Abstract
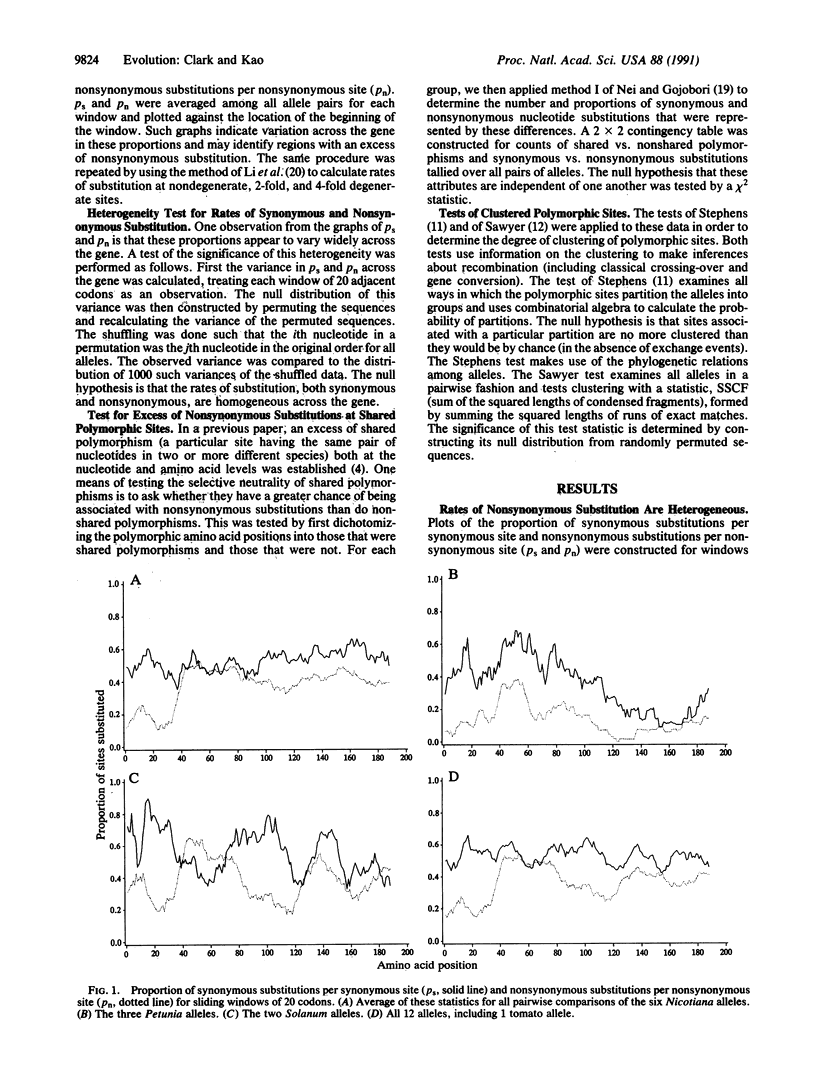
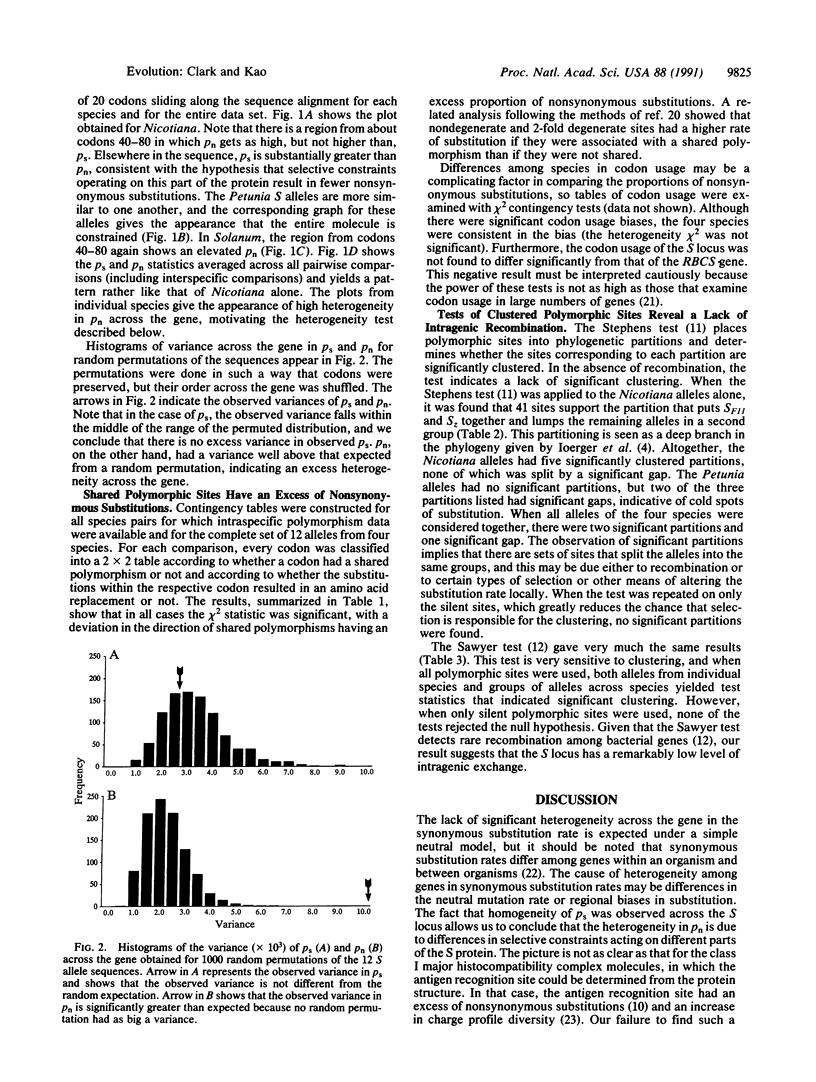
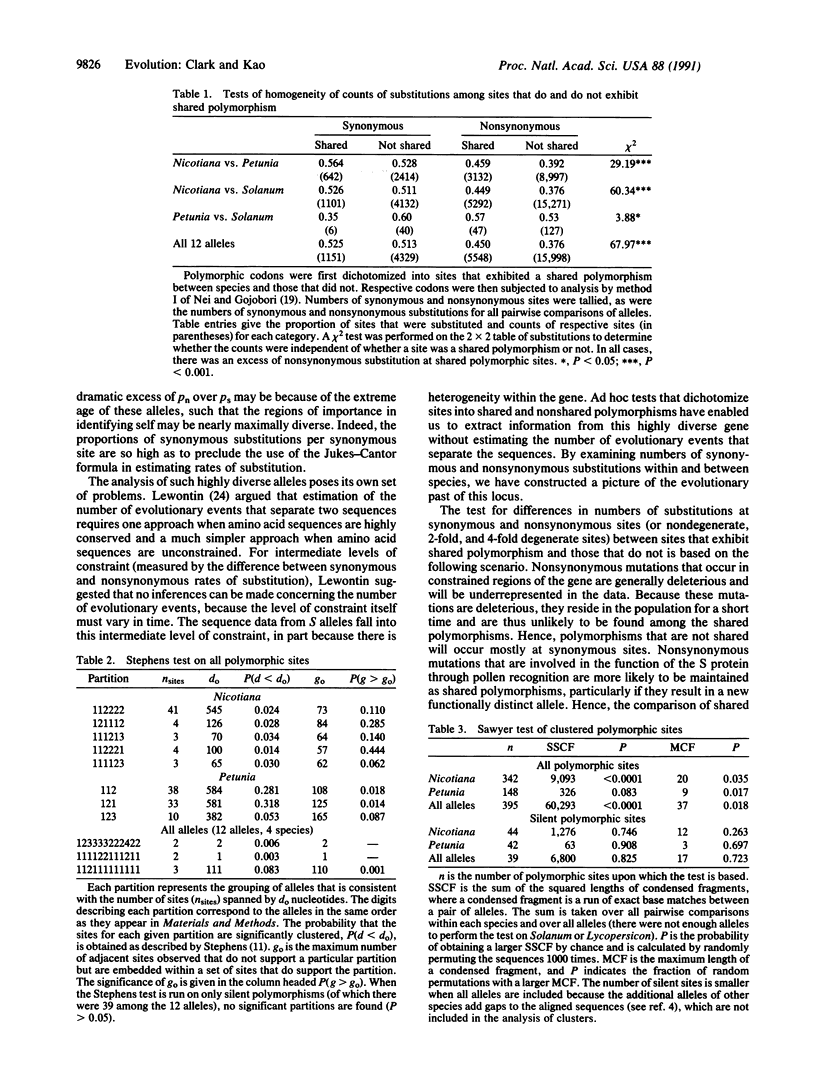
The function of the self-incompatibility locus (S locus) of many plant species dictates that natural selection will favor high levels of protein diversity. Pairwise sequence comparisons between S alleles from four species of Solanaceae reveal remarkably high sequence diversity and evidence for shared polymorphism. The level of amino acid constraint was found to be significantly heterogeneous among different regions of the gene, with some regions being highly constrained and others appearing to be virtually unconstrained. In some regions of the protein, there was an excess of nonsynonymous over synonymous substitution, consistent with the strong diversifying selection that must operate on this locus. These hypervariable regions are candidates for the sites that determine functional allelic identity. Simple contingency table tests show that sites that have polymorphism shared between species have more nonsynonymous substitution than polymorphic sites that do not exhibit shared polymorphism. This is consistent with the idea that adaptive evolution favoring amino acid replacement is occurring at sites with shared polymorphism. Tests of clustered polymorphism reveal that an unusually low rate of recombination must be occurring in this locus, allowing very ancient alleles to preserve their identity.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson M. A., McFadden G. I., Bernatzky R., Atkinson A., Orpin T., Dedman H., Tregear G., Fernley R., Clarke A. E. Sequence variability of three alleles of the self-incompatibility gene of Nicotiana alata. Plant Cell. 1989 May;1(5):483–491. doi: 10.1105/tpc.1.5.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K. R., Okuley J. J., Collins P. D., Sims T. L. Sequence variability and developmental expression of S-alleles in self-incompatible and pseudo-self-compatible petunia. Plant Cell. 1990 Aug;2(8):815–826. doi: 10.1105/tpc.2.8.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert P. R., Anderson M. A., Bernatzky R., Altschuler M., Clarke A. E. Genetic polymorphism of self-incompatibility in flowering plants. Cell. 1989 Jan 27;56(2):255–262. doi: 10.1016/0092-8674(89)90899-4. [DOI] [PubMed] [Google Scholar]
- Glass N. L., Vollmer S. J., Staben C., Grotelueschen J., Metzenberg R. L., Yanofsky C. DNAs of the two mating-type alleles of Neurospora crassa are highly dissimilar. Science. 1988 Jul 29;241(4865):570–573. doi: 10.1126/science.2840740. [DOI] [PubMed] [Google Scholar]
- Haring V., Gray J. E., McClure B. A., Anderson M. A., Clarke A. E. Self-incompatibility: a self-recognition system in plants. Science. 1990 Nov 16;250(4983):937–941. doi: 10.1126/science.2237440. [DOI] [PubMed] [Google Scholar]
- Hughes A. L., Nei M. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature. 1988 Sep 8;335(6186):167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- Hughes A. L., Ota T., Nei M. Positive Darwinian selection promotes charge profile diversity in the antigen-binding cleft of class I major-histocompatibility-complex molecules. Mol Biol Evol. 1990 Nov;7(6):515–524. doi: 10.1093/oxfordjournals.molbev.a040626. [DOI] [PubMed] [Google Scholar]
- Ioerger T. R., Clark A. G., Kao T. H. Polymorphism at the self-incompatibility locus in Solanaceae predates speciation. Proc Natl Acad Sci U S A. 1990 Dec;87(24):9732–9735. doi: 10.1073/pnas.87.24.9732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin R. C. Inferring the number of evolutionary events from DNA coding sequence differences. Mol Biol Evol. 1989 Jan;6(1):15–32. doi: 10.1093/oxfordjournals.molbev.a040532. [DOI] [PubMed] [Google Scholar]
- Li W. H., Wu C. I., Luo C. C. A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Mol Biol Evol. 1985 Mar;2(2):150–174. doi: 10.1093/oxfordjournals.molbev.a040343. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Altschul S. F., Kececioglu J. D. A tool for multiple sequence alignment. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4412–4415. doi: 10.1073/pnas.86.12.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T., Nei M. Genetic variability maintained by mutation and overdominant selection in finite populations. Genetics. 1981 Jun;98(2):441–459. doi: 10.1093/genetics/98.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986 Sep;3(5):418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987 Jul;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sawyer S. Statistical tests for detecting gene conversion. Mol Biol Evol. 1989 Sep;6(5):526–538. doi: 10.1093/oxfordjournals.molbev.a040567. [DOI] [PubMed] [Google Scholar]
- Schulz B., Banuett F., Dahl M., Schlesinger R., Schäfer W., Martin T., Herskowitz I., Kahmann R. The b alleles of U. maydis, whose combinations program pathogenic development, code for polypeptides containing a homeodomain-related motif. Cell. 1990 Jan 26;60(2):295–306. doi: 10.1016/0092-8674(90)90744-y. [DOI] [PubMed] [Google Scholar]
- Sharp P. M., Cowe E., Higgins D. G., Shields D. C., Wolfe K. H., Wright F. Codon usage patterns in Escherichia coli, Bacillus subtilis, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster and Homo sapiens; a review of the considerable within-species diversity. Nucleic Acids Res. 1988 Sep 12;16(17):8207–8211. doi: 10.1093/nar/16.17.8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens J. C. Statistical methods of DNA sequence analysis: detection of intragenic recombination or gene conversion. Mol Biol Evol. 1985 Nov;2(6):539–556. doi: 10.1093/oxfordjournals.molbev.a040371. [DOI] [PubMed] [Google Scholar]
- Takahata N. A simple genealogical structure of strongly balanced allelic lines and trans-species evolution of polymorphism. Proc Natl Acad Sci U S A. 1990 Apr;87(7):2419–2423. doi: 10.1073/pnas.87.7.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata N., Nei M. Allelic genealogy under overdominant and frequency-dependent selection and polymorphism of major histocompatibility complex loci. Genetics. 1990 Apr;124(4):967–978. doi: 10.1093/genetics/124.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyenoyama M. K. On the evolution of genetic incompatibility systems. VI. A three-locus modifier model for the origin of gametophytic self-incompatibility. Genetics. 1991 Jun;128(2):453–469. doi: 10.1093/genetics/128.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. The Distribution of Self-Sterility Alleles in Populations. Genetics. 1939 Jun;24(4):538–552. doi: 10.1093/genetics/24.4.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B. B., Mu J. H., Nevins D. L., Grun P., Kao T. H. Cloning and sequencing of cDNAs encoding two self-incompatibility associated proteins in Solanum chacoense. Mol Gen Genet. 1990 Dec;224(3):341–346. doi: 10.1007/BF00262427. [DOI] [PubMed] [Google Scholar]



