Abstract
Microbial communities contain diverse bacteria that play important functional roles in every environment. Advances in sequencing and computational methodologies over the past decades have illuminated the phylogenetic and functional diversity of microbial communities from diverse habitats. Among the functions encoded in microbiomes are the abilities to synthesize and resist small molecules, yielding antimicrobial activity. These functions are of particular interest when viewed in light of the public health emergency posed by the increase in clinical antimicrobial resistance and the dwindling antimicrobial discovery and approval pipeline, and given the intimate ecological and evolutionary relationship between antimicrobial biosynthesis and resistance. Here, we review genomic and functional methods that have been developed for accessing the antimicrobial biosynthesis and resistance capacity of microbiomes and highlight outstanding examples of their applications.
Keywords: antimicrobials, antimicrobial resistance, microbiome, metagenomics
Introduction
Nobel laureate Joshua Lederberg was the first to propose the term “microbiome,” which he defined as “the ecological community of commensal, symbiotic, and pathogenic microorganisms that literally share our body space.”1 While Lederberg coined the term in reference to the microorganisms that inhabit human body sites, microbial communities can be associated with any environmental, clinical, or engineered habitat. Concurrent with the expansion in sequencing capacity and the widespread adoption of “omics” technologies, the term microbiome was redefined as the collective genetic material of a microbial community, while “microbiota” is generally used to refer to the microorganisms themselves.2 Initial studies of microbial communities attempted to characterize the phylogenetic and functional diversity by culturing the organisms and performing targeted functional assays. However, culture-based assays are inherently biased, as a large fraction of microbes from most habitats are not readily cultured using standard methods.3 Fortunately, metagenomic sequencing of microbial communities, facilitated by the revolution in genomics over the past several decades, has enabled increased investigation and appreciation of the breadth and depth of functions encoded in diverse microbiomes.
The study of antimicrobial resistance and biosynthesis in microbial communities is motivated by the ubiquity and diversity of microbes in wide-ranging habitats, the evolutionary and ecological origins of antimicrobial resistance, and the global health crisis posed by increasing antimicrobial resistance. The human commensal microbiota harbors approximately 10 trillion microbes,4,5 while the diversity of the soil microbiota has been estimated to be as high as 10,000 unique species per gram.6 Both culture-based and culture-independent analyses of these and other microbial communities have revealed biosynthetic pathways for small molecules with antimicrobial activity,7,8 as well as abundant and diverse antimicrobial resistance genes.9–11 The collection of resistance genes in a given microbiome, known as the antimicrobial resistome, consists of acquired, intrinsic, proto, and silent resistance genes.12,13 It is important to characterize resistomes and biosynthetic pathways from varied microbial communities in order to better understand and combat the rise of clinical antimicrobial resistance.10 This is ideally achieved by pairing functional and sequence-based genomic technologies with traditional culture-based analyses.
This review is intended to provide an overview of the functional and sequence-based methods that have been developed for discovery of antimicrobial biosynthesis pathways and resistance genes from microbial communities. We will critically evaluate these methods and highlight outstanding examples of their applications, with a specific focus on the advantages and disadvantages that each method provides.
Rise in antimicrobial resistance and decline in antimicrobial discovery
The World Health Organization has described antimicrobial resistance as the single greatest challenge in infectious disease today, posing a serious public health threat on a global scale.14 Clinical and environmental antimicrobial resistance have been increasing since the widespread anthropogenic deployment of antimicrobials. Indeed, clinical resistance is now frequently observed within a year of deployment of a new antimicrobial15,16 (Fig. 1). Furthermore, polymerase chain reaction (PCR)–based surveys of archived soils have revealed that the prevalence of environmental antimicrobial resistance genes has been on the rise since 1940.3,17 In the United States alone, 2 million people acquire serious infections that are resistant to the antimicrobials designed to treat those infections, resulting in an estimated 23,000 deaths.18 Incidences of multidrug resistance in human pathogens are also increasing.18 The proportion of clinical isolates that are resistant to antimicrobials, including deadly methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant Enterococcus (VRE), and fluoroquinolone-resistant Pseudomonas aeruginosa (FQRP) strains, has been steadily increasing since the 1980s.19 The economic burden of antimicrobial resistance is also tremendous, with an estimated 8 million additional hospital days resulting in $20 billion in direct healthcare costs and $35 billion in lost productivity in the United States alone.18, 20 If the current trajectory is not altered, it is estimated that worldwide deaths from antimicrobial resistance could climb to 10 million per year by 2050.21
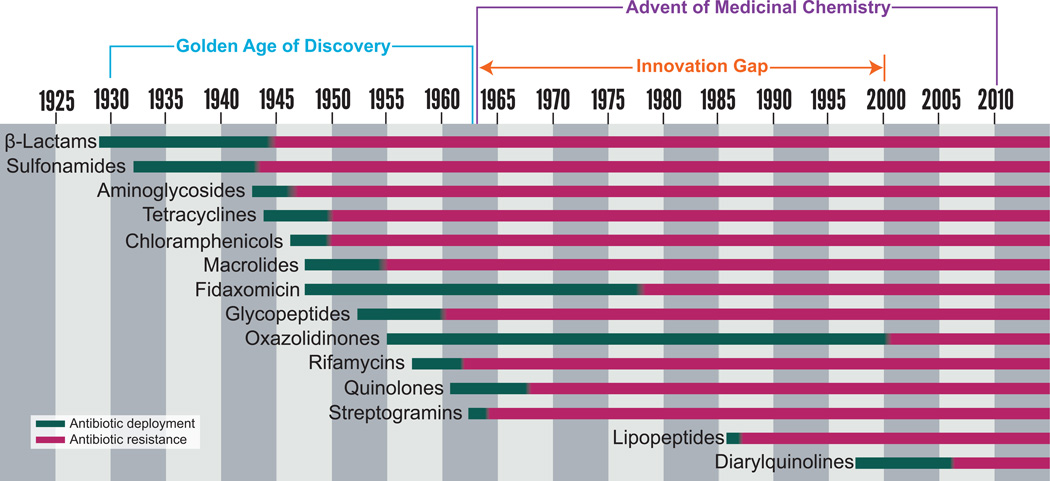
Figure 1.
The introduction of an antimicrobial to the clinic (green) is rapidly followed by the first observation of resistance to that antimicrobial (red).15,16
Compounding the problem posed by antimicrobial resistance, the golden age of antimicrobial discovery (ca. 1940–1960) was followed by a substantial innovation gap spanning 1962–2000 in which no new classes of antimicrobials were introduced (Fig. 1).15,22 Only three new antimicrobials were approved for human use during the first decade of the 21st century.23 The decrease in antimicrobial discovery can be partially attributed to low incentives for research and development of new drug classes.14 While most antimicrobial products currently in the pipeline are active against at least some of the pathogens identified by the U.S. Food and Drug Administration (FDA) as posing a “serious threat to public health,” barely a third (16 drugs) show significant activity against multidrug-resistant Gram-negative species.14 The issue is complicated by the collateral damage from excessive antimicrobial use: when an antimicrobial is administered clinically or agriculturally, it provides selective pressure for increased antimicrobial resistance in all microbes in those habitats, not simply the targeted pathogen(s), and thus has a negative societal impact.24 To counter the withdrawal of pharmaceutical companies from the antimicrobial market, the U.S. Congress is considering legislation to create a streamlined antimicrobial approval pathway. This would approve drugs to treat life-threatening infections on the basis of smaller (and hence faster) clinical trials, provided that they meet the same phase I safety and efficacy standards as other drugs.25 While these measures are being considered, it is imperative that we continue to better understand the ecological and evolutionary dimensions of antimicrobial production and resistance to support the improved stewardship of both our current and future arsenals of antimicrobial compounds.
Brief history of antimicrobials
Antimicrobials are defined as chemicals that either kill (bacteriocidal) or inhibit the growth (bacteriostatic) of bacteria at defined concentrations, and function by targeting systems critical to bacterial physiology. The five major modes of antimicrobial action are (1) inhibition of cell wall synthesis, (2) inhibition of protein synthesis via the bacterial ribosome, (3) inhibition of DNA or RNA synthesis, (4) inhibition of the folic acid pathway of nucleic acid synthesis, and (5) disruption of cell membrane integrity.15,26–29
Antimicrobials have likely been naturally produced by environmental microbes as a means of communication and defense for billions of years.30–32 Residues of antimicrobials were discovered in human skeletal remains dating from 350–550 CE, suggesting that humans have been exposed to antimicrobials for thousands of years.33–35 However, it was not until the 20th century that natural-product antimicrobials were deliberately repurposed for human use, revolutionizing our treatment of infectious diseases. Up until the 1920s, infectious diseases were the leading cause of death, with pneumonia, influenza, gastrointestinal infections, and tuberculosis accounting for a combined 540 per 100,000 deaths in the United States during the first decade of the 20th century.36 Ehrlich’s discovery of arsphenamine, an organoarsenic drug that could cure syphilis-infected rabbits35,37 and Fleming’s subsequent serendipitous discovery of penicillin, a β-lactam,38 signaled the advent of the antimicrobial era. The golden age of antimicrobial discovery began in the 1940s and lasted through the 1960s, fueled by Selman Waksman’s rapid antimicrobial discovery platform.16,39 Waksman was awarded the Nobel Prize for the development of this method,40 and the drugs developed during the golden age of antimicrobial discovery substantially improved the treatment of infectious disease.
Many natural-product antimicrobials were found to have pharmacological or toxicological drawbacks or eventually became obsolete owing to evolution and spread of resistance.41 Synthetically altering natural-product antimicrobials to improve pharmacology or evade resistance launched a new era of medicinal chemistry. Many antimicrobials were synthesized by modifying molecular scaffolds of antimicrobials previously discovered during the golden era. As a result, the amphenicol, tetracycline, aminoglycoside, macrolide, glycopeptide, and quinolone classes were introduced or expanded, further improving our treatment of infectious diseases.16,22,35,42,43
Brief history of antimicrobial resistance
The introduction of antimicrobials to the clinic resulted in a precipitous drop in worldwide mortality from infectious disease.44 However, the selective pressure placed on bacterial systems targeted by antimicrobials owing to the extensive use of antimicrobials in clinical and agricultural settings soon led to the steady evolution and increase of antimicrobial resistance, systematically compromising each of the antimicrobials in our arsenal (Fig 1).27,45–47
The strategies that bacteria use to resist antimicrobials can be divided into four general categories: (1) drug efflux, (2) reducing the permeability of the cell wall or membrane, (3) target overexpression, modification, or protection, and (4) enzymatic inactivation of the drug.27,48 In the first resistance mechanism of drug efflux, bacteria pump out the antimicrobial, keeping the intracellular concentration low and preventing the drug from reaching inhibitory concentrations. This mechanism encompasses efflux pumps that are either specific to single antimicrobials or classes, such as the tetracycline efflux pumps,49 or nonspecific, multidrug resistance efflux pumps.50–53 The second resistance strategy, thematically related to the first, involves reducing the permeability of the cell wall or cell membrane to the antimicrobial, thereby preventing it from reaching its target. This can occur via reduction of porin expression or expressions of a more selective porin variant.54,55 The third resistance strategy is overexpression, modification, or protection of the drug target in the bacterium, enabling survival. This mechanism is used by MRSA, which expresses PBP2a, a redundant and methicillin-insensitive version of the native PBP2 penicillin-binding protein encoded by the mecA gene.56 The final (fourth) resistance strategy involves enzymatic inactivation of the drug, by either degrading or modifying the antimicrobial compound such that it no longer has activity. Examples of this mechanism include β-lactamases, which degrade β-lactam antimicrobials,57,58 tetracycline destructases, such as Tet(X),59,60 and chloramphenicol acetyltransferases.61
While we primarily evaluate antimicrobial resistance in terms of its effects on the clinical use of antimicrobials, the capacity to resist such compounds is in fact a natural and ancient feature of all microbial communities.62,63 It is important to consider that the genes that we refer to as “resistance genes” may have had functions unrelated to clinical antimicrobial resistance in their original context, but could be repurposed for resisting antimicrobials when they are encountered in a new context. The original functions of such genes may include cell wall biosynthesis (e.g., β-lactamases), export of signaling molecules, metabolic intermediates, or plant-produced compounds (e.g., multidrug efflux pumps), or detoxifying the original host from the antimicrobial that it is producing (e.g., the tetracycline efflux pump induced just in time by its biosynthetic precursor anhydrotetracycline64).65 The discovery of β-lactam, tetracycline, and vancomycin resistance genes in 30,000-year-old Beringian permafrost sediments confirms that genes that confer resistance to antimicrobials preceded the clinical use of antimicrobials by several thousand years.62 Furthermore, while the first enzymatic degradation of penicillin was described in 1940, when researchers noted that extracts of certain bacterial cells were capable of inactivating penicillin,66 it is estimated that environmental β-lactamases have existed for over 2 billion years.67,68 Intensive human use of antimicrobials in clinical, agricultural, and industrial settings over the past 80 years have led to the fairly recent exaptation of such genes with resistance-conferring potential for surviving in the presence of antimicrobials.69,70
Since the introduction of antimicrobials to the clinic, the study of antimicrobial resistance has largely focused on isolates of human pathogens and has relied on information gathered from culturing these organisms.71 However, it is thought that most clinical antimicrobial resistance has its evolutionary origins in the aforementioned genes, with resistance-conferring potential encoded by benign environmental and human-associated bacteria. Furthermore, a recent study identified numerous resistance genes from benign soil bacteria with 100% nucleotide identity to resistance genes that were present in diverse human pathogens, suggesting the relatively recent sharing of resistance genes via horizontal gene transfer.72 This finding highlights the need to study antimicrobial resistance in benign as well as pathogenic bacteria and in the context of microbial communities as well as isolates, as horizontal gene transfer is the mechanism by which most pathogens become resistant to antimicrobials.73
Culture-based methods underestimate the functional diversity of microbiomes
Discovering novel antimicrobials and resistance from microbial communities through culture-based methods is challenging and limited to the cultivable minority of bacteria.74 Genomic technologies offer the promise of substantial performance improvements for characterizing the functional capacity of microbiota, which has implications in both the basic science and translational realms.74,75 For example, characterizing antimicrobial resistance using current culture-based methods can take approximately 1–3 weeks, versus the proposed less than 12 hours using genomic technologies.74 Whole-genome sequencing has the potential to reduce exposure to ineffective drugs and the evolution of resistance by reducing diagnosis to days instead of weeks, as seen in the case of drug-resistant Mycobacterium tuberculosis.75,76
Depending on the habitat, traditional culturing efforts may capture as little as 1% of a microbial community.3 The other 99% includes species that are recalcitrant to culture by standard methods.77 This severe undersampling has motivated considerable recent advancements in culturing methods to enable improvements in the proportion of microbial communities that can be cultured. However, these methods, which may leverage microfluidic technologies,78,79 clever simulations of natural growth conditions,79,80 or exhaustive sampling of culture conditions,81 can be technically challenging and are not yet part of standard culturing workflows. Culture-based methods are further hindered by the possible unreliable identification of the cultured organisms, which is dependent on the expertise of the researcher. Culture-based methods are also riddled with irreproducibility on non-selective media.82 It is often difficult to recapitulate the phenotype of resistance of a targeted bacterium from selective media to non-selective media.
Advancements in genomic and computational technologies and the concomitant dramatic reductions in sequencing costs over the past decades have allowed for an explosion of microbial genome and metagenome sequencing, illuminating the diversity of microbial communities and the breadth and depth of functions encoded therein.74,83–89 Genomic technologies can overcome culture-based limitations and facilitate the discovery of novel antimicrobials and resistance determinants. The remaining review details the functional and sequencing-based methods that can be employed to quantitatively interrogate microbiomes and resistomes (Fig. 2).
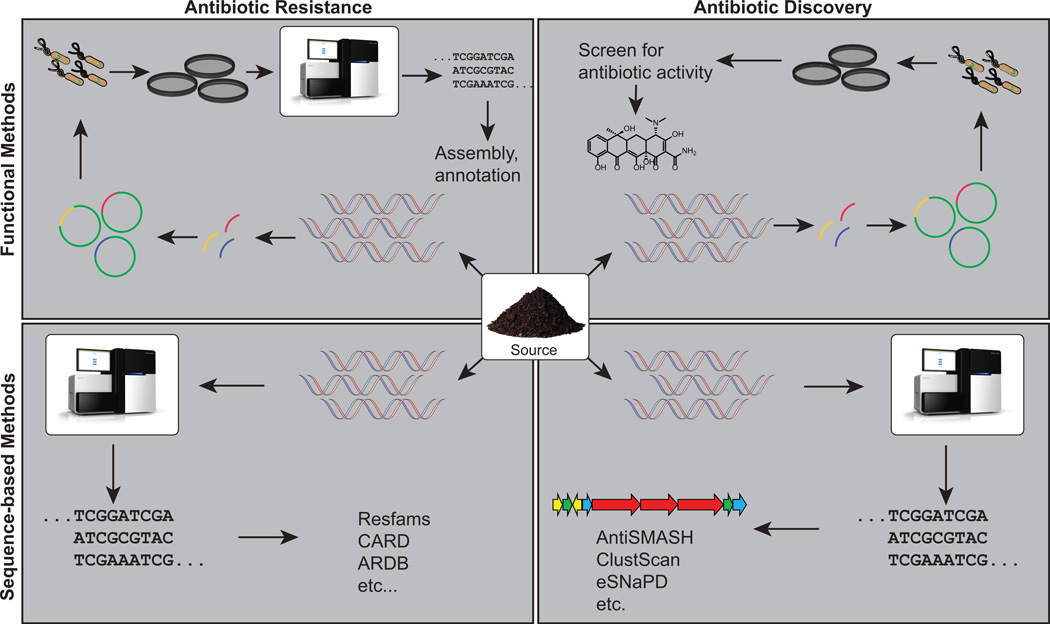
Figure 2.
An overview of function- and sequence-based methods for the discovery of antimicrobials and antimicrobial resistance from microbiomes. DNA is isolated from source material (center), which can then be mined for antimicrobial resistance genes (left) or antimicrobial biosynthetic pathways (right). Functional metagenomic methods (top) typically entail shotgun cloning metagenomic DNA into an expression vector and selecting for a desired phenotype (e.g., antimicrobial resistance or antimicrobial activity). Sequence-based methods (bottom) customarily involve sequencing metagenomic DNA and annotating sequences using general or function-specific databases.
Function-based methods for resistance discovery
Functional metagenomics
Functional metagenomic methods for the discovery of antimicrobial resistance genes sit at the intersection of culture-based and genomic techniques.90 The general functional metagenomic pipeline consists of extracting total metagenomic DNA from a microbial community, shearing the metagenomic DNA to a target size distribution (e.g., 1–5 kb), and shotgun cloning these fragments into an expression vector. The expression library is then transformed into a suitable heterologous host, such as Escherichia coli. A phenotype, such as antimicrobial resistance, is selected by plating a metagenomic library on media supplemented with antimicrobials.71,91 The combination of functional metagenomics with next-generation sequencing and improved computational sequence-assembly algorithms has dramatically increased the throughput of functional metagenomics while also decreasing the cost.72 This method is particularly well suited for discovering novel genes with antimicrobial resistance function, including those that are less than 65% identical at the amino acid level to known resistance genes.81 Functional metagenomics vastly outperforms methods that rely solely on alignment to reference databases to predict resistance functionality because it is independent of prior knowledge of resistance function.72,90,92–96 However, a limitation of this method is that the identification of novel resistance genes depends on functionality of genes in the heterologous host.90 It is thus best to use a range of heterologous hosts in order to truly capture all phenotypic resistance.90,97 Additionally, some combinatorial antimicrobial resistance mechanisms require the presence of multiple genes, which may not be captured within a functional metagenomic library depending on the insert size.98
One of the primary strengths of functional metagenomics is that it enables the identification of novel resistance-conferring genes from diverse microbiomes.71,72,96 For example, a recent study used functional metagenomics to identify 1100 genes encoding resistance functions, 121 of which were unique, from 96 fecal and environmental samples from rural areas in El Salvador and a peri-urban shanty town in Lima, Peru.96 By complementing this functional metagenomic profiling with deep metagenomic shotgun sequencing of 285 fecal and environmental samples from these same habitats (representing 344 Gb), researchers identified chicken coops and wastewater treatment systems as hot-spots of resistance gene exchange and provided a new framework for risk assessment of resistance gene exchange across interconnected habitats. In addition to clearly identifying novel antimicrobial-resistance determinants, functional metagenomics can also be used to predict horizontal gene transfer of antimicrobial-resistance genes by identifying co-localized mobility elements.99 For instance, the aforementioned cross-habitat resistome study in El Salvador and Peru identified a single β-lactamase (TEM-1) in over 25 different genomic contexts, highlighting its high genomic mobility.96
Functional metagenomic selections in phylogenetically diverse expression hosts are powerful tools for expanding our knowledge of novel, functional resistance determinants in the face of rising clinical and environmental resistance. Going forward, it is important that functional methods be used to identify novel resistance genes for existing antimicrobials and to quantify the prevalence of different resistance mechanisms across habitats. Furthermore, new antimicrobial lead compounds should be screened for potentially cryptic resistance genes from diverse environments using functional metagenomic assays. This would allow us to anticipate the specific mechanism of inevitable evolution and dissemination of antimicrobial resistance to next-generation antimicrobials before the widespread emergence of these resistance genes in clinical and agricultural settings. Such information would enable design of preemptive molecular surveillance and diagnostics of these resistance determinants, as well as potential targets for design of resistance gene inhibitors.
Hybridization/PCR-based methods
Another method that integrates culturing and sequencing for resistance characterization is a new technology that utilizes molecular padlock probes.100 In this method, which shows particular clinical promise in predicting the antimicrobial susceptibility profile of patient samples, the microbial community is exposed to the antimicrobial of interest for a short period of time, after which metagenomic DNA is extracted.100 The metagenomic DNA is then mixed with complementary biotinylated oligonucleotides of targeted rRNA gene sequences and streptavidin-coated magnetic beads. This is to ensure the capture of the relevant rRNA gene sequences. The mixture is then hybridized and ligated to the designed padlock probes, usually designed to detect certain positions in the 16S rRNA gene.100 The probes are then amplified through rolling-circle amplification.100 After amplification, the final product is fluorescently labeled, and DNA copy number is quantified using a high-performance fluorescence detector.100 An antimicrobial susceptibility profile can be determined using the DNA copy number of the sample without antimicrobials and comparing it with the DNA copy number of the sample after exposure to antimicrobials.100 If there is an increase in DNA copy number after exposure to antimicrobials, this indicates resistance. If there is no significant growth after exposure to antimicrobials, this indicates susceptibility.100 This method has been used to characterize the antimicrobial susceptibility profile of uropathogens from complex urinary tract infection samples.100 Researchers accurately predicted the antimicrobial susceptibility profile for 55 of 56 samples, highlighting the clinical utility of this method.100 An advantage that the padlock probe method presents is short turnaround time, leveraging high throughput to give results in a matter of hours and help clinicians prescribe the correct antimicrobials to patients with bacterial infections. However, the probes that are used in the assays have to be designed for individual bacterial species and antimicrobials that are targeted to be used in the assay and cannot characterize novel resistance genes.
Sequence-based methods for resistance discovery
While function-based methods have been valuable in identifying novel resistance determinants from diverse microbiomes, they are limited in throughput and are ideally complemented by sequence-based methods, which we will review here. The ability to analyze bacterial communities through culture-independent shotgun metagenomic sequencing has revolutionized our ability to characterize resistomes.96,101–103 These analyses have benefited from databasing projects, including the 35,000+ public metagenomes that have been uploaded to the Metagenomics Rapid Annotations using Subsystems Technology server (MG-RAST).104 Shotgun metagenomic sequencing involves extracting and sequencing total metagenomic DNA from a microbial community. Once sequenced, a specific subset of informative marker genes from the metagenome can be used to infer phylogeny and functions that are present in the microbial community of interest.105 To predict antimicrobial resistance from shotgun sequencing, most studies have relied on pair-wise comparisons (e.g., by Basic Local Alignment Search Tool (BLAST)106) of sequences to databases of known or predicted resistance genes.107–111 Many antimicrobial resistance databases are highly biased toward human-associated organisms; therefore, environmental antimicrobial resistance is comparatively ignored.112 Prediction algorithms based on hidden Markov models (HMMs) trained on functionally validated resistance genes from diverse environments have proven to be superior to traditional pairwise annotators in both precision and accuracy of resistance gene annotation, since HMM profiles allow for the discovery of highly diverse and understudied antimicrobial resistance genes.112
Antimicrobial resistance gene databases
A number of databases that store nucleotide and protein sequence information for antimicrobial resistance genes have been developed. Query sequences are aligned to sequences of known antimicrobial resistance determinants in the databases by BLAST,106 which performs a strict pairwise DNA or protein sequence alignment, to catalog the antimicrobial resistance gene content of a genome or metagenome. These databases have dramatically advanced the field of genomic and metagenomic analyses of antimicrobial resistance by allowing the sequence-based identification of known resistance determinants.113
The Antibiotic Resistance Gene Database (ARDB) (http://ardb.cbcb.umd.edu/) is a manually curated antimicrobial resistance database.114 The ARDB provides a comprehensive ontology which creates a resistance profile by matching specific genes and mechanisms of action. It uses BLAST to identify and annotate antimicrobial resistance genes. Many studies have used the ARDB as their sole method to identify antimicrobial resistance genes115–117 and have revealed that a majority of antimicrobial resistance genes cluster based on ecology.107,116,118 Although this database was the first of its kind, it has not been updated since 2009.
The Comprehensive Antibiotic Resistance Database (CARD) (https://card.mcmaster.ca/) is a collection of known resistance determinants and associated antimicrobials.119 It is designed to predict known antimicrobial resistance from genome sequence data using a BLAST-based pairwise sequence alignment to known antimicrobial resistance genes in the database. The database is organized on the basis of ontologies, linking genes to their function. This is an ideal database to use if the antimicrobial resistance mechanism of interest has been thoroughly researched. However, it is not suited for the detection of point mutations in chromosomal target genes known to be associated with antimicrobial resistance genes.120 Many studies have annotated antimicrobial resistance using the CARD alone or in combination with the ARDB.110,111,121,122 Unlike the ARDB, the CARD is actively managed and periodically updated.
The Bush-Jacoby β-lactamase list (http://www.lahey.org/Studies/) is a curated database that matches β-lactamase gene sequences to resistance to β-lactam antimicrobials. This database will soon be hosted on the National Center for Biotechnology Information (NCBI) website. This database was originally focused heavily on TEM, SHV, and OXA type β-lactamases, only a subset of β-lactam antimicrobial resistance genes. Other β-lactamases that are now included in the database are CTX-M, CMY, AmpC, CARB, IMP, VIM, KPC, GES, PER, and VEB.113
The Lactamase Engineering Database (LacED) (http://www.laced.uni-stuttgart.de/)123 is an extension of the Bush-Jacoby β-lactamase list. This database includes a way to predict TEM, SHV, and class B enzymes by merging the information from the both NCBI peptide database and the TEM mutation table. The LacED builds protein families from these databases and integrates protein sequence and structure information using DWARF.124
The Repository of Antibiotic Resistance Cassettes (RAC) (http://rac.aihi.mq.edu.au/rac/) takes advantage of the fact that resistance genes are often syntenic with mobile genetic elements, forming resistance cassettes. This database was the first to simultaneously automate the identification of antimicrobial resistance genes and place the genes in their broader genetic context.125
ResFinder (https://cge.cbs.dtu.dk//services/ResFinder/)126 combines the Bush-Jacoby β-lactamase list with the ARDB and other published antimicrobial resistance gene sequences. Similar to other resistance databases, query sequences are matched to the sequences in the database via BLAST. The difference between ResFinder and most other resistance gene databases is the ability for the user to tailor the identity and length coverage thresholds. This allows the user to specify settings that are suited for the depth and quality of sequencing for each project. The current challenge in using ResFinder and the previously described databases is the inability to classify chromosomal mutations that can lead to antimicrobial resistance or to classify novel resistance genes, and it is thus limited to resistance genes commonly acquired through horizontal gene transfer.120
The Antibiotic Resistance Gene-Annotation (ARG-ANNOT) (http://www.mediterranee-infection.com/article.php?laref=282&titer=arg-annot)120 uses a BLAST-enabled search on a curated antimicrobial resistance gene database compiled from the Bush-Jacoby β-lactamase list (http://www.lahey.org/Studies/), Resfinder,126 the ARDB,114 and others. A main difference between this program and others is the ability to additionally predict resistance function on the basis of chromosomal point mutations. ARG-ANNOT contains a curated database focused on mutations found in many chromosomal genes (i.e., rpoB, gyrA1, gyrA2, parC) that can confer resistance to antimicrobials.
It is difficult to identify novel resistance genes when using pairwise comparisons (e.g., by BLAST) to resistance databases, which are largely based on resistance genes from cultured bacterial isolates, and are especially biased towards human associated pathogens. Resfams (http://www.dantaslab.org/resfams/) implements an alternative method in order to identify known and novel resistance genes with high precision and accuracy.112 Resfams is a curated database of profile HMMs built on resistance proteins compiled from CARD,119 LacED,123 and Jacoby and Bush’s collection of curated β-lactamases, based on the gene ontology from the CARD database. Additionally, the Resfams profile HMMs incorporate antimicrobial resistance genes from environmental, human commensal, and pathogenic bacteria that have been discovered through culture-independent functional metagenomic selections. The HMM approach is superior to pairwise annotation approaches in both precision and accuracy. Pairwise BLAST searches against resistance gene databases were unable to identify over 60% of resistance genes identified by Resfams from a functionally validated resistance gene test set.112 Perhaps more importantly, differences in HMM-based versus BLAST-based pairwise resistome analyses of the same data can lead to significantly different (and sometimes completely contradictory) biological conclusions. For instance, comparison of resistome predictions from over 6000 bacterial genomes by Resfams versus predictions by pairwise alignment of resistance gene databases revealed that pairwise methods are inaccurately biased in both their phylogenetic and ecological distributions toward the relatively oversampled human-associated environments that they represent.112
These myriad resistance gene databases are clearly important tools for interrogating microbiomes for antimicrobial resistance genes, but some challenges in their use and opportunities for their improvement exist. An important limitation to both HMM- and BLAST-based searches is the implicit requirement for users to determine the accuracy of the predicted resistance gene function, especially when the exact sequence being annotated has not previously been functionally assayed. Additionally, many of these databases do not incorporate information on the host-specific functionality of some antimicrobial resistance genes.127 For example, most Gram-negative bacteria are intrinsically resistant to important classes of Gram-positive–specific drugs (e.g., vancomycin, linezolid, and macrolides) because they cannot cross the Gram-negative outer membrane.48 Accordingly, it is likely inappropriate to annotate homologs of genes that provide resistance to these antimicrobials in Gram-positive bacteria when they are identified in Gram-negative bacteria. An important future goal in these databases is a species-dependent risk estimator for horizontal gene transfer to quantify the likelihood of resistance gene dissemination between species and habitats. Additionally, it is important that environmental or nonclinical resistance genes, which are currently underrepresented these databases, be included.112 Because most clinical resistance originated in the environment, this will facilitate the surveillance of emerging resistance mechanisms as they disseminate beyond benign organisms. Despite the inherent limitations in ascribing resistance function based on sequence alone, antimicrobial resistance databases play a key role in curating sequences of known resistance determinants. It is crucial that these databases are regularly updated and simultaneously cross-validated to maintain accuracy and relevance. Continued maintenance and improvement of these databases, including the ongoing incorporation of novel, functionally validated resistance determinants and associated metadata from diverse habitats, will ensure that they remain valuable resources for the antimicrobial resistance community for years to come.
It is likely that the selective pressure that drove the evolution of most antimicrobial resistance genes is the incredible biosynthetic potential of microbial communities to synthesize natural products with antimicrobial activity. This motivates the need to understand antimicrobial resistance in microbial communities in the context of antimicrobial biosynthesis, and vice versa. Paralleling recent improvements in methods to characterize resistomes, there have also been significant advances in functional and sequence-based methods for discovering novel antimicrobial biosynthesis machinery, which we will review below.
Function-based methods for antimicrobial discovery from microbiomes
Function-based methods for the discovery of antimicrobials from microbiomes present a clear advantage over sequence-based methods in that no a priori knowledge of the sequence of a natural product biosynthetic pathway is required. This facilitates the discovery of novel natural products that are produced by biosynthetic pathways that are divergent from previously characterized pathways and provides access to novel chemical diversity. This unbiased approach has been successfully used for the discovery of natural products with antimicrobial activity.128,129 The method depends on shotgun cloning of metagenomic DNA from a given environment into an expression vector, heterologous expression of pathways encoded in the metagenomic DNA in an appropriate host, and screening for production of bioactive natural products.130
Despite the advantages that this unbiased approach provides, there are several challenges that have hindered the discovery of natural-product antimicrobials using functional metagenomics. The main disadvantage is that construction and heterologous expression of large insert libraries is technically challenging.131 Strategies to improve the heterologous expression of biosynthetic pathways, including overexpression of alternative sigma factors in E. coli, have proven successful for heterologous production of polyketides and hold promise for the development of improved metagenomic screening hosts.132 Additionally, nontraditional heterologous hosts, such as Streptomyces lividans, have demonstrated utility in the discovery of bioactive natural products from metagenomic libraries.133 A final challenge inherent in functional metagenomic antimicrobial discovery lies in the fact that antimicrobial production is a more complicated phenotype to select for than antimicrobial resistance, for reasons that we will detail below. This necessitates the use of clever screening strategies to identify and isolate clones of interest.131
One screening strategy that successfully enables the discovery of antimicrobial biosynthesis from metagenomic libraries is to directly assay for antimicrobial activity against an indicator strain using a double–agar layer method.134 In this method, the heterologous host of the metagenomic library is allowed to grow on plates, after which top agar containing the indicator strain is overlaid. This method was used to discover two clones from a 113,500 member fosmid library representing 4 Gb constructed from a soil metagenome that were able to inhibit the growth of Bacillus subtilis. The clones encoded a biosynthetic pathway that produced indigo and indirubicin, pigments that were confirmed to be responsible for the observed antibacterial activity.128 Another application of this method resulted in the identification of 65 clones with antibacterial activity from a 700,000-member soil library. A small molecule from the clone with the highest apparent bioactivity was purified and identified as a long chain N-acyl amino acid. This represented the first report of the antibacterial activity of long-chain N-acyl amino acids, as well as the first identification of genes involved in long-chain N-acyl amino acid biosynthesis, highlighting two key advantages of the functional metagenomic approach to antimicrobial discovery.129
An alternative approach that extends traditional culturing methods uses the innovative iChip, a multichannel, microfluidic diffusion growth chamber.79 This device was recently used to discover a novel non-ribosomal peptide with antimicrobial activity from a soil microbial community.135 The iChip requires that input microbial community samples are diluted such that each channel receives approximately one bacterial cell. The device is sealed with a semipermeable membrane and returned to the source so that nutrients and growth factors from the natural environment of the source organisms can diffuse into the chamber, increasing the likelihood of culturing fastidious organisms.79 This high-throughput method was used to culture 10,000 diverse isolates from a soil sample, which were predicted to be recalcitrant to culture using traditional methods. Screening of extracts from these isolates for antimicrobial activity revealed the presence of a non-ribosomal peptide named teixobactin, synthesized by a previously uncultured betaproteobacterial species. Teixobactin was isolated and characterized and found to have activity against multidrug-resistant Gram-positive pathogens in vitro, as well as in animal models for MRSA and Streptococcus pneumoniae.135 Although this method relied on classical antimicrobial screening platforms, innovative culture methods facilitated the discovery of a novel antimicrobial and revealed the biosynthetic potential of the uncultivable fraction of microbial communities.
Sequence-based methods for antimicrobial discovery from microbiomes
Sequence-based methods for the discovery of antimicrobial biosynthetic pathways from microbiomes depend on homology between the query sequence and known biosynthetic pathways. A clear advantage of this method is throughput. Advancements in annotation and prediction algorithms coupled with the abundance of microbial genome and metagenome sequences have made feasible the analysis of sequence data for the identification of biosynthetic gene clusters that may encode novel bioactive natural products. A disadvantage of this strategy is that prediction of the structure of a small molecule from the sequence of a biosynthetic gene cluster is nontrivial, as is prediction of a small molecule’s activity from its structure.
One of the first attempts to identify biosynthetic pathways from an uncultured community relied on PCR amplification of ketosynthase genes from Streptomyces isolates, as well as from metagenomic DNA isolated from soil.136 The amplicons were purified, cloned into an expression vector, and sequenced. Of 20 randomly selected transformants that were sequenced, only two had similarity to any known ketosynthase gene products. Four of the novel ketosynthases were shown to be functional when expressed in a Streptomyces lividans or Streptomyces glaucescens host as part of a hybrid polyketide synthase pathway (that is, the native ketosynthase was replaced with the newly discovered ketosynthase). The polyketide products of the hybrid biosynthetic pathways were purified and characterized to be novel octaketide and decaketide natural products. This highlights the utility of culture-free methods for identifying novel, sequence-divergent genes involved in biosynthetic pathways to expand access to novel natural products with diverse bioactivities, including potential antimicrobial activity.
Tools for identifying genes involved in natural-product biosynthesis from sequence data are a significant advancement over targeted PCR-based studies. One of the resources developed for mining genomic data for biosynthetic gene clusters is antiSMASH.137 AntiSMASH makes use of the ClusterFinder algorithm,138 which is an HMM-based algorithm for the discovery of biosynthetic gene clusters from genomic data. The ClusterFinder algorithm comprises a four-step prediction pipeline that can identify biosynthetic gene clusters in genomic data. The use of an HMM-based, probabilistic algorithm allows identification of biosynthetic gene clusters that lack high similarity in domain structure to any known biosynthetic gene clusters, circumventing the problem that had previously been posed in identification of biosynthetic gene clusters from sequence data. AntiSMASH also includes an active site finder module that identifies conserved amino acid motifs in the active sites of key biosynthetic enzymes, including those involved in polyketide biosynthesis. A final, key feature of antiSMASH is an algorithm that predicts the structure of a natural product synthesized by the annotated biosynthetic gene cluster. Furthermore, the development of a web-based server (http://antismash.secondarymetabolites.org/) has democratized the process of identifying biosynthetic gene clusters, because it is available to those without access to high-throughput computing facilities.137
The ClusterFinder/antiSMASH workflow was successfully used to identify a natural product with antimicrobial activity from metagenomic sequencing data generated as part of the National Institutes of Health Human Microbiome Project. Genomes of 2430 members of the human microbiota were queried using ClusterFinder to identify over 14,000 putative biosynthetic gene clusters. Metagenomic reads from 752 samples representing five body sites of healthy subjects were mapped to the 14,000 predicted biosynthetic gene clusters. Among the natural products predicted was a thiopeptide from the vaginal commensal Lactobacillus gasseri. The thiopeptide, named lactocillin, was purified and characterized. Lactocillin was found to have antimicrobial activity against Gram-positive bacteria, including Enterococus faecalis, Gardnerella vaginalis, Staphylococcus aureus, and Corynebacterium aurimucosum. The discovery and characterization of the structure and bioactivity of lactocillin from human microbiome metagenomic sequencing data demonstrates the potential that diverse metagenomes hold for encoding natural products with antimicrobial activity.8
A current shortcoming of the software available to identify biosynthetic gene clusters from metagenomic sequencing data is that assembly of short-read data into genomes or large contigs is required. Although there have been recent advances in metagenomic assemblies of this type,139–141 it remains a challenge. One way to circumvent the requirement for metagenomic assembly is the use of ShortBRED,142 which finds short, unique amino acid markers from reference proteins of interest and maps metagenomic sequencing reads to these markers. This tool has been successfully implemented for the identification of antimicrobial resistance genes101 and has potential for the identification of genes involved in natural product biosynthesis directly from short-read metagenomic sequencing data.
An alternative method that can be used to access the biochemical diversity of microbiomes without the necessity for metagenomic assembly uses short DNA sequences that are conserved between biosynthetic gene clusters. PCR amplification of these conserved biosynthetic motifs from a metagenomic source using degenerate primers followed by amplicon sequencing can provide an overview of the biosynthetic capabilities of a given microbiome and highlight promising leads for heterologous expression and further characterization.143 A bioinformatics platform for mapping short DNA sequence tags to known biosynthetic gene clusters, termed Environmental Surveyor of Natural Product Diversity (eSNaPD), has been developed to facilitate analysis of amplicon sequencing data in the context of natural-product biosynthesis.144 This strategy has been successfully used in the identification of novel bioactive small molecules, including some with antimicrobial activity.143,145
A final, promising method for sequence-based identification of biosynthetic gene clusters from microbiomes involves assembly of complete or near-complete genomes using single-molecule, real-time sequencing (SMRT),146 followed by annotation with antiSMASH or a similar annotation pipeline. This approach was successfully applied in tandem with short-read data reference-independent assembly of a high-quality Corynebacterium simulans genome de novo from a skin microbiota sample. This previously uncharacterized genome was annotated with antiSMASH, revealing predicted nonribosomal peptide synthetases, type I polyketide synthetases, terpene synthases, and bacteriocin production genes.147 Interestingly, the bacteriocin production locus was homologous to the locus for biosynthesis of lactococcin, a bacteriocin that is known to be bactericidal.148 Although the natural products whose biosynthetic gene clusters were identified in this study were not isolated or confirmed for bioactivity, the authors demonstrate the power of metagenomic assembly in the identification of biosynthetic gene clusters from microbiomes.
Future directions in antimicrobial and antimicrobial resistance discovery
The increasing threat to human health from multidrug-resistant pathogens underscores the need to develop tools to interrogate microbiomes for novel antimicrobials and to understand and mitigate the evolution and transmission of resistance genes. A more complete understanding of antimicrobial resistance in the context of microbial communities will advance our treatment of antimicrobial-resistant infections and help us choose therapies that will reduce selection for resistance. Additionally, novel antimicrobials discovered within microbial communities will lay the groundwork for narrow-spectrum, next-generation antimicrobials that reduce collateral microbiome damage. Given that most clinical antimicrobial resistance genes originated in environmental microbes, and most antimicrobials are natural products or derivatives thereof, it is critical that these two microbial functions be studied in tandem and in the context of microbial communities in order to understand the dynamics governing the evolution and transmission of antimicrobial resistance and biosynthetic capacities.
Advances in sequencing technology will facilitate the discovery of complex functions from metagenomes on the basis of sequence alone. For example, long-read sequencing technologies, such as the NanoPore Minion and the PacBio SMRT, will help to overcome the challenges currently posed by metagenomic assembly from short-read data, providing greater information about functions encoded in large operons, such as biosynthetic and multidrug-resistance gene clusters.140,149,150 Furthermore, cheaper sequencing will allow for deeper sequencing, improving our ability to assemble metagenomes and profile antimicrobial resistance and biosynthetic capacities.83 Single-molecule sequencing, particularly when combined with short-read shotgun metagenomic data, may also improve metagenomic assembly and allow for better identification of biosynthetic gene clusters.147 In addition to progress resulting from improvements to DNA reading, advancements in DNA writing will likely benefit the field. Dramatic reductions in the cost and efficiency of DNA synthesis may soon make synthesis and recoding of entire biosynthetic gene clusters feasible, simplifying heterologous expression in genetically tractable hosts and downstream characterization of bioactive natural products.151,152 An integration of functional and genomic techniques, aided by these and other inevitable technical and computational developments, will help us better understand the delicate interplay between antimicrobial biosynthesis and resistance in microbiomes.
Acknowledgments
This work was supported in part by grants to G.D. from the NIH Director’s New Innovator Award (http://commonfund.nih.gov/newinnovator/), the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK: http://www.niddk.nih.gov/), the National Institute of General Medical Sciences (NIGMS: http://www.nigms.nih.gov/), and the National Institute of Allergy and Infectious Diseases (NIAID: https://www.niaid.nih.gov/) of the National Institutes of Health (NIH) under Award Numbers DP2DK098089, R01GM099538, and R01AI123394. B.A. is a National Science Foundation graduate research fellow (Award Number DGE-1143945). A.J.G. received support from a NIGMS training grant through Award Number T32 GM007067. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Footnotes
Conflicts of interest
The authors declare no conflicts of interest.
References
- 1.Lederberg J, McCray A. The Scientist : 'Ome Sweet 'Omics-- A Genealogical Treasury of Words. The Scientist. 2001;17 [Google Scholar]
- 2.Turnbaugh PJ, Ley RE, Hamady M, et al. The Human Microbiome Project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen HK, Donato J, Wang HH, et al. Call of the wild: antibiotic resistance genes in natural environments. Nat Rev Microbiol. 2010;8:251–259. doi: 10.1038/nrmicro2312. [DOI] [PubMed] [Google Scholar]
- 4.Sender R, Fuchs S, Milo R. Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans. Cell. 2016;164:337–340. doi: 10.1016/j.cell.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 5.Ursell LK, Metcalf JL, Parfrey LW, et al. Defining the human microbiome. Nutr Rev. 2012;70(Suppl 1):S38–S44. doi: 10.1111/j.1753-4887.2012.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roesch LFW, Fulthorpe RR, Riva A, et al. Pyrosequencing enumerates and contrasts soil microbial diversity. Isme j. 2007;1:283–290. doi: 10.1038/ismej.2007.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waldor MK, Tyson G, Borenstein E, et al. Where next for microbiome research? PLoS Biol. 2015;13:e1002050. doi: 10.1371/journal.pbio.1002050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donia MS, Cimermancic P, Schulze CJ, et al. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell. 2014;158:1402–1414. doi: 10.1016/j.cell.2014.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sommer MO, Church GM, Dantas G. The human microbiome harbors a diverse reservoir of antibiotic resistance genes. Virulence. 2010;1:299–303. doi: 10.4161/viru.1.4.12010. [DOI] [PubMed] [Google Scholar]
- 10.Perry JA, Westman EL, Wright GD. The antibiotic resistome: what's new? Curr Opin Microbiol. 2014;21:45–50. doi: 10.1016/j.mib.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 11.Sommer MO, Dantas G. Antibiotics and the resistant microbiome. Curr Opin Microbiol. 2011;14:556–563. doi: 10.1016/j.mib.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 12.D'Costa VM, McGrann KM, Hughes DW, et al. Sampling the antibiotic resistome. Science. 2006;311:374–377. doi: 10.1126/science.1120800. [DOI] [PubMed] [Google Scholar]
- 13.Wright GD. The antibiotic resistome: the nexus of chemical and genetic diversity. Nat Rev Micro. 2007;5:175–186. doi: 10.1038/nrmicro1614. [DOI] [PubMed] [Google Scholar]
- 14.O'Neill J. Securing new drugs for future generations: the pipeline of antibiotics. [Accesssed August 22, 2016]; http://amr-review.org/sites/default/files/SECURING%20NEW%20DRUGS%20FOR%20FUTURE%20GENERATIONS%20FINAL%20WEB_0.pdf. [Google Scholar]
- 15.Walsh CT, Wencewicz TA. Prospects for new antibiotics: a molecule-centered perspective. J Antibiot (Tokyo) 2014;67:7–22. doi: 10.1038/ja.2013.49. [DOI] [PubMed] [Google Scholar]
- 16.Lewis K. Platforms for antibiotic discovery. Nat Rev Drug Discov. 2013;12:371–387. doi: 10.1038/nrd3975. [DOI] [PubMed] [Google Scholar]
- 17.Knapp CW, Dolfing J, Ehlert PA, et al. Evidence of increasing antibiotic resistance gene abundances in archived soils since 1940. Environ Sci Technol. 2010;44:580–587. doi: 10.1021/es901221x. [DOI] [PubMed] [Google Scholar]
- 18.Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States, 2013. [Accessed August 22, 2016]; http://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf.
- 19.Cooper MA, Shlaes D. Fix the antibiotics pipeline. Nature. 2011;472:32. doi: 10.1038/472032a. [DOI] [PubMed] [Google Scholar]
- 20.Roberts RR, Hota B, Ahmad I, et al. Hospital and societal costs of antimicrobial-resistant infections in a Chicago teaching hospital: implications for antibiotic stewardship. Clin Infect Dis. 2009;49:1175–1184. doi: 10.1086/605630. [DOI] [PubMed] [Google Scholar]
- 21.O'Neill J. Tackling drug-resistant infections globally: Final report and recommendations [Google Scholar]
- 22.Fischbach MA, Walsh CT. Antibiotics for emerging pathogens. Science. 2009;325:1089–1093. doi: 10.1126/science.1176667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kinch MS, Patridge E, Plummer M, et al. An analysis of FDA-approved drugs for infectious disease: antibacterial agents. Drug Discov Today. 2014;19:1283–1287. doi: 10.1016/j.drudis.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 24.Howard DH, Scott RD. The Economic Burden of Drug Resistance. Clinical Infectious Diseases. 2005;41:S283–S286. doi: 10.1086/430792. [DOI] [PubMed] [Google Scholar]
- 25.Miller A. Antibacterial Development: a Changing Landscape. [Accessed August 22, 2016];Microbe Magazine. http://www.asmscience.org/content/journal/microbe/10.1128/microbe.11.111.1. [Google Scholar]
- 26.Tenover FC. Mechanisms of antimicrobial resistance in bacteria. Am J Infect Control. 2006;34:S3–S10. doi: 10.1016/j.ajic.2006.05.219. discussion S64–73. [DOI] [PubMed] [Google Scholar]
- 27.Walsh C. Molecular mechanisms that confer antibacterial drug resistance. Nature. 2000;406:775–781. doi: 10.1038/35021219. [DOI] [PubMed] [Google Scholar]
- 28.Alekshun MN, Levy SB. Molecular mechanisms of antibacterial multidrug resistance. Cell. 2007;128:1037–1050. doi: 10.1016/j.cell.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 29.MacLean RC, Hall AR, Perron GG, et al. The population genetics of antibiotic resistance: integrating molecular mechanisms and treatment contexts. Nat Rev Genet. 2010;11:405–414. doi: 10.1038/nrg2778. [DOI] [PubMed] [Google Scholar]
- 30.Baltz RH. Genomics and the ancient origins of the daptomycin biosynthetic gene cluster. J Antibiot (Tokyo) 2010;63:506–511. doi: 10.1038/ja.2010.82. [DOI] [PubMed] [Google Scholar]
- 31.Fajardo A, Martinez JL. Antibiotics as signals that trigger specific bacterial responses. Curr Opin Microbiol. 2008;11:161–167. doi: 10.1016/j.mib.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 32.Clardy J, Fischbach MA, Currie CR. The natural history of antibiotics. Curr Biol. 2009;19:R437–R441. doi: 10.1016/j.cub.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nelson ML, Dinardo A, Hochberg J, et al. Brief communication: Mass spectroscopic characterization of tetracycline in the skeletal remains of an ancient population from Sudanese Nubia 350–550 CE. Am J Phys Anthropol. 2010;143:151–154. doi: 10.1002/ajpa.21340. [DOI] [PubMed] [Google Scholar]
- 34.Bassett EJ, Keith MS, Armelagos GJ, et al. Tetracycline-labeled human bone from ancient Sudanese Nubia (A.D. 350) Science. 1980;209:1532–1534. doi: 10.1126/science.7001623. [DOI] [PubMed] [Google Scholar]
- 35.Aminov RI. A brief history of the antibiotic era: lessons learned and challenges for the future. Front Microbiol. 2010;1:134. doi: 10.3389/fmicb.2010.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones DS, Podolsky SH, Greene JA. The burden of disease and the changing task of medicine. N Engl J Med. 2012;366:2333–2338. doi: 10.1056/NEJMp1113569. [DOI] [PubMed] [Google Scholar]
- 37.Elsner HL. The new treatment of syphilis (ehrlich-hata): Observations and results. Journal of the American Medical Association. 1910;55:2052–2057. [Google Scholar]
- 38.Fleming A. On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzae. 1929. Bull World Health Organ. 2001;79:780–790. [PMC free article] [PubMed] [Google Scholar]
- 39.Schatz A, Bugie E, Waksman SA. Streptomycin, a substance exhibiting antibiotic activity against gram-positive and gram-negative bacteria. 1944. Clin Orthop Relat Res. 2005:3–6. doi: 10.1097/01.blo.0000175887.98112.fe. [DOI] [PubMed] [Google Scholar]
- 40.Zetterstrom R. Selman A. Waksman (1888–1973) Nobel Prize in 1952 for the discovery of streptomycin, the first antibiotic effective against tuberculosis. Acta Paediatr. 2007;96:317–319. doi: 10.1111/j.1651-2227.2007.00182.x. [DOI] [PubMed] [Google Scholar]
- 41.Brown ED, Wright GD. Antibacterial drug discovery in the resistance era. Nature. 2016;529:336–343. doi: 10.1038/nature17042. [DOI] [PubMed] [Google Scholar]
- 42.Pelaez F. The historical delivery of antibiotics from microbial natural products--can history repeat? Biochem Pharmacol. 2006;71:981–990. doi: 10.1016/j.bcp.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 43.Fernandes P. Antibacterial discovery and development--the failure of success? Nat Biotechnol. 2006;24:1497–1503. doi: 10.1038/nbt1206-1497. [DOI] [PubMed] [Google Scholar]
- 44.Zaffiri L, Gardner J, Toledo-Pereyra LH. History of antibiotics. From salvarsan to cephalosporins. J Invest Surg. 2012;25:67–77. doi: 10.3109/08941939.2012.664099. [DOI] [PubMed] [Google Scholar]
- 45.Walsh C. Antibiotics : actions, origins, resistance. Washington, D.C: ASM Press; 2003. [Google Scholar]
- 46.Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de la Cruz F, Davies J. Horizontal gene transfer and the origin of species: lessons from bacteria. Trends Microbiol. 2000;8:128–133. doi: 10.1016/s0966-842x(00)01703-0. [DOI] [PubMed] [Google Scholar]
- 48.Blair JM, Webber MA, Baylay AJ, et al. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol. 2015;13:42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- 49.Roberts MC. Tetracycline resistance determinants: mechanisms of action, regulation of expression, genetic mobility, and distribution. FEMS Microbiol Rev. 1996;19:1–24. doi: 10.1111/j.1574-6976.1996.tb00251.x. [DOI] [PubMed] [Google Scholar]
- 50.Ogawa W, Onishi M, Ni R, et al. Functional study of the novel multidrug efflux pump KexD from Klebsiella pneumoniae. Gene. 2012;498:177–182. doi: 10.1016/j.gene.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 51.Ruggerone P, Murakami S, Pos KM, et al. RND efflux pumps: structural information translated into function and inhibition mechanisms. Curr Top Med Chem. 2013;13:3079–3100. doi: 10.2174/15680266113136660220. [DOI] [PubMed] [Google Scholar]
- 52.Hinchliffe P, Symmons MF, Hughes C, et al. Structure and operation of bacterial tripartite pumps. Annu Rev Microbiol. 2013;67:221–242. doi: 10.1146/annurev-micro-092412-155718. [DOI] [PubMed] [Google Scholar]
- 53.Piddock LJ. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin Microbiol Rev. 2006;19:382–402. doi: 10.1128/CMR.19.2.382-402.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tamber S, Hancock RE. On the mechanism of solute uptake in Pseudomonas. Front Biosci. 2003;8:s472–s483. doi: 10.2741/1075. [DOI] [PubMed] [Google Scholar]
- 55.Poulou A, Voulgari E, Vrioni G, et al. Outbreak caused by an ertapenem-resistant, CTX-M-15-producing Klebsiella pneumoniae sequence type 101 clone carrying an OmpK36 porin variant. J Clin Microbiol. 2013;51:3176–3182. doi: 10.1128/JCM.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Katayama Y, Ito T, Hiramatsu K. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2000;44:1549–1555. doi: 10.1128/aac.44.6.1549-1555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Neu HC. Effect of beta-lactamase location in Escherichia coli on penicillin synergy. Appl Microbiol. 1969;17:783–786. doi: 10.1128/am.17.6.783-786.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bush K. Bench-to-bedside review: The role of beta-lactamases in antibiotic-resistant Gram-negative infections. Crit Care. 2010;14:224. doi: 10.1186/cc8892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang W, Moore IF, Koteva KP, et al. TetX is a flavin-dependent monooxygenase conferring resistance to tetracycline antibiotics. J Biol Chem. 2004;279:52346–52352. doi: 10.1074/jbc.M409573200. [DOI] [PubMed] [Google Scholar]
- 60.Forsberg KJ, Patel S, Wencewicz TA, et al. The Tetracycline Destructases: A Novel Family of Tetracycline-Inactivating Enzymes. Chem Biol. 2015;22:888–897. doi: 10.1016/j.chembiol.2015.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shaw WV, Packman LC, Burleigh BD, et al. Primary structure of a chloramphenicol acetyltransferase specified by R plasmids. Nature. 1979;282:870–872. doi: 10.1038/282870a0. [DOI] [PubMed] [Google Scholar]
- 62.D'Costa VM, King CE, Kalan L, et al. Antibiotic resistance is ancient. Nature. 2011;477:457–461. doi: 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]
- 63.Bhullar K, Waglechner N, Pawlowski A, et al. Antibiotic resistance is prevalent in an isolated cave microbiome. PLoS One. 2012;7:e34953. doi: 10.1371/journal.pone.0034953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Palmer AC, Angelino E, Kishony R. Chemical decay of an antibiotic inverts selection for resistance. Nat Chem Biol. 2010;6:105–107. doi: 10.1038/nchembio.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martinez JL. Natural antibiotic resistance and contamination by antibiotic resistance determinants: the two ages in the evolution of resistance to antimicrobials. Front Microbiol. 2012;3:1. doi: 10.3389/fmicb.2012.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abraham EP, Chain E. An enzyme from bacteria able to destroy penicillin. 1940. Rev Infect Dis. 1988;10:677–678. [PubMed] [Google Scholar]
- 67.Hall BG, Salipante SJ, Barlow M. Independent origins of subgroup Bl + B2 and subgroup B3 metallo-beta-lactamases. J Mol Evol. 2004;59:133–141. doi: 10.1007/s00239-003-2572-9. [DOI] [PubMed] [Google Scholar]
- 68.Hall BG, Barlow M. Evolution of the serine beta-lactamases: past, present and future. Drug Resist Updat. 2004;7:111–123. doi: 10.1016/j.drup.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 69.Stokes HW, Gillings MR. Gene flow, mobile genetic elements and the recruitment of antibiotic resistance genes into Gram-negative pathogens. FEMS Microbiol Rev. 2011;35:790–819. doi: 10.1111/j.1574-6976.2011.00273.x. [DOI] [PubMed] [Google Scholar]
- 70.Gould SJ, Vrba ES. Exaptation—a Missing Term in the Science of Form. Paleobiology. 1982;8:4–15. [Google Scholar]
- 71.Sommer MO, Dantas G, Church GM. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science. 2009;325:1128–1131. doi: 10.1126/science.1176950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Forsberg KJ, Reyes A, Wang B, et al. The shared antibiotic resistome of soil bacteria and human pathogens. Science. 2012;337:1107–1111. doi: 10.1126/science.1220761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.D'Costa VM, Griffiths E, Wright GD. Expanding the soil antibiotic resistome: exploring environmental diversity. Curr Opin Microbiol. 2007;10:481–489. doi: 10.1016/j.mib.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 74.Didelot X, Bowden R, Wilson DJ, et al. Transforming clinical microbiology with bacterial genome sequencing. Nat Rev Genet. 2012;13:601–612. doi: 10.1038/nrg3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kwong JC, McCallum N, Sintchenko V, et al. Whole genome sequencing in clinical and public health microbiology. Pathology. 2015;47:199–210. doi: 10.1097/PAT.0000000000000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Koser CU, Bryant JM, Becq J, et al. Whole-genome sequencing for rapid susceptibility testing of M. tuberculosis. N Engl J Med. 2013;369:290–292. doi: 10.1056/NEJMc1215305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li L, Mendis N, Trigui H, et al. The importance of the viable but non-culturable state in human bacterial pathogens. Front Microbiol. 2014;5:258. doi: 10.3389/fmicb.2014.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ma L, Kim J, Hatzenpichler R, et al. Gene-targeted microfluidic cultivation validated by isolation of a gut bacterium listed in Human Microbiome Project's Most Wanted taxa. Proc Natl Acad Sci U S A. 2014;111:9768–9773. doi: 10.1073/pnas.1404753111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nichols D, Cahoon N, Trakhtenberg EM, et al. Use of Ichip for High-Throughput In Situ Cultivation of “Uncultivable” Microbial Species. Appl Environ Microbiol. 2010;76:2445–2450. doi: 10.1128/AEM.01754-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kaeberlein T, Lewis K, Epstein SS. Isolating "uncultivable" microorganisms in pure culture in a simulated natural environment. Science. 2002;296:1127–1129. doi: 10.1126/science.1070633. [DOI] [PubMed] [Google Scholar]
- 81.Lagier JC, Armougom F, Million M, et al. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect. 2012;18:1185–1193. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- 82.McLain JE, Cytryn E, Durso LM, et al. Culture-based Methods for Detection of Antibiotic Resistance in Agroecosystems: Advantages, Challenges, and Gaps in Knowledge. J Environ Qual. 2016;45:432–440. doi: 10.2134/jeq2015.06.0317. [DOI] [PubMed] [Google Scholar]
- 83.Wetterstrand KA. DNA Sequencing Costs: Data from the NHGRI Genome Sequencing Program (GSP) [Accessed August 22, 2016]; www.genome.gov/sequencingcostsdata.
- 84.Pettersson E, Lundeberg J, Ahmadian A. Generations of sequencing technologies. Genomics. 2009;93:105–111. doi: 10.1016/j.ygeno.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 85.Bonetta L. Whole-genome sequencing breaks the cost barrier. Cell. 2010;141:917–919. doi: 10.1016/j.cell.2010.05.034. [DOI] [PubMed] [Google Scholar]
- 86.Niedringhaus TP, Milanova D, Kerby MB, et al. Landscape of next-generation sequencing technologies. Anal Chem. 2011;83:4327–4341. doi: 10.1021/ac2010857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Christensen KD, Dukhovny D, Siebert U, et al. Assessing the Costs and Cost-Effectiveness of Genomic Sequencing. J Pers Med. 2015;5:470–486. doi: 10.3390/jpm5040470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mardis ER. Next-generation DNA sequencing methods. Annu Rev Genomics Hum Genet. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- 89.Koboldt DC, Steinberg KM, Larson DE, et al. The next-generation sequencing revolution and its impact on genomics. Cell. 2013;155:27–38. doi: 10.1016/j.cell.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pehrsson EC, Forsberg KJ, Gibson MK, et al. Novel resistance functions uncovered using functional metagenomic investigations of resistance reservoirs. Front Microbiol. 2013;4:145. doi: 10.3389/fmicb.2013.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Allen HK, Moe LA, Rodbumrer J, et al. Functional metagenomics reveals diverse beta-lactamases in a remote Alaskan soil. Isme j. 2009;3:243–251. doi: 10.1038/ismej.2008.86. [DOI] [PubMed] [Google Scholar]
- 92.Moore AM, Patel S, Forsberg KJ, et al. Pediatric fecal microbiota harbor diverse and novel antibiotic resistance genes. PLoS One. 2013;8:e78822. doi: 10.1371/journal.pone.0078822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Moore AM, Munck C, Sommer MO, et al. Functional metagenomic investigations of the human intestinal microbiota. Front Microbiol. 2011;2:188. doi: 10.3389/fmicb.2011.00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Perron GG, Whyte L, Turnbaugh PJ, et al. Functional characterization of bacteria isolated from ancient arctic soil exposes diverse resistance mechanisms to modern antibiotics. PLoS One. 2015;10:e0069533. doi: 10.1371/journal.pone.0069533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Clemente JC, Pehrsson EC, Blaser MJ, et al. The microbiome of uncontacted Amerindians. Sci Adv. 2015;1 doi: 10.1126/sciadv.1500183. pii: e1500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pehrsson EC, Tsukayama P, Patel S, et al. Interconnected microbiomes and resistomes in low-income human habitats. Nature. 2016;533:212–216. doi: 10.1038/nature17672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lam KN, Cheng J, Engel K, et al. Current and future resources for functional metagenomics. Front Microbiol. 2015;6:1196. doi: 10.3389/fmicb.2015.01196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schmieder R, Edwards R. Insights into antibiotic resistance through metagenomic approaches. Future Microbiol. 2012;7:73–89. doi: 10.2217/fmb.11.135. [DOI] [PubMed] [Google Scholar]
- 99.Forsberg KJ, Patel S, Gibson MK, et al. Bacterial phylogeny structures soil resistomes across habitats. Nature. 2014;509:612–616. doi: 10.1038/nature13377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mezger A, Gullberg E, Goransson J, et al. A general method for rapid determination of antibiotic susceptibility and species in bacterial infections. J Clin Microbiol. 2015;53:425–432. doi: 10.1128/JCM.02434-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gibson MK, Wang B, Ahmadi S, et al. Developmental dynamics of the preterm infant gut microbiota and antibiotic resistome. Nature Microbiology. 2016;1:16024. doi: 10.1038/nmicrobiol.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Buelow E, Gonzalez TB, Versluis D, et al. Effects of selective digestive decontamination (SDD) on the gut resistome. J Antimicrob Chemother. 2014;69:2215–2223. doi: 10.1093/jac/dku092. [DOI] [PubMed] [Google Scholar]
- 103.Bengtsson-Palme J, Boulund F, Fick J, et al. Shotgun metagenomics reveals a wide array of antibiotic resistance genes and mobile elements in a polluted lake in India. Front Microbiol. 2014;5:648. doi: 10.3389/fmicb.2014.00648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Meyer F, Paarmann D, D'Souza M, et al. The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Segata N, Waldron L, Ballarini A, et al. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods. 2012;9:811–814. doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 107.Hu Y, Yang X, Qin J, et al. Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota. Nat Commun. 2013;4:2151. doi: 10.1038/ncomms3151. [DOI] [PubMed] [Google Scholar]
- 108.Forslund K, Sunagawa S, Kultima JR, et al. Country-specific antibiotic use practices impact the human gut resistome. Genome Res. 2013;23:1163–1169. doi: 10.1101/gr.155465.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Durso LM, Harhay GP, Bono JL, et al. Virulence-associated and antibiotic resistance genes of microbial populations in cattle feces analyzed using a metagenomic approach. J Microbiol Methods. 2011;84:278–282. doi: 10.1016/j.mimet.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 110.Ma L, Li B, Zhang T. Abundant rifampin resistance genes and significant correlations of antibiotic resistance genes and plasmids in various environments revealed by metagenomic analysis. Appl Microbiol Biotechnol. 2014;98:5195–5204. doi: 10.1007/s00253-014-5511-3. [DOI] [PubMed] [Google Scholar]
- 111.Abeles SR, Ly M, Santiago-Rodriguez TM, et al. Effects of Long Term Antibiotic Therapy on Human Oral and Fecal Viromes. PLoS One. 2015;10:e0134941. doi: 10.1371/journal.pone.0134941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gibson MK, Forsberg KJ, Dantas G. Improved annotation of antibiotic resistance determinants reveals microbial resistomes cluster by ecology. ISME J. 2015;9:207–216. doi: 10.1038/ismej.2014.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Xavier BB, Das AJ, Cochrane G, et al. Consolidating and Exploring Antibiotic Resistance Gene Data Resources. J Clin Microbiol. 2016;54:851–859. doi: 10.1128/JCM.02717-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liu B, Pop M. ARDB--Antibiotic Resistance Genes Database. Nucleic Acids Res. 2009;37:D443–D447. doi: 10.1093/nar/gkn656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Looft T, Johnson TA, Allen HK, et al. In-feed antibiotic effects on the swine intestinal microbiome. Proc Natl Acad Sci U S A. 2012;109:1691–1696. doi: 10.1073/pnas.1120238109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nesme J, Cecillon S, Delmont TO, et al. Large-scale metagenomic-based study of antibiotic resistance in the environment. Curr Biol. 2014;24:1096–1100. doi: 10.1016/j.cub.2014.03.036. [DOI] [PubMed] [Google Scholar]
- 117.Modi SR, Lee HH, Spina CS, et al. Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature. 2013;499:219–222. doi: 10.1038/nature12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Smillie CS, Smith MB, Friedman J, et al. Ecology drives a global network of gene exchange connecting the human microbiome. Nature. 2011;480:241–244. doi: 10.1038/nature10571. [DOI] [PubMed] [Google Scholar]
- 119.McArthur AG, Waglechner N, Nizam F, et al. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother. 2013;57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gupta SK, Padmanabhan BR, Diene SM, et al. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother. 2014;58:212–220. doi: 10.1128/AAC.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yamashita A, Sekizuka T, Kuroda M. Characterization of Antimicrobial Resistance Dissemination across Plasmid Communities Classified by Network Analysis. Pathogens. 2014;3:356–376. doi: 10.3390/pathogens3020356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.McCall CA, Bent E, Jorgensen TS, et al. Metagenomic Comparison of Antibiotic Resistance Genes Associated with Liquid and Dewatered Biosolids. J Environ Qual. 2016;45:463–470. doi: 10.2134/jeq2015.05.0255. [DOI] [PubMed] [Google Scholar]
- 123.Thai QK, Bos F, Pleiss J. The Lactamase Engineering Database: a critical survey of TEM sequences in public databases. BMC Genomics. 2009;10:390. doi: 10.1186/1471-2164-10-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fischer M, Thai QK, Grieb M, et al. DWARF--a data warehouse system for analyzing protein families. BMC Bioinformatics. 2006;7:495. doi: 10.1186/1471-2105-7-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tsafnat G, Copty J, Partridge SR. RAC: Repository of Antibiotic resistance Cassettes. Database (Oxford) 2011;2011:bar054. doi: 10.1093/database/bar054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zankari E, Hasman H, Cosentino S, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.McArthur AG, Wright GD. Bioinformatics of antimicrobial resistance in the age of molecular epidemiology. Curr Opin Microbiol. 2015;27:45–50. doi: 10.1016/j.mib.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 128.Lim HK, Chung EJ, Kim JC, et al. Characterization of a forest soil metagenome clone that confers indirubin and indigo production on Escherichia coli. Appl Environ Microbiol. 2005;71:7768–7777. doi: 10.1128/AEM.71.12.7768-7777.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Brady SF, Clardy J. Long-Chain N-Acyl Amino Acid Antibiotics Isolated from Heterologously Expressed Environmental DNA. Journal of the American Chemical Society. 2000;122:12903–12904. [Google Scholar]
- 130.Banik JJ, Brady SF. Recent application of metagenomic approaches toward the discovery of antimicrobials and other bioactive small molecules. Curr Opin Microbiol. 2010;13:603–609. doi: 10.1016/j.mib.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Katz M, Hover BM, Brady SF. Culture-independent discovery of natural products from soil metagenomes. J Ind Microbiol Biotechnol. 2016;43:129–141. doi: 10.1007/s10295-015-1706-6. [DOI] [PubMed] [Google Scholar]
- 132.Stevens DC, Conway KR, Pearce N, et al. Alternative Sigma Factor Over-Expression Enables Heterologous Expression of a Type II Polyketide Biosynthetic Pathway in Escherichia coli. PLoS One. 2013;8:e64858. doi: 10.1371/journal.pone.0064858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.McMahon MD, Guan C, Handelsman J, et al. Metagenomic analysis of Streptomyces lividans reveals host-dependent functional expression. Appl Environ Microbiol. 2012;78:3622–3629. doi: 10.1128/AEM.00044-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lee MH, Lee SW. Bioprospecting potential of the soil metagenome: novel enzymes and bioactivities. Genomics Inform. 2013;11:114–120. doi: 10.5808/GI.2013.11.3.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ling LL, Schneider T, Peoples AJ, et al. A new antibiotic kills pathogens without detectable resistance. Nature. 2015;517:455–459. doi: 10.1038/nature14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Seow KT, Meurer G, Gerlitz M, et al. A study of iterative type II polyketide synthases, using bacterial genes cloned from soil DNA: a means to access and use genes from uncultured microorganisms. J Bacteriol. 1997;179:7360–7368. doi: 10.1128/jb.179.23.7360-7368.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Weber T, Blin K, Duddela S, et al. antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015;43:W237–W243. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Cimermancic P, Medema MH, Claesen J, et al. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell. 2014;158:412–421. doi: 10.1016/j.cell.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Raveh-Sadka T, Thomas BC, Singh A, et al. Gut bacteria are rarely shared by co-hospitalized premature infants, regardless of necrotizing enterocolitis development. Elife. 2015;4 doi: 10.7554/eLife.05477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Frank JA, Pan Y, Tooming-Klunderud A, et al. Improved metagenome assemblies and taxonomic binning using long-read circular consensus sequence data. Sci Rep. 2016;6:25373. doi: 10.1038/srep25373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sangwan N, Xia F, Gilbert JA. Recovering complete and draft population genomes from metagenome datasets. Microbiome. 2016;4:8. doi: 10.1186/s40168-016-0154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kaminski J, Gibson MK, Franzosa EA, et al. High-Specificity Targeted Functional Profiling in Microbial Communities with ShortBRED. PLoS Comput Biol. 2015;11:e1004557. doi: 10.1371/journal.pcbi.1004557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Owen JG, Charlop-Powers Z, Smith AG, et al. Multiplexed metagenome mining using short DNA sequence tags facilitates targeted discovery of epoxyketone proteasome inhibitors. Proc Natl Acad Sci U S A. 2015;112:4221–4226. doi: 10.1073/pnas.1501124112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Reddy BV, Milshteyn A, Charlop-Powers Z, et al. eSNaPD: a versatile, web-based bioinformatics platform for surveying and mining natural product biosynthetic diversity from metagenomes. Chem Biol. 2014;21:1023–1033. doi: 10.1016/j.chembiol.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Reddy BV, Kallifidas D, Kim JH, et al. Natural product biosynthetic gene diversity in geographically distinct soil microbiomes. Appl Environ Microbiol. 2012;78:3744–3752. doi: 10.1128/AEM.00102-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Eid J, Fehr A, Gray J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 147.Tsai YC, Conlan S, Deming C, et al. Resolving the Complexity of Human Skin Metagenomes Using Single-Molecule Sequencing. MBio. 2016;7:e01948–e01915. doi: 10.1128/mBio.01948-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Oppegard C, Rogne P, Emanuelsen L, et al. The two-peptide class II bacteriocins: structure, production, and mode of action. J Mol Microbiol Biotechnol. 2007;13:210–219. doi: 10.1159/000104750. [DOI] [PubMed] [Google Scholar]
- 149.Koren S, Harhay GP, Smith TP, et al. Reducing assembly complexity of microbial genomes with single-molecule sequencing. Genome Biol. 2013;14:R101. doi: 10.1186/gb-2013-14-9-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gomez-Escribano JP, Alt S, Bibb MJ. Next Generation Sequencing of Actinobacteria for the Discovery of Novel Natural Products. Mar Drugs. 2016;14 doi: 10.3390/md14040078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Meyer QC, Burton SG, Cowan DA. Subtractive hybridization magnetic bead capture: a new technique for the recovery of full-length ORFs from the metagenome. Biotechnol J. 2007;2:36–40. doi: 10.1002/biot.200600156. [DOI] [PubMed] [Google Scholar]
- 152.Kosuri S, Church GM. Large-scale de novo DNA synthesis: technologies and applications. Nat Methods. 2014;11:499–507. doi: 10.1038/nmeth.2918. [DOI] [PMC free article] [PubMed] [Google Scholar]