morrison SJ: Deep imaging of bone marrow shows non-dividing stem cells are mainly perisinusoidal. Nature 526: 126–130, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reddy V, Bhandari S, Seymour AM: Myocardial function, energy provision, and carnitine deficiency in experimental uremia. J Am Soc Nephrol 18: 84–92, 2007 [DOI] [PubMed] [Google Scholar]
- 27.Cullen-McEwen LA, Kett MM, Dowling J, Anderson WP, Bertram JF: Nephron number, renal function, and arterial pressure in aged GDNF heterozygous mice. Hypertension 41: 335–340, 2003 [DOI] [PubMed] [Google Scholar]
- 28.Heilmann M, Neudecker S, Wolf I, Gubhaju L, Sticht C, Schock-Kusch D, Kriz W, Bertram JF, Schad LR, Gretz N: Quantification of glomerular number and size distribution in normal rat kidneys using magnetic resonance imaging. Nephrol Dial Transplant 27: 100–107, 2012 [DOI] [PubMed] [Google Scholar]
- 29.Kriz W, LeHir M: Pathways to nephron loss starting from glomerular diseases-insights from animal models. Kidney Int 67: 404–419, 2005 [DOI] [PubMed] [Google Scholar]
- 30.Hochheiser K, Engel DR, Hammerich L, Heymann F, Knolle PA, Panzer U, Kurts C: Kidney dendritic cells become pathogenic during crescentic glomerulonephritis with proteinuria. J Am Soc Nephrol 22: 306–316, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scholz J, Lukacs-Kornek V, Engel DR, Specht S, Kiss E, Eitner F, Floege J, Groene HJ, Kurts C: Renal dendritic cells stimulate IL-10 production and attenuate nephrotoxic nephritis. J Am Soc Nephrol 19: 527–537, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jung S, Aliberti J, Graemmel P, Sunshine MJ, Kreutzberg GW, Sher A, Littman DR: Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol Cell Biol 20: 4106–4114, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hasenberg A, Hasenberg M, Männ L, Neumann F, Borkenstein L, Stecher M, Kraus A, Engel DR, Klingberg A, Seddigh P, Abdullah Z, Klebow S, Engelmann S, Reinhold A, Brandau S, Seeling M, Waisman A, Schraven B, Göthert JR, Nimmerjahn F, Gunzer M: Catchup: A mouse model for imaging-based tracking and modulation of neutrophil granulocytes. Nat Methods 12: 445–452, 2015 [DOI] [PubMed] [Google Scholar]
- 34.Becker K, Jährling N, Kramer ER, Schnorrer F, Dodt HU: Ultramicroscopy: 3D reconstruction of large microscopical specimens. J Biophotonics 1: 36–42, 2008 [DOI] [PubMed] [Google Scholar]
- 35.Bohm G, Zech G: Introduction to Statistics and Data Analysis for Physicists, Hamburg, Germany, Verlag Deutsches Elektronen-Synchrotron, 2010 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.