Abstract
The obligate bitrophic fungus Puccinia striiformis f. sp. tritici (Pst) causes stripe (yellow) rust on wheat worldwide. Here, we report a novel fungal strain able to hyperparasitize Pst. The strain was isolated from gray-colored rust pustules, and was identified as Alternaria alternata (Fr.: Fr.) keissler based on a combination of morphological characteristics and multi-locus (ITS, GAPDH, and RPB2) phylogeny. Upon artificial inoculation, the hyperparasite reduced the production and viability of urediniospores, and produced a typical gray-colored rust pustule symptom. Scanning electron microscopy demonstrated that the strain could efficiently penetrate and colonize Pst urediniospores. This study first demonstrates that A. alternata could parasitize Pst and indicates its potential application in the biological control of wheat stripe rust disease.
Keywords: wheat stripe rust, Puccinia striiformis, hyperparasite, Alternaria alternata, biological control
Introduction
Strip rust (yellow rust), caused by Puccinia striiformis Westend f. sp. tritici Erikss. (Pst), is one of the most important diseases of wheat in many regions of world (Saari and Prescott, 1985; Stubbs, 1985; Chen, 2005). Because the Pst urediniospores could be dispersed over long distances by the wind, the fungal pathogen is able to cause large-scale epidemics and severe yield losses under conducive environmental conditions (Brown and Hovmøller, 2002; Wan et al., 2007; Zhao et al., 2008). In China, the devastating epidemics occurring in 1950, 1964, 1990, and 2002, has caused up to 6.0 × 109, 3.0 × 109, 2.6 × 109, and 1.0 × 109 kg of yield losses, respectively (Wan et al., 2004; Chen et al., 2009; Gao et al., 2015). So far, cultivation of resistant varieties is the most effective way to control wheat stripe rust. Nevertheless, most resistant varieties were bred for major gene resistance and rapidly lost their resistance within 3–6 years after field cultivation (Cheng et al., 2014; Han et al., 2015). In addition, the constant and indiscriminate use of fungicides poses serious environmental problems and health hazards to animals and humans. Biological control strategy is thus attractive for the potential to achieve effective disease management with minimal environmental cost.
Puccinia striiformis f. sp. tritici (Pst) is an obligate biotrophic fungus, which normally forms yellow to orange urediniospores on leaf blade surfaces during disease progression (Hovmøller et al., 2011). However, we have observed that the color appearance of uredinia (urediniospore mass) occasionally turn dark gray overtime during greenhouse propagation, especially under high humidity conditions. The color shift takes place gradually and becomes increasingly common in frequency, which finally causes the cessation of uredinia sporulation. Our previous study has demonstrated that such discoloration and sporulation cessation could be associated with hyperparasite infection (Zhan et al., 2014).
Hyperparasitism is common in filamentous fungi, and could be developed into a useful alternative to chemical fungicides for effective control of plant fungal diseases (Hijwegen and Buchenauer, 1984; Blakeman, 1992; McLaren et al., 1996; De Cal et al., 2008; Adhikari et al., 2014; Zhong et al., 2016). Previous studies showed that approximately 30 genera of fungi can hyperparasitize rust pathogens, which included Tuberculina spp. (Mijušković and Vučinić, 2001), Darluca filum (Yuan et al., 1999), Fusarium spp. (Kapooria and Sinha, 1969), Scytalidium uredinicola (Tsuneda et al., 2011), Aphanocladium album (Koç and Défago, 2008) and Cladosporium spp. (Moricca et al., 2001). However, so far only four species, Cladosporium cladosporioides, Lecanicillium lecanii, Microdochium nivale, and Typhula idahoensis, have been reported to infect uredinia and urediniospores of Pst (Littlefield, 1981; Zhan et al., 2014).
Here we describe the discovery of a novel Pst hyperparasite. Morphological observations and phylogenetic analysis demonstrated that the fungus belongs to the species of Alternaria alternata (Fr.: Fr.) keissler, which has never been reported to parasitize Pst or any other fungal organisms. Pathogenicity test and microscopic examination showed that the obtained A. alternata strain is able to reduce Pst urediniospore production and viability, which indicates a biological control potential of this novel mycoparasite against wheat stripe rust disease.
Materials and Methods
Isolation and Purification of the Mycoparasite
The hyperparasitic strain CPA001 was isolated from Pst urediniospores in Northwest A&F University, Yangling, Shaanxi, China. Firstly, urediniospores of Pst were propagated on the susceptible wheat cultivar Mingxian 169 as previously described (Cao et al., 2008). Wheat seedlings inoculated with urediniospores of Pst were kept in a growth chamber at about 16°C and 80–90% relative humidity (Lu et al., 2011). In total, 151 seedling plants were inoculated. Fourteen to twenty days after inoculation, more than half of the uredinia changed color from fresh yellow orange to gray or dark gray. Gray urediniospores were then transferred onto potato dextrose agar (PDA) medium with a sterilized needle. After incubation at 25° for 3 days, mycelia from the colony margins were transferred to fresh PDA plate and singe-spore purified to obtain a pure culture. The purified culture was stored on PDA slants at 4–8°C.
Morphological Observation
For the morphological observation, mycelial disks of 5 mm in diameter were taken from the growing margins of 3-day old PDA culture, transferred to potato carrot agar (PCA) plates and incubated at 25°C in a 12-h photoperiod for 1 week to induce conidia production. In addition, microscope slide cultures were prepared by placing a small amount of mycelia on PDA medium blocks (5 mm diameter) overlaid by a cover slip (Wang et al., 2015). Examination of the morphological characteristics of hyphae, conidiophores and conidia were conducted using an Olympus BX51T-32P01 optical microscope.
To further observe the ultrastructure of the parasitic fungus, wheat leaves bearing uredinia with abnormal colors were cut into pieces approximately 0.5 cm × 0.5 cm in size for scanning electronic microscope (SEM). Samples were immersed in 4.0% glutaraldehyde (pH 6.8) and fixed at 4°C for 4 h. Then samples were washed four times with 0.1 M phosphate buffer for 15 min each. Subsequently, samples were dehydrated for 30 min each in 30, 50, 70, 80, and 90% ethanol series, and finally 3 repeats in 100% ethanol. Samples were dried in a CO2 vacuum, and sputter coated with gold (E-1045, Hitachi, Japan) for SEM examination (S-4800, Hitachi, Japan).
Pathogenicity Test to Confirm Hyperparasitism
The susceptible wheat cultivar Mingxian 169 was used for propagating Pst urediniospores. When the first leaf had successfully expanded after 10 days, seedlings were inoculated with CYR32, a predominant race of Pst in China. Approximately 14 days later, the diseased leaves bearing urediniospores were inoculated with the mycoparasite A. alternata strain CPA001. The pure culture of CPA001 was formulated into spore suspension (1.0 × 106 spores/mL in concentration) for spray inoculation. Healthy wheat leaves receiving A. alternata inoculation represented control check1 (CK1). Wheat leaves infected by Pst but not treated with the A. alternata conidia suspension represented CK2. Each treatment was carried out with wheat seedlings growing in three independent pots, with each pot containing about 24 plants. All treatments were placed in the same growth chamber, and observation of the symptoms was performed at the same time.
Simultaneously, Pst urediniaspores were directly inoculated with the A. alternata strain CPA001. The A. alternata conidia suspension (1.0 × 106 spores/mL in concentration) was mixed with the Pst urediniospores, the spore mixture was sprayed on PDA medium, and incubated at 25° for 24 h. During co-cultivation, samples were collected to observe the dynamic infection process with SEM. Ultrastructural sample treatments were the same as ones described above.
Uredinia Quantification
The phenotype of disease was quantitively assessed by counting the number of uredinia pustules within a 5 cm2 area at 9 days post CPA001 inoculation, using IMAGEJ1. To avoid bias among leaf samples, 35 random leaves were selected for each treatment and the entire experiment was repeated for three times.
Germination Rate of Pst
Freshly collected urediniospores were cultured on sterile water at 9°C for 6 h, then placed on slides to count the numbers of germinated urediniospores using an Olympus BX51T-32P01 optical microscope. A germ tube length up to the one-half spore diameter was defined as germination. The germination rate was expressed as a percentage based on 100 urediniospores. One hundred urediniospores were selected randomly, and all experiments were performed at least three times.
Molecular Characterization
DNA Extraction
The strain CPA001 was cultured on cellophane placed on top of PDA medium and incubated at 25°C for 7 days, the mycelia were then harvested for DNA extraction. Genomic DNA was extracted with CTAB method described by Wang et al. (2015). DNA concentration was measured with a spectrophotometry (Nanodrop 2000, Thermo Fisher Scientific, Wilmington, DE, USA). The DNA was stored at -20°C and diluted to 100 ng/μL as the working solution for polymerase chain reaction (PCR) amplification.
PCR Amplification and Sequencing
Three representative genes [the complete rDNA-ITS (ITS) region, glyceraldephyde-3-phosphate dehydrogenase (GAPDH) and the second largest subunit of RNA polymerase II (RPB2)] of A. alternata were amplified using gene-specific PCR primers of the V9G (5′-TTACGTCCCTGCCCTTTGTA-3′) (Hoog and Gerrits van den Ende, 1998) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) (White et al., 1990), gpd1 (5′-CAACGGCTTCGGTCGCATTG-3′) and gpd2 (5′-GCCAAGCAGTTGGTTGTGC-3′) (Berbee et al., 1999) and RPB2-5F2 (5′-GGGGWGAYCAGAAGAAGGC-3′) (Sung et al., 2007) and fRPB2-7cR (5′-CCCATRGCTTGTYYRCCCAT-3′) (Liu et al., 1999), respectively. Conditions for PCR amplification of the three genes were as follows: initial denaturing at 94°C for 5 min; 35 cycles of denaturing (each cycle at 94°C for 30 s), annealing at 55°C for 30 s, and extension at 72°C for 1 min; and then a final extension at 72°C for 10 min. PCR products were detected by 1.0% agarose gel electrophoresis, purified using a PCR Purification Kit (Bio-tek Co., Ltd, China) according to the manufacture’s protocol. The amplified products were cloned into pMD20-T vector (Takara) for Sanger sequencing. All sequences were deposited in GenBank under accession numbers KX976465, KX976466, and KX976467, respectively.
Phylogenetic Analysis
Reference sequences from other Alternaria spp. were retrieved from GenBank (Table 1). Sequences were aligned with Clustal X (Thompson et al., 1997), and the final alignment was inspected with BioEdit 5.0.9.1 (Saitou and Nei, 1987). On the basis of the aligned sequences, a phylogenetic tree was constructed with the Maximum Likelihood (ML) method in the Molecular Evolutionary Genetics Analysis (MEGA) software version 6.0 with 1000 bootstrap replicates (Efron et al., 1996; Tamura et al., 2013).
Table 1.
Sources of Alternaria spp. strains with GenBank accession numbers.
| Species name | Strain number | Gene and GenBank accession no. |
Substrate/Host | Origin/Locality | ||
|---|---|---|---|---|---|---|
| ITSa | GAPDHb | RPB2c | ||||
| A. alternantherae | CBS124392 | KC584179 | KC584096 | KC584374 | Solanum melongena | China |
| A. iridiaustralis | CBS118404 | KP124434 | KP124283 | KP124904 | Iris sp. | New Zealand |
| CBS118486 | KP124435 | KP124284 | KP124905 | Iris sp. | Australia | |
| CBS118487 | KP124436 | KP124285 | KP124906 | Iris sp. | Australia | |
| A. betae-kenyensis | CBS118810 | KP124419 | KP124270 | KP124888 | Beta vulgaris var. cicla | Kenya |
| A. eichhorniae | CBS489.92 | KC146356 | KP124276 | KP124895 | Eichhornia crassipes | India |
| A. burnsii | CBS107.38 | KP124420 | JQ646305 | KP124889 | Cuminum cyminum | India |
| CBS110.50 | KP124421 | KP124271 | KP124890 | Gossypium sp. | Mozambique | |
| CBS118816 | KP124423 | KP124273 | KP124892 | Rhizophora mucronata | India | |
| A. tomato | CBS103.30 | KP124445 | KP124294 | KP124915 | Solanum lycopersicum | Unknown |
| CBS114.35 | KP124446 | KP124295 | KP124916 | Solanum lycopersicum | Unknown | |
| A. jacinthicola | CBS878.95 | KP124437 | KP124286 | KP124907 | Arachis hypogaea | Mauritius |
| CBS133751 | KP124438 | KP124287 | KP124908 | Eichhornia crassipes | Mali | |
| CPC25267 | KP124439 | KP124288 | KP124909 | Cucumis melo var. indorus | Unknown | |
| A. alternata | CBS102599 | KP124330 | KP124185 | KP124798 | Minneola tangelo | Turkey |
| CBS107.53 | KP124305 | KP124162 | KP124774 | Pyrus pyrifolia | Japan | |
| CBS115200 | KP124352 | KP124206 | KP124820 | Minneola tangelo | South Africa | |
| CBS115616 | AF347031 | AY278808 | KC584375 | Arachis hypogaea | India | |
| CBS117143 | KP124355 | KP124209 | KP124823 | Capsicum annuum | Italy | |
| CBS118812 | KC584193 | KC584112 | KC584393 | Daucus carota | USA | |
| CBS118814 | KP124357 | KP124211 | KP124825 | Solanum lycopersicum | USA | |
| CBS118815 | KP124358 | KP124212 | KP124826 | Solanum lycopersicum | USA | |
| CBS121348 | KP124367 | KP124219 | KP124836 | Platycodon grandiflorus | China | |
| CBS127671 | KP124381 | KP124233 | KP124851 | Stanleya pinnata | USA | |
| CBS127334 | KP124380 | KP124232 | KP124850 | soil | USA | |
| CBS121456 | KP124369 | KP124221 | KP124839 | Sanguisorba officinalis | China | |
| CBS126910 | KP124379 | KP124231 | KP124849 | Stanleya pinnata | USA | |
| CBS795.72 | KP124309 | KP124166 | KP125085 | Plantago aristida | USA | |
| CBS620.83 | KP124315 | KP124171 | KP124783 | Nicotiana tabacum | USA | |
| A. alternata | CPA001d | KX976465 | KX976466 | KX976467 | Puccinia striiformis | China |
| CBS102600 | KP124331 | KP124186 | KP124799 | Citrus reticulata | USA | |
| CBS115069 | KP124347 | KP124201 | KP124815 | Malus domestica | South Africa | |
| CBS119543 | KP124363 | KP124215 | KP124831 | Citrus paradisi | USA | |
| A. gaisen | CBS632.93 | KC584197 | KC584116 | KC584399 | Pyrus pyrifolia | Japan |
| CBS118488 | KP124427 | KP124278 | KP124897 | Pyrus pyrifolia | Japan | |
| CPC25268 | KP124428 | KP124279 | KP123976 | Unknown | Portugal | |
| A. arborescens SC | CBS101.13 | KP124392 | KP124244 | KP124862 | Peat soil | Switzerland |
| CBS105.24 | KP124393 | KP124245 | KP124863 | Solanum tuberosum | Unknown | |
| CBS116329 | KP124405 | KP124257 | KP124875 | Malus domestica | Germany | |
| CBS105.49 | KP124396 | KP124248 | KP124866 | Contaminant blood culture | Italy | |
| CBS126.60 | KP124397 | KP124249 | KP124867 | Wood | UK | |
| CBS109730 | KP124399 | KP124251 | KP124869 | Solanum lycopersicum | USA | |
| CBS112749 | KP124401 | KP124253 | KP124871 | Malus domestica | South Africa | |
| CBS112633 | KP124400 | KP124252 | KP124870 | Malus domestica | South Africa | |
| A. arborescens SC | CBS117587 | KP124406 | KP124258 | KP124876 | Brassica sp. | Netherlands |
| CBS118389 | KP124407 | KP124259 | KP124877 | Pyrus pyrifolia | Japan | |
| CBS123266 | KP124411 | KP124262 | KP124881 | Human toenail | Denmark | |
| CBS127263 | KP124417 | KP124268 | KP124886 | Human nasal infection | Mexico | |
| CBS115516 | KP124403 | KP124255 | KP124873 | Malus domestica | South Africa | |
aITS complete rDNA-ITS region; bGAPDH glyceraldephyde-3-phosphate dehydrogenase; cRPB2 the second largest subunit of RNA polymerase II; dSequences from isolates of A. alternatea from Puccinia striiformis.
Results
Isolation of an Alternaria alternata Strain from Pst Uredinia Showing Mycoparasitic Symptom
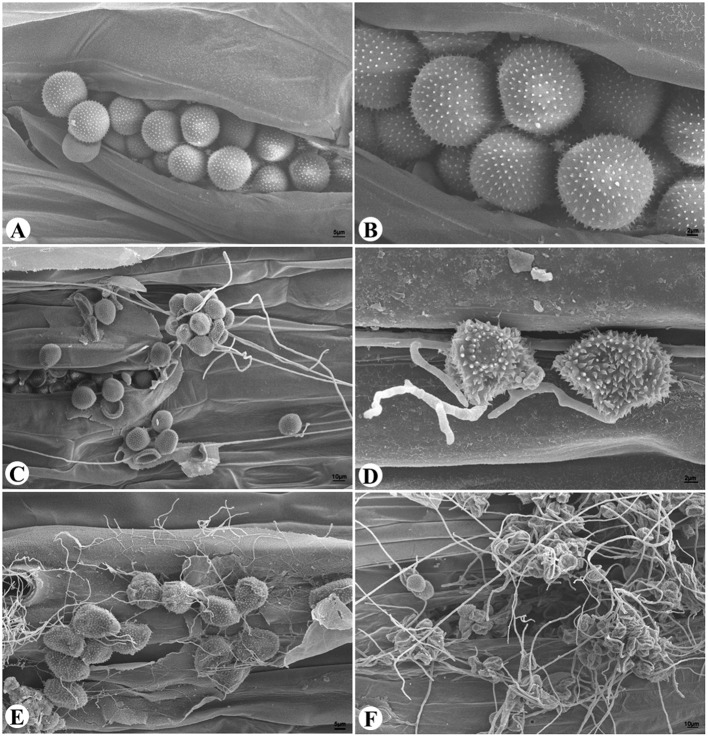
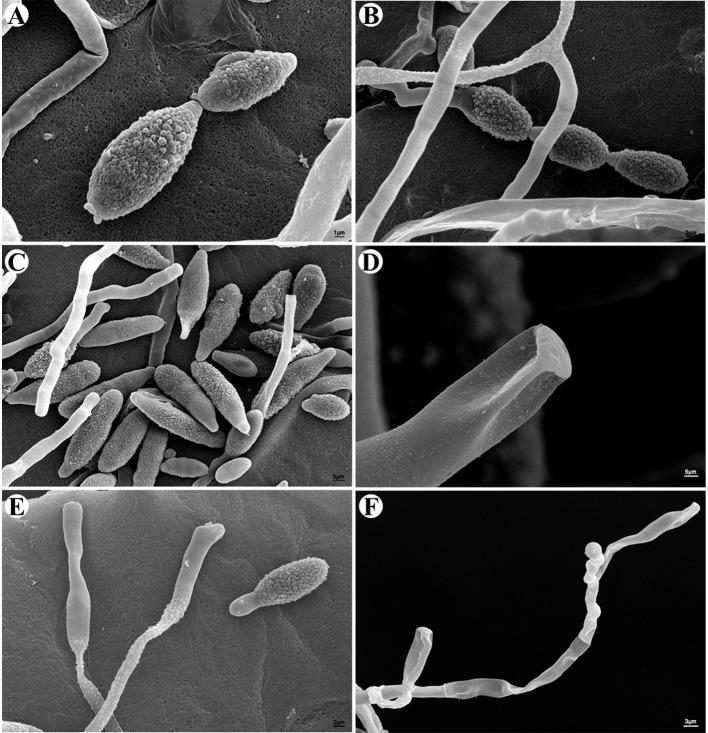
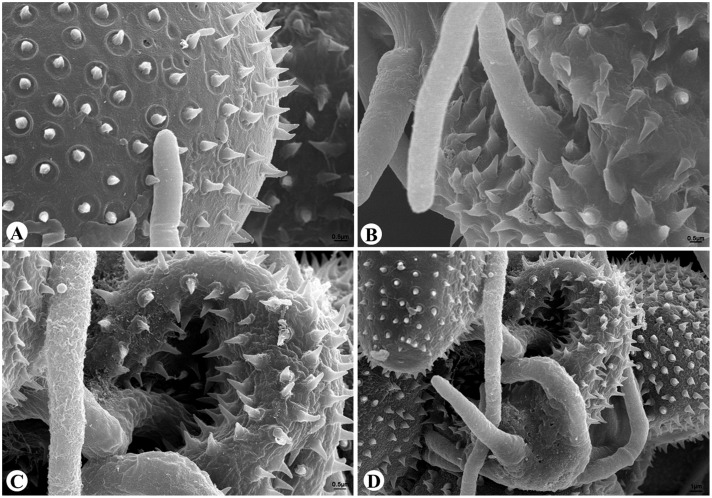
Wheat leaves bearing normal yellow-colored uredinia and leaves bearing gray-colored uredinia resembling hyperparasite infection were examined under SEM (Figure 1). Yellow-colored uredinia contained round-shaped urediniospores, and contained no mycelium or spore of other fungal organism (Figures 1A,B). On the other hand, gray-colored uredinia were made up of shriveled or ruptured urediniospores, these spores were intertwined by dense filamentous hyphae, which become increasingly prevalent over time (Figures 1C–F). The dramatic shape change of urediniospores indicated their loss of cell viability. The prevalence of intertwined hyphae strongly suggested hyperparasitic colonization events.
FIGURE 1.
The hyperparasitic colonization of Pst uredinia and urediniospores. (A,B) Uredinium and urediniospores in normal shape. (C) Early stage of hyperparasitic infection, note the shriveled urediniospores. (D,E) Mid infection stage. (F) Late infection stage.
Through in vitro culture, several candidate mycoparasitic fungal strains were obtained. Most strains resembled the previously reported C. cladosporioides in morphological appearance (Zhan et al., 2014), which were not characterized further. One strain, named CPA001, was characterized further and reported here. On PCA medium, the CPA001 culture initially developed light-gray colony and the center turned dark gray after 7 days (Figure 2A). The vegetative hyphae were brown, branched, septate, and 4 μm in diameter (Figure 2D). Conidia were typically obpyriform, dark brown, 20.2 to 35.2 μm × 8.0 to 12.6 μm in size, with 1–3 transverse and 0–1 longitudinal septate. Most conidia also had a short beak with a dimension of 1.6 to 9.4 μm × 2.9 to 5.0 μm (Figures 2B–D). SEM observation obtained more detailed morphological characteristics of the conidia and conidiophores (Figures 3A–F). SEM observations indicated that the fungus produced abundant conidia in long chains on short conidiophores (Figures 3A–C). Conidiophores were single, straight or slightly curved, and ranged from 12.3 to 60.6 μm × 2.2 to 4.0 μm (Figures 3E,F). These morphological features resembled Alternaria spp.
FIGURE 2.
Morphological characterizations of A. alternata cultured on PCA medium. (A) Colony morphology grown at 25°C for 7 days. (B–D) Conidia and vegetative hyphae morphology.
FIGURE 3.
Morphological characterizations of A. alternata under SEM. (A–C) Conidia. (D) Scars on a secondary conidium. (E,F) Conidiophores.
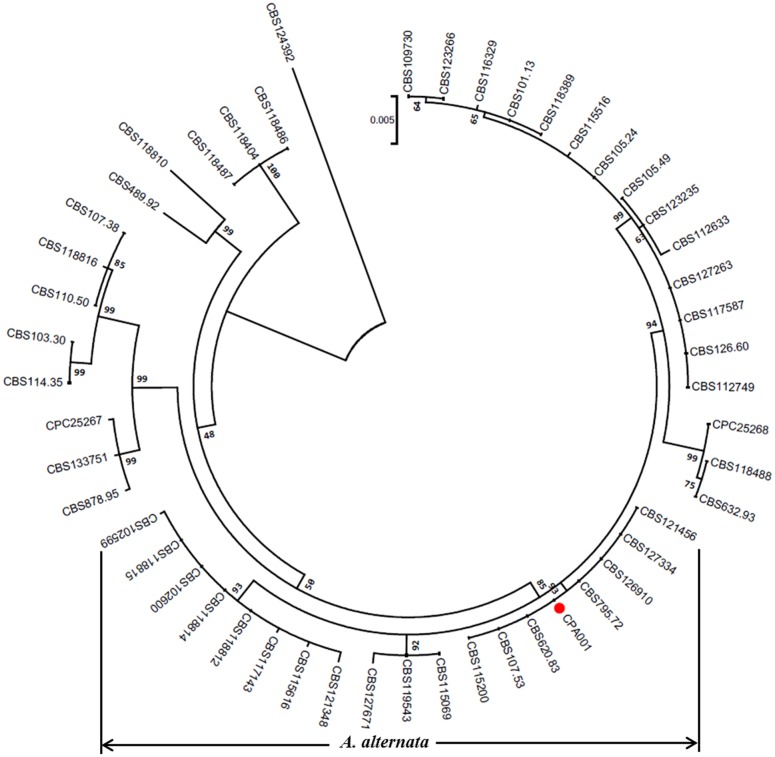
CPA001 was further identified to be A. alternata based on phylogenetic analysis with the ITS, GAPDH and RPB2 genetic markers (Figure 4). A range of Alternaria spp. reference isolates were selected for the phylogenetic tree construction. CPA001 was found to be most closely related to CBS121456, CBS127334, CBS126910,CBS795.72, CBS620.83, CBS107.53 and CBS115200, all belonging to the A. alternata species. The entire A. alternata clade was well-separated from other clades with a bootstrap value of 85%.
FIGURE 4.
A phylogenetic tree constructed based on the three genes (ITS, GAPDH and RPB2) of the members in Alternaria genus using the maximum likelihood (ML) method with 1000 bootstrap replicates. The red circle represents the A. alternata isolate characterized in the present study.
Confirmation of Hyperparasitism
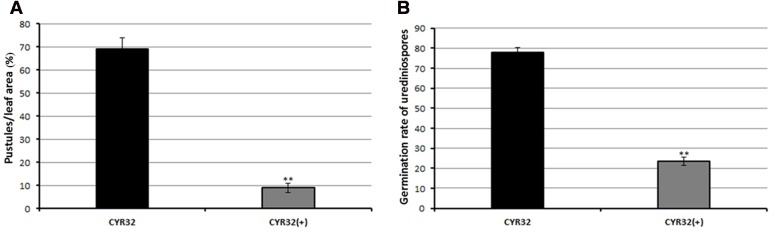
Pathogenicity testing showed that the obtained CPA001 strain could efficiently hyperparasitize Pst (Figure 5). Wheat leaves inoculated with Pst alone produced abundant orange-colored uredinia after 21 days post inoculation (Figure 5B). On the other hand, Pst pre-inoculated wheat leaves receiving subsequent A. alternata treatment showed a typical sign of mycoparasitic colonization, namely fewer rust pustule formation and abundant gray-colored hyphae covering the uredinia (Figures 5C–F). Such gray-colored hyphae were never observed with wheat leaves treated with A. alternata conidia suspension alone (CK1, Figure 5A). Reisolated strain from the parasitized uredinia showed the same morphological characteristics CPA001. At 9 days post CPA001 inoculation, the frequency of Pst pustule formation was merely 10% whereas that of the control treatment was 70% (Figure 6A). Urediniospores collected from CPA001-treated pustules also showed dramatically reduced viability (∼25% vs. 80%), indicated by germination rate (Figure 6B).
FIGURE 5.
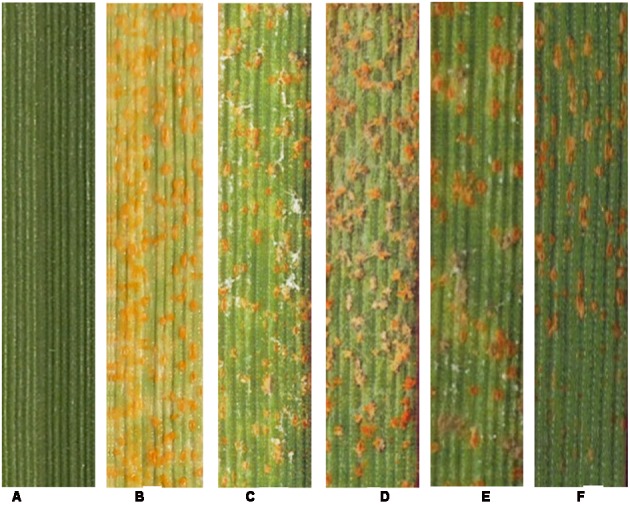
Pathogenicity test to confirm that A. alternata could hyperparasite Pst. (A) CK1, wheat leaves inoculated with the spore suspension of A. alternata, 20 dpi, without any symptom; (B) CK2, wheat leaves only inoculated with CYR32, 20 dpi; (C–F) Wheat leaves inoculated with CYR32 for 14 days prior to inoculating with the spore suspension of A. alternata. (C–F) are symptoms at 3, 5, 7, and 9 d after A. alternata inoculation respectively.
FIGURE 6.
Quantification of the percentage of leaf area covered by Pst pustules (A) and germination rate of Pst urediniospores (B). CYR32 (+) means mycoparasite infected. Values represent mean ± standard errors of three independent assays, and the statistical analysis was assessed by using Student’s t-tests. Double asterisks indicate P < 0.01.
Scanning electronic microscope observations further confirmed that the A. alternata CPA001 strain could efficiently parasitize Pst. The A. alternata germ tube contacted with and penetrated into Pst urediniospores at 24 hpi, and caused complete urediniospore collapse at 36–48 hpi (Figures 7A–D).
FIGURE 7.
Scanning electronic microscope observations of Pst urediniospore infection by A. alternata. (A) 24 h after inoculation, the A. alternata germ tube contacted with Pst urediniospore; (B) 36 h after inoculation, an A. alternata germ tube penetrated into a urediniospore; (C,D) 48 h after inoculation, the hyphae of A. alternata directly penetrated through the urediniospore.
Discussion
Characterization of newly isolated mycoparasites has contributed to a better understanding of the diversity of hyperparasites, and will lead to the discoveries of novel fungal species and the development of novel biocontrol agents (Vandermeer et al., 2009; Baiswar et al., 2014; Wang et al., 2015; Zhong et al., 2016). The present study revealed a novel mycoparasite infecting Pst, the causal agent of wheat stripe rust. In addition, the hyperparasite could reduce the production and viability of urediniospores, indicating its potential application in the biological control of Pst.
Conidial morphology and size used to be important features used in Alternaria taxonomy. However, these phenotypes are plastic, showing considerable variations under different environmental and culture conditions, making it difficult to identify species based on phenotype alone (Rotem, 1994). Although ITS region of nuclear ribosomal DNA (nrDNA) is a universal marker used for the identification of fungal species (Seena et al., 2010; Schoch et al., 2012), it is ineffective in distinguishing closely related fungal species (Pryor and Gilbertson, 2002; Kiss, 2012). Currently, multiple gene-based phylogeny has been widely used in the classification of species in the genus Alternaria (Andrew and Pryor, 2008; Lawrence et al., 2013; Woudenberg et al., 2015). In the present study, a combination of three markers identified the obtained mycoparasite to be A. alternata. To our knowledge, this is the first report that A. alternata could hyperparasitize Pst, and also the first report of A. alternata as a hyperparasite.
Resistance breeding is critical for wheat rust disease control. Nevertheless, most resistant genes used for breeding are ones with major effect, which tend to lose their resistance rapidly upon field release (Line and Qayoum, 1992; Li and Zeng, 2000). So far, mycoparasitism has been reported as an effective measure for controlling several diseases (Zhong et al., 2016). For example, Trichoderma spp. has been successfully used to minimize the effect of Fusarium oxysporum pathogen on tomato plants (Adhikari et al., 2014). Ampelomyces quisqualis is in commercial use for biocontrol of powdery mildew on grapes and other crops (Sullivan and Maddock, 2000). However, there have been little known attempts to control Pst with hyperparasites.
The A. alternata strain CPA001 obtained in the present study can colonize Pst urediniospores in an aggressive manner. CPA001 treatment dramatically reduces uredinial pustule formation and the viability of ureniniospores. Moreover, our observation indicated that CPA001 can colonize a broad range of Pst isolates being different in virulence profile (physiological race) and geographic origin. These facts make CPA001 a good candidate for further characterization efforts to develop novel Pst biocontrol agent. But now, we are unclear about the mycoparasitism spectrum of CPA001 at a broader level (e.g., its hypoparasitic potential against other rust pathogens), and whether and how environmental factors affect the survival ability and hypoparasitic potential of CPA001. We also do not know by which strategies CPA001 kills and colonizes Pst urediniospores and whether these strategies are CPA001-unique or are general features of the A. alternata species. In the near future, it is important to study the hypoparasitic characteristics of CPA001 in more detail in the laboratory, so as to understand its parasitism spectrum, its hypoparasitic mechanisms, and the potential environmental and ecological impacts upon massive release. Key factors impacting mycoparasitism efficiency should be identified and controlled field test should be performed to determine the disease control effect.
Urediniospores are important inoculation materials for rust disease research. Based on our experience, mycoparasitic infection of Pst is common in the greenhouse, which could pose a great challenge to research activities such as spore propagation. In the near future, we are planning to further characterize the biological characteristics of the obtained mycoparasitic isolate, such as the experimental host range, the spore type specificity, and the effects of environmental conditions (e.g., humidity, moisture) on the final parasitic infection outcome. These efforts will offer important principle guidelines for the field application and greenhouse control of Pst mycoparasites.
Author Contributions
ZK designed experiments; LZ performed the experiments; JZ and XL analyzed the data; GZ and SJ joined the discussion and gave the original ideas; LZ wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Key Basic Research Program of China (2013CB127700), National ‘Twelfth Five-Year’ Plan for Science and Technology Support (2012BAD19B04-12), Modern Agro-industry Technology Research System in China (CARS-3-1-11) and the 111 Project of the Ministry of Education of China (No. B07049), and the China Postdoctoral Science Foundation (2016M592845).
Footnotes
References
- Adhikari A., Nandi S., Dutta S., Bhattacharya I., Mandal T. (2014). Study of morphology and mycoparasitism of some antagonists of Trichoderma sp from West Bengal. India. Int. J. Res. 1 593–605. [Google Scholar]
- Andrew M., Pryor B. M. (2008). An expanded multilocus phylogeny does not resolve morphological species within the small-spored Alternaria species complex. Mycologia 101 95–109. 10.3852/08-135 [DOI] [PubMed] [Google Scholar]
- Baiswar P., Ngachan S., Chandra S. (2014). Ramularia coleosporii, a hyperparasite on Coleosporium plumeriae, in India. J. Phytopathol. 163 407–410. 10.1111/jph.12286 [DOI] [Google Scholar]
- Berbee M. L., Pirseyedi M., Hubbard S. (1999). Cochliobolus phylogenetics and the origin of known, highly virulent pathogens, inferred from ITS and glyceraldehyde-3-phosphate dehydrogenase gene sequences. Mycologia 91 964–977. 10.2307/3761627 [DOI] [Google Scholar]
- Blakeman J. P. (1992). Potential for biological of plant diseases on the phyllosphere. Annu. Rev. Phytopathol. 20 167–192. 10.1146/annurev.py.20.090182.001123 [DOI] [Google Scholar]
- Brown J. K. M., Hovmøller M. S. (2002). Aerial dispersal of pathogens on the global and continental scales and its impact on plant disease. Science 297 537–541. 10.1126/science.1072678 [DOI] [PubMed] [Google Scholar]
- Cao L. H., Xu S. C., Chen W. Q., Liu T. G., Chen W. Q. (2008). Early molecular diagnosis and detection of Puccinia striiformis f. sp. tritici in China. Lett. Appl. Microbiol. 46 501–506. 10.1111/j.1472-765X.2007.@02313.x [DOI] [PubMed] [Google Scholar]
- Chen W. Q., Wu L. R., Liu T. G., Xu S. C. (2009). Race dynamics, diversity, and virulence evolution in Puccinia striiformis f. sp. tritici, the causal agent of wheat stripe rust in China from 2003 to 2007. Plant Dis. 93 1093–1101. 10.1094/PDIS-93-11-1093 [DOI] [PubMed] [Google Scholar]
- Chen X. M. (2005). Epidemiology and control of stripe rust [Puccinia striiformis f. sp. tritici] on wheat. Can. J. Plant Pathol. 27 314–337. 10.1080/07060660509507230 [DOI] [Google Scholar]
- Cheng P., Xu L. S., Wang M. N., See D. R., Chen X. M. (2014). Molecular mapping of genes Yr64 and Yr65 for stripe rust resistance in hexaploid derivatives of durum wheat accessions PI 331260 and PI 480016. Theor. Appl. Genet. 127 2267–2277. 10.1007/s00122-014-2378-8 [DOI] [PubMed] [Google Scholar]
- De Cal A., Redondo C., Sztejnberg A., Melgarejo P. (2008). Biocontrol of powdery mildew by Penicillium oxalicum in open-field nurseries of strawberries. Biol. Control 47 103–107. 10.1016/j.biocontrol.2008.07.010 [DOI] [Google Scholar]
- Efron B., Halloran E., Holmes S. (1996). Bootstrap confidence levels for phylogenetic trees. Proc. Natl. Acad. Sci. U.S.A. 93 13429–13434. 10.1073/pnas.93.14.7085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L., Yu H. X., Kang X. H., Shen H. M., Li C., Liu T. G., et al. (2015). Development of SCAR markers and an SYBR green assay to detect Puccinia striiformis f. sp. tritici in infected wheat leaves. Plant Dis. 100 1840–1847. 10.1094/PDIS-06-15-0693-RE [DOI] [PubMed] [Google Scholar]
- Han D. J., Wang Q. L., Chen X. M., Zeng Q. D., Wu J. H., Xue W. B., et al. (2015). Emerging Yr26 – virulent races of Puccinia striiformis f. tritici are threatening wheat production in the Sichuan basin. China. Plant Dis. 99 754–760. 10.1094/PDIS-08-14-0865-RE [DOI] [PubMed] [Google Scholar]
- Hijwegen T., Buchenauer H. (1984). Isolation and identification of hyperparasitic fungi associated with erysiphaceae. Neth. J. Plant Pathol. 90 79–83. 10.1007/BF01999956 [DOI] [Google Scholar]
- Hoog G. S. D., Gerrits van den Ende A. H. G. (1998). Molecular diagnostics of clinical strains of filamentous basidiomycetes. Mycoses 41 183–189. 10.1111/j.1439-0507.1998.tb00321.x [DOI] [PubMed] [Google Scholar]
- Hovmøller M. S., Sørensen C. K., Walter S., Justesen A. F. (2011). Diversity of Puccinia striiformis on cereals and grasses. Annu. Rev. Phytopathol. 49 197–217. 10.1146/annurev-phyto-072910-095230 [DOI] [PubMed] [Google Scholar]
- Kapooria R. G., Sinha S. (1969). Phylloplane mycoflora of pearl millet and its influence on the development of Puccinia penniseti. Trans. Br. Mycol. Soc. 53 153–155. 10.1016/S0007-1536(69)80024-0 [DOI] [Google Scholar]
- Kiss L. (2012). Limits of nuclear ribosomal DNA internal transcribed spacer (ITS) sequences as species barcodes for Fungi. Proc. Natl. Acad. Sci. U.S.A. 109 10741–10742. 10.1073/pnas.1207143109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koç N. K., Défago G. (2008). Studies on the host range of the hyperparasite Aphanocladium album. J. Phytopathol. 107 214–218. [Google Scholar]
- Lawrence D. P., Gannibal P. B., Peever T. L., Pryor B. M. (2013). The sections of Alternaria: formalizing species-group concepts. Mycologia 105 530–546. 10.3852/12-249 [DOI] [PubMed] [Google Scholar]
- Li Z. Q., Zeng S. M. (2000). Wheat Rust in China. Beijing: China Agricultural Press. [Google Scholar]
- Line R. F., Qayoum A. (1992). Virulence, aggressiveness, evolution and distribution of races of Puccinia striiformis (the cause of stripe rust of wheat) in North America. Tech. Bull. 1968–1987. [Google Scholar]
- Littlefield L. J. (1981). Biology of the Plant Rust: An Introduction. Ames, IA: Iowa State University Press, 103. [Google Scholar]
- Liu Y. J., Whelen S., Hall B. D. (1999). Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 16 1799–1808. 10.1093/oxfordjournals.molbev.a026092 [DOI] [PubMed] [Google Scholar]
- Lu N. H., Wang J. F., Chen X. M., Zhan G. M., Chen C. Q., Huang L. L., et al. (2011). Spatial genetic diversity and interregional spread of Puccinia striiformis f. sp. tritici in Northwest China. Eur. J. Plant Pathol. 131 685–693. 10.1007/s10658-011-9842-y [DOI] [Google Scholar]
- McLaren D. L., Huang H. C., Rimmer S. R. (1996). Control of apothecial production of Sclerotinia sclerotiorum by coniothyrium minitans and talaromyces flavus. Plant Dis. 80 1373–1378. 10.1094/PD-80-1373 [DOI] [Google Scholar]
- Mijušković M., Vučinić Z. (2001). Hyperparasitic Fungi in Mycopopulation of Montenegro. Podgorica: Montenegrien Mycological Center. [Google Scholar]
- Moricca S., Ragazzi A., Mitchelson K. R., Assante G. (2001). Antagonism of the two-needle pine stem rust fungi Cronartium flaccidum and Peridermium pini by Cladosporium tenuissimum in vitro and in planta. Phytopathology 91 457–468. 10.1094/PHYTO.2001.91.5.457 [DOI] [PubMed] [Google Scholar]
- Pryor B. M., Gilbertson R. L. (2002). Relationships and taxonomic status of alternaria radicina, A. carotiincultae, and A. petroselini based upon morphological, biochemical, and molecular characteristics. Mycologia 94 49–61. [PubMed] [Google Scholar]
- Rotem J. (1994). The Genus Alternaria: Biology, Epidemiology and Pathogenicity. St Paul, MN: American Phytopathological Society Press. [Google Scholar]
- Saari E. E., Prescott J. M. (1985). World distribution in relation to economic losses. Cereal Rusts 2 260–298. [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 406–425. [DOI] [PubMed] [Google Scholar]
- Schoch C. L., Seifert K. A., Huhndorf S., Robert V., Spouge J. L., Levesque C. A., et al. (2012). Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. U.S.A. 109 6241–6246. 10.1073/pnas.1117018109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seena S., Pascoal C., Marvanová L., Cássio F., Hyde K. D. (2010). DNA barcoding of fungi: a case study using its sequences for identifying aquatic hyphomycete species. Fungal Divers. 44 77–87. 10.1007/s13225-010-0056-y [DOI] [Google Scholar]
- Stubbs R. W. (1985). Stripe rust. The Cereal Rusts 2 61–101. [Google Scholar]
- Sullivan S. M., Maddock J. R. (2000). Bacterial sporulation: pole-to-pole protein oscillation. Curr. Biol. 10 159–161. 10.1016/S0960-9822(00)00331-6 [DOI] [PubMed] [Google Scholar]
- Sung G. H., Sung J. M., Hywel-Jones N. L., Spatafora J. W. (2007). A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): identification of localized incongruence using a combinational bootstrap approach. Mol. Phylogenet. Evol. 44 1204–1223. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. D., Gibson T. J., Plewniak F., Higgins D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25 4876–4882. 10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuneda A., Hiratsuka Y., Maruyama P. J. (2011). Hyperparasitism of Scytalidium uredinicola on western gall rust, Endocronartium harknessii. Can. J. Bot. 58 1154–1159. 10.1139/b80-143 [DOI] [Google Scholar]
- Vandermeer J., Perfecto I., Liere H. (2009). Evidence for hyperparasitism of coffee rust (Hemileia vastatrix) by the entomogenous fungus, Lecanicillium lecanii, through a complex ecological web. Plant Pathol. 58 636–641. [Google Scholar]
- Wan A. M., Chen X. M., He Z. H. (2007). Wheat stripe rust in China. Aust. J. Agr. Res. 58 605–619. 10.1071/AR06142 [DOI] [Google Scholar]
- Wan A. M., Zhao Z. H., Chen X. M., He Z. H., Jin S. L., Jia Q. Z., et al. (2004). Wheat stripe rust 1139 epidemic and virulence of Puccinia striiformis f. sp. tritici in China in 2002. Plant Dis. 88 896–904. 10.1094/PDIS.2004.88.8.896 [DOI] [PubMed] [Google Scholar]
- Wang Y. B., Yu H., Dai Y. D., Wu C. K., Zeng W. B., Yuan F., et al. (2015). Polycephalomyces agaricus, a new hyperparasite of Ophiocordyceps, sp. infecting melolonthid larvae in southwestern China. Mycol. Prog. 14 1–9. 10.1007/s11557-015-1090-7 [DOI] [Google Scholar]
- White T. J., Bruns T., Lee S., Taylor J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide to Methods and Applications, eds Innis M. A., Gelfand D. H., Sninsky J. J., White T. J. (San Diego, CA: Academic Press; ), 315–322. [Google Scholar]
- Woudenberg J. H. C., Seidl M. F., Groenewald J. Z., Vries M. D., Stielow J. B., Thomma B. P. H. J., et al. (2015). Alternaria section Alternaria: species, formae speciales, or pathotypes? Stud. Mycol. 82 1–21. 10.1016/j.simyco.2015.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z. M., Hunter T., Ruiz C., Royle D. J. (1999). Pathogenicity to willow rust, melampsora epitea, of the mycoparasite sphaerellopsis filum from different sources. Mycol. Res. 103 509–512. 10.1017/S0953756298007448 [DOI] [Google Scholar]
- Zhan G. M., Tian Y., Wang F. P., Chen X. M., Guo J., Jiao M., et al. (2014). A novel fungal hyperparasite of Puccinia striiformis f. sp. tritici, the causal agent of wheat stripe rust. PLoS ONE 9:e111484 10.1371/journal.pone.0111484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Wang X. J., Chen C. Q., Huang L. L., Kang Z. S. (2008). A PCR-based assay for detection of Puccinia striiformis f. sp. tritici in wheat. Plant Dis. 91 1669–1674. 10.1094/PDIS-91-12-1669 [DOI] [PubMed] [Google Scholar]
- Zhong X., Li S. S., Peng Q. Y., Zhang J. S., Kan X. T., Zhang G. R., et al. (2016). A polycephalomyces, hyperparasite of ophiocordyceps sinensis, leads to shortened duration of production and reduced numbers of host ascospores. Fungal Ecol. 21 24–31. 10.1016/j.funeco.2016.03.002 [DOI] [Google Scholar]