Abstract
Rhazya stricta is a unique medicinal plant source for many indole alkaloids, non-alkaloids, flavonoids, triterpenes and other unknown molecules with tremendous potential for therapeutic applications against many diseases. In the present article, we generated computational data on predictive properties and activity across two key therapeutic areas of cancer and obesity, and corresponding cheminformatics studies were carried out to examine druggable properties of these alkaloids. Computed physiochemical properties of the 78 indole alkaloids from R. stricta plant using industry-standard scientific molecular modeling software and their predictive anti-cancer activities from reliable web-source technologies indicate their plausible therapeutic applications. Their predictive ADME properties are further indicative of their drug-like-ness. We believe that the top-ranked molecules with anti-cancer activity are clearly amenable to chemical modifications for creating potent, safe and efficacious compounds with the feasibility of generating new chemical entities after pre-clinical and clinical studies.
Keywords: Rhazya stricta, Alkaloids, Physiochemical properties, Druggability, Anticancer molecules, Anti-obesity molecules
Background
Rhazya stricta Decsne (Apocynaceae family), a traditional herbal medicinal plant from Western and South Asia, has been shown to have multiple pharmacological effects due to the presence of over 100 alkaloids [1–3]. The chemical constituents of this plant (R. stricta) may possess biological activities of antifungal, antimicrobial, antioxidant, CNS, hypertension, metabolic, and inflammatory disorders. Rhazimine, an alkaloid isolated from R. stricta leaves, was shown to affect arachidonic acid metabolism in human blood [4]. This alkaloid was shown to be a dual and selective inhibitor of platelet activating factor (PAF)-induced platelet aggregation and arachidonic acid metabolism. Other effects of the lyophilized extract of R. stricta include an antispasmodic effect in rat muscles [5]. In another study, antioxidant effects were observed at higher doses, and it reduced the hepatic and renal concentrations of glutathione (GSH) and increased the ascorbic acid levels, whereas the degree of lipid peroxidation was reduced [6]. A recent study has shown that the basic alkaloid fraction from R. stricta significantly induces one of the chemopreventive enzyme-Nqo 1, through an Nrf 2-dependent mechanism, thereby establishing its role as an anti-tumor agent [7]. In another pharmacological study, the biochemical parameters including blood lipid profile concentrations, liver enzyme activities and kidney functions were analyzed in rats [8]. It was also found that aqueous extract of R. stricta and indole alkaloids caused a significant increase in serum adiponectin levels and resulted in significant improvements in insulin resistance [9]. In another follow up study, we observed indole-alkaloids of R. stricta improved not only the lipid profile and liver function but also led to improvements in the insulin levels in rats, most likely via modulating insulin resistance [10]. Indole-alkaloids of R. stricta had been reported to have anticancer properties [11]. Other studies by our departmental colleagues showed that alkaloid extract of R. stricta leaves inhibited proliferation, colony formation and anchorage-independent growth in various cancer cell lines such as colon cancer, breast cancer and lung cancer [12–14].
Understanding the chemical structure, physiochemical, and chemical-informatic properties of these natural product compounds will give clues for further modifications required in their structures responsible for their biological activities. Even though, there have been about 100 chemical entities of indole-based alkaloid constituents of R. stricta which have been reported but their chemical structures are yet to be clustered and identified, and moreover the pharmacological application of any one of these constituents towards human health is yet to be identified. Understanding qualitative correlation of structures to their chemical druggability, IP potential, and their applicability towards a therapeutic area would be worth exploring prior to pre-clinical studies. Availability of this plant (R. stricta), thus its phytochemical constituents largely in Arabian and South Asian region makes it worth studying through computational, synthetic, and biological view point. Indole based alkaloids such as vinblastine and vincristine are well known for their anti-cancer properties. From systematically generated informatics data analysis, one would be able to evaluate the physiochemical properties of the potential therapeutic compounds. These promising molecules which have “desirable pharmacophores” may provide obvious extension to a better targeted therapeutic benefit. Conventional drugs obey set of rules such as Lipinski’s Rule-of-Five (RO5) [15], wherein all orally administered molecules need to have certain physiochemical properties. Calculation of these cheminformatic properties has thus become essential for all projects of new drug discovery which go through oral route of administration. Along with RO5, the new molecules also have to adhere to certain parameters which yield favorable ADMET outcome of an oral drug. We further evaluated these molecules for therapeutic activity, including anticancer, anti-obesity, anti-inflammatory, and anti-bacterial properties. Although these predictions are indicative only, the value of predictions in various target classes and therapeutic areas would be very useful for future experimental studies. Moreover, their metabolic fate with key enzymes such as P450’s is also predicted for probable drug–drug and drug-target (P450) interactions (reviewed in [16, 17]).
Methods
For prediction of various therapeutic potential of these molecules, commercially and publicly available technologies as below were utilized.
PharmaExpert (http://www.pharmaexpert.ru)—PASS [18]
Superpred (http://prediction.charite.de)—Predictive Targets [19]
SwissTargetPrediction (http://www.swisstargetprediction.ch)—Predictive Target [20]
CDRUG (http://bsb.kiz.ac.cn/CDRUG)—Anti-cancer activity [21]
Schrodinger [22], a scientific software that predicts drug-like properties and liabilities (viz. HERG and CNS), and ACD/Labs [23] for physiochemical and cheminformatics studies were utilized. Details of the molecules, names, structures were obtained from the literature, commercial sources, and knowledge-based web sources. Tables 1 and 2 gives the details of these molecules together with their 2D SMILES notation, respectively.
Table 1.
Chemical structures and names of Rhazya stricta compounds
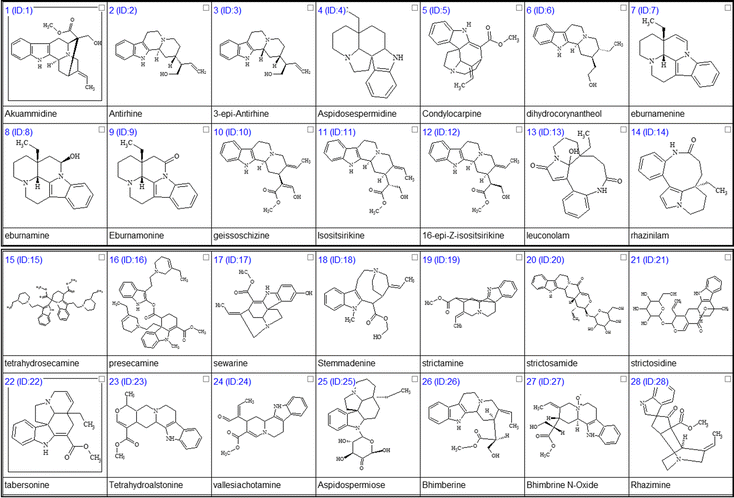
| |
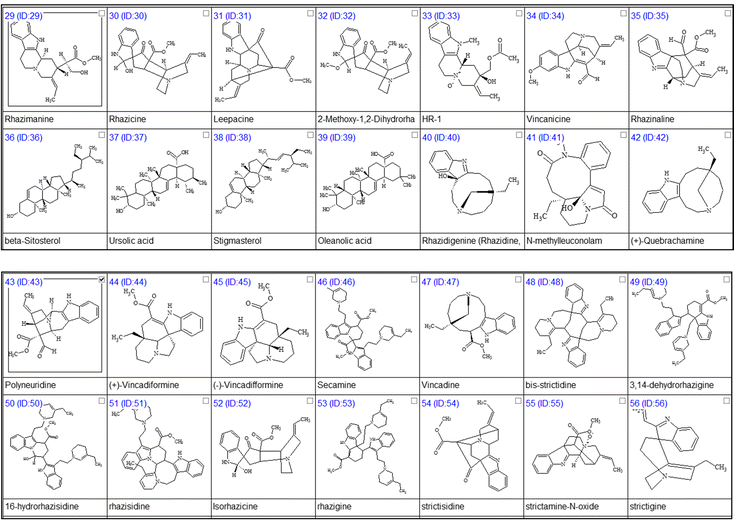
| |
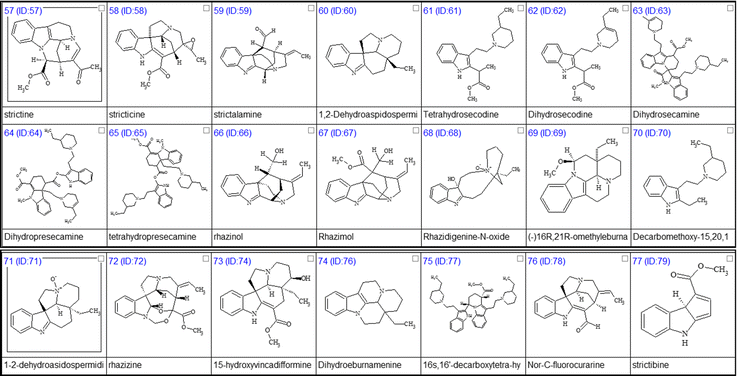
|
Table 2.
SMILES codes for Rhazya stricta compounds
| MOL ID | Name | SMILES code |
|---|---|---|
| M1 | Akummidine | COC(=O)C1(CO)C2CC3=C([NH]C4=C3C=CC=C4)C5CC1\C(CN25)=C/C |
| M2 | Antirhine | OCC(C=C)C1CCN2CCC3=C([NH]C4=C3C=CC=C4)C2C1 |
| M3 | 3-Epi-antirhine | OCC(C=C)C1CCN2CCC3=C([NH]C4=C3C=CC=C4)C2C1 |
| M4 | Aspidosespermidine | CCC12CCCN3CCC4(C(CC1)NC5=C4C=CC=C5)C23 |
| M5 | Condylocarpine | COC(=O)C1=C2NC3=CC=CC=C3C24CCN5CCC1\C(=C\C)C45 |
| M6 | Dihydrocorynantheol | CCC1CN2CCC3=C([NH]C4=CC=CC=C34)C2CC1CCO |
| M7 | Eburnamenine | CCC12CCCN3CCC4=C(C13)[N](C=C2)C5=CC=CC=C45 |
| M8 | Eburnamine | CCC12CCCN3CCC4C(C13)[N](C(O)C2)C5=CC=CC=C45 |
| M9 | Eburnamonine | CCC12CCCN3CCC4=C(C13)[N](C(=O)C2)C5=CC=CC=C45 |
| M10 | Geissoschizine | COC(=O)\C(=C/O)C\1CC2N(CCC3=C2[NH]C4=CC=CC=C34)CC1=C\C |
| M11 | Isositsirikine | COC(=O)C(CO)C\1CC2N(CCC3=C2[NH]C4=CC=CC=C34)CC1=C/C |
| M12 | 16-Epi-Z-isositsirikine | COC(=O)C(CO)C\1CC2N(CCC3=C2[NH]C4=CC=CC=C34)CC1=C\C |
| M13 | Leuconalm | CCC12CCCN3C(=O)C=C(C4=CC=CC=C4NC(=O)CC1)C23O |
| M14 | Rhazinliam | CCC12CCC[N]3C=CC(=C13)C4=CC=CC=C4NC(=O)CC2 |
| M15 | Tetrahydrosecamine | CCC1CCCN(CCC2=C([NH]C3=CC=CC=C23)C4(CCC(C(=O)OC)C5=C(CCN6CCCC(CC)C6)C7=C C=CC=C7[N]45)C(=O)OC)C1 |
| M16 | Presecamine | CCC1=CCCN(CCC2=C([NH]C3=CC=CC=C23)OC(=O)C4CCC(=C5N(C)C6=C C=CC=C6C45C CN7CCC=C(CC)C7)C(=O)OC)C1 |
| M17 | Sewarine | COC(=O)C1=C2NC3=C(C=C(O)C=C3)C24CCN5C\C(=C\C)C1CC45 |
| M18 | Stemmadenine | C\C=C1/CN2CCC1C(C(=O)OCO)C3=C(CC2)C4=CC=CC=C4[N]3C |
| M19 | Strictamine | COC(=O)C1C\2CC3N(CCC14C3=NC5=CC=CC=C45)CC2=C\C |
| M20 | Strictosamide | OCC1OC(OC2OC=C3C(CC4N(CCC5=C4[NH]C6=CC=CC=C56)C3=O)C2C=C)C(O)C(O)C1O |
| M21 | Strictosidine | COC(=O)C1=COC(OC2OC(CO)C(O)C(O)C2O)C(C=C)C1CC3NCCC4=C3[NH]C5=CC=CC=C45 |
| M22 | Taberonine | CCC12CC(=C3NC4=CC=CC=C4C35CCN(CC=C1)C25)C(=O)OC |
| M23 | Tetrahydrlstonine | COC(=O)C1=COC(C)C2CN3CCC4=C([NH]C5=CC=CC=C45)C3CC12 |
| M24 | Vallesiachotamine | COC(=O)C1=CN2CCC3=C([NH]C4=CC=CC=C34)C2CC1\C(=C/C)C=O |
| M25 | Aspidospermoise | CCC12CCCN3CCC4(C(CC1)N(C5OC(O)C(=O)C(O)C5O)C6=CC=CC=C46)C23 |
| M26 | Bhimbrine | COC(=O)C(CO)C\1CC2N(CCC3=C2[NH]C4=C3C=CC=C4)CC1=C/C |
| M27 | Bhimbrine N-oxide | COC(=O)C(CO)C\1CC2C3=C(CC[N+]2([O-])CC1=C/C)C4=C([NH]3)C=CC=C4 |
| M28 | Rhazimine | COC(=O)C12C(CC3(C=NC4=CC=CC=C34)C1=O)N5CCC2\C(C5)=C/C |
| M29 | Rhazimanine | COC(=O)C(CO)C\1CC2N(CCC3=C2[NH]C4=CC=CC=C34)CC1=C\C |
| M30 | Rhazicine | COC(=O)C12C(CC3(C(O)NC4=CC=CC=C34)C1=O)N5CCC2\C(C5)=C\C |
| M31 | Leepacine | COC(=O)C12C3CC4(C(NC5=CC=CC=C45)C6CC1\C(CN36)=C/C)C2=O |
| M32 | 2-Methoxy 1-2,dihydrorhazamine | COC1NC2=CC=CC=C2C13CC4N5CCC(\C(C5)=C/C)C4(C(=O)OC)C3=O |
| M33 | HR-1 | C\C=C1\C[N+]2([O-])CCC3=C(C2CC1(O)COC(C)=O)[N](C)C4=CC=CC=C34 |
| M34 | Vincanicine | COC1=CC=C2C(=C1)NC3=C(C=O)C\4CC5N(CCC235)CC4=C\C |
| M35 | Rhazinaline | COC(=O)C1(C=O)C\2CC3N(CCC14C3=NC5=CC=CC=C45)CC2=C/C |
| M36 | Beta-sitosterol | CCC(CCC(C)C1CCC2C3CC=C4CC(O)CCC4(C)C3CCC12C)C(C)C |
| M37 | Ursolic acid | CC1CCC2(CCC3(C)C(=CCC4C5(C)CCC(O)C(C)(C)C5CCC34C)C2C1C)C(O)=O |
| M38 | Stigmasterol | CCC(\C=C\C(C)C1CCC2C3CC=C4CC(O)CCC4(C)C3CCC12C)C(C)C |
| M39 | Olenaolic acid | CC1(C)CCC2(CCC3(C)C(=CCC4C5(C)CCC(O)C(C)(C)C5CCC34C)C2C1)C(O)=O |
| M40 | Rhazidigenine (rhazidine) | CCC12CCCN(CCC3(O)C(=NC4=CC=CC=C34)CC1)C2 |
| M41 | N-methylleuconolam | CCC12CCCN3C(=O)C=C(C4=CC=CC=C4N(C)C(=O)CC1)C23O |
| M42 | (+)-Quebranchamine | CCC12CCCN(CCC3=C(CC1)[NH]C4=CC=CC=C34)C2 |
| M43 | Polyneuridine | COC(=O)C1(C=O)C2CC3=C([NH]C4=CC=CC=C34)C5CC1\C(CN25)=C\C |
| M44 | (+)-Vincadiformine | CCC12CCCN3CCC4(C13)C(=C(C2)C(=O)OC)NC5=CC=CC=C45 |
| M45 | (−)-Vincadiformine | CCC12CCCN3CCC4(C13)C(=C(C2)C(=O)OC)NC5=CC=CC=C45 |
| M46 | Secamine | CCC1=CCCN(CCC2=C([NH]C3=C2C=CC=C3)C4(CCC(C(=O)OC)C5=C (CCN6CCC=C(CC) C6)C7=CC=CC=C7[N]45)C(=O)OC)C1 |
| M47 | Vincadine | CCC12CCCN(CCC3=C([NH]C4=CC=CC=C34)C(C1)C(=O)OC)C2 |
| M48 | Bis-strictidine | CCC1=C2C3CC4(CCN2CCC1)C5C=CC=CC5 N=C4C6CC7(CCN8CCCC(=C68) CC)C3=NC9=C7C=CC=C9 |
| M49 | 3,14-Dehydrorhazigine | CCC1=CN(CCC1)CCC2C(=NC3=C2C=CC=C3)C4CCC(=C5NC6=C(C=CC=C6) C45C CN7CC(=CC=C7)CC)C(=O)OC |
| M50 | 16-Hydrorhazisidine | CCC1=CCCN(CCC2=C3C(CC(C(O)[N]3C4=C2C=CC=C4)C5=C(CCN6CCCC(=C6)CC) C7=C([NH]5)C=CC=C7)C(=O)OC)C1 |
| M51 | Rhazisidine | CCC1=CCCN(CCC2=C3C(CC4C([N]3C5=C2C=CC=C5)C6=C(CC)C=C CN6C CC7=C4[NH]C8=C7C=CC=C8)C(=O)OC)C1 |
| M52 | Isorhazicine | COC(=O)C12C(CC3(C(O)NC4=C3C=CC=C4)C1=O)N5CCC2\C(C5)=C\C |
| M53 | Rhazigine | CCC1=CCCN(CCC2=C([NH]C3=C2C=CC=C3)C4CCC(=C5NC6=C(C=CC=C6) C45C CN7CCC=C(CC)C7)C(=O)OC)C1 |
| M54 | Strictisidine | COC(=O)C12C3CC4(C1=O)C(=NC5=C4C=CC=C5)C6CC2\C(CN36)=C\C |
| M55 | Strictamine-N-oxide | COC(=O)C1C\2CC3C4=NC5=CC=CC=C5C14CC[N +]3([O-])CC2=C/C |
| M56 | Strictigine | CCC1=C2CCN(CCC23C(=NC4=CC=CC=C34)C=C)C1 |
| M57 | Strictine | COC(=O)C1C2CC3 N(CCC4=C3[N]1C5=CC=CC=C45)C=C2C(C)=O |
| M58 | Stricticine | COC(=O)C1=C2NC3=CC=CC=C3C24CCN5CC6(OC6C)C1CC45 |
| M59 | Strictalamine | C\C=C1/CN2CCC34C(C=O)C1CC2C3=NC5=CC=CC=C45 |
| M60 | 1,2-Dehydroaspidospermine | CCC12CCCN3CCC4(C13)C(=NC5=CC=CC=C45)CC2 |
| M61 | Tetrahydrosecodine | CCC1CCCN(CCC2=C([NH]C3=CC=CC=C23)C(C)C(=O)OC)C1 |
| M62 | Dihydrosecodine | CCC1=CCCN(CCC2=C([NH]C3=CC=CC=C23)C(C)C(=O)OC)C1 |
| M63 | Dihydrosecamine | CCC1CCCN(CCC2=C([NH]C3=C2C=CC=C3)C4(CCC(C(=O)OC)C5=C (CCN6CC C=C(CC)C6)C7=CC=CC=C7[N]45)C(=O)OC)C1 |
| M64 | Dihydropresecamine | CCC1CCCN(CCC2=C([NH]C3=CC=CC=C23)OC(=O)C4CCC(=C5 N© C6=CC=C C=C6C45CCN7CCC=C(CC)C7)C(=O)OC)C1 |
| M65 | Tetrahydropresecamine | CCC1CCCN(CCC2=C([NH]C3=CC=CC=C23)OC(=O)C4CCC(=C5 N© C6=CC=C C=C6C45CCN7CCCC(CC)C7)C(=O)OC)C1 |
| M66 | Rhazinol | C\C=C1\CN2CCC34C(CO)C1CC2C3=NC5=CC=CC=C45 |
| M67 | Rhazimol | COC(=O)C1(CO)C\2CC3N(CCC14C3=NC5=CC=CC=C45)CC2=C/C |
| M68 | Rhazidigenine-N-oxide | CCC12CCC[N+]([O-])(CCC3(O)C(=NC4=CC=CC=C34)CC1)C2 |
| M69 | (−)-16R,21R-Omethyleburmanine | CCC12CCCN3CCC4=C(C13)[N](C(C2)OC)C5=CC=CC=C45 |
| M70 | Decarbomethoxy-15,20,16,17-tetrahydrosecodine | CCC1CCCN(CCC2=C(CC)[NH]C3=CC=CC=C23)C1 |
| M71 | 1,2-Dehydroaspidospermidine-N-oxide | CCC12CCC[N+]3([O–])CCC4(C13)C(=NC5=CC=CC=C45)CC2 |
| M72 | Rhazizine | COC(=O)C12OCN3C(O1)C4(CCN5C\C(=C\C)C2CC45)C6=CC=CC=C36 |
| M73 | 15-Hydroxyvincadifformine | CCC12CC(=C3NC4=CC=CC=C4C35CCN(CCC1O)C25)C(=O)OC |
| M74 | Dihydroburnamenine | CCC12CCCN3CCC4=C(C13)[N](CC2)C5=CC=CC=C45 |
| M75 | 16s,16′-Decarboxytetrahydrosecamine | CCC1CCCN(CCC2=C([NH]C3=C2C=CC=C3)C4CCC(C(=O)OC)C5=C (CCN6CCCC(CC)C6)C7=C(C=CC=C7)[N]45)C1 |
| M76 | Nor-C-fluorocuraine | C\C=C1\CN2CCC34C2CC1C(=C3NC5=CC=CC=C45)C=O |
| M77 | Strictibine | COC(=O)C1=CC=C2NC3=CC=CC=C3C12 |
Results and discussion
Physiochemical and cheminformatic studies
ACD/Laboratories informatics modules generated physiochemical and cheminformatics data of R. stricta indole and non-indole alkaloids. For all the selected 78 molecules in this study, it was observed that less than 20% of the molecules are having molecular weights >450, while most molecules range around 300–350, indicating their viability for additional medicinal chemistry amenable nature. Most of these molecules are also moderately to highly soluble—mainly due to the high value of pKa (leading to solubility at neutral pH). Additionally, many of these indole/non-indole molecules are also less lipophilic (~75% of them have logP ~3 to 4). Alkaloids that violate Lipinski’s Rule-of-5 are either due to molecular weight or logP, are tetrahydrosecamine; presecamine; beta-sitosterol; ursolic acid; stigmasterol; oleanolic acid; secamine; bis-strictidine; 3,14-dehydrorhazigine; 16-hydroxyrhazisidine; rhazisidine; rhazigine; dihydrosecamine; dihydropresecamine; tetrahydropresecamine; decarbomethoxy-15,17-tetrahydrosecodine;16s,16′-decarboxytetrahydro-secamine. Figures 1 and 2 give the plots of molecular weight and LogP (lipophilicity) of individual compounds, accordingly. Since most of the molecules have a basic nitrogen and sometimes, may be more than one, leading to a larger pKa at physiological pH—thus leading most molecules are highly to moderately soluble at physiological pH. Very few compounds and non-indole alkaloids have no basic nitrogen leading to highly insoluble compounds in water at physiological pH. As the acidity goes up (leading towards pH 1), most compounds become largely soluble. A qualitative and quantitative (computational) estimate of solubility of these compounds are given in Tables 3 and 4, respectively.
Fig. 1.
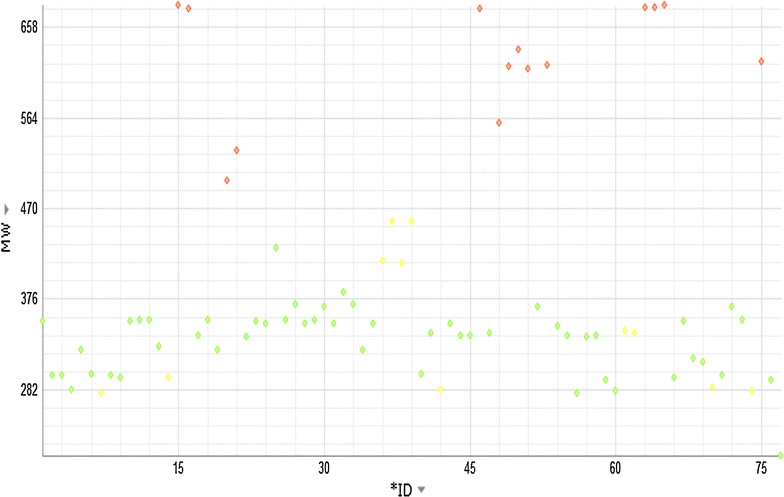
Variation of Molecular weight of compounds of Rhazya stricta
Fig. 2.

Variation of LogP of compounds of Rhazya stricta
Table 3.
Qualitative assessment of Rhazya stricta compounds with respect to Lipinski’s Rule-of-5 and solubility
| ID | Name | LogP | MW | HBD | HBA | #RotB | Rings | Rule-of-5 | Leadlike | Solubility |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Akummidine | Optimal | Good | Good | Good | Good | Bad | Good | Good | Soluble |
| 2 | Antirhine | Optimal | Good | Good | Good | Good | Good | Good | Good | Soluble |
| 3 | 3-epi-Antirhine | Optimal | Good | Good | Good | Good | Good | Good | Good | Soluble |
| 4 | Aspidosespermidine | Optimal | Good | Good | Good | Good | Bad | Good | Good | Soluble |
| 5 | Condylocarpine | Optimal | Good | Good | Good | Good | Bad | Good | Good | Soluble |
| 6 | Dihydrocorynantheol | Optimal | Good | Good | Good | Good | Good | Good | Good | Soluble |
| 7 | Eburnamenine | Lipophilic | Good | Good | Good | Good | Bad | Good | Moderate | Soluble |
| 8 | Eburnamine | Optimal | Good | Good | Good | Good | Bad | Good | Good | Soluble |
| 9 | Eburnamonine | Optimal | Good | Good | Good | Good | Bad | Good | Good | Soluble |
| 10 | Geissoschizine | Optimal | Good | Good | Good | Good | Good | Good | Good | Insoluble |
| 11 | Isositsirikine | Optimal | Good | Good | Good | Good | Good | Good | Good | Soluble |
| 12 | 16-Epi-Z-isositsirikine | Optimal | Good | Good | Good | Good | Good | Good | Good | Soluble |
| 13 | Leuconalm | Optimal | Good | Good | Good | Good | Good | Good | Good | Soluble |
| 14 | Rhazinliam | Lipophilic | Good | Good | Good | Good | Good | Good | Moderate | Highly insoluble |
| 15 | Tetrahydrosecamine | Very lipophilic | Bad | Good | Good | Bad | Bad | Bad | Bad | Soluble |
| 16 | Presecamine | Very lipophilic | Bad | Good | Good | Bad | Bad | Bad | Bad | Soluble |
| 17 | Sewarine | Optimal | Good | Good | Good | Good | Bad | Good | Good | Soluble |
| 18 | Stemmadenine | Optimal | Good | Good | Good | Good | Bad | Good | Good | Soluble |
| 19 | Strictamine | Optimal | Good | Good | Good | Good | Bad | Good | Good | Insoluble |
| 20 | Strictosamide | Optimal | Moderate | Good | Good | Good | Bad | Good | Bad | Soluble |
| 21 | Strictosidine | Optimal | Bad | Bad | Bad | Good | Bad | Bad | Bad | Soluble |
| 22 | Taberonine | Optimal | Good | Good | Good | Good | Bad | Good | Good | Soluble |
| 23 | Tetrahydrlstonine | Optimal | Good | Good | Good | Good | Bad | Good | Good | Soluble |
| 24 | Vallesiachotamine | Optimal | Good | Good | Good | Good | Good | Good | Good | Highly insoluble |
| 25 | Aspidospermoise | Optimal | Good | Good | Good | Good | Bad | Good | Good | Soluble |
| 26 | Bhimbrine | Optimal | Good | Good | Good | Good | Good | Good | Good | Soluble |
| 27 | Bhimbrine N-oxide | Optimal | Good | Good | Good | Good | Good | Good | Good | Soluble |
| 28 | Rhazimine | Optimal | Good | Good | Good | Good | Bad | Good | Good | Soluble |
| 29 | Rhazimanine | Optimal | Good | Good | Good | Good | Good | Good | Good | Soluble |
| 30 | Rhazicine | Optimal | Good | Good | Good | Good | Bad | Good | Good | Soluble |
| 31 | Leepacine | Optimal | Good | Good | Good | Good | Bad | Good | Good | Soluble |
| 32 | 2-Methoxy,1-2,dihydro rhazamine | Optimal | Good | Good | Good | Good | Bad | Good | Good | Soluble |
| 33 | HR-1 | Optimal | Good | Good | Good | Good | Good | Good | Good | Soluble |
| 34 | Vincanicine | Optimal | Good | Good | Good | Good | Bad | Good | Good | Soluble |
| 35 | Rhazinaline | Optimal | Good | Good | Good | Good | Bad | Good | Good | Insoluble |
| 36 | Beta-sitosterol | Very lipophilic | Lipophilic | Good | Good | Good | Good | Good | Moderate | Moderate |
| 37 | Ursolic acid | Very lipophilic | Lipophilic | Good | Good | Good | Good | Bad | Moderate | Highly insoluble |
| 38 | Stigmasterol | Lipophilic | Good | Good | Good | Good | Good | Moderate | Moderate | Insoluble |
| 39 | Olenaolic acid | Very lipop | Lipophilic | Good | Good | Good | Good | Bad | Moderate | Highly insoluble |
| 40 | Rhazidigenine (rhazidine) | Optimal | Optimal | Good | Good | Good | Good | Good | Good | Good |
| 41 | N-methylleuconolam | Optimal | Good | Good | Good | Good | Good | Good | Good | Soluble |
| 42 | (+)-Quebranchamine | Lipophilic | Good | Good | Good | Good | Good | Good | Moderate | Soluble |
| 43 | Polyneuridine | Optimal | Good | Good | Good | Good | Bad | Good | Good | Soluble |
| 44 | (+)-Vincadiformine | Optimal | Good | Good | Good | Good | Bad | Good | Good | Soluble |
| 45 | (−)-Vincadiformine | Optimal | Good | Good | Good | Good | Bad | Good | Good | Soluble |
| 46 | Secamine | Very | Lipop | Bad | Good | Good | Bad | Bad | Bad | Bad |
| 47 | Vincadine | Optimal | Good | Good | Good | Good | Good | Good | Good | Soluble |
| 48 | Bis-strictidine | Very lipop | Bad | Good | Good | Good | Bad | Bad | Bad | Insoluble |
| 49 | 3,14-Dehydrorhazigine | Very lipop | Bad | Good | Good | Bad | Bad | Bad | Bad | Highly insoluble |
| 50 | 16-Hydrorhazisidine | Very lipop | Bad | Good | Good | Bad | Bad | Bad | Bad | Soluble |
| 51 | Rhazisidine | Very lipop | Bad | Good | Good | Good | Bad | Bad | Bad | Insoluble |
| 52 | Isorhazicine | Optimal | Good | Good | Good | Good | Bad | Good | Good | Soluble |
| 53 | Rhazigine | Very lipop | Bad | Good | Good | Bad | Bad | Bad | Bad | Soluble |
| 54 | Strictisidine | Optimal | Good | Good | Good | Good | Bad | Good | Good | Soluble |
| 55 | Strictamine-N-oxide | Optimal | Good | Good | Good | Good | Bad | Good | Good | Soluble |
| 56 | Strictigine | Optimal | Good | Good | Good | Good | Bad | Good | Good | Soluble |
| 57 | Strictine | Optimal | Good | Good | Good | Good | Bad | Good | Good | Highly insoluble |
| 58 | Stricticine | Optimal | Good | Good | Good | Good | Bad | Good | Good | Soluble |
| 59 | Strictalamine | Optimal | Good | Good | Good | Good | Bad | Good | Good | Insoluble |
| 60 | 1,2-Dehydro-aspidospermine | Optimal | Good | Good | Good | Good | Bad | Good | Good | Soluble |
| 61 | Tetrahydrosecodine | Lipophilichilic | Good | Good | Good | Good | Good | Good | Moderate | Soluble |
| 62 | Dihydrosecodine | Lipophilichilic | Good | Good | Good | Good | Good | Good | Moderate | Soluble |
| 63 | Dihydrosecamine | Very lipophilic | Bad | Good | Good | Bad | Bad | Bad | Bad | Soluble |
| 64 | Dihydropresecamine | Very lipophilic | Bad | Good | Good | Bad | Bad | Bad | Bad | Soluble |
| 65 | Tetrahydropresecamine | Very lipop | Bad | Good | Good | Bad | Bad | Bad | Bad | Soluble |
| 66 | Rhazinol | Optimal | Good | Good | Good | Good | Bad | Good | Good | Insoluble |
| 67 | Rhazimol | Optimal | Good | Good | Good | Good | Bad | Good | Good | Insoluble |
| 68 | Rhazidigenine-N-oxide | Optimal | Good | Good | Good | Good | Good | Good | Good | Soluble |
| 69 | (−)-16R,21R-Omethyleburmanine | Optimal | Good | Good | Good | Good | Bad | Good | Good | Soluble |
| 70 | Decarbomethoxy-15,20,16,17-tetrahydrosecodine | Very lipophilic | Good | Good | Good | Good | Good | Moderate | Moderate | Soluble |
| 71 | 1,2-Dehydroaspidosper midine-N-oxide | Optimal | Good | Good | Good | Good | Bad | Good | Good | Soluble |
| 72 | Rhazizine | Optimal | Good | Good | Good | Good | Bad | Good | Good | Soluble |
| 73 | 15-Hydroxyvincadiffor mine | Optimal | Good | Good | Good | Good | Bad | Good | Good | Soluble |
| 74 | Dihydroburnamenine | Lipophilic | Good | Good | Good | Good | Bad | Good | Moderate | Soluble |
| 75 | 16s,16′-Decarboxytetra hydrosecamine | Very lipop | Bad | Good | Good | Bad | Bad | Bad | Bad | Soluble |
| 76 | Nor-C-fluorocuraine | Optimal | Good | Good | Good | Good | Bad | Good | Good | Soluble |
| 77 | Strictibine | Optimal | Good | Good | Good | Good | Good | Good | Good | Insoluble |
LogP partition-coefficient, MW molecular weight, HBD hydrogen bond donor, HBA hydrogen bond acceptors, #RotB number of rotatable bonds, Rings # of ideally acceptable rings, Rule-of-5 Lipinski’s rule of five, Leadlike leadlikeness, Solubility solubility classification
Table 4.
Predicted solubility and pKa (acid and base) of various Rhazya stricta compounds
| ID | Name | Solubility | LogSW/LogSw | LogSw/pH | pKa (acid) | pKa (base) |
|---|---|---|---|---|---|---|
| 1 | Akuammidine | Soluble | −3.32 | 8.85 | 14.79 | 6.88 |
| 2 | Antirhine | Soluble | −4.08 | 9.49 | 14.72 | 9.24 |
| 3 | 3-Epi-antirhine | Soluble | −4.08 | 9.49 | 14.72 | 9.24 |
| 4 | Aspidosespermidine | Soluble | −2.34 | 10.82 | 9.94 | |
| 5 | Condylocarpine | Soluble | −3.13 | 9.36 | 7.98 | |
| 6 | Dihydrocorynantheol | Soluble | −4.04 | 9.57 | 15.08 | 9.37 |
| 7 | Eburnamenine | Soluble | −4.6 | 8.92 | 8.61 | |
| 8 | Eburnamine | Soluble | −4.39 | 9.15 | 14.3 | |
| 9 | Eburnamonine | Soluble | −4.4 | 8.82 | 8.13 | |
| 10 | Geissoschizine | Insoluble | −3.64 | 6.59 | 4.73 | 8.25 |
| 11 | Isositsirikine | Soluble | −4.1 | 9.16 | 14.29 | 8.49 |
| 12 | 16-Epi-Z-isositsirikine | Soluble | −4.1 | 9.16 | 14.29 | 8.49 |
| 13 | leuconolam | Soluble | −1.83 | 6.71 | 11.76 | 0.36 |
| 14 | Rhazinilam | Highly insoluble | −4.47 | 7 | 1.21 | |
| 15 | Tetrahydrosecamine | Soluble | −3.67 | 8.07 | 17.43 | 9.4 |
| 16 | Presecamine | Soluble | −5.27 | 8.48 | 15.79 | 8.54 |
| 17 | Sewarine | Soluble | −2.98 | 9.17 | 11.08 | 1.95 |
| 18 | Stemmadenine | Soluble | −3.63 | 9.21 | 11.84 | 8.08 |
| 19 | Strictamine | Insoluble | −4.47 | 7.7 | 5.74 | |
| 20 | Strictosamide | Soluble | −3.26 | 7 | 12.79 | −1.64 |
| 21 | Strictosidine | Soluble | −2.73 | 10.83 | 12.81 | 10.62 |
| 22 | Tabersonine | Soluble | −2.99 | 9.25 | 7.64 | |
| 23 | Tetrahydroalstonine | Soluble | −4.4 | 8.89 | 18.03 | 8.27 |
| 24 | Vallesiachotamine | Highly insoluble | −5.21 | 7.45 | 17.46 | 6.08 |
| 25 | Aspidospermiose | Soluble | −0.19 | 9.81 | 10.11 | 9.88 |
| 26 | Bhimberine | Soluble | −4.1 | 9.16 | 14.29 | 8.49 |
| 27 | Bhimbhrine N-oxide | Soluble | 0.4 | 9.66 | 14.2 | 5.17 |
| 28 | Rhazimine | Soluble | −2.89 | 8.9 | 6.51 | |
| 29 | Rhazimanine | Soluble | −4.1 | 9.16 | 14.29 | 8.49 |
| 30 | Rhazicine | Soluble | −1.6 | 8.94 | 11.3 | 6.36 |
| 31 | Leepacine | Soluble | −1.84 | 9.43 | 6.69 | |
| 32 | 2-Methoxy-1,2-dihydrorhazimine | Soluble | −2.18 | 9.15 | 6.3 | |
| 33 | HR-1 | Soluble | 0.43 | 8.55 | 12.69 | 4.6 |
| 34 | Vincanicine | Soluble | −2.67 | 9.67 | 8.16 | |
| 35 | Rhazinaline | Insoluble | −4.14 | 7.47 | 5.03 | |
| 36 | Beta-sitosterol | Highly insoluble | −7.6 | 7 | 15.03 | |
| 37 | Ursolic acid | Highly insoluble | −6 | 6.01 | 15.18 | |
| 38 | Stigmasterol | Highly insoluble | −7.52 | 7 | 15.03 | |
| 39 | Oleanolic acid | Highly insoluble | −6.02 | 6.04 | 15.18 | |
| 40 | Rhazidigenine | Soluble | −3.2 | 9.92 | 12.43 | 8.82 |
| 41 | N-methylleuconolam | Soluble | −1.52 | 6.55 | 11.62 | 0.09 |
| 42 | (+)-Quebrachamine | Soluble | −4.15 | 9.55 | 17.84 | 9.74 |
| 43 | Polyneuridine | Soluble | −3.2 | 8.46 | 17.19 | 6.11 |
| 44 | (+)-Vincadiformine | Soluble | −3.06 | 10.04 | 9.33 | |
| 45 | (−)-Vincadifformine | Soluble | −3.06 | 10.04 | 9.33 | |
| 46 | Secamine | Soluble | −5.12 | 8.22 | 17.34 | 8.71 |
| 47 | Vincadine | Soluble | −4.23 | 9.28 | 16.98 | 9.11 |
| 48 | Bis-strictidine | Insoluble | −6.11 | 7.79 | 7.57 | |
| 49 | 3,14-Dehydrorhazigine | Highly insoluble | −5.89 | 8.12 | 10.62 | |
| 50 | 16-Hydrorhazisidine | Soluble | −5.05 | 8.28 | 13.98 | 10.8 |
| 51 | Rhazisidine | Insoluble | −5.56 | 8.2 | 17.47 | 8.76 |
| 52 | Isorhazicine | Soluble | −1.6 | 8.94 | 11.3 | 6.36 |
| 53 | Rhazigine | Soluble | −4.44 | 7.7 | 17.45 | 8.89 |
| 54 | Strictisidine | Soluble | −2.18 | 8.18 | 4.27 | |
| 55 | Strictamine-N-oxide | Soluble | −0.67 | 8.73 | 4.17 | |
| 56 | Strictigine | Soluble | −4.07 | 8.83 | 7.71 | |
| 57 | Strictine | Highly insoluble | −4.79 | 7.36 | 5.41 | |
| 58 | Stricticine | Soluble | −3.68 | 9.33 | 8.43 | |
| 59 | Strictalamine | Insoluble | −3.94 | 8.04 | 5.87 | |
| 60 | 1,2-Dehydroaspidospermidine(eburenine) | Soluble | −2.84 | 10.23 | 9.38 | |
| 61 | Tetrahydrosecodine | Soluble | −3.85 | 9.67 | 16.75 | 9.33 |
| 62 | Dihydrosecodine | Soluble | −3.84 | 9.44 | 16.66 | 8.73 |
| 63 | Dihydrosecamine | Soluble | −4.61 | 8.3 | 17.43 | 9.4 |
| 64 | Dihydropresecamine | Soluble | −4.78 | 8.28 | 15.88 | 9.16 |
| 65 | Tetrahydropresecamine | Soluble | −3.89 | 8.23 | 15.88 | 9.65 |
| 66 | Rhazinol | Insoluble | −4.1 | 8.25 | 14.53 | 6.3 |
| 67 | Rhazimol | Insoluble | −4.24 | 7.67 | 14.53 | 5.45 |
| 68 | Rhazidigenine-N-oxide | Soluble | 0.5 | 8.35 | 11.98 | 49.2 |
| 69 | (−)16R,21R-omethyleburnamine | Soluble | −4.93 | 8.73 | 8.66 | |
| 70 | Decarbomethoxy-15,20,16,17-tetrahydros | Soluble | −3.79 | 9.81 | 17.83 | 9.46 |
| 71 | 1-2-Dehydroasidospermidine-N-oxide | Soluble | −1.2 | 8.95 | 4.82 | |
| 72 | Rhazizine | Soluble | −2.61 | 9.2 | 7.31 | |
| 73 | 15-Hydroxyvincadifformine | Soluble | −2.36 | 9.88 | 14.4 | 8.46 |
| 74 | Dihydroeburnamenine | Soluble | −4.72 | 9.06 | 9.41 | |
| 75 | 16s,16’-Decarboxytetra-hydrosecamine | Soluble | −3.5 | 7.88 | 17.43 | 9.4 |
| 76 | Nor-C-fluorocurarine | Soluble | −2.4 | 9.8 | 8.14 | |
| 77 | Strictibine | Insoluble | −3.7 | 7 | 1.06 |
Solubility solubility classifications, LogSW/LogSw ratio of solubility in water vs. intrinsic solubility, LogSw/pH solubility in water at pH 7.0, pKa (acid) pKa in acidic pH, pKa(base) pKa in basic pH
QUIKPROP calculations
Predicted Quikprop properties for potential cardiac liabilities such as HERG, and CNS liabilities (Blood–Brain-Barrier) and drug-like nature of these molecules indicate that many of these molecules are well within the boundaries of accepted hit-, and lead-like nature. QuikProp calculations were performed using Schrodinger’s Maestro for various alkaloids of R. stricta. These predictions not only give Rule-of-5 data, but also predict the cardiotoxicity predictions (HERG) and CNS penetration potential (logBBB) properties. More importantly, it also gives the prediction regarding cell-permeability (Caco2). All these models are well validated in literature, and most of them perform well within the reproducible results for training datasets. Results indicate that many of the molecules have decent permeation through Caco2 cell lines (>300), while the polar surface area (PSA) is not too high (>120) for oral absorption. For HERG toxicity prediction, below −5 (i.e. −6, −7 etc.) is not considered to be safe. Hence, those molecules whose logHERG values are well below -5 (such as geissoschizine, presecamine, tetrahydrosecamine) may exhibit cardioliability. The human intestinal absorption is also predicted, and it appears for most molecules, these values are larger. Any %HIA prediction >90% is expected to be well absorbed, and their polar surface area (PSA) is also a direct correlation to it. Those molecules whose molecular weights are >500 exhibit rule-of-5 violation and this violation goes beyond 1 to a maximum of 3. Those molecules appear structurally much larger and like dimers. Table 5 gives computed Quikprop computed values of various alkaloids of R. stricta. Table 6 also indicates various other physiochemical parameters including surface tension, parachor etc. of R. stricta indole and non-indole analogs.
Table 5.
Quikprop calculation (for physiochemical properties) of Rhazya stricta compounds
| Title | Name | M.W | HBD | HBA | QP logP | QP logS | QP logHERG | QP Caco2 | QP logBB | %HOA | PSA | RO5v |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M1 | Akummidine | 352.432 | 1 | 5 | 3.2 | −3.5 | −5.1 | 410.4 | 0.1 | 93 | 63 | 0 |
| M2 | Antirhine | 296.411 | 2 | 4 | 3.1 | −3.3 | −5.6 | 583.1 | 0.1 | 95 | 40 | 0 |
| M3 | 3-Epi-antirhine | 296.411 | 2 | 4 | 3.1 | −3.3 | −5.6 | 583.1 | 0.1 | 95 | 40 | 0 |
| M4 | Aspidosespermidine | 282.428 | 1 | 3 | 2.8 | −1.9 | −5.2 | 382.4 | 1.1 | 90 | 18 | 0 |
| M5 | Condylocarpine | 322.406 | 0 | 3 | 4 | −4.5 | −5.5 | 735.5 | 0.4 | 100 | 48 | 0 |
| M6 | Dihydrocorynantheol | 298.427 | 2 | 4 | 3.2 | −3.6 | −5.7 | 521.2 | 0.1 | 95 | 40 | 0 |
| M7 | Eburnamenine | 278.396 | 0 | 2 | 4.1 | −3.7 | −5.2 | 2375.6 | 0.9 | 100 | 7 | 0 |
| M8 | Eburnamine | 296.411 | 1 | 4 | 3.2 | −3.1 | −4.9 | 1159.1 | 0.5 | 100 | 27 | 0 |
| M9 | Eburnamonine | 294.396 | 0 | 5 | 2.4 | −2.1 | −4.8 | 1051.6 | 0.6 | 95 | 32 | 0 |
| M10 | Geissoschizine | 352.432 | 1 | 6 | 3 | −4.4 | −6.2 | 202.7 | −0.4 | 86 | 79 | 0 |
| M11 | Isositsirikine | 354.448 | 1 | 5 | 3.6 | −3.9 | −5.5 | 348.3 | −0.1 | 94 | 68 | 0 |
| M12 | 16-Epi-Z-isositsirikine | 354.448 | 1 | 5 | 3.7 | −4.6 | −6.1 | 305.6 | −0.2 | 93 | 71 | 0 |
| M13 | Leuconalm | 326.394 | 2 | 6 | 2 | −3.3 | −3.7 | 600.6 | −0.6 | 88 | 82 | 0 |
| M14 | Rhazinliam | 294.396 | 1 | 3 | 4.1 | −4.6 | −4.2 | 3342.3 | 0.1 | 100 | 36 | 0 |
| M15 | Tetrahydrosecamine | 680.929 | 0 | 7 | 8.5 | −8.1 | −8.1 | 198.9 | 0.2 | 92 | 75 | 2 |
| M16 | Presecamine | 676.897 | 0 | 8 | 7.5 | −5.7 | −7.1 | 134.1 | 0 | 83 | 79 | 2 |
| M17 | Sewarine | 338.405 | 1 | 4 | 3.3 | −4.1 | −5.3 | 305.7 | 0 | 91 | 69 | 0 |
| M18 | Stemmadenine | 354.448 | 0 | 5 | 3.3 | −3.4 | −5.2 | 363.8 | 0 | 92 | 57 | 0 |
| M19 | Strictamine | 322.406 | 0 | 6 | 2.4 | −2.3 | −4.7 | 624.1 | 0.4 | 91 | 47 | 0 |
| M20 | Strictosamide | 498.532 | 5 | 15 | 0.6 | −4 | −5.9 | 94.5 | −2.1 | 66 | 147 | 0 |
| M21 | Strictosidine | 530.574 | 6 | 15 | 0.5 | −2.6 | −6.4 | 34.7 | −1.7 | 19 | 164 | 3 |
| M22 | Taberonine | 336.433 | 0 | 3 | 4 | −4.1 | −5.3 | 617.2 | 0.3 | 100 | 51 | 0 |
| M23 | Tetrahydrlstonine | 352.432 | 1 | 6 | 3.2 | −4.3 | −6.1 | 573.5 | 0.3 | 95 | 59 | 0 |
| M24 | Vallesiachotamine | 350.416 | 1 | 6 | 3.4 | −5 | −5.1 | 932 | −0.6 | 100 | 81 | 0 |
| M25 | Aspidospermoise | 428.527 | 3 | 12 | 0.2 | −1.3 | −5.9 | 16.8 | −0.4 | 50 | 102 | 0 |
| M26 | Bhimbrine | 354.448 | 1 | 5 | 3.6 | −3.9 | −5.5 | 370.7 | −0.1 | 94 | 69 | 0 |
| M27 | Bhimbrine N-oxide | 370.447 | 1 | 6 | 3.7 | −3.2 | −5.1 | 917.7 | −0.7 | 100 | 79 | 0 |
| M28 | Rhazimine | 350.416 | 0 | 8 | 2.4 | −3.5 | −6.7 | 333.6 | 0 | 86 | 67 | 0 |
| M29 | Rhazimanine | 354.448 | 1 | 5 | 3.7 | −4.6 | −6.1 | 305.6 | −0.2 | 93 | 71 | 0 |
| M30 | Rhazicine | 368.432 | 0 | 7 | 1.6 | −1 | −5.5 | 56.9 | 0.3 | 68 | 88 | 0 |
| M31 | Leepacine | 350.416 | 1 | 7 | 1.6 | −1.7 | −5.9 | 103.1 | 0.6 | 72 | 74 | 0 |
| M32 | 2-Methoxy 1-2,dihydrorhazamine | 382.458 | 0 | 8 | 1.8 | −1.1 | −5.7 | 113.2 | 0.6 | 74 | 72 | 0 |
| M33 | HR-1 | 370.447 | 1 | 6 | 3.8 | −3.3 | −5 | 1346.8 | −0.5 | 100 | 70 | 0 |
| M34 | Vincanicine | 322.406 | 0 | 4 | 3.1 | −3.1 | −4.7 | 454.4 | 0.2 | 93 | 62 | 0 |
| M35 | Rhazinaline | 350.416 | 0 | 8 | 1.5 | −1.3 | −4.7 | 337 | 0.1 | 81 | 68 | 0 |
| M36 | beta-Sitosterol | 414.713 | 1 | 2 | 7.5 | −8.2 | −4.4 | 4119.2 | −0.2 | 100 | 21 | 1 |
| M37 | Ursolic acid | 456.707 | 2 | 4 | 6.1 | −6.8 | −1.7 | 304.5 | −0.4 | 94 | 60 | 1 |
| M38 | Stigmasterol | 412.698 | 1 | 2 | 7.4 | −8.1 | −4.3 | 4119.2 | −0.2 | 100 | 21 | 1 |
| M39 | Olenaolic acid | 456.707 | 2 | 4 | 6.2 | −7 | −1.8 | 306 | −0.4 | 95 | 60 | 1 |
| M40 | Rhazidigenine (rhazidine) | 298.427 | 1 | 4 | 3.1 | −3.1 | −4.8 | 849.1 | 0.4 | 100 | 34 | 0 |
| M41 | N-methylleuconolam | 340.421 | 1 | 7 | 2.4 | −3.4 | −3.8 | 1336.6 | −0.3 | 100 | 66 | 0 |
| M42 | (+)-Quebranchamine | 282.428 | 1 | 2 | 4.1 | −4 | −5 | 1678.5 | 0.7 | 100 | 15 | 0 |
| M43 | Polyneuridine | 350.416 | 1 | 6 | 2.4 | −3.1 | −5 | 299.1 | 0 | 85 | 75 | 0 |
| M44 | (+)-Vincadiformine | 338.449 | 0 | 3 | 4.1 | −4.3 | −5.2 | 655.8 | 0.3 | 100 | 49 | 0 |
| M45 | (−)-Vincadiformine | 338.449 | 0 | 3 | 4.1 | −4.3 | −5.2 | 713 | 0.3 | 100 | 49 | 0 |
| M46 | Secamine | 676.897 | 0 | 7 | 8.6 | −8.3 | −8.5 | 200.4 | 0.2 | 92 | 76 | 2 |
| M47 | Vincadine | 340.464 | 0 | 3 | 4.6 | −5.4 | −6 | 637.6 | 0.2 | 100 | 46 | 0 |
| M48 | Bis-strictidine | 560.824 | 2 | 3 | 7.9 | −7.7 | −5.8 | 1941.9 | 0.7 | 100 | 24 | 2 |
| M51 | Rhazisidine | 614.829 | 1 | 5 | 8.8 | −9.3 | −7.6 | 1208.3 | 0.2 | 100 | 50 | 2 |
| M52 | Isorhazicine | 368.432 | 0 | 7 | 1.6 | −1.1 | −5.7 | 49 | 0.2 | 66 | 88 | 0 |
| M53 | Rhazigine | 618.861 | 1 | 5 | 8.9 | −9.2 | −8.6 | 153.6 | 0.1 | 92 | 65 | 2 |
| M54 | Strictisidine | 348.401 | 0 | 8 | 1.5 | −1.7 | −5.1 | 270.9 | 0 | 79 | 74 | 0 |
| M55 | Strictamine-N-oxide | 338.405 | 0 | 7 | 2.3 | −0.9 | −3.8 | 1371.3 | −0.2 | 97 | 58 | 0 |
| M56 | Strictigine | 278.396 | 0 | 4 | 3.1 | −2.5 | −4.7 | 1380.1 | 0.6 | 100 | 19 | 0 |
| M57 | Strictine | 336.39 | 0 | 6 | 3.1 | −3.3 | −4.1 | 2156.2 | −0.2 | 100 | 59 | 0 |
| M58 | Stricticine | 338.405 | 0 | 5 | 2.9 | −2.8 | −4.9 | 836.2 | 0.5 | 96 | 65 | 0 |
| M59 | Strictalamine | 292.38 | 0 | 6 | 1.6 | −1.7 | −4.7 | 602.9 | 0.4 | 86 | 50 | 0 |
| M60 | 1,2-Dehydro-aspido-spermine | 280.412 | 0 | 4 | 3.2 | −2.7 | −4.6 | 1558 | 0.7 | 100 | 15 | 0 |
| M61 | Tetrahydrosecodine | 342.48 | 0 | 3 | 5 | −5.1 | −5.9 | 687.8 | 0 | 100 | 50 | 0 |
| M62 | Dihydrosecodine | 340.464 | 0 | 3 | 5 | −5.5 | −6.5 | 632.6 | −0.1 | 100 | 51 | 0 |
| M63 | Dihydrosecamine | 678.913 | 0 | 7 | 8.6 | −8 | −8.1 | 214.4 | 0.2 | 93 | 75 | 2 |
| M64 | Dihydropresecamine | 678.913 | 0 | 8 | 7.8 | −6 | −7 | 155.8 | 0.1 | 86 | 78 | 2 |
| M65 | Tetrahydropresecamine | 680.929 | 0 | 8 | 7.8 | −6.5 | −7.3 | 144.9 | 0 | 85 | 80 | 2 |
| M66 | Rhazinol | 294.396 | 1 | 5 | 1.2 | −1.8 | −4.6 | 581 | 0.3 | 83 | 40 | 0 |
| M67 | Rhazimol | 352.432 | 0 | 6 | 2.4 | −2 | −4.8 | 572.5 | 0.2 | 90 | 60 | 0 |
| M68 | Rhazidigenine-N-oxide | 314.427 | 1 | 5 | 3.1 | −1.9 | −4 | 1964.2 | −0.2 | 100 | 46 | 0 |
| M69 | (−)-16R,21R-Omethyleburmanine | 310.438 | 0 | 4 | 3.3 | −3.3 | −4.9 | 2470.6 | −0.7 | 100 | 13 | 0 |
| M70 | Decarbomethoxy-15,20,16,17-tetrahydrosecodine | 284.444 | 1 | 2 | 4.7 | −4.6 | −5.8 | 1672.7 | 0.5 | 100 | 18 | 0 |
| M71 | 1,2-Dehydro-aspidospermidine-N-oxide | 296.411 | 0 | 5 | 3.1 | −1.2 | −3.5 | 4109.3 | 0.2 | 100 | 29 | 0 |
| M72 | Rhazizine | 368.432 | 0 | 7 | 2.5 | −2.1 | −4.5 | 1005.1 | 0.6 | 95 | 53 | 0 |
| M73 | 15-Hydroxy-vincadifformine | 354.448 | 1 | 5 | 3.2 | −3.8 | −5.3 | 302.4 | −0.1 | 90 | 67 | 0 |
| M74 | Dihydroburnamenine | 280.412 | 0 | 2 | 3.9 | −3.6 | −4.8 | 2470.6 | 0 | 100 | 5 | 0 |
| M75 | 16s,16’-Decarboxy-tetrahydrosecamine | 622.892 | 1 | 6 | 7.6 | −6.1 | −6.8 | 295.2 | 0.5 | 90 | 45 | 2 |
| M76 | Nor-C-fluorocuraine | 292.38 | 0 | 3 | 2.8 | −2.3 | −3.8 | 512 | 0.4 | 92 | 52 | 0 |
| M77 | Strictibine | 213.235 | 1 | 2 | 2.5 | −3.3 | −4.7 | 1789.3 | −0.2 | 100 | 49 | 0 |
MW molecular weight, HBD hydrogen bond donors, HBA hydrogen bond acceptors, QPlogP predicted octanol/water partition coefficient, QPlogS predicted aqueous solubility, QPlogHERG predicted IC50 value for blockage of HERG K+ channels, QPCaco2 predicted Caco-2 cell permeability, QPlogBB predicted brain/blood partition coefficient, %HOA percentage of human oral absorption, PSA polar surface area, RO5v number of violations of Lipinski’s Rule of Five
Table 6.
Surface related and ring-related properties of Rhazya stricta compounds
| ID | Name | CR | NR | NOR | HetR | #R | Para | Ind.Ref | Sur.Ten | Density | Polar. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Akuammidine | 0.81 | 0.08 | 0.19 | 0.19 | 6 | 743.43 | 1.68 | 65.34 | 1.35 | 39.32 |
| 2 | Antirhine | 0.86 | 0.09 | 0.14 | 0.14 | 4 | 676.25 | 1.65 | 56.53 | 1.2 | 35.76 |
| 3 | 3-Epi-antirhine | 0.86 | 0.09 | 0.14 | 0.14 | 4 | 676.25 | 1.65 | 56.53 | 1.2 | 35.76 |
| 4 | Aspidosespermidine | 0.9 | 0.1 | 0.1 | 0.1 | 5 | 647.87 | 1.63 | 50.04 | 1.16 | 34.2 |
| 5 | Condylocarpine | 0.83 | 0.08 | 0.17 | 0.17 | 5 | 681.18 | 1.66 | 56.36 | 1.3 | 36.43 |
| 6 | Dihydrocorynantheol | 0.86 | 0.09 | 0.14 | 0.14 | 4 | 687.1 | 1.64 | 55.86 | 1.19 | 35.85 |
| 7 | Eburnamenine | 0.9 | 0.1 | 0.1 | 0.1 | 5 | 589.57 | 1.7 | 49.78 | 1.25 | 33.94 |
| 8 | Eburnamine | 0.86 | 0.09 | 0.14 | 0.14 | 5 | 595.24 | 1.72 | 54.34 | 1.35 | 34.28 |
| 9 | Eburnamonine | 0.86 | 0.09 | 0.14 | 0.14 | 5 | 595.24 | 1.72 | 54.34 | 1.34 | 34.28 |
| 10 | Geissoschizine | 0.81 | 0.08 | 0.19 | 0.19 | 4 | 762.54 | 1.66 | 61.38 | 1.29 | 40.01 |
| 11 | Isositsirikine | 0.81 | 0.08 | 0.19 | 0.19 | 4 | 776.63 | 1.64 | 59.3 | 1.27 | 40.13 |
| 12 | 16-Epi-Z-isositsirikine | 0.81 | 0.08 | 0.19 | 0.19 | 4 | 776.63 | 1.64 | 59.3 | 1.27 | 40.13 |
| 13 | Leuconolam | 0.79 | 0.08 | 0.21 | 0.21 | 4 | 692.66 | 1.65 | 63.34 | 1.33 | 35.61 |
| 14 | Rhazinilam | 0.86 | 0.09 | 0.14 | 0.14 | 4 | 635.67 | 1.65 | 47.86 | 1.22 | 34.93 |
| 15 | Tetrahydrosecamine | 0.84 | 0.08 | 0.16 | 0.16 | 7 | 1449.04 | 1.63 | 46.81 | 1.23 | 78.28 |
| 16 | Presecamine | 0.84 | 0.08 | 0.16 | 0.16 | 7 | 1516.34 | 1.65 | 60.13 | 1.24 | 78.73 |
| 17 | Sewarine | 0.8 | 0.08 | 0.2 | 0.2 | 5 | 696.4 | 1.69 | 64.76 | 1.38 | 37.04 |
| 18 | Stemmadenine | 0.81 | 0.08 | 0.19 | 0.19 | 5 | 729.69 | 1.64 | 47.88 | 1.28 | 39.55 |
| 19 | Strictamine | 0.83 | 0.08 | 0.17 | 0.17 | 5 | 631.14 | 1.71 | 52.17 | 1.37 | 36.23 |
| 20 | Strictosamide | 0.72 | 0.06 | 0.28 | 0.28 | 6 | 986.67 | 1.72 | 84.28 | 1.53 | 50.75 |
| 21 | Strictosidine | 0.71 | 0.05 | 0.29 | 0.29 | 5 | 1078.5 | 1.66 | 74.07 | 1.44 | 54 |
| 22 | Tabersonine | 0.84 | 0.08 | 0.16 | 0.16 | 5 | 723.31 | 1.65 | 55.72 | 1.27 | 38.37 |
| 23 | Tetrahydroalstonine | 0.81 | 0.08 | 0.19 | 0.19 | 5 | 748.43 | 1.66 | 58.69 | 1.3 | 39.39 |
| 24 | Vallesiachotamine | 0.81 | 0.08 | 0.19 | 0.19 | 4 | 754.43 | 1.65 | 59.07 | 1.29 | 39.54 |
| 25 | Aspidospermiose | 0.77 | 0.06 | 0.23 | 0.23 | 6 | 885.22 | 1.68 | 74 | 1.42 | 45.19 |
| 26 | Bhimberine | 0.81 | 0.08 | 0.19 | 0.19 | 4 | 776.63 | 1.64 | 59.3 | 1.27 | 40.13 |
| 27 | Bhimbhrine N-oxide | 0.78 | 0.07 | 0.22 | 0.22 | 4 | 45.12 | ||||
| 28 | Rhazimine | 0.81 | 0.08 | 0.19 | 0.19 | 6 | 690.3 | 1.69 | 54.96 | 1.38 | 38.6 |
| 29 | Rhazimanine | 0.81 | 0.08 | 0.19 | 0.19 | 4 | 776.63 | 1.64 | 59.3 | 1.27 | 40.13 |
| 30 | Rhazicine | 0.78 | 0.07 | 0.22 | 0.22 | 6 | 757.54 | 1.66 | 64.83 | 1.38 | 39.13 |
| 31 | Leepacine | 0.81 | 0.08 | 0.19 | 0.19 | 7 | 709 | 1.68 | 63.18 | 1.39 | 37.7 |
| 32 | 2-Methoxy-1,2-dihydrorhazimine | 0.79 | 0.07 | 0.21 | 0.21 | 6 | 800.93 | 1.63 | 56.72 | 1.31 | 41.05 |
| 33 | HR-1 | 0.78 | 0.07 | 0.22 | 0.22 | 4 | |||||
| 34 | Vincanicine | 0.83 | 0.08 | 0.17 | 0.17 | 5 | 683.12 | 1.66 | 57.29 | 1.3 | 36.52 |
| 35 | Rhazinaline | 0.81 | 0.08 | 0.19 | 0.19 | 5 | 690.3 | 1.69 | 54.96 | 1.38 | 38.6 |
| 36 | Beta-sitosterol | 0.97 | 0 | 0.03 | 0.03 | 4 | 1051.02 | 1.52 | 37.64 | 0.98 | 51.22 |
| 37 | Ursolic acid | 0.91 | 0 | 0.09 | 0.09 | 5 | 1076.71 | 1.56 | 45 | 1.1 | 52.93 |
| 38 | Stigmasterol | 0.97 | 0 | 0.03 | 0.03 | 4 | 1038.63 | 1.53 | 38.25 | 0.99 | 51.19 |
| 39 | Oleanolic acid | 0.91 | 0 | 0.09 | 0.09 | 5 | 1077.07 | 1.56 | 45.41 | 1.1 | 52.95 |
| 40 | Rhazidigenine | 0.86 | 0.09 | 0.14 | 0.14 | 4 | 650.55 | 1.64 | 48.09 | 1.21 | 35.15 |
| 41 | N-methylleuconolam | 0.8 | 0.08 | 0.2 | 0.2 | 4 | 730.79 | 1.65 | 61.9 | 1.31 | 37.53 |
| 42 | (+)-Quebrachamine | 0.9 | 0.1 | 0.1 | 0.1 | 4 | 672.49 | 1.62 | 50.29 | 1.12 | 35.27 |
| 43 | Polyneuridine | 0.81 | 0.08 | 0.19 | 0.19 | 6 | 735.31 | 1.67 | 62.83 | 1.34 | 38.85 |
| 44 | (+)-Vincadiformine | 0.84 | 0.08 | 0.16 | 0.16 | 5 | 735.7 | 1.63 | 53.98 | 1.25 | 38.4 |
| 45 | (-)-Vincadifformine | 0.84 | 0.08 | 0.16 | 0.16 | 5 | 735.7 | 1.63 | 53.98 | 1.25 | 38.4 |
| 46 | Secamine | 0.84 | 0.08 | 0.16 | 0.16 | 7 | 1449.04 | 1.63 | 46.81 | 1.22 | 78.28 |
| 47 | Vincadine | 0.84 | 0.08 | 0.16 | 0.16 | 4 | 776.11 | 1.61 | 52.34 | 1.18 | 39.67 |
| 48 | Bis-strictidine | 0.9 | 0.1 | 0.1 | 0.1 | 9 | 1150.88 | 1.73 | 52.95 | 1.31 | 67.18 |
| 49 | 3,14-Dehydrorhazigine | 0.87 | 0.09 | 0.13 | 0.13 | 7 | 1340.13 | 1.64 | 46.53 | 1.2 | 73.46 |
| 50 | 16-Hydrorhazisidine | 0.85 | 0.09 | 0.15 | 0.15 | 7 | 1345.28 | 1.65 | 48.02 | 1.24 | 73.94 |
| 51 | Rhazisidine | 0.87 | 0.09 | 0.13 | 0.13 | 8 | 1284.82 | 1.68 | 49.12 | 1.27 | 72.59 |
| 52 | Isorhazicine | 0.78 | 0.07 | 0.22 | 0.22 | 6 | 757.54 | 1.66 | 64.83 | 1.38 | 39.13 |
| 53 | Rhazigine | 0.87 | 0.09 | 0.13 | 0.13 | 7 | 1412.62 | 1.65 | 58.36 | 1.21 | 74.23 |
| 54 | Strictisidine | 0.81 | 0.08 | 0.19 | 0.19 | 7 | 635.5 | 1.78 | 63.63 | 1.55 | 37.59 |
| 55 | Strictamine-N-oxide | 0.8 | 0.08 | 0.2 | 0.2 | 5 | |||||
| 56 | Strictigine | 0.9 | 0.1 | 0.1 | 0.1 | 5 | 622.49 | 1.63 | 42.69 | 1.14 | 34.52 |
| 57 | Strictine | 0.8 | 0.08 | 0.2 | 0.2 | 5 | 636.29 | 1.73 | 55.79 | 1.44 | 36.71 |
| 58 | Stricticine | 0.8 | 0.08 | 0.2 | 0.2 | 6 | 682.41 | 1.68 | 61.46 | 1.39 | 36.43 |
| 59 | Strictalamine | 0.86 | 0.09 | 0.14 | 0.14 | 5 | 580.88 | 1.74 | 55.15 | 1.37 | 33.92 |
| 60 | 1,2-Dehydroaspidospermidine | 0.9 | 0.1 | 0.1 | 0.1 | 5 | 590.09 | 1.7 | 50.6 | 1.27 | 33.8 |
| 61 | Tetrahydrosecodine | 0.84 | 0.08 | 0.16 | 0.16 | 3 | 807.26 | 1.56 | 42.67 | 1.08 | 40.69 |
| 62 | Dihydrosecodine | 0.84 | 0.08 | 0.16 | 0.16 | 3 | 793.18 | 1.58 | 44.47 | 1.11 | 40.53 |
| 63 | Dihydrosecamine | 0.84 | 0.08 | 0.16 | 0.16 | 7 | 1449.04 | 1.63 | 46.81 | 1.23 | 78.28 |
| 64 | Dihydropresecamine | 0.84 | 0.08 | 0.16 | 0.16 | 7 | 1530.43 | 1.64 | 59.1 | 1.23 | 78.84 |
| 65 | Tetrahydropresecamine | 0.84 | 0.08 | 0.16 | 0.16 | 7 | 1544.52 | 1.63 | 58.11 | 1.22 | 78.96 |
| 66 | Rhazinol | 0.86 | 0.09 | 0.14 | 0.14 | 5 | 580.88 | 1.74 | 55.15 | 1.38 | 33.92 |
| 67 | Rhazimol | 0.81 | 0.08 | 0.19 | 0.19 | 5 | 690.3 | 1.69 | 54.96 | 1.39 | 38.6 |
| 68 | Rhazidigenine-N-oxide | 0.83 | 0.09 | 0.17 | 0.17 | 4 | |||||
| 69 | (-)16R,21R-omethyleburnamine | 0.87 | 0.09 | 0.13 | 0.13 | 5 | 639.83 | 1.67 | 47.55 | 1.27 | 36.25 |
| 70 | Decarbomethoxy-15,20,16,17-tetrahydros | 0.9 | 0.1 | 0.1 | 0.1 | 3 | 703.65 | 1.57 | 40.68 | 1.02 | 36.31 |
| 71 | 1-2-Dehydroasidospermidine-N-oxide | 0.86 | 0.09 | 0.14 | 0.14 | 5 | |||||
| 72 | Rhazizine | 0.78 | 0.07 | 0.22 | 0.22 | 6 | 744.62 | 1.67 | 62.43 | 1.39 | 39.14 |
| 73 | 15-Hydroxyvincadifformine | 0.81 | 0.08 | 0.19 | 0.19 | 5 | 750.68 | 1.65 | 60.33 | 1.32 | 39 |
| 74 | Dihydroeburnamenine | 0.9 | 0.1 | 0.1 | 0.1 | 5 | 589.57 | 1.7 | 49.78 | 1.26 | 33.94 |
| 75 | 16s,16′-Decarboxytetra-hydrosecamine | 0.87 | 0.09 | 0.13 | 0.13 | 7 | 1339.61 | 1.64 | 46.2 | 1.21 | 73.6 |
| 76 | Nor-C-fluorocurarine | 0.86 | 0.09 | 0.14 | 0.14 | 5 | 624.5 | 1.68 | 57.83 | 1.29 | 33.99 |
| 77 | Strictibine | 0.81 | 0.06 | 0.19 | 0.19 | 3 | 442.8 | 1.65 | 51.74 | 1.29 | 23.76 |
Ind Ref refractive index, Para parachor, Sur ten surface tension, Polar polarizability, #R number of rings, CR ratio of carbons, NR ratio of nitrogens, NOR ratio of oxygens, HetR ratio of heteroatoms
Predicted therapeutic area applications
PASS—prediction of activity spectra for substances
This web-based predictive server from Way2Drug, has variety of annotators of substances for their probability of active or inactive towards few targets. Out of all services and products of them, we utilized PASS method of predictions. More than 100 activities are predicted with their probability of activities and in-activities. Some of them include kinase inhibitors, GPCR antagonists, and some specific targets like adrenergic receptors, and their kinase inhibitors. We considered the probability of active (Pa) >0.3 (i.e. >30%), and should be greater than probability of inactive (Pi). Given these conditions, we observed many alkaloids have indicated Pa >0.8 in certain conditions (such as, anthrine has predicted Pa at 90% towards β-adrenergic receptor kinase inhibitor, 5-HTA release stimulant). Majority of them also is predicted to be substrate to CYP3A4 and CYP2D6 indicating their metabolic instability (Pa ~ 0.5, 0.4, respectively). Several such predictions for all 78 alkaloids has been computed—leaving predictions to be validated, experimentally. Similarly, dihydrocorynantheol and corynantheol were also predicted to be 5-HT release stimulants, and have been projected to be chemosensitizers. Eburnamenine is predicted to be a Nootropic agent at 90% Pa, while eburnamine is predicted to be a CNS (anti-depressant and mood disorder management agent at >96% Pa). Strictosidine is predicted to be an antiprotozoal at 86% Pa, β-sitosterol is anti-hypercholesterolemic agent with Pa ~98%, rhazidigenine (rhazidine) is an antidyskinetic at 60% Pa, secamine is a H1F1A expression inhibitor at 83% Pa (but a non-pharmaceutically acceptable molecule due to high MW and many RO5 violations). A similar observations is also made for 16-hydrorhazisidine (72% Pa for H1F1A expression inhibitor). Strictamine is predicted to be gluconate 2-dehydrogenase acceptor with 70% Pa, and 1,2-dehydroaspidospermine (which is a small molecule) has been predicted to be analeptic with 77% Pa. Dihydrosecamine is predicted to be a H1F1A expression inhibitor with 77% Pa, and rhazidigenine-N-oxide is predicted to be a cognition disorder agent with 64% Pa. Decarbomethoxy-15,20,16,17-tetrahydrosecodine is a small molecule with ~70% Pa for antidyskinetic and antineuronic agent, 1,2-dehydrospidospermidine-N-oxide is predicted to be 87% as analeptic.
Anticancer activity through CDRUG
This set of predictions using the structures and SMILES codes of the alkaloids, annotates the anti-cancer activity by predicting “Mean logGI50”. Most molecules that have Mean LogGI50 values lower than −5 are considered to have anti-cancer activity. It is interesting to know that all the molecules of R. stricta alkaloids (indole/non-indole) have predicted mean logGI50 values ranging between −4.95 and −6.50—indicating they all may have anti-cancer activities. There are about 10 compounds that have predicted logGI50 values less than −6, which indicate strong anti-cancer activity. Table 7 shows the predicted mean LogGI50 values of all the compounds considered in the present study.
Table 7.
Predicted mean LogGI50 of Rhazya stricta compounds whose values lower than −6.0 are highlighted in italics may exhibit anti-cancer activity
| MOL ID | Name | Mean LogGI50 CDRUG |
|---|---|---|
| M1 | Akummidine | −5.408 |
| M2 | Antirhine | −5.408 |
| M3 | 3-Epi-antirhine | −5.408 |
| M4 | Aspidosespermidine | −5.726 |
| M5 | Condylocarpine | −5.726 |
| M6 | Dihydrocorynantheol | −5.408 |
| M7 | Eburnamenine | −5.096 |
| M8 | Eburnamine | −5.096 |
| M9 | Eburnamonine | −5.096 |
| M10 | Geissoschizine | −5.048 |
| M11 | Isositsirikine | −5.408 |
| M12 | 16-Epi-Z-isositsirikine | −5.408 |
| M13 | Leuconalm | −5.154 |
| M14 | Rhazinliam | −5.096 |
| M15 | Tetrahydrosecamine | −4.975 |
| M16 | Presecamine | −5.726 |
| M17 | Sewarine | −5.726 |
| M18 | Stemmadenine | −5.408 |
| M19 | Strictamine | −5.726 |
| M20 | Strictosamide | −5.256 |
| M21 | Strictosidine | −5.937 |
| M22 | Taberonine | −5.726 |
| M23 | Tetrahydrlstonine | −5.408 |
| M24 | Vallesiachotamine | −5.408 |
| M25 | Aspidospermoise | −5.726 |
| M26 | Bhimbrine | −5.408 |
| M27 | Bhimbrine N-oxide | −5.408 |
| M28 | Rhazimine | −5.726 |
| M29 | Rhazimanine | −5.408 |
| M30 | Rhazicine | −5.726 |
| M31 | Leepacine | −5.726 |
| M32 | 2-Methoxy 1-2,dihydrorhazamine | −5.726 |
| M33 | HR-1 | −5.096 |
| M34 | Vincanicine | −5.726 |
| M35 | Rhazinaline | −5.726 |
| M36 | Beta-sitosterol | −5.918 |
| M37 | Ursolic acid | −5.124 |
| M38 | Stigmasterol | −5.918 |
| M39 | Olenaolic acid | −5.124 |
| M40 | Rhazidigenine (rhazidine) | − 6.327 |
| M41 | N-methylleuconolam | −5.154 |
| M42 | (+)-Quebranchamine | −5.861 |
| M43 | Polyneuridine | −5.408 |
| M44 | (+)-Vincadiformine | −5.726 |
| M45 | (−)-Vincadiformine | −5.726 |
| M46 | Secamine | − 6.298 |
| M47 | Vincadine | −5.486 |
| M48 | Bis-strictidine | −5.409 |
| M49 | 3,14-Dehydrorhazigine | −5.726 |
| M50 | 16-Hydrorhazisidine | − 6.298 |
| M51 | Rhazisidine | −5.406 |
| M52 | Isorhazicine | −5.726 |
| M53 | Rhazigine | −5.726 |
| M54 | Strictisidine | −5.726 |
| M55 | Strictamine-N-oxide | −5.726 |
| M56 | Strictigine | −5.726 |
| M57 | Strictine | −5.096 |
| M58 | Stricticine | −5.726 |
| M59 | Strictalamine | − 6.327 |
| M60 | 1,2-Dehydroaspidospermine | − 6.327 |
| M61 | Tetrahydrosecodine | −5.783 |
| M62 | Dihydrosecodine | −5.408 |
| M63 | Dihydrosecamine | − 6.298 |
| M64 | Dihydropresecamine | −5.726 |
| M65 | Tetrahydropresecamine | −5.726 |
| M66 | Rhazinol | −5.726 |
| M67 | Rhazimol | −5.726 |
| M68 | Rhazidigenine-N-oxide | − 6.327 |
| M69 | (−)-16R,21R-Omethyleburmanine | −5.096 |
| M70 | Decarbomethoxy-15,20,16,17-tetrahydrosecodine | − 6.471 |
| M71 | 1,2-Dehydroaspidospermidine-N-oxide | − 6.327 |
| M72 | Rhazizine | −4.878 |
| M73 | 15-Hydroxyvincadifformine | −5.726 |
| M74 | Dihydroburnamenine | −5.096 |
| M75 | 16s,16′-Decarboxytetrahydrosecamine | −4.975 |
| M76 | Nor-C-fluorocuraine | −5.726 |
| M77 | Strictibine | −5.785 |
SuperPred—predicted target interactions
From this server studies on R. stricta alkaloids, we observed that many of these molecules may interact with CYP2D6 or CYP3A4 as substrates. The indication of these results mean that their target may be unknown, but they do modify the drug metabolism, and affect drug–drug interactions.
SwissTarget prediction
While predictions from this web-server may suggest each molecule have certain target activity, they almost correlate well with the PASS server prediction—which gives additional probability of prediction for each molecule to be active or inactive against the target of interest.
Overall from the calculated cheminformatics studies and web-server predictions, we understand that few molecules like anthrine, condylocarpine, dihydrocorynantheol etc. have predicted GIC50 values in sub µM concentrations, while they also have predicted drug–drug activity towards CYP3A4, and CYP2D6 enzymes. Most molecules turnout to be modulators of membrane receptor ligands while some have predicted cholinesterase, CNS (5HT2x), adenosine (A2A/A2B) activity. Moreover, all molecules have predicted activity towards certain targets (Pa > 30%).
Conclusions
Table 8 indicates the top 10-best naturally occurring indole alkaloids of R. stricta that were predicted to be having decent anti-cancer activity and other good physiochemical properties together with cheminformatics properties—these molecules are antirhine, 3-epi-antirhine, condylocarpine, eburnamine, eburnamonine, taberonine, ursolic acid, stigmasterol, olenaolic acid, (+)-vincadiformine, (−)-vincadiformine, (−)-16R,21R-omethyleburmanine, 15-hydroxy-vincadifformine, and dihydroburnamenine.
Table 8.
Key details of top molecules with predicted targets for anti-cancer and anti-obesity, probable rule-of-5, predicted LogGI50 with predicted H-, and p values
| SI. No | Mol. name | Mol. wt | Predicted | ||||
|---|---|---|---|---|---|---|---|
| LogG150/H-/p val | Target | RO5 violations | Liability | Comment | |||
| Anti-cancer | Anti-obesity | Druggability | Hepatic | HERG, renal issues | |||
| M2 | Antirhine | 296.411 | −5.41/0.39/0.05 | 5HT2A,BC | Good | CYP2D6 | None predicted |
| M3 | 3-Epi-antirhine | 296.411 | −5.41/0.39/0.05 | 5HT2A,B | Good | CYP2D6 | None predicted |
| M5 | Condylocarpine | 322.406 | −5.73/0.42/0.03 | Negative | Good | None | None predicted |
| M8 | Eburnamine | 296.411 | −5.10/0.74/0.01 | 5HT2A,BC | Good | 2D6,3A4 | None predicted |
| M9 | Eburnamonine | 294.396 | −5.10/1.00/0.01 | 5HT2A,BC | Good | 2D6,3A4 | None predicted |
| M22 | Taberonine | 336.433 | −5.73/0.67/0.01 | Negative | Good | None | None predicted |
| M37 | Ursolic acid | 456.707 | −5.12/1.00/0.00 | Negative | Moderate (LogP) | None | Highly hydrophobic |
| M38 | Stigmasterol | 412.698 | −5.92/0.93/0.04 | Negative | Moderate (LogP) | CYP17A1 | Highly hydrophobic |
| M39 | Olenaolic acid | 456.707 | −5.12/0.71/0.07 | Negative | Moderate (LogP) | None | Highly hydrophobic |
| M44 | (+)-Vincadiformine | 338.449 | −5.73/0.56/0.02 | 5HT3A | Good | None | None predicted |
| M45 | (−)-Vincadiformine | 338.449 | −5.73/0.56/0.02 | 5HT3A | Good | None | None predicted |
| M69 | (−)-16R,21R-Omethyleburma nine | 310.438 | −5.10/0.55/0.02 | 5HT2A,BC | Good | CYP2D6 | None predicted |
| M73 | 15-Hydroxy-vincadifformine | 354.448 | −5.73/0.56/0.02 | 5HT2A,BC | Good | None | None predicted |
| M74 | Dihydroburnamenine | 280.412 | −5.10/0.63/0.01 | Negative | Good | 2D6,3A4 | None predicted |
Authors’ contribution
AYO, SV, RSB were involved in generation of computational data on predictive properties of various Rhazya stricta’s alkaloids; NHH and AMSO participated in data acquisition. SV, JSMS and KSS were involved in overall research planning & supervision, data analysis and manuscript writing. All authors read and approved the final manuscript.
Acknowledgements
The authors gratefully acknowledge the financial support from KAU Vice President for Educational Affairs Prof. Dr. Abdulrahman O Alyoubi, The Deanship of Scientific Research (DSR) at King Abdulaziz University (KAU), Jeddah, Saudi Arabia, represented by the Unit of Strategic Technologies Research through the Project number (D 008/431) for the Project entitled: “Identification and Isolation of Salt and Heat-Tolerance Genes of R. stricta and Detection of Metabolites and their Therapeutic Effects via Cheminformatics”.
Competing interests
The authors declare that they have no competing interests.
Contributor Information
Abdullah Y. Obaid, Email: aobaid@kau.edu.sa
Sreedhara Voleti, Email: sreedhara.voleti@gmail.com.
Roop Singh Bora, Email: roopbora@yahoo.co.in.
Nahid H. Hajrah, Email: nhajrah260@gmail.com
Abdulkader M. Shaikh Omer, Email: ak_shaikh@yahoo.com.
Jamal S. M. Sabir, Email: jsabir2622@gmail.com
Kulvinder Singh Saini, Email: ksaini@kau.edu.sa.
References
- 1.Marwat SK, Fazal-ur-Rehman, Usman K, Shah SS, Anwar N, Ullah I. A review of phytochemistry, bioactivities and ethno medicinal uses of Rhazya stricta Decsne (Apocynaceae) Afr J Microbiol Res. 2012;6(8):1629–1641. [Google Scholar]
- 2.Ahmad Y, Fatima K, Le Quesne PPW, Atta-ur-Rahman Further alkaloidal constituents of the leaves of Rhazya stricta. Phytochemistry. 1983;22:1017–1019. doi: 10.1016/0031-9422(83)85045-6. [DOI] [Google Scholar]
- 3.Atta-ur-Rahman, Zaman K, Habib-ur-Rehman, Malik S. Studies on alkaloids of Rhazya stricta. J Nat Prod. 1986;49:1138–1139. doi: 10.1021/np50048a033. [DOI] [Google Scholar]
- 4.Saeed SA, Simjee RU, Mahmood F, Sultana N, Atta-ur-Rahman Rhazimine from Rhazya stricta: a dual inhibitor of arachidonic acid metabolism and platelet activating factor-induced platelet aggregation. Planta Med. 1993;59(6):566–568. doi: 10.1055/s-2006-959765. [DOI] [PubMed] [Google Scholar]
- 5.Tanira MO, Ali BH, Bashir AK, Wasfi IA, Chandranath I. Evaluation of the relaxant activity of some United Arab Emirates plants on intestinal smooth muscles. J Pharm Pharmacol. 1996;47:457–459. doi: 10.1111/j.2042-7158.1996.tb05971.x. [DOI] [PubMed] [Google Scholar]
- 6.Ali BH, Bashir AK, Tanira MO. The anti-oxidant action of Rhazya stricta in rats. Phytothera Res. 2000;14:469–471. doi: 10.1002/1099-1573(200009)14:6<469::AID-PTR612>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 7.El Gendy MAM, Ali BH, Michail K, Siraki AG, El Kadi AOS. Induction of quinine oxidoreductase 1 enzyme by R. stricta through Nrf2-dependent mechanisms. J Ethnopharmacol. 2012;144:416–424. doi: 10.1016/j.jep.2012.09.032. [DOI] [PubMed] [Google Scholar]
- 8.Baeshen NA, Lari SA, Aldoghaither HA, Elkady AI. Biochemical evaluation of the effect of Rhazya stricta aqueous leaves extract in the liver and kidney functions in rats. Nat Sci. 2010;8:136–142. [Google Scholar]
- 9.Baeshen N, Lari S, Al Doghaither HA, Ramadan HAI. Effect of Rhazya stricta extract on rat adiponectin gene and insulin resistance. J American Sci. 2010;6:1237–1245. [Google Scholar]
- 10.Baeshina NA, Yaghmoor SS, Ashmaouia HM, Kumosani TA, Saini KS. The indole-alkaloid fraction of Rhazya stricta improves key biochemical parameters associated with metabolic syndrome in rats. Bothalia J. 2014;44:358–371. [Google Scholar]
- 11.Mukhopadhayay S, Handy GA, Funayama S, Cordell GA. Anticancer indole alkaloids of Rhazya stricta. J Nat Prod. 1981;44:696–700. doi: 10.1021/np50018a014. [DOI] [PubMed] [Google Scholar]
- 12.Baeshen NA, Elkady AI, AbuZinadah OA, Mutwakil MH. Potential anticancer activity of the medicinal herb, Rhazya stricta, against human breast cancer. Af J Biotechnol. 2012;11(37):8960–8972. doi: 10.5897/AJB12.570. [DOI] [Google Scholar]
- 13.Elkady AI. Crude alkaloid extract of Rhazya stricta inhibits cell growth and sensitizes human lung cancer cells to cisplatin through induction of apoptosis. Gen Mol Biol. 2013;36(1):12–21. doi: 10.1590/S1415-47572013005000009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elkady AI, Hussein RAH, Abu-Zinadah OA. Differential control of growth, apoptotic activity and gene expression in human colon cancer cells by extracts derived from medicinal herbs, Rhazya stricta and Zingiber officinale and their combination. World J Gastroenterol. 2014;20(41):15275–15288. doi: 10.3748/wjg.v20.i41.15275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46(1–3):3–26. doi: 10.1016/S0169-409X(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 16.Purnapatre K, Khattar SK, Saini KS. Cytochrome P450s in the development of target-based anticancer drugs. Cancer Lett. 2008;259:1–15. doi: 10.1016/j.canlet.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 17.Singh D, Kashyap A, Pandey RV, Saini KS. Novel advances in cytochrome P450 research. Drug Discovery Today. 2011;16:793–799. doi: 10.1016/j.drudis.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 18.Filimonov DA, Lagunin AA, Gloriozova TA, Rudik AV, Druzhilovskii DS, Pogodin PV, Poroikov VV. Prediction of the biological activity spectra of organic compounds using the PASS Online web resource. Chem Heterocycl Compd. 2014;50(3):444–457. doi: 10.1007/s10593-014-1496-1. [DOI] [Google Scholar]
- 19.Nickel J, Gohlke BO, Erehman J, Banerjee P, Rong WW, Goede A, Dunkel M, Preissner R. SuperPred: update on drug classification and target prediction. Nucleic Acids Res. 2014;42:26–31. doi: 10.1093/nar/gku477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gfeller D, Grosdidier A, Wirth M, Daina A, Michielin O, Zoete V. Swiss Target prediction: a web server for target prediction of bioactive small molecules. Nucleic Acids Res. 2014;42(1–5):W32–W38. doi: 10.1093/nar/gku293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li GH, Huang JF. CDRUG: a web server for predicting anticancer activity of chemical compounds. Bioinformatics. 2012;28(24):3334–3335. doi: 10.1093/bioinformatics/bts625. [DOI] [PubMed] [Google Scholar]
- 22.Schrödinger Release 2015-4: Maestro, version 10.4, Schrödinger, LLC, New York, 2015
- 23.http://www.acdlabs.com/products/enterprise


