Abstract
The emergence of carbapenem-resistant Enterobacteriaceae (CRE) has become a major public health concern worldwide and a new challenge in the treatment of infectious diseases. The molecular characteristics of Enterobacter cloacae in Ningxia China are unknown. In this study, we reported 10 carbapenem-resistant E. cloacae isolates from the General Hospital of Ningxia Medical University, the largest university hospital in Ningxia between January 2012 and December 2013. Bacteria isolates were identified by Vitek2 compact and the identity of non-duplicate E. cloacae isolates was further confirmed by PCR and sequencing. The drug susceptibility and phenotype identification of these isolates were analyzed by agar dilution method, modified Hodge test (MHT), and EDTA synergy test. Beta-lactamase (bla) genes blaNDM−1 was found in 8 out of 10 isolates. Most isolates harbored multiple resistance genes including blaESBL, blaAmpC, quinolones, aminoglycosides, and disinfectant resistance genes. Pulsed field gel electrophoresis (PFGE) showed that these E. cloacae isolates were grouped into 6 clusters based on a cutoff of 80% genetic similarity. In conjugative assay, 9 out of 10 isolates transferred carbapenem-resistant genes to Escherichia coli. Our study has revealed that NDM-1-producing isolates are the most prevalent carbapenem-resistant E. cloacae in Ningxia. These isolates also carry several other carbapenem-resistant genes and can transfer these genes to other bacteria through conjugation. These findings highlight an urgent need to monitor these isolates to prevent their further spread in this region.
Keywords: Enterobacteriaceae, carbapenem resistance, antibiotic susceptibility, New Delhi metallo-beta-lactamase-1, genetic homologeneity
Introduction
The Enterobacteriaceae are a large family of Gram-negative bacteria that may cause several infectious diseases such as respiratory tract infection, intra-abdominal infection, urinary tract infection, etc. (Wang J. T. et al., 2015). Carbapenem has been the main treatment for severe infections associated with Enterobacteriaceae (Kaniga et al., 2010). Nevertheless, carbapenem-resistant Enterobacteriaceae (CRE) has gradually emerged and becomes one of the serious public health concerns worldwide (Alam et al., 2014; Ling et al., 2015; Xu et al., 2015; Iredell et al., 2016). According to the 2012 CHINET report on CRE in China, the domestic incidence of CRE has increased 2.2-fold between 2010 and 2012 (Hu et al., 2012). Carbapenem resistance in Enterobacteriaceae is mainly mediated by the production of carbapenemases, a form of β-lactamase, that cleave the β-lactam ring, an essential component of β-lactam antibiotics such as cephalosporins and carbapenems (Queenan and Bush, 2007).
Enterobacter cloacae, a species of the Enterobacteriaceae family, is an important nosocomial pathogen that may cause infections in wound, urinary tract, lower respiratory tract, skin and soft tissues. While CRE infections are primarily caused by Klebsiella pneumoniae and Escherichia coli (Goodman et al., 2016), carbapenem-resistant E. cloacae has only occasionally been reported (Bennett et al., 2009; Heller et al., 2012). In this study, we reported 10 carbapenem-resistant E. cloacae isolated in the largest university hospital in Ningxia province, China and investigated their molecular characteristics.
Materials and methods
Reagents
Imipenem, meropenem, ceftazidime, and tigecycline was purchased from Pfizer. MH agar was purchased from OXOID. Primers were synthesized by Sangon Biotech Shanghai Co. Ltd. Premix Taq was purchased from Takara Biotech Dalian Co. Ltd. Xba I restriction enzymes were purchased from Thermo.
Sample collection, bacterial identification, and reference strains
Bacterial isolates were collected from the General Hospital of Ningxia Medical University, a large-sized hospital in Ningxia province, China, between January 2012 and December 2013. Species identification and initial drug susceptibility were analyzed by Vitek2 compact (bioMerieux, France). E. coli ATCC25922, Klebsiella pneumonia ATCC BAA-1705, and K. pneumonia ATCC BAA-1706 were used as quality control strains.
Identification of E. cloacae by PCR and sequencing
The identity of all E. cloacae isolates was further confirmed by PCR and sequencing. Briefly, the isolates were cultured by standard methods. To isolate chromosomal DNA, one freshly grown colony was scraped into an Eppendorf tube and resuspended in 200 μL of sterile water. The bacterial suspension was boiled at 100°C for 10 min to release the genomic DNA. The DNA was then used as the template to amplify the 16S rRNA gene segment (1465 bp) by PCR. The sequences of primers: 27F: AGA GTT TGA TCM TGG CTC AG, and 1492R: TAC GGY TAC CTT GTT ACG ACT T. The PCR programs were as follows: denaturation at 94°C 5 min, followed by 30 cycles of 94°C 30 s, 55°C 40 s, and 72°C 1 min, and extension at 72°C 5 min. The PCR products were sequenced by BGI Genomics (Beijing, China).
Drug susceptibility test and phenotype identification
The antibiotic susceptibilities of the E. cloacae isolates were tested by measuring the minimal inhibitory concentration (MIC) for imipenem, meropenem and tigecycline using the agar dilution method. Imipenem and meropenem results were interpreted according to the Clinical Laboratory Standards Institute (CLSI) guidelines (2013). Freshly prepared tigecycline (<12 h old) was used, and results were interpreted based on the FDA criteria (R:MIC ≥ 8, I:MIC = 4, S:MIC ≤ 2). All E. cloacae isolates were screened for carbapenemase production by the modified Hodge test (MHT) and EDTA synergy test according to the CLSI guidelines. In EDTA synergy test, 0.5M units bacterial sample was spread on MH agar plate. Two meropenem disks were placed on the plate with a distance of 15–20 mm. Aliquots of EDTA (10 μL of 0.5 mol/L solution) was added onto one of the disks, and the plate was incubated at 35°C for 18–24 h. A positive result was scored when the diameter of the inhibition zone around EDTA disk was 5 mm larger than that around the other disk. Both MHT and EDTA synergy test were performed in triplicates.
Detection of resistance genes
The DNA was extracted from each carbapenem-resistant E. cloacae isolate. Resistance genes were detected by PCR, including extended-spectrum β-lactamases (ESBL: blaTEM, blaSHV, blaCTX, blaVEB, blaPER, and blaSFO), AmpC enzyme (blaDHA, blaEBC, blaACC, blaCIT, and blaMOX), carbapenemase (blaKPC, blaNDM−1, blaSME, blaIMP, blaVIM, blaOXA−23, and blaOXA−48), class 1 and 2 integron, and other resistant genes (qnrA, qnrB, qnrS, acc-Ib, qacΔE, and sul). The reaction system (25 μL) was prepared: Premix Taq 12.5 μL, template 2 μL, each primer 0.2 μL (~10 μM), and sterile water 10.1 μL. Reaction program was as follows: 94°C denaturation for 5 min, followed by 30 cycles of 94°C denaturation for 45 s, 52–58°C annealing for 45 s, 72°C extension for 1 min, 72°C final extension for 10 min. Positive amplification products were sequenced and the sequencing results were compared by BLAST.
Genetic homologeity analysis
All carbapenem-resistant E. cloacae were analyzed by pulsed field gel electrophoresis (PFGE) to determine their genetic homologeity. The bacterial DNA embedded in small plastic pieces was treated with proteinase K and cleaved by the restriction enzyme XbaI at 37°C for 12–14 h. The XbaI-digested DNA was analyzed by electrophoresis for 21 h at 6 V/cm, with a pulse angle of 120° and pulse times from 2.16 to 63.8 s. PFGE was performed in triplicates. Comparison of the PFGE patterns was performed with Bionumerics software based on the Tenover's criteria (Tenover et al., 1995). Isolates were allocated into genetic similarity clusters using a cut-off value of 80%.
Plasmid conjugation test
The conjugation test was performed by the broth mating method using carbapenem-resistant E. cloacae as the donor strain and rifampin-resistant E. coli EC600 as the recipient strain. The donor and recipient strain were inoculated respectively in 5 mL LB, and was incubated in a thermostatic shaker at 37°C for 18–24 h. The donor strain (60 μL) and recipient strain (90 μL) were inoculated into 20 ml of LB medium and incubated 37°C for 18–24 h without any shaking. Transconjugants were selected on MH agar plates containing 30 μg/mL ceftazidime + 400 μg/mL rifampicin and plates with 1 μg/mL imipenem + 400 μg/mL rifampicin. The donor and recipient strains were used as negative controls. The transconjugants strains were identified by PCR and sequencing. The MIC for imipenem, meropenem, and tigecycline was measured by the agar dilution method. The conjugation test was performed in triplicates
Results
Phenotype and genotype of carbapenem-resistant E. cloacae isolates
A total of 18 non-duplicate CRE isolates were identified from 2012 to 2013, 10 of which were E. cloacae. Eight and nine positive E. cloacae isolates were detected in the MHT and EDTA double disk synergy test, respectively. PCR amplification showed that these E. cloacae isolates carried multiple resistant genes. Integrons and insertion sequence common regions (ISCR) were present in 8 and 9 out of 10 E. cloacae isolates, respectively. As shown in Table 1, the carbapenem-resistant gene was primarily New Delhi metallo-beta-lactamase-1 (blaNDM−1, 8/10), followed by K. pneumoniae carbapenemase (blaKPC, 6/10). ESBL, AmpC, quinolones, aminoglycosides and disinfectant resistance gene was found in 7, 8, 8, 4, and 8 out of 10 isolates, respectively.
Table 1.
Summary of resistant genes detected in the carbapenem-resistant E. cloacae isolates.
| Isolate ID | MHT/EDTA double-disk synergy test | Associated resistance determinants | |
|---|---|---|---|
| β-lactamase gene | Others | ||
| 3 | +/+ | blaTEM, blaKPC, blaNDM−1, blaIMP, blaVIM | qacΔE, IntI, ISCR |
| 9 | +/+ | blaCTX−1, blaEBC, blaKPC, blaNDM−1, blaVIM | acc-Ib, qacΔE, sul, IntI, ISCR |
| 11 | +/+ | blaACC, blaEBC, blaKPC, blaNDM−1 | qnrA, qacΔE, IntI, ISCR |
| 18 | +/+ | blaCTX−1, blaEBC, blaKPC, blaNDM−1 | qnrA, qacΔE, sul, IntI, ISCR |
| 40 | ± | blaTEM, blaSHV, blaACC, blaEBC, blaNDM−1, blaVIM | acc-Ib, qacΔE, sul, IntI, ISCR |
| 42 | +/+ | blaTEM, blaKPC, blaNDM−1, blaVIM | qnrS, acc-Ib, qacΔE, sul, IntI, ISCR |
| 55 | +/+ | blaIMP | – |
| 56 | +/+ | blaTEM, blaDHA, blaEBC, blaNDM−1 | acc-Ib, qacΔE, sul, IntI, ISCR |
| 02 | −/− | blaACC, blaEBC | ISCR |
| 03 | +/+ | blaCTX−1, blaDHA, blaEBC, blaKPC, blaNDM−1, blaVIM | qnrA, qacΔE, sul, IntI, ISCR |
Molecular epidemiology
Based on a cutoff of 80% genetic similarity, these isolates were grouped into A (isolate No: 9, 11, 18, 03), B (isolate No: 33), C (isolate No: 42), D (isolate No: 55), E (isolate No: 40, 56), and F (isolate No: 02) clusters (Figure 1).
Figure 1.
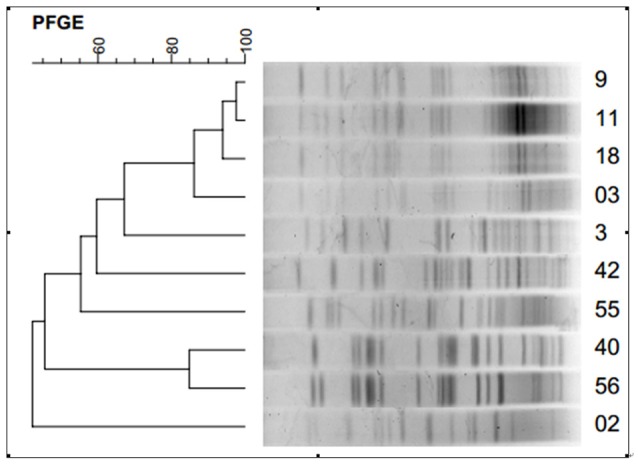
Dendrogram showing pulsed-field gel electrophoresis (PFGE) analysis of the 10 carbapenem-resistant E. cloacae isolates.
Plasmid conjugation test
The plasmid transconjugant test was performed for each carbapenem-resistant E. cloacae isolate. The transconjugants were analyzed by PCR and sequencing. Results showed that a total of 9 transconjugants were E. coli, indicating the successful transfer of resistant plasmid from E. cloacae isolate to E. coli. The MIC of the 9 transconjugants for imipenem, meropenem and tigecycline was measured by the agar dilution method (Table 2).
Table 2.
Minimum inhibitory concentrations (MIC) for imipenem, meropenem and tigecycline of the carbapenem-resistant E. cloacae isolates and their transconjugants.
| Isolate | S/F | IPM | TC-IPM | MEM | TC-MEM | Tigecycline | TC-Tigecycline |
|---|---|---|---|---|---|---|---|
| 3 | S | 4 | 4 | 8 | 8 | 1 | 1 |
| 9 | S | 64 | 4 | 32 | 1 | 1 | ≤0.125 |
| 11 | S | ≥128 | 4 | ≥128 | 4 | 0.5 | 1 |
| 18 | S | 64 | 2 | 32 | 8 | 8 | 1 |
| 40 | S | 64 | 32 | 32 | 32 | 8 | 8 |
| 42 | S | 8 | 4 | 8 | 16 | 8 | 2 |
| 55 | S | 4 | 4 | 4 | 4 | 32 | 8 |
| 56 | S | 32 | 32 | 32 | 16 | 8 | 4 |
| 02 | F | – | – | – | – | – | – |
| 03 | S | 16 | 4 | 8 | 4 | 0.5 | 1 |
| EC600 | – | 0.5 | – | ≤0.125 | – | ≤0.125 | – |
S/F, success/failure; IMP, impipenem; MEM, meropenem; TC, transconjugant strain; and EC600, recipient strain.
Discussion
Carbapenem-resistant E. cloacae can markedly prolong the hospital length of stay, increase the medical cost, and most importantly cause high mortality rate. With the wide spread of CRE and associated resistant genes around the word (Castanheira et al., 2013; Poirel et al., 2014; Zweigner et al., 2014; Jakobsen et al., 2015), it has become urgent to investigate the epidemiology and molecular characteristics of carbapenem-resistant E. cloacae, which may provide a solid basis for an effective control of carbapenem-resistant E. cloacae.
In this study, 55.6% of the carbapenem-resistant E. cloacae isolated in our hospital can produce carbapenemase, which is substantially different from the CHINET report (Ling et al., 2015). The finding has reflected the distinct distribution of the bacteria in this region, which might be related to the antibiotic-prescribing habits of local doctors. After NDM-1 was first reported by Kumarasamy in 2010 (Wang et al., 2011), NDM-1-producing E. cloacae have been identified throughout the world (Gupta et al., 2011; Ho et al., 2012). In China, NDM-1 was initially detected in Ningxia province and has been later reported in other regions (Ho et al., 2012; Huang et al., 2015; Zhang et al., 2015). In this study, the carbapenem-resistant gene is primarily blaNDM−1, and other carbapenemase genes are also detected including blaKPC, imipenem-hydrolizing-lactamase gene (blaIMP), and Verona imipenemase gene (blaVIM). Inconsistent with our results, blaKPC−2 and blaVIM−1 were identified as the primary carbapenem-resistant genes of CRE isolates (Levy Hara et al., 2013; Li et al., 2014; Villa et al., 2014). More importantly, in our study, non-carbapenemase-producing resistant genes such as β-lactamase, quinolones, aminoglycosides, etc. were detected. The gene of integration, ISCR and other genetic elements were also identified in these isolates, suggesting the possibility of exogenous gene transfer. Two of the isolates even carry six β-lactamases and some other resistance genes. Our results have revealed a diversity of resistance genes in our isolates, indicating that these isolates might have achieved resistance through different mechanisms.
Ningxia province, a relatively less developed region in the northwest of China, has a much lower risk of bacterial transmission compared with other regions (Hu et al., 2014; Ling et al., 2015). In this study, the patients carrying the carbapenem-resistant E. cloacae isolates had no prior travel or living in any epidemic areas, indicating that these isolates might have acquired the carbapenem resistance gene through a unusual way. Moreover, Acinetobacter baumannii had been previously isolated from 6 of these carrying patients, suggesting the probable transmission of resistant genes between different bacterial species. Such cross-species gene transmissions have been previously detected (Bogaerts et al., 2013; Wang X. et al., 2015).
MHT is recommended for the carbapenemas phenotype identification test for by CLSI. Studies have shown that the sensitivity of MHT for some strains is much higher compared with others containing IMP, VIM, and NDM (a sensitivity of 50%; Girlich et al., 2012; Ramana et al., 2013). In this study, the positive rate of MHT is 8/10, indicating the occurrence of false negative, which might be associated with the production of IMP, VIM, and NDM in some isolates. EDTA can inactivate the type B carbapenemas by chelating Zn ions (Miriagou et al., 2013), resulting in the expansion of the inhibition zone in EDTA synergy test. In this study, the positive rate of EDTA synergy test is 9/10, and most of the positive strains produce the type B carbapenemas. Although, Azimi et al. have suggested that false negative results are associated with the test method instead of the type of carbapenemases (Azimi et al., 2016), Wang et al. have reported a false positive rate of 3.3% in MHT due to the low ertapenem hydrolysis capacity of ESBL, especially CTX-M (Wang et al., 2011). Therefore, a combination examination of carbapenemas phenotype by MHT and EDTA synergy test can improve the sensitivity and specificity of carbapenemas gene detection. Moreover, PCR method remains the gold standard for carbapenemas phenotype identification (Rastegar Lari et al., 2013). Consistently, the positive rate of PCR in our study is 10/10.
In this study, based on a cutoff of 80% genetic similarity, PFGE analysis revealed a high degree of genetic homologeity among 5 isolates, suggesting that small-scale epidemic spread might have occurred. Further transconjugation assay has detected a positive rate of 9/10, which is markedly higher than that in a previous study (Fu et al., 2012). The finding suggests that our carbapenemase-producing E. cloacae can transfer the resistant genes to susceptible bacterium, leading to horizontal transfer of resistant genes, and thus an infection outbreak of the pathogens. Furthermore, our study also showed that the MIC of most transconjugates for imipenem, meropenem, and tigecycline was lower compared with their donor bacteria, indicating that the bacterial resistance is dependent on several factors. The mechanism of clinical transmission among different bacteria needs further investigation.
In summary, our study demonstrated that NDM-1-producing isolates were the most prevalent carbapenem-resistant E. cloacae in Ningxia province, China. These isolates can transfer the resistant gene to other bacteria through conjugation. To our best knowledge, this is the first report on the prevalence and molecular characteristics of carbapenem-resistant E. cloacae in, China. Early detection and surveillance of these E. cloacae isolates are urgently needed to prevent their further spread.
Author contributions
WJ designed and supervise the study. ZS performed the experiments, analyzed the data, and wrote the paper. HZ helped with experiments. GL helped with the writing of the paper.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Alam M. Z., Aqil F., Ahmad I., Ahmad S. (2014). Incidence and transferability of antibiotic resistance in the enteric bacteria isolated from hospital wastewater. Braz. J. Microbiol. 44, 799–806. 10.1590/S1517-83822013000300021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimi L., Talebi M., Owlia P., Pourshafie M. R., Najafi M., Lari E. R., et al. (2016). Tracing of false negative results in phenotypic methods for identification of carbapenemase by Real-time PCR. Gene 576, 166–170. 10.1016/j.gene.2015.10.008 [DOI] [PubMed] [Google Scholar]
- Bennett J. W., Herrera M. L., Lewis J. S., Wickes B. W., Jorgensen J. H. (2009). KPC-2-producing Enterobacter cloacae and pseudomonas putida coinfection in a liver transplant recipient. Antimicrob. Agents Chemother. 53, 292–294. 10.1128/AAC.00931-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogaerts P., Huang T. D., Rezende de Castro R., Bouchahrouf W., Glupczynski Y. (2013). Could Acinetobacter pittii act as an NDM-1 reservoir for Enterobacteriaceae? J. Antimicrob. Chemother. 68, 2414–2415. 10.1093/jac/dkt201 [DOI] [PubMed] [Google Scholar]
- Castanheira M., Farrell S. E., Wanger A., Rolston K. V., Jones R. N., Mendes R. E. (2013). Rapid expansion of KPC-2-producing Klebsiella pneumoniae isolates in two Texas hospitals due to clonal spread of ST258 and ST307 lineages. Microb. Drug Resist. 19, 295–297. 10.1089/mdr.2012.0238 [DOI] [PubMed] [Google Scholar]
- Fu Y., Du X., Ji J., Chen Y., Jiang Y., Yu Y. (2012). Epidemiological characteristics and genetic structure of blaNDM-1 in non-baumannii Acinetobacter spp. in China. J. Antimicrob. Chemother. 67, 2114–2122. 10.1093/jac/dks192 [DOI] [PubMed] [Google Scholar]
- Girlich D., Poirel L., Nordmann P. (2012). Value of the modified Hodge test for detection of emerging carbapenemases in Enterobacteriaceae. J. Clin. Microbiol. 50, 477–479. 10.1128/JCM.05247-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman K. E., Simner P. J., Tamma P. D., Milstone A. M. (2016). Infection control implications of heterogeneous resistance mechanisms in carbapenem-resistant Enterobacteriaceae (CRE). Expert Rev. Anti Infect. Ther. 14, 95–108. 10.1586/14787210.2016.1106940 [DOI] [PubMed] [Google Scholar]
- Gupta N., Limbago B. M., Patel J. B., Kallen A. J. (2011). Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin. Infect. Dis. 53, 60–67. 10.1093/cid/cir202 [DOI] [PubMed] [Google Scholar]
- Heller I., Grif K., Orth D. (2012). Emergence of IM-1-carbapenemase-producing Enterobacter cloacae in Tyrol, Austria. J. Med. Microbiol. 61, 567–571. 10.1099/jmm.0.038646-0 [DOI] [PubMed] [Google Scholar]
- Ho P. L., Li Z., Lai E. L., Chiu S. S., Cheng V. C. (2012). Emergence of NDM-1-producing Enterobacteriaceae in China. J. Antimicrob. Chemother. 67, 1553–1555. 10.1093/jac/dks095 [DOI] [PubMed] [Google Scholar]
- Hu P. F., Zhu D., Wang F., Yu Y. S., Lin J., Hu Y. J., et al. (2012). CHINET report on the distribution and drug resistance of carbapenem-resistant Enterobacteriaceae in China. Chin. J. Infect. Chemother. 2014, 382–386. [Google Scholar]
- Hu P. F., Zhu D. M., Wang F., Jiang X. F., Xu Y. C., Zhang X. J., et al. (2014). CHINET report on the bacterial drug reisstance in China. Chin. J. Infect. Chemother. 2015, 401–410. [Google Scholar]
- Huang Y. M., Zhong L. L., Zhang X. F., Hu H. T., Li Y. Q., Yang X. R., et al. (2015). NDM-1-producing Citrobacter freundii, Escherichia coli, and Acinetobacter baumannii identified from a single patient in China. Antimicrob. Agents Chemother. 59, 5073–5077. 10.1128/AAC.04682-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iredell J., Brown J., Tagg K. (2016). Antibiotic resistance in Enterobacteriaceae: mechanisms and clinical implications. BMJ 352:h6420. 10.1136/bmj.h6420 [DOI] [PubMed] [Google Scholar]
- Jakobsen L., Hansen F., Stegger M., Andersen P. S., Hammerum A. M., Fuglsang-Damgaard D. (2015). Use of whole-genome sequencing for detection of the spread of VIM-4-producing Escherichia coli between two patients in Denmark. Int. J. Antimicrob. Agents 45, 327–329. 10.1016/j.ijantimicag.2014.12.003 [DOI] [PubMed] [Google Scholar]
- Kaniga K., Flamm R., Tong S. Y., Lee M., Friedland I., Redman R. (2010). Worldwide experience with the use of doripenem against extended-spectrum-β-lactamase-producing and ciprofloxacin-resistant Enterobacteriaceae: analysis of six phase 3 clinical studies. Antimicrob. Agents Chemother. 54, 2119–2124. 10.1128/AAC.01450-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy Hara G., Gould I., Endimiani A., Pardo P. R., Daikos G., Hsueh P. R., et al. (2013). Detection, treatment, and prevention of carbapenemase-producing Enterobacteriaceae: recommendations from an international working group. J. Chemother. 25, 129–140. 10.1179/1973947812Y.0000000062 [DOI] [PubMed] [Google Scholar]
- Li H., Zhang J., Liu Y., Zheng R., Chen H., Wang X., et al. (2014). Molecular characteristics of carbapenemase-producing Enterobacteriaceae in China from 2008 to 2011: predominance of KPC-2 enzyme. Diagn. Microbiol. Infect. Dis. 78, 63–65. 10.1016/j.diagmicrobio.2013.10.002 [DOI] [PubMed] [Google Scholar]
- Ling M. L., Tee Y. M., Tan S. G., Amin I. M., How K. B., Tan K. Y., et al. (2015). Risk factors for acquisition of carbapenem resistant Enterobacteriaceae in an acute tertiary care hospital in Singapore. Antimicrob. Resist. Infect. Control. 4, 26. 10.1186/s13756-015-0066-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miriagou V., Tzelepi E., Kotsakis S. D., Daikos G. L., Bou Casals J., Tzouvelekis L. S. (2013). Combined disc methods for the detection of KPC- and/or VIM-positive Klebsiella pneumoniae: improving reliability for the double carbapenemase producers. Clin. Microb. Infect. 19, E412–E4125. 10.1111/1469-0691 [DOI] [PubMed] [Google Scholar]
- Poirel L., Yilmaz M., Istanbullu A., Arslan F., Mert A., Bernabeu S., et al. (2014). Spread of NDM-1-producing Enterobacteriaceae in a neonatal intensive care unit in Istanbul, Turkey. Antimicrob. Agents Chemother. 58, 2929–2933. 10.1128/AAC.02047-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queenan A. M., Bush K. (2007). Carbapenemases: the versatile β-lactamases. Clin. Microbiol. Rev. 20, 440–458. 10.1128/CMR.00001-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramana K. V., Rao R., Sharada Ch V., Kareem M., Reddy L. R., Ratna Mani M. (2013). Modified hodge test: a useful and the low-cost phenotypic method for detection of carbapenemase producers in Enterobacteriaceae members. J. Nat. Sci. Biol. Med. 4, 346–348. 10.4103/0976-9668.117009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastegar Lari A., Azimi L., Rahbar M., Fallah F., Alaghehbandan R. (2013). Phenotypic detection of Klebsiella pneumoniae carbapenemase among burns patients: first report from Iran. Burns 39, 174–176. 10.1016/j.burns.2012.02.025 [DOI] [PubMed] [Google Scholar]
- Tenover F. C., Arbeit R. D., Goering R. V., Mickelsen P. A., Murray B. E., Persing D. H., et al. (1995). Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33, 2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa J., Viedma E., Branas P., Orellana M. A., Otero J. R., Chaves F. (2014). Multiclonal spread of VIM-1-producing Enterobacter cloacae isolates associated with In624 and In488 integrons located in an IncHI2 plasmid. Int. J. Antimicrob. Agents 43, 451–455. 10.1016/j.ijantimicag.2014.02.006 [DOI] [PubMed] [Google Scholar]
- Wang J. T., Wu U. I., Lauderdale T. L., Chen M. C., Li S. Y., Hsu L. Y., et al. (2015). Carbapenem-nonsusceptible Enterobacteriaceae in Taiwan. PLoS ONE 10:e0121668. 10.1371/journal.pone.0121668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Chen S., Guo Y., Xiong Z., Hu F., Zhu D., et al. (2011). Occurrence of false positive results for the detection of carbapenemases in carbapenemase-negative Escherichia coli and Klebsiella pneumoniae isolates. PLoS ONE 6:e26356. 10.1371/journal.pone.0026356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Chen G., Wu X., Wang L., Cai J., Chan E. W., et al. (2015). Increased prevalence of carbapenem resistant Enterobacteriaceae in hospital setting due to cross-species transmission of the bla NDM-1 element and clonal spread of progenitor resistant strains. Front. Microbiol. 6:595. 10.3389/fmicb.2015.00595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Gu B., Huang M., Liu H., Xu T., Xia W., et al. (2015). Epidemiology of carbapenem resistant Enterobacteriaceae (CRE) during 2000-2012 in Asia. J. Thorac. Dis. 7, 376–385. 10.3978/j.issn.2072-1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Li X., Wang M., Yue H., Li P., Liu Y., et al. (2015). Outbreak of NDM-1-producing Klebsiella pneumoniae causing neonatal infection in a teaching hospital in mainland China. Antimicrob. Agents Chemother. 59, 4349–4351. 10.1128/AAC.03868-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweigner J., Gastmeier P., Kola A., Klefisch F. R., Schweizer C., Hummel M. A. (2014). carbapenem-resistant Klebsiella pneumoniae outbreak following bronchoscopy. Am. J. Infect. Control. 42, 936–937. 10.1016/j.ajic.2014.04.022 [DOI] [PubMed] [Google Scholar]


