Abstract
Mechanism of inactivation of bio-particles exposed to non-thermal plasma (NTP), namely, dielectric barrier discharge (DBD), and plasma jet (PJ), has been studied using E. coli, B. subtilis spore, S. cerevisiae and bacteriophages. States of different biological components were monitored during the course of inactivation. Analysis of green fluorescent protein, GFP, introduced into E. coli. or B. subtiles spore cells proved that radicals generated by NTP penetrate into microbes, destroying the cell membrane and finally damage the genes. We have evaluated the damage of the bacteriophages. Bacteriophage λ having double stranded DNA was exposed to DBD, then DNA was purified and subjected to in vitro DNA packaging reactions. The re-packaged phages consist of the DNA from discharged phages and brand-new coat proteins were proved to be active, indicating that the damage of coat proteins is responsible for inactivation. M13 phages having single stranded DNA were also examined with the same manner. In this case, damage to the DNA was as severe as that of the coat proteins. For practical applications, DBD showed very intense sterilization ability for B. Subtilis spore with the D-value of less than 10 s. This result indicates a possibility of application of NTP for quick sterilization.
Keywords: non-thermal plasma, sterilization, dielectric barrier discharge, plasma jet, radical
Introduction
Influenza, avian flu (H5N1) or swine flu (H1N1), and drug resistant microbes are of concern for public health. Non-thermal atmospheric pressure plasma (NTP), is effective for dealing with such menaces caused by bio-particles (BPs) because, in principle, it can decontaminate both the surface of materials and room air at room temperature without using bactericides.(1,2) Though the study of plasma decontamination is expanding, the mechanism of inactivation is still to be studied.(3,4) In this study, we report several aspects to evaluate damages due to radicals generated by NTP on E. coli, S. cerevisiae, B. subtilis and bacteriophages.(5–9)
Collection and Destruction of BPs by Electrostatic Precipitation
BPs are commonly existing around us, and electrostatic precipitation can collect suspended BPs in air with high efficiency and low pressure drop. At the meantime, corona discharges used in electrostatic precipitators, ESPs, generate radicals and other oxidative particles.(10) Those are effective to destroy BPs. Therefore, ESPs are potentially an important device to control airborne BPs, especially in hospitals and in public spaces.
Previous experimental work indicated effective destruction of yeast cell on the collection electrode of a corona discharge system.(2) We have tested the destruction effect of corona discharge generated in the needle-plate system. Bacteriophage MS2 can also be destroyed, with improved efficiency when the phages are in wet condition.
Fig. 1a illustrates the ESP used to collect airborne bio-particles. Film or culture media can be used as the collecting electrode. The needle electrode with DC high voltage was placed above the yeast extract peptone dextrose (YPD) medium, and corona discharge was generated between them. Distance between the needle electrode and YPD medium was 40 mm. The ESP was placed in a box and a fan was set at the top of the box to introduce the air from the upper side and the BPs are collected on the YPD medium. Different voltage was tested (0~30 kV) on each fresh YPD plate. Collected samples were incubated at 30°C for 48 h for the colony forming. Fig. 1b shows the effect of ESP on the BPs collection. Particles in room air were collected on YPD medium for 1 min. Positive and negative voltages were examined in this experiment. The result shows that, for both polarities, the number of collected bacteria increased with applied voltage. However, the positive corona discharge shows the higher efficiency than the negative corona. The highest collection efficiency was observed with applied voltage of 25 kV. This efficiency suggesting that collection of bacteria was enhanced by about 50 times compared with non-applied discharge BPs amount. In addition, influence of applied voltage and distance of electrodes on collection profiles of BPs was noticed, as shown in Fig. 2. In this case, the distance between the needle electrode and the surface of YDP media was 10 mm. This small distance was selected to measure the effect of radial distance more clearly. Relative density of the colonies was measured for comparison. This was calculated by measuring the colonies in a concentric circle of 9 mm width, and divided by total number of colonies. Close to the center of the plate, just under the high voltage electrode, the relative colony density increased. However, above a certain applied voltage, the relative colony density at the center decreased. The total number of colonies (Fig. 1b) also decreased with very high voltage. At 30 kV, the number of collected bacteria decreased. Due to ionization of electrical discharge, reactive atoms and radicals are generated,(11) and not only short-lived atoms and radicals, long lived oxidative species are generated. From mass spectroscopy, it is known that corona discharge generates stable neutral O3 molecules, and ions such as O3−(H2O)n, NO3−(H2O)n, O2−(H2O)n, in negative corona and NH4+(H2O)n in positive corona.(12) The decrease in colonies is due to lethal effect of these species generated by the corona discharge. It should be noted that, with the silver needle, ozone generation can be reduced in positive corona by at least 1/3 compared with that in negative corona.(13) This is the reason for larger number of colonies in the positive corona.
Fig. 1.
Collection of suspended bio-particles in air using electrostatic precipitation. (a) An electrostatic precipitator for collecting bio-particles in air. (b) Number of colony due to collected bio-particles using the ESP.
Fig. 2.
Radial distribution of the colonies collected by the electrostatic precipitation (electrode separation: 10 mm). (a) Positive corona. (b) Negative corona.
Experimental Apparatus and Samples
Dielectric barrier discharge and plasma jet for studying the mechanism of BP destruction
Fig. 3a illustrates the dielectric barrier discharge (DBD) reactor(14–17) used in this study. Stainless steel mesh (diameter: 50 mm, 20 mesh (separation between adjacent wire of the mesh = 0.127 mm) and aluminum plate were used as high voltage and GND electrodes respectively. Teflon sheet (2 mm-thick) was set on the high voltage electrode as a dielectric barrier. A high voltage AC power supply (Kasuga-denki AGF-010) was used to generate the uniform filamentous streamers in the atmospheric air gap (3 mm). All of the discharge experiments in this report were done in a fixed electrical condition except operating time. The peak to peak voltage, the frequency of the applied voltage and the input power were 20 kV peak-to-peak value (p-p), 2 kHz and the power density of 1.2 W/cm2, respectively. A piece of polyethylene terephthalate (PET) film (90 mm × 30 mm, 0.1 mm thickness) was soaked in 0.1% gelatin for 5 min and air dried for 24 h under UV light.
Fig. 3.
Electrical discharging devices used in the experiment. (a) Dielectric barrier discharge. (b) Plasma jet.
20 µl of sample solution was spotted and widely spread to 3~4 mm2 on the PET film and immediately applied the atmospheric plasma for intended time. After the DBD application, the sample solution was recovered into a microtube with additional washing by 100 µl of distilled water on the surface of the PET film. The recovered samples were rendered for experiments of survival estimation, fluorescence measurement, protein analysis, DNA analysis and microscopic observation.
The schematic of the plasma jet generator is shown in Fig. 3b.(18–20) The plasma jet generator was constructed with a glass tube, a stainless wire, and a stainless mesh. The stainless wire (0.2 mm in diameter) was fixed as a high voltage electrode coaxially inside the glass tube. The Stainless mesh (10 mm width) was wrapped around the end of the glass tube. This serves as a ground electrode. Dielectric-barrier discharge (DBD) is generated between both electrodes. Pulsed voltage of 12 kV0-P, frequency of 2.5 kHz, and the pulse width of 2.8 µs was used. Helium (or Argon) gas was used for the plasma torch with the flow rate of 2.0 L/min.
Samples and evaluation
To analyze the plasma inactivation process, we have genetically constructed a series of Bacillus subtilis strains whose proteins of spore layer are tagged with GFP (Green Fluorescent Protein).(21–23) Location of GFP fluorescence in each spore strain is limited to one of the layer of the spore (Fig. 4). Using these spore collections, relation between cell death and inactivation of GFP was investigated.
Fig. 4.
Bacillus subtilis spores differentially labeled with GFP.
Green Fluorescent Protein, GFP, coding plasmid pGLO (5.4 kb in size) was purchased from Bio-Rad Co., Inc. and transfected to E. coli MV1184. The transformants, E. coli MV1184 (pGLO), were propagated by shaking at 37°C for over night in 20 ml of LB medium supplemented with 5 mM l-arabinose which induces and accumulates GFP in the cells. The cells were harvested by centrifugation at 5,000 × g for 5 min, washed and finally suspended in 3 ml of 100 mM Tris·HCl (pH 8.0).
In order to study the stress of NTP on microbes, we have constructed a yeast-based genotoxicity test system using reporter assay linked to DNA damage-inducible promoter.(7) When damage is introduced to chromosomal DNA, the damage works as a signal which order to induce high level of ribonucleotide reductase subunit (RNR2) gene expression. RNR2 is involved in cellular DNA repair reactions. Therefore, on the reporter plasmid, lacZ gene under the control of RNR2 promoter is induced by chromosomal DNA damage. The extent of DNA damage is evaluated by the reporter β-glactosidase activity in the plasma treated yeast cells. Our yeast test system responded to many type of carcinogenic reagents specifically, H2O2 and UV irradiation.
Bacteriophage lambda (λCI857Sam7) was induced from λ lisogen, E. coli M65. The E. coli cells were inoculated to 200 ml of LB medium in a 1 L flask and shaken at 32°C. When OD650 (optical density at 650 nm wavelength) reached to 0.5, cultivation temperature was quickly shifted to 42°C and shaken for 20 min and then shaken for 3 h at 40°C. The cells were harvested by centrifugation at 8,000 × g for 5 min and resuspended with 10 ml of SM buffer. The cells were lysed by adding 0.1 ml of chloroform and 10 µl of 2 mg/ml of pancreatic DNase and gentle shaking at 37°C for 20 min. The cell lysate was centrifuged at 10,000 × g for 10 min and recovered the supernatant. Further purification of the λ phage in the lysate by stepwise CsCl density gradient centrifugation and CsCl equilibrium density gradient centrifugation was performed according to the literature protocol except that the phage was finally dialyzed against 100 mM Tris·HCl (pH 8.0), 1 mM MgCl2.
Purified DNA from E. coli MV1184 (pGLO) was obtained by serial extraction of the cells with phenol, phenol-chloroform, and chloroform. After precipitation with ethanol, the DNA was dissolved in 100 mM Tris·HCl (pH 8.0), 1 mM EDTA.
The DBD treated cells were serially diluted with SM buffer and plated on LB plates. After incubation at 37°C for 30 h, colonies on the plates were counted.
Titration of the DBD treated λ phage was done as follows. As indicator cells, E. coli Y-mel was cultivated in LB medium at 32°C for over night. 0.1 ml of the serially diluted λ phage solution was mixed with 0.1 ml of the indicator cell suspension and kept at 37°C for 10 min. 3 ml of LB soft agar kept at 45°C was added to the mixture of infection and poured on a LB plate. Phage plaques on the plates were counted after incubation at 40°C for 30 h.
φX174 phages were also used. A 50 µl aliquot of φX174 phages sample solution which is suspended in TE buffer (pH 7.5) to an appropriate density (1012 phages/ml) was spotted and widely spread to 3–4 cm2 on a PET film and the DBD was immediately applied for the intended time. On the other hand, dry φX174 phages sample which was dried up 3 h after a 50 µl aliquot of φX174 phages sample solution was spread on a PET firm also examined. After that the samples were recovered by additional 50 µl for wet sample or 100 µl for dry sample of the distilled water on the surface of the PET film. For the measurement of phage activity, the recovered phage solution was mixed with E. coli ATCC13706 and poured on an agar media plate. Phage plaques on the plates were counted after incubation at 37°C for 24 h. On the other hand the activity of DNA extracted from DBD-treated φX174 phages were measured by transfection, which is a method to form infectious φX174 phage particle by mixing purified DNA with competent E. coli cells. Transfection allows measurement of DNA damage only without being affected by the damages of coat proteins. Damages of coat proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
All of the survival measurement experiments were duplicated independently and the mean value was adopted.
Results and Discussion
Sterilization of Bacillus subtilis spore
Time-lapse change of the effect of DBD on sterilization of the dry and wet samples was measured using the apparatus shown in Fig. 3a. AC voltage of 20 kVp-p, and 2 kHz was used with the power density of the plasma of 1.2 W/cm2. The spacing between the sample and the upper electrode (bottom of the ceramic plate) was 2.0 mm.
The sample with 106 CFU/100 µl density was applied on an aluminum plate and dried (the dry sample) or to the agarose gel for the wet sample.
Fig. 5 shows the experimental results. The dry sample was sterilized to be less than 10−6 with 10 s. This value is very quick, and is indicating possibility of practical applications.
Fig. 5.
Sterilization using dielectric barrier discharge. (a) Dry sample. (b) Wet sample.
The result of the wet condition is also shown. In this condition, effect of the sterilization is dependent on pH value of the sample. When the gel was made using H2O, pH value quickly decreased to become acid. The survival rate decreased to 10−6 in 20 s. With TBE buffer, the pH value was kept at around 7. In this case, about 60 s was necessary to decrease the survival rate to 10−6. With the wet sample, radicals are injected to the gel, and should migrate to the spore to destroy. This process is affected by the pH value of the medium. It should be noted that, the electric field and the UV associated with the DBD had negligible effect of sterilization (data not shown).
States of the spore membrane was monitored during the course of the exposure using the GFP-tagged spore. Location of GFP fluorescence in each spore strain is limited to one of the layer of the spore as shown in Fig. 4 and 6. Using these spore collections, relation between cell death and inactivation of GFP was investigated. DBD was used with the samples in wet condition. The power density was decreased to prolong the period of sterilization (viability reduced to 10−6). The results are shown in Fig. 6. Bleaching of the GFP initially occurred from outermost coat labeled GFP, and proceeded to inner positions of cortex, inner membrane, and finally reached to core. The timing of cell death and bleaching of the fused GFP positioned in inner membrane was overlapped. The results suggest that the damage of inner membrane or germination receptor proteins located on the membrane is closely related to the plasma inactivation of Bacillus subtilis spores in liquids.
Fig. 6.
Plasma bleaching of fused-GFP in the spores.
Destruction of E. coli
E. coli cells in water solution (wet samples) were exposed to DBD.(3) The sample solution was spread on the PET film widely, keeping the thickness of the water layer less than 0.1 mm. This wet-samples did not evaporate significantly during the DBD application which suggested the cells were kept in low and harmless temperature. All the wet samples for DBD application contained 100 mM Tris·HCl (pH 8.0) to avoid rapid dropping down the initial neutral pH. The phenomena of increasing acidity of the samples may be mainly caused by NOx gas produced in the air field of the discharge. NOx will form nitric acid or nitrous acid after dissolving in water. In our buffering conditions, neutral pH of the samples were guaranteed until 30 s discharge, but 40 s discharge led the sample solution to acidic around pH 4.
Fig. 7 shows the survival curve of DBD treated E. coli MV1184 (pGLO) cells. The power density of the DBD was 0.4 W/cm2. Two decimal reduction was seen for 30 s DBD treatment and 5 decimal reduction was achieved for 40 s of the treatment. The curve showed a characteristic of multi-slope lines. The D value of the first slope (0~10 s), the second slope (10~30 s) and the third slope (30~40 s) were about 10, 35 and 4 s, respectively. The steep decrease between 30~40 s might be due to acidification of the solution, generating H-O-O radicals from O2−.(19)
Fig. 7.
Survival rate and the GFP fluorescence of the E. coli after the DBD treatment (Power density: 0.4 W/cm2).
Fluorescent images of E. coli cells which produce GFP are shown in Fig. 8. Originally the fluorescence was very bright, and the cells were clearly observed. With the exposure to DBD, the GFP fluorescence of the cells rapidly changed into faint images because of the bleaching of GFP (Fig. 8a 10~20 s). As shown in Fig. 8d, at 30 s after the DBD exposure, YOYO-1 {1,1'-[(4,4,7,7-Tetramethyl)-4,7-diazaundecamethylene]bis-4-[3-methyl-2,3-dihydro(benzo-1,3-oxazole)-2-methylidene]quinolinium tetraiodide} was added. This is a fluorescent dye binding specifically to double stranded DNA. The restored blight images at 30 s means that YOYO-1 penetrated through the cell membrane into the cell body, and that considerable amount of chromosomal DNA was remaining inside the cell without severe degradation; this is consistent with the results from DNA analysis by electrophoresis. The cells over-sterilized with 80 s discharge (Fig. 8e) showed tight aggregation. Those cell walls might have been destroyed and interconnected with each other.
Fig. 8.
Fluorescent image of the E. coli producing GFP. 0–20 s fluorescent of GFP, 30–80 s fluorescent of DNA by adding YOYO-1. Bar: 20 µm.
The picture at 0 s shows the YOYO-1 stained host MV1184 cells which do not produce GFP. Only a fraction of the cells were strongly stained and other major fractions were stained weakly. The bright cells seem to be dead ones because YOYO-1 does not intrude easily inside the living healthy cells. Increasing of the rate of brightly stained cells by DBD treatment after 30 s strongly suggests that cell wall and cell membrane are damaged and destroyed, at least locally or in a small manner, during the sterilization.
Fig. 7 also indicates the fluorescent intensity of GFP in E. coli MV1184 (pGLO) cells after time-lapse treatment with DBD. The intensity decreased with increasing the time of DBD treatment and complete inactivation of the GFP function was seen at 40 s of the treatment. The exhibited results recall some correlation to the cell survival. Pouring alkali solution to the bleached samples did not improve their fluorescence though normal (non-discharged) GFP inactivated in acid solution exhibited full recovery of the fluorescence by neutralization with alkali. It was suggested that the bleaching of GFP by DBD treatment seemed to be caused by its irreversible denaturation or chemical modification or degradation.
Fig. 9 shows the SDS-PAGE of E. coli MV1184 (pGLO) subjected to the atmospheric DBD.
Fig. 9.
SDS polyacrylamide gel electrophoresis of E. coli MV1184 (pGLO) exposed to the DBD. Cells were lysed and fractionated in a 14% gel before staining with Coomassie Brilliant Blue (CBB). Lane M is a protein standards marker, lane 1 is purified GFP, lane 2 is non-induced E. coli MV1184 (pGLO), lanes 3–8 represent the cells treated with DBD for 0, 5, 10, 20, 30 and 40 s, respectively. The arrow indicates GFP.
The thick band (indicated by the arrow) of the lane 3~8 corresponds to GFP. GFP and many other intrinsic proteins of E. coli were not degraded significantly. Even in the 40 s discharge treated sample (lane 8), considerable amount of full length GFP was remaining though its fluorescence was completely lost (Fig. 8c). These results suggest that the cause of the GFP bleaching by DBD treatment was irreversible denaturation of its tertiary structure or chemical modification by oxidation or reduction but not the degradation of the peptide bonds.
Fig. 10a and b shows the analysis of DNA from the DBD treated cells by agarose gel electrophoresis. The chromosome DNA was slightly cut by sharing force of mixing before separation with electrophoresis. In Fig. 10a, the chromosomal DNA was separated into two different positions on the 0.3% agarose gel. One stayed near the sample well which did not enter the gel matrix and the other migrated to the position slightly larger than the λ phage DNA (48.5 kb). The pattern of the chromosomal DNA bands did not change until 30 s of the DBD treatment. Some degradation was seen in the 40 s treated sample which seems to be caused by increased acidity of the sample because DNA is labile to acid. It is concluded that the chromosomal DNA did not degraded remarkably during inactivation of E. coli cells by DBD treatment. Bright materials at the bottom of the gel are ribosomal RNA. Similarly to the DNA, influence of the DBD treatment to the ribosomal RNA was not detected. Plasmid DNA is a good reporter molecule about DNA degradation because only a nick introduction changes its topological structure from super-coiled to relaxed form. Fig. 10b shows the plasmid DNA (pGLO) fraction separated from the DBD treated cells on 0.8% agarose gel electrophoresis. The bands at lower, upper and middle positions correspond to super-coiled, relaxed (open circular) and linear form of the plasmid molecules respectively. The appearance of three types of molecular form represents some degradation of the plasmid DNA has occurred, but it seems to be caused by cellular intrinsic DNase during recovery of the samples because non-discharged sample (0 s sample) also contained the three types of the DNA (Fig. 10b, lane 1 control). The pattern of the plasmid DNA bands did not change until 30 s of the DBD treatment which means that the nick caused by the discharge was introduced scarcely until 30 s of the DBD treatment to the cells (Fig. 10b, lane 2~5). The degradation of the plasmid seen in lane 6 may be caused by acidic effect. Fig. 10c shows the plasmid DNA fractions from the purified E. coli DNA solution (naked DNA) exposed to the DBD. With the elapsed time, super-coiled molecules have decreased and relaxed molecules have increased (Fig. 10c, lane 2~6). The rational change of the isomeric form of the plasmid DNA indicates the nick introducing activity was present in the DBD treatment to the naked DNA. The inertness of the DBD for the DNA inside the bacterial cells (Fig. 10b) may be caused by the protective effect of gram negative bacterial cell envelope (inner membrane, outer membrane, periplasm and cell wall). In any case, DNA destroying activity of the DBD treatment was very small, though it should be reminded that subtle changes in DNA may largely affect to cell viability.
Fig. 10.
Agarose gel electrophoresis of the cellular DNA from E. coli MV1184 (pGLO) exposed to the DBD. (a) DNA was extracted and separated in 0.3% gel before staining with ethidium bromide. Lane M1 is Hind III digests of λDNA, lane M2 is monomeric λDNA. Lanes 1–6 represent the DNA from the cells treated with DBD for 0, 5, 10, 20, 30 and 40 s, respectively. Arrows indicate chromosome DNA. (b) Plasmid DNA fractionated in 0.8% gel. Lane M represents the purified pGLO DNA. Lanes 1–6 represent the plasmid DNA fraction from the cells treated with DBD for 0, 5, 10, 20, 30 and 40 s, respectively. sc, oc and l represent super-coiled, open circler, and linear form, respectively. (c) Plasmid DNA fractionated in 0.8% gel from the purified DNA subjected to DBD. Lane M represents the purified pGLO DNA. Lanes 1–6 represent the naked DNA samples treated with DBD for 0, 5, 10, 20, 30 and 40 s, respectively.
Protective effect of cell membrane on the exposure to plasma jet
Using the Plasma Jet, protective effect of the cell membrane was estimated. Suspension of GFP-expressing E. coli and solution of extracted GFP were exposed to plasma jet. This is to evaluate inhibiting effect of cell membrane on inactivation of GFP. Bactericidal effect by NTP can be enhanced in acidic condition. Superoxide anion radical (O2•−) is one of reactive oxidant generated by NTP. Because of its negative charge, O2•− cannot penetrate cell membrane. By protonation, O2•− changes to hydroperoxy radical (HO2•) which has no charge. Neutral species could easily penetrate cell membrane, then turn to superoxide anion radical inside the cell.(19,20) To confirm this hypothesis, inhibiting effect of cell membrane in neutral condition was evaluated as follows.
The GFP-expressing E. coli was used, with concentration of about 105 cells/ml. GFP concentration was adjusted to 2.4 mg/ml. GFP was extracted from GFP-expressing E. coli and purified by hydrophobic affinity column chromatography and Sephadex G100 gel filtration column chromatography, and dissolved in the buffer similar to E. coli suspension sample. A 100 µl drop of the suspension or the solution were put on a 20 mm piece of PET film, and placed on the grounded aluminum plate at 40 mm from tip of the plasma jet generator as shown in Fig. 3b. Pulsed voltage of 30 kVp-p, 2.8 µs width and 3.0 kHz frequency was applied, and Helium gas (2 L/min flow rate) was used. The discharge power was 8 W. The treatment times were 5, 10, 15, 20, 25 and 30 min. For correction of the sample concentration, distilled water was added up to the original sample volume after every 5 min exposure. After the treatment, 100 µl of PBS was added to the sample to facilitate the collection. Fluorescent intensity was measured by fluorescent microplate reader. For the E. coli suspension sample, concentration of living cell was measured by standard plate count method. Each value was normalized by the result without plasma on (only gas flow). Fig. 11 shows the relative fluorescent intensity plotted against treatment time. Both fluorescent intensity and survival rate are decreased by the plasma jet (PJ) treatment. E. coli was inactivated more rapidly than GFP in it. Fluorescent intensity of GFP in E. coli was inactivated more slowly than purified GFP. Those are due to protective effect of cell membrane and organic-rich cytoplasm.
Fig. 11.
Decrease in fluorescent intensity and survival rate of the GFP-expressing E. coli by the exposure to plasma jet.
Effect of growth phase on the exposure to plasma jet
Fig. 12 shows the effect of E. coli growth phase on the bacterial inactivation by PJ exposure. E. coli strain ATCC13706 was used for the experiment. After a certain lag, E. coli increases exponentially until the stationary phase.(24) In this experiment, a single colony of E. coli from a LB agar medium was inoculated into 20 ml LB liquid medium and pre-incubated. 500 µl of the bacterial suspension in stationary phase was added to 100 ml fresh LB nutrient broth in a 1.0 L flask. This new culture was incubated at 37°C while shaking for aeration. Once in every hour, a portion of the bacterial suspension was harvested. Data were collected this way for 8 h. Before plasma jet treatment, the bacterial cells were collected by centrifugation, and resuspended into phosphate buffer saline (PBS). The cell density was adjusted to 106 cell/ml at each plotted time. At each cultivation time, the harvested bacterial suspension was exposed to plasma jet for 5 min. After that, viable cell concentration was measured by standard plate count. The survival rates after the plasma jet treatment were calculated and plotted against the cultivation time. The cell concentrations of the culture before the adjustment were also shown in Fig. 12. In practice, growth phases are determined by cell concentration. Substantially, the cell growth is caused by cellular adaptation to environment. In lag phase, microbes adapt themselves to the new environment. If the change in the cell environment is small, this phase becomes short, as in our case, and hard to notice. In the exponential phase, cells continue to divide. Therefore, the growth rate is proportional to the cell concentration at the time. The logarithmic plot of cell concentration in this phase is a linear function of the incubation time. The proportionality constant of the function indicates the doubling time. The transition to the stationary phase occurs by restriction of growth through nutrient depletion, accumulation of metabolites, and congestion. Previously, the stationary phase was considered as equilibrium between proliferation and cell death. In recent years, controlled transition of gene expression is demonstrated by metabolomic analysis. This has changed the interpretation of stationary phase to include the controlled adaptation to high environmental stress factor.
Fig. 12.
Cell density and E. coli survival rate with cultivation time from early exponential phase, through transition phase (6 h) to stationary phase, after the plasma jet exposure.
From the results, two distinct peaks can be observed of the survival rate. The first peak is after around 1 h of cultivation and the second peak is at 6 h of cultivation time. For 1 h cultivation, the survival rate was around 90%, though it dropped to about 20% for 3, 4 and 5 h. For 6 h cultivation, the cell viability rose to 30%, then dropped again to about 15% after 8 h. These peaks of plasma resistance coincidently correspond to two of the known state transitions of E. coli. Specifically, from Fig. 12, the 1 h cultivation time and the 6 h cultivation time are considered as corresponding to the transition from the lag phase to the early exponential phase, and the exponential phase to the early stationary phase, respectively. The resistance of E. coli to plasma treatment increased at the transitions of growth phase. Cellular adaptation during these transitions is considered as the reason. Generally, when cells confront stresses, such as change of environment, they express genes for repair from damage, protection, and adaptation. This could result in the increase in the resistance to PJ, or other stresses. For example, reports show that proteins with the highest rate of synthesis during the lag phase are not detectably produced at 37°C.(25)
From these considerations, the first peak in cell viability (Fig. 12) for plasma treatment after one hour of cultivation is likely caused by the transition of E. coli from the lag phase (not shown) to the exponential phase.
The second peak, at the transition to stationary phase, can be due to an increase of cyclopropane fatty acids (CFA) content occurred in the cell membrane.(26) Cell membrane of E. coli is composed of phospholipid bilayer. The composition of the fatty acid chains in phospholipids is one of the factors that affect the properties of the membrane. CFA synthesis in E. coli is related to the gene transcription factor RpoS. RpoS plays the dominant role at the beginning of the stationary phase. CFA concentration in cell membrane has its peak at the transition between the exponential phase to the stationary phase. Increase of CFA in cell membrane reduces the fluidity of the lipid bilayer, i.e., the fluidity of the cell membrane. This leads to a decrease in membrane permeability, as a defense mechanism to cell environmental shock. This cell hardening mechanism has been previously explained in detail.(26)
In summary, in early exponential phase and early stationary phase, the E. coli resistance to plasma treatment increased. These effects could be attributed to change of fatty acid composition, as a cell adaptation mechanism.
Yeast reporter assay system
Fig. 13 shows the concept of the genotoxicity test system using reporter assay with yeast DF5 strain. Using Yep365 vector, RNR2 or heat-shock protein 26 (HSP26) promoter connected with lacZ reporter genes were inserted. When damage is introduced to chromosomal DNA, RNR2 involved in cellular DNA repair is activated, then, the reporter plasmid, lacZ gene is activated to produce β-glactosidase, which corresponds to the stress and can be measured by light absorption. The yeast reporter assay system responded to many type of carcinogenic reagents, specifically H2O2 and UV irradiation.
Fig. 13.
The yeast reporter assay system.
Fig. 14 shows the results of plasma jet application to the yeast reporter assay system, DF5(YEp365) (without the reporter gene), DF5(YEp365)-RNR2-lacZ, and DF5(YEp365)-HSP26-lacZ. The survival rates are also indicated.
Fig. 14.
Response of the yeast reporter assay ((a)–(f) fold induction) and the survival rate of the yeast vs exposure time of the plasma jet (g). The plasma jet was used with the voltage of 16 kVp-p, 3.5 kHz with Ar or He of 2 L/min gas flow rate. The sample was set in a 96-well container. Distance between the nozzle of the PJ and the surface of the sample was 25 mm. (a) MMS (methyl methanesulfonate), (b) H2O2, (c) UV lamp, (d) Heat shock (40°C), (e) Ar plasma jet, (f) He plasma jet, (g) Survival rate.
High levels of RNR2-lacZ expression occurred by the Ar or He plasma jet exposure. Another stress response promoter HSP26 which responds to heat shock did not respond to the plasma. Because emission of UV-C region did not occur significantly in the plasma jet, the DNA damage seems to be brought by the radicals injected by the plasma jet. The results indicate that the Ar and He plasma jets have strong genotoxicity. The remarkable genotoxic (and carcinogenic) property of atmospheric pressure NTP has an important and essential role when the plasma is used as medicine.
Inactivation of φX174 by DBD
Fig. 15a and b shows the inactivation profile of the wet φX174 phage after DBD treatment (with 0.4 W/cm2 power density), as well as the activity of DNA extracted from DBD-treated phages obtained from the transfection.(9) These profiles were normalized with each control sample (0 s samples) to give the plaque forming unit (PFU) value of 100. The profile exhibited a decrease in activity of φX174 phage. The activity of DNA also decreased in the first 10 s. However, they were retained at long time from 20 s to 40 s. These results show that the predominant factor in the inactivation of φX174 phage is not DNA damage.
Fig. 15.
Inactivation of φX174 phage by the exposure to DBD. (a, b) Wet sample. (c, d) Dry sample. PFU: plaque forming unit. (a) Wet φX174 phage and φX174 DNA. (b) Wet φX174 phage coat protein (SDS-PAGE). (c) Dry φX174 phage and φX174 DNA. (d) Dry φX174 phage coat protein (SDS-PAGE).
Fig. 15b shows the analysis of coat proteins from DBD-treated φX174 phage in the wet condition by SDS-PAGE. These coat proteins degraded with the treatment time. In addition, band shifts with the treatment time showed that some modifications of proteins exist. These results show that despite φX174 phage activity decrease with treatment time, the activity of DNA extracted from DBD-treated phage was retained. Moreover damages of coat proteins obtained from DBD-treated φX174 phage exist. Therefore the cause of inactivation of wet φX174 phage by DBD exposure should be mainly due to the damages of coat proteins.
Fig. 15c shows the inactivation profile of the dry φX174 phage after DBD treatment, as well as the activity of DNA extracted from DBD-treated phages obtained from the transfection. The profile exhibited a decrease in activity of φX174 phage. The activity of DNA also decreased. The activity of DNA extracted from treated phage, however, decreased more gradually compared to the activity of treated phage. These results show that the predominant factor in the inactivation of φX174 phage is not DNA damage.
Fig. 15d shows the analysis of coat proteins from DBD-treated φX174 phage in the dry condition by SDS-PAGE. These coat proteins degraded with the treatment time. In addition, band shifts with the treatment time showed that some modifications of proteins exist. Therefore, coat proteins were damaged by the DBD treatment in dry condition as well as in wet condition.
Inactivation of bacteriophage λ
Inactivation of λ phage with the DBD exposure is as rapid as that for E. coli. Fig. 16a shows the analysis of proteins from the DBD treated λ phages by SDS-PAGE. λ phage has about 20 genes of coat proteins, but only two major proteins were detectable on the gel (Fig. 16a lane 1~4). These proteins degraded rapidly comparing to the E. coli cellular proteins and could not detect in the sample with 30 s exposed to the DBD of 0.4 W/cm2.
Fig. 16.
Electrophoresis of the protein and the extracted DNA from λ phage treated by the DBD. (a) SDS gel electrophoresis λ phage. Lane M: protein marker, lane 0: large amount of purified λ phage, lanes 1–6 represent the proteins from the phages treated with the DBD for 0, 5, 10, 20, 30 and 40 s. (b) Electrophoresis of the extracted DNA. Lane M1: monomeric DNA, lane M2: Hind III digests of λ DNA, lanes 1–6: DNA from the phages treated with the DBD for 0, 5, 10, 20, 30 and 40 s.
Though the degradation rate of the proteins was slow compare with the inactivation rate of the phage, time of the disappearance of the protein and completion of the inactivation coincided. Inactivation of the phage proteins involved in binding to the cell surface receptor may contribute largely to the decrease of infectious phage.
Fig. 16b shows the analysis of DNA from the DBD treated λ phages by 0.3% agarose gel electrophoresis. The phage DNA degraded faster comparing to the E. coli cellular DNA and could not detect in the 40 s discharged sample, but the degradation was slow when compared to the inactivation rate of the phage. Here also, the protective function of the cellular membrane from the attack of discharge to the DNA was suggested.
Fig. 17 illustrates the method to estimate the DNA-specific damage in plasma applied λ phages. Putative damage is introduced in both protein and DNA of the plasma treated phages. DNA is extracted from the plasma treated phage and re-packaged in vitro to form newly packaged phages. Newly packaged phages do not have protein damage and carries only the plasma exposed DNA. Therefore, if activity of the re-packaged phages is low, the damage should be brought from the plasma treated phages. Because re-packaging procedure of λ phage usually decreases the efficiency of infection, absolute number of active phages (phage titer) in the plasma treated phages and the re-packaged phage cannot be compared directly. Comparison of the normalized survival curves for these phages enables to evaluate the DNA damage and the protein damage.
Fig. 17.
Method to identify the damage of bacteriophage by NTP exposure.
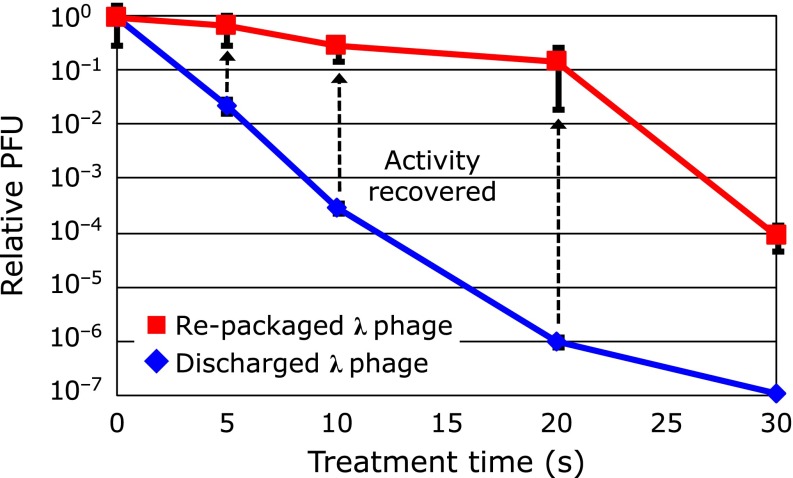
Fig. 18 shows the relative PFU curves obtained from the re-packaged λ phages and the plasma treated phages. These PFU curves were normalized with each control sample (0 s samples) to give the PFU value of 100. The profile exhibited the characteristic of a single slope curve until 20 s discharge and the D value was about 25 s. Large D value of the re-packaged phages means very slow decrease of infective phages until 20 s. The results indicate that the DNA damage introduced by plasma was very small and did not accumulate prominently with increase of the discharge time. It seems that all of the initial damage for inactivation in phages introduced by plasma was protein damage, and DNA damage was marked on the phages already inactivated by protein damage. It is concluded that inactivation of λ phage by atmospheric DBD was attributed to the damage of coat proteins. The damage of λDNA was negligible in the early stage of the inactivation.
Fig. 18.
Relative PFU (plaque forming unit) curves obtained from the re-packaged λ phages and the discharged (plasma treated) phages.
The damage responsible for phage inactivation can be recognized only when the assay of phage viability was carried out. Therefore, the damage for inactivation may not be directly correlated to the amount of the molecular damage. For example, molecular damage introduced in the binding protein which interacts with phage receptors on E. coli cell surface may largely affect to the inactivation. Moreover, it should be counted that cells have a highly developed DNA repair system. Molecular damage of DNA might have been repaired effectively than that of proteins.
The bio-assay of DNA damage has enough reproducibility when the strains of phage and host cell were fixed. This assay can be applied to not only plasma inactivation but also any type of inactivation step of bacteriophages. Application of the DNA damage assay to other plasma sources or different sets of bacteriophages and host cells may bring valuable insights into the mechanism of plasma inactivation.
For comparison, the damage to M13 phages was evaluated. M13 has a single stranded DNA. After the exposure to DBD, the DNAs were extracted, and were transfected to E. coli to measure the infection rate. The result indicated that the infection ability did not recover. The comparison indicated that double-stranded DNA is stronger against the exposure to DBD, as the DNA repair system of the host cell could fix single strand breaks of the λ phage having double-stranded DNA.
Application of NTP for destruction of bio-particles, including Bacillus subtilis spore, E. coli, S. cerevisiae, and bacteriophages, was investigated, and summarized as follows.
The bio-particles examined were inactivated effectively by NTP. The DBD exposure shows very quick destruction of B. Subtilis spore. Turbidity of the spore suspension quickly decreases with the DBD exposure. This could be due to surface modification by radicals.
With the GFP-fused spores, penetration of radicals can be visualized. In the reported experimental condition, radicals injected by the DBD penetrated into the spore with the velocity of about 1 µm/min, and when the protein damage reached to the inner membrane, the spore is destroyed.
E. coli with GFP showed the DBD exposure triggers breakdown of the membrane to penetrate the radicals. Then DNAs are damaged. The protective function of the cellular membrane from the attack of discharge to the biological materials inside the cell was suggested.
Some proteins (GFP, λ coat protein) were irreversibly inactivated quickly by denaturation or chemical modification such as oxidation or reduction but not degradation. Not only the membrane destruction or DNA damage, but the quick inactivation of proteins may have a central role in sterilization by DBD.
The bio-assay to separate the damages on DNA and coat proteins of phages has been developed. This assay has enough reproducibility when the strains of phage and host cell were fixed. Inactivation of λ phage by the DBD exposure was attributed to the damage of coat proteins. The damage of DNA was negligible in the early stage of the inactivation.
Acknowledgments
The author is grateful for the support and discussions given to this work by Mr. Hachiro Yasuda, Dr. Hirofumi Kurita, Dr. Kazunori Takashima, Prof. Toshihiko Eki, Dr. Tomoko Nakajima, Mr. Yoshimasa Tanaka, Mr. Takuya Miura, Dr. Masudur Rahman, Mr. Hiroki Yamaguchi, Mr. Yasuaki Tanino, Mr. Mai Hashimoto, Mr. Masakazu Tanino of Toyohashi Univ. of Technology, Prof. Shinji Katsura of Gunma University, Dr. Jun-Seok Oh of Meijo Univ. Prof. Hiromu Takamatsu and Prof. Kazuhito Watabe of Setsunan Univ., Prof. M. Abdel-Salam of Assiut Univ. This work was partly supported by a Grant-in-Aid for Scientific Research (KAKENHI 24108005) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan.
Conflict of Interest
No potential conflicts of interest were disclosed.
References
- 1.Mizuno A. Industrial applications of atmospheric non-thermal plasma in environmental remediation. Plasma Physics and Controlled Fusion. 2007;49:A1–A15. [Google Scholar]
- 2.Mizuno A, Yamazaki Y, Ito H, Yoshida H. Ac energized ferroelectric pellet bed gas cleaner. IEEE Trans Ind Appl. 1992;28:535–540. [Google Scholar]
- 3.Yasuda H, Hashimoto M, Rahman M, Takashima K, Mizuno A. States of biological components in bacteria and bacteriophages during inactivation by atmospheric dielectric barrier discharges. Plasma Process Polym. 2008;5:615–621. [Google Scholar]
- 4.Fridman G, Brooks AD, Balasubramanian M, et al. Comparison of direct and indirect effects of non-thermal atmospheric-pressure plasma on bacteria. Plasma Process Polym. 2007;4:370–375. [Google Scholar]
- 5.Hashimoto M, Rahman MM, Tanino M, et al. Cell destruction by dielectric barrier discharge for real-time monitoring of bio-particles. Int J Plasma Environ Sci Technol. 2007;1:146–150. [Google Scholar]
- 6.Yasuda H, Miura T, Kurita H, Takashima K, Mizuno A. Biological evaluation of DNA damage in bacteriophages inactivated by atmospheric pressure cold plasma. Plasma Process Polym. 2010;7:301–308. [Google Scholar]
- 7.Yamaguchi H, Yasuda H, Eki T, Kurita H, Takashima K, Mizuno A. Detection of DNA damages in Saccharomyces cerevisiae caused by non-thermal atmospheric pressure plasmas. J Inst Electrostat Jpn. 2011;35:8–13. [Google Scholar]
- 8.Mizuno A, Yasuda H. Damages of biological components in bacteria and bacteriophages exposed to atmospheric non-thermal plasma. In: Machala Z, Hensel K, Akishev Y, editors. Plasma for Bio-Decontamination, Medicine and Food Security (NATO Science for Peace and Security Series A: Chemistry and Biology) Netherlands: Springer; 2012. pp. 79–92. [Google Scholar]
- 9.Tanaka Y, Yasuda H, Kurita H, Takashima K, Mizuno A. Analysis of the inactivation mechanism of bacteriophage φX174 by atmospheric pressure discharge plasma. IEEE Trans Ind Appl. 2014;50:1397–1401. [Google Scholar]
- 10.Abdel-Salam M, Nakano M, Tanino M, Mizuno A. Culturing of cells as influenced by exposure to AC and DC fields. J Phys: Conference Series. 2008;142:012051. [Google Scholar]
- 11.Ono R, Oda T. Optical diagnosis of pulsed streamer discharge under atmospheric pressure. Int J Plasma Environ Sci Technol. 2008;1:123–129. [Google Scholar]
- 12.Nagato K. Chemical composition of atmospheric ions. J Inst Electrostat Jpn. 1999;23:37–43. [Google Scholar]
- 13.Yehia A, Abdel-Salam M, Mizuno A. On assessment of ozone generation in dc coronas. J Phys D: Appl Phys. 2000;33:831–835. [Google Scholar]
- 14.Eliasson B, Kogelschatz U. Nonequilibrium volume plasma chemical processing. IEEE Trans Plasma Sci. 1991;19:1063–1077. [Google Scholar]
- 15.Tanaka M, Kuzumoto M, Tamita T, Inanaga Y. In: Barrier Discharge. Yagi S, editor. Tokyo: Asakura Publishing; 2012. pp. 52–128. (in Japanese) [Google Scholar]
- 16.Kirkpatrick M, Dodet B, Odic E. Atmospheric pressure humid argon DBD plasma for the application of sterilization - measurement and simulation of hydrogen, oxygen, and hydrogen peroxide formation. Int J Plasma Environ Sci Technol. 2007;1:96–101. [Google Scholar]
- 17.Tanino M, Mizuno A. Sterilization using dielectric barrier discharge at atmospheric pressure. Int J Plasma Environ Sci Technol. 2007;1:108–113. [Google Scholar]
- 18.Kurita H, Nakajima T, Yasuda H, et al. Single-molecule measurement of strand breaks on large DNA induced by atmospheric pressure plasma jet. Appl Phys Lett. 2011;99:191504. [Google Scholar]
- 19.Ikawa S, Kitano K, Hamaguchi S. Effects of pH on bacterial inactivation in aqueous solutions due to low-temperature atmospheric pressure plasma application. Plasma Process Polym. 2010;7:33–42. [Google Scholar]
- 20.Jun-Seok O, Endre JS, Nishtha G, et al. How to assess the plasma delivery of RONS into tissue fluid and tissue. J Phys D: Appl Phys. 2016;49:304005. [Google Scholar]
- 21.Imamura D, Kuwana R, Takamatsu H, Watabe K. Localization of proteins to different layers and regions of Bacillus subtilis spore coats. J Bacteriol. 2010;192:518–524. doi: 10.1128/JB.01103-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ichikawa K, Eki T. A novel yeast-based reporter assay system for the sensitive detection of genotoxic agents mediated by a DNA damage-inducible LexA-GAL4 protein. J Biochem. 2006;139:105–112. doi: 10.1093/jb/mvj011. [DOI] [PubMed] [Google Scholar]
- 23.Ochi Y, Sugawara H, Iwami M, Tanaka M, Eki T. Sensitive detection of chemical-induced genotoxicity by the Cypridina secretory luciferase reporter assay, using DNA repair-deficient strains of Saccharomyces cerevisiae. Yeast. 2011;28:265–278. doi: 10.1002/yea.1837. [DOI] [PubMed] [Google Scholar]
- 24.Ali Azam T, Iwata A, Nishimura A, Ueda S, Ishihama A. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J Bacteriol. 1999;181:6361–6370. doi: 10.1128/jb.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones TH, Murray A, Johns M, Gill CO, McMullen LM. Differential expression of proteins in cold-adapted log-phase cultures of Escherichia coli incubated at 8, 6 or 2°C. Int J Food Microbiol. 2006;107:12–19. doi: 10.1016/j.ijfoodmicro.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 26.Chang YY, Eichel J, Cronan JE, Jr. Metabolic instability of Escherichia coli cyclopropane fatty acid synthase is due to RpoH-dependent proteolysis. J Bacteriol. 2000;182:4288–4294. doi: 10.1128/jb.182.15.4288-4294.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]