Abstract
The photosynthetic capabilities of cyanobacteria make them interesting candidates for industrial bioproduction. One obstacle to large-scale implementation of cyanobacteria is their limited growth rates as compared to industrial mainstays. Synechococcus UTEX 2973, a strain closely related to Synechococcus PCC 7942, was recently identified as having the fastest measured growth rate among cyanobacteria. To facilitate the development of 2973 as a model organism we developed in this study the genome-scale metabolic model iSyu683. Experimental data were used to define CO2 uptake rates as well as the biomass compositions for each strain. The inclusion of constraints based on experimental measurements of CO2 uptake resulted in a ratio of the growth rates of Synechococcus 2973 to Synechococcus 7942 of 2.03, which nearly recapitulates the in vivo growth rate ratio of 2.13. This identified the difference in carbon uptake rate as the main factor contributing to the divergent growth rates. Additionally four SNPs were identified as possible contributors to modified kinetic parameters of metabolic enzymes and candidates for further study. Comparisons against more established cyanobacterial strains identified a number of differences between the strains along with a correlation between the number of cytochrome c oxidase operons and heterotrophic or diazotrophic capabilities.
Cyanobacteria are a morphologically and geographically diverse group of photosynthetic prokaryotes that have adapted to a wide variety of environmental niches including fresh, sea, and wastewater1,2,3. These organisms therefore do not compete for arable land with food and energy crops4. Cyanobacteria have been explored as candidates for numerous applications, including carbon dioxide sequestration5,6 and as a food source7,8. In recent years, cyanobacteria have become the targets of synthetic biology for use as platforms for the production of a variety of industrially relevant chemicals9,10. These include both biochemically active products11,12, as well as a number of biofuel products such as hydrogen13, butanol14 and polymer precursors including 2,3-butanediol15,16 and isoprene17.
Despite the inherent advantage of not requiring a reduced carbon substrate, cyanobacteria are still limited by growth rates lower than industrial mainstays such as E. coli and Saccharomyces cerevisiae. The model cyanobacterium Synechocystis PCC 6803 has a maximum doubling time of 7–10 hours under optimal conditions18. However a recently discovered cyanobacterium, Synechococcus UTEX 2973 has been shown to have the lowest doubling time among cyanobacteria at 1.9 hours19. This organism is closely related to Synechococcus PCC 7942, with a total of 55 single nucleotide polymorphisms and insertion-deletions, including a 188.6 kb inversion and a deletion of six open reading frames that are present in 794219. Despite the small differences, the growth rate for 2973 is 2.13 times greater than that of 7942 under their optimal illumination conditions at 38C19. This rapid growth rate and recent development of additional genetic tools20 makes Synechococcus 2973 an attractive candidate for bioproduction. Developing Synechococcus 2973 as a platform requires a more thorough understanding of its metabolic network and capabilities.
Genome-scale metabolic (GSM) models provide a blueprint for tracing all possible routes for utilizing carbon substrates in the context of the overall metabolism. They contain the set of metabolic reactions catalyzed within the organism, the gene-protein-reaction (GPR) relationships, and reaction directionalities21,22. These models also have a biomass equation, which contains the experimentally measured stoichiometric amounts of biomass precursors for the specific organism. GSM models are created using the sequenced genome for the organism along with available literature evidence. Semi-automated model generation balances the significant time investment required for de novo model development with the risk of losing details specific to certain organisms associated with completely automated development23. The workflow introduced and implemented in Mueller et al. uses an available curated model and reviewed gene annotations to expedite the development of a GSM model23. The GSM models required for semi-automated model development of cyanobacteria strains already exist, as models have been developed for a number of cyanobacteria including Synechocystis PCC 680324,25, Cyanothece ATCC 5114224, and Synechococcus PCC 700226.
Once a GSM model is created, flux balance analysis (FBA) can be used to estimate the maximum theoretical biomass yield by maximizing the flux through the biomass equation27. Flux variability analysis (FVA) can then be used to determine the feasible ranges for each reaction flux at maximum growth conditions. GSM models can also be used to predict differences in pathways between organisms23, product yields28 and the effect of genetic interventions29. GPR relationships enable the modeling of gene knockout phenotypes. This can be used not only for model refinement30 but also to facilitate organism design by suggesting genetic manipulations to reapportion fluxes to obtain a desired phenotype29.
In this paper we describe the development of a composite GSM model for both Synechococcus 7942 and Synechococcus 2973 using a previously defined workflow23, the sequenced genomes, and a curated reference GSM model24. The iSyn731 GSM model of Synechocystis PCC 6803 was chosen as the reference model following the identification of similar percent identity between previously modeled cyanobacteria24,26 and the Synechococcus strains (see Materials and Methods). These two strains are represented via a composite model given that the minimal genomic differences between the two strains do not result in any changes to the set of reactions in each strain. The model is combined with additional experimental measurements; specifically carbon dioxide uptake rates and biomass composition, to further interrogate what factors contribute to these differences in growth rates. When the two strains were evaluated, the same reaction network was used with carbon dioxide uptake rates and a biomass reaction specific to each strain. Additional model refinement was performed by comparing the model against available gene essentiality data31. Using experimental measurements for CO2 uptake as constraints on the model, the in silico ratio of the growth rate of Synechococcus 2973 to Synechococcus 7942 of 2.03 is close to the in vivo ratio of 2.13, identifying differing carbon uptake rates as the main contributing factor to the fast-growth phenotype. Specific SNPs and their possible effects on metabolism and the fast-growth growth phenotype are discussed. Genomic comparisons between the Synechococcus strains and several subsets of organisms identify genes and functionalities exclusive to the two strains.
Results and Discussion
Model Generation and Testing
This work resulted in the creation of a composite GSM model for two closely related cyanobacteria strains, Synechococcus PCC 7942 and Synechococcus UTEX 2973 (hereafter referred to as Synechococcus 7942 and Synechococcus 2973). The composite model contains 1,178 reactions and 1,028 metabolites. GPRs were developed for both strains, with the Synechococcus 2973 GPRs containing 683 genes and the Synechococcus 7942 GPRs containing 687 genes. This difference in gene number is due to four pairs of Synechococcus 7942 genes that each map to a single Synechococcus 2973 gene. The composite model contains 150 reactions not present in iSyn73124, the original Synechocystis model used in the model generation workflow. The energetic parameters from iSyn731 (i.e. non-growth and growth associated maintenance energy costs) were also used in the Synechococcus model. 145 reactions were not transferred from iSyn731 to the composite model. 93 of these reactions have a GPR and did not have the necessary homologs to transfer.
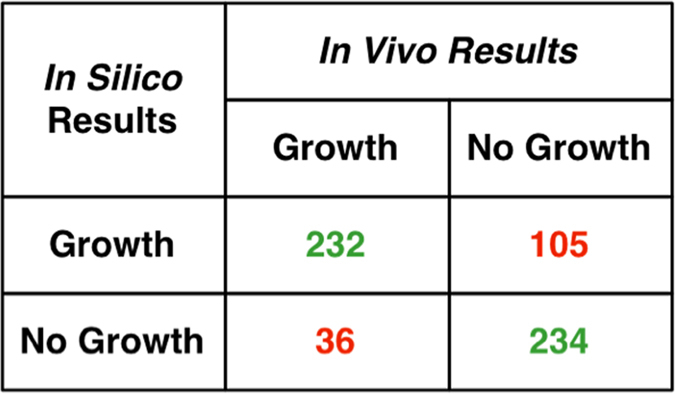
607 of the 687 genes in iSyu683 have in vivo essential/nonessential verdicts from the transposon library by Rubin et al.31. In silico predictions of these gene knockouts correctly predicted 232 of 268 non-essential genes and 234 of 395 essential genes (Fig. 1). All refinements made to the Synechococcus 7942 GPR were mapped to the Synechococcus 2973 GPR as well. Currently only one gene in the set of Synechococcus 2973 GPR relationships, zwf, has been knocked out. Both in vivo and in silico results indicate that zwf, which codes for glucose-6-phosphate 1-dehydrogenase in the oxidative pentose phosphate pathway, is not an essential gene for growth under autotrophic conditions (Abernathy et al., in review).
Figure 1. Categorization of in silico gene knockout predictions in Synechococcus 7942 as compared to in vivo data.
Composition and Genomic Differences Between Strains
Measurements for each strain were obtained for amino acids, lipids, glycogen, and chlorophyll (Abernathy et al., in review) and used in the formulation of strain specific biomass equations (see Materials and Methods for a full description of biomass equation development). The most pronounced difference between the strains is in the glycogen levels, with Synechococcus 7942 generating 8.3 times the amount produced by Synechococcus 2973 during the exponential growth phase (Abernathy et al., in review). This supports the observations made by Yu et al. on the number of electron-dense bodies between the two strains19. Another significant difference is between amino acid levels, with 53.0% of Synechococcus 2973 biomass by weight composed of amino acids, as compared to 40.9% for Synechococcus 7942. Both strains have similar weight percentages of lipids, although measurements for C18:0 and C18:1 fatty acid chains are an order of magnitude higher in Synechococcus 7942. The weight percentage of chlorophyll a in the two strains is also quite similar, with 2973 having only 1.05 times the amount of the pigment found in 7942. These measured components sum to 755.46 and 743.89 mg/gDW for Synechococcus 7942 and Synechococcus 2973, respectively. Therefore, the unmeasured biomass components (e.g. nucleic acids) account for similar fraction of biomass.
Yu et al. compared the Synechococcus 7942 and 2973 genomes and identified 26 SNPs in 21 genes and six open reading frames (ORFs) present in Synechococcus 7942 that were not found in Synechococcus 2973. The six ORFs not found in Synechococcus 2973 are five hypothetical proteins and a chromophore lyase, none of which are present within the Synechococcus 7942 GPR. Of the 21 genes with SNPs, 11 are found within the Synechococcus 7942 GPR (Supplementary File S4). Seven of these genes are essential for carrying out 22 different reactions within the model. 12 of these reactions are associated with long-chain fatty acid CoA ligase. Nine of the ten remaining reactions can carry flux at experimental biomass, two of which are in nucleic acid synthesis and salvage pathways, two are ATP synthase reactions located at the thylakoid lumen and periplasm respectively, and five are in amino acid biosynthesis pathways (Table 1). SNPs in these genes could result in an increase to the kinetic parameters associated with the enzyme and contribute to a higher production of corresponding biomass components. When examining flux ranges standardized by carbon uptake for both strains at experimental growth rates, four ORFs with SNPs (M744_01335, M744_06650, M744_06850, and M744_12285) have associated reactions with higher achievable fluxes in Synechococcus 2973. This suggests that these locii code for enzymes that may have different kinetic constants between the two strains. This calls for enzymology studies of the enzymes with SNPs and kinetic modeling to assess the effect of kinetic parameter tune-up on growth rate.
Table 1. Compilation of ORFs with SNPs that are essential to GSM model reactions.
| Locus Tag | Protein Annotation | Number of Reactions Gene is Essential For | Pathway containing reaction (s) |
|---|---|---|---|
| M744_01335 | ATP synthase F0F1 subunit alpha | 2 | ATP synthesis |
| M744_03965 | ABC-transporter substrate-binding protein | 1 | Adenine salvage |
| M744_06650 | CTP synthetase | 2 | CTP synthesis |
| M744_06850 | Chorismate mutase | 1 | Phenylalanine/Tyrosine biosynthesis |
| M744_11685 | Anthranilate synthase, component I | 2 | Tryptophan biosynthesis |
| M744_12130 | Long-chain-fatty-acid CoA ligase | 1 | Fatty acid metabolism |
| M744_12285 | Glutamate synthase | 1 | Glutamate biosynthesis |
The remaining 10 genes with SNPs are not present in the GSM model and are associated with a diverse group of Gene Ontology (GO) categories32. Several SNPs imply potential effects that cannot be captured with a GSM model. Specifically, several SNP’s in RNA polymerase 23 S ribosomal RNA could result in higher rates of translation, which may explain the higher amino acid content in Synechococcus 2973 biomass as compared to Synechococcus 7942 (Fig. 2). Another SNP is in M744_13540, annotated as the Photosystem 1 assembly protein Ycf4. The ycf4 knockout has been shown to be essential for PSI activity in Chlamydomonas reinhardtii33 and inactivation of an ycf4 homolog in Synechocystis 6803 increases the PSII-PSI ratio34. Comparing the flux through the photosystem reactions under limiting light and maximum biomass production, Synechococcus 7942 has a higher PSII-PSI ratio than Synechococcus 2973, a difference that could correlate to an increased activity from M744_13540. While the effects of these changes cannot be captured with stoichiometric modeling they provide a starting point for further exploration with models that capture in detail more biological processes.
Figure 2. Mass fractions of the main constituents of biomass for the two Synechococcus strains.
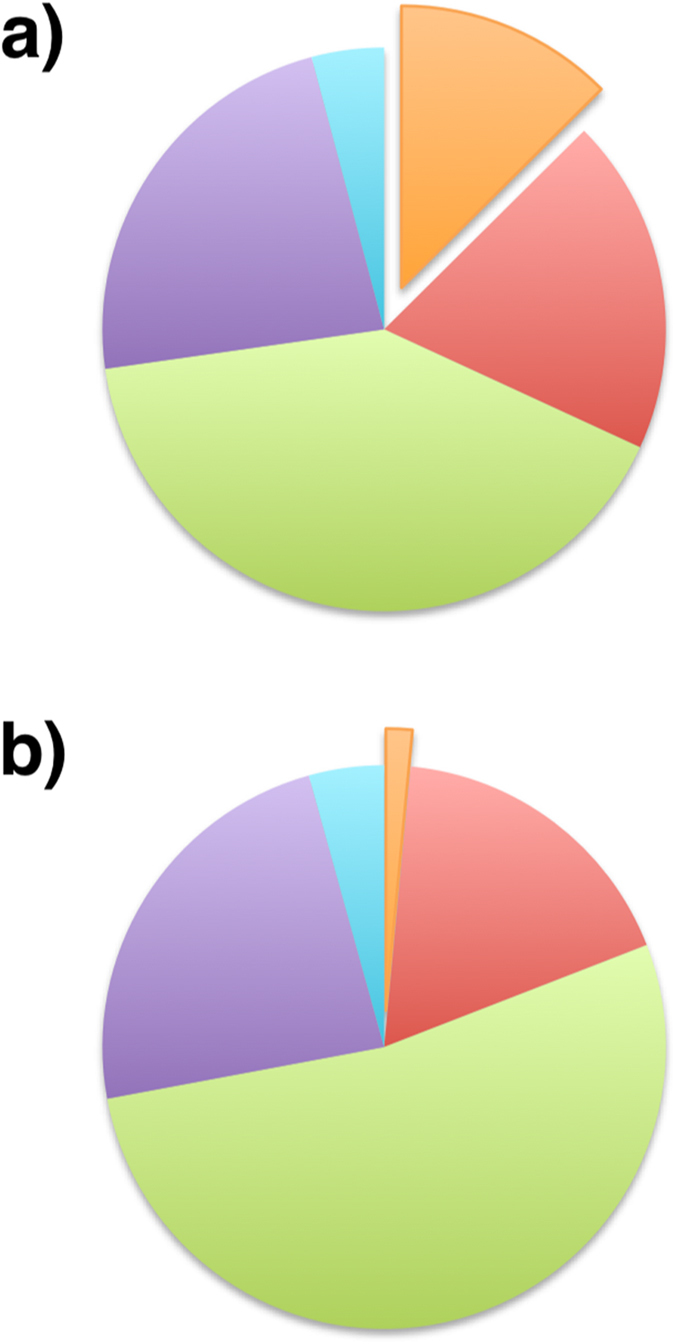
(a) Mass fractions for Synechococcus 7942 (b) Mass fractions for Synechococcus 2973. Color-Category relationship: orange – glycogen, red – lipids, green – amino acids, purple – nucleic acids, blue – other.
Identification of Causes for Fast-Growth Phenotype
Under their optimal illumination conditions at 38 C, the growth rate ratio of Synechococcus 2973: Synechococcus 7942 is 2.1319. The GSM model can aid in the identification of a set of contributing factors to this difference. The varying biomass composition for the two strains and constant growth, transport, and maintenance energy requirements implies different biomass yields per mmol of CO2 taken up. The required moles of carbon for one theoretical mole of biomass were calculated by multiplying biomass stoichiometric coefficients by the number of carbon atoms in each precursor. Synechococcus 2973 requires 0.502 g carbon per g biomass as compared to 0.497 for Synechococcus 7942. When supplied equal amounts of CO2 the ratio of biomass yields per mol of carbon of Synechococcus 2973 to Synechococcus 7942 is 0.98 implying that Synechococcus 2973 requires slightly more carbon per gDW of biomass. Experimental measurements have shown that under their optimal conditions, Synechococcus 2973 takes up CO2 at a rate 2.06 times that of 7942 (Supplementary File S5). Using these measurements as a constraint on the exchange fluxes on the model resulted in an in silico growth rate ratio of 2.03. This is close to recapitulating the experimental ratio of 2.13. Therefore, due to the carbon limitation of the models, the difference in the rates of CO2 uptake is a primary contributing factor to the different growth rates seen for the two Synechoccocus strain models. The minimum required photons for both strains were calculated by minimizing the sum of photons taken up for both photosystems at maximum biomass. Flux variability analysis (FVA) was used to calculate the flux ranges for both strains at max biomass with minimum required photons. When the flux ranges are standardized by the amount of CO2 taken up the reactions with non-overlapping ranges are dictated by the biomass composition, with 2973 having a higher flux through the amino acid biosynthetic pathways and 7942 having a higher flux through the biosynthesis pathways for glycogen as well as C18:0 and 18:1 lipids (Fig. 3). Other notable differences include the aforementioned ratio of photon use between photosystems 1 and 2, and a higher flux through photorespiration in Synechococcus 2973. Photorespiration, the photosynthetic oxygenation of ribulose 1,5-bisphosphate (RuBP), competes with the carboxylation of RuBP and generates 2-phosphoglycolate (2PG). Eisenhut et al. previously showed that 2PG metabolism is essential for organisms such as cyanobacteria that perform oxygenic photosynthesis35, and similar requirements for flux through the photorespiration reaction at optimal growth rates has been seen in other in silico models36. The higher flux through photorespiration in Synechococcus 2973 could be due to the production of glyoxylate, a precursor to glycine, serine, and cysteine, from the photorespiration pathway and the increased requirements for amino acids in Synechococcus 2973.
Figure 3. Fraction of select pathways with non-overlapping flux ranges between the two Synechococcus strains.
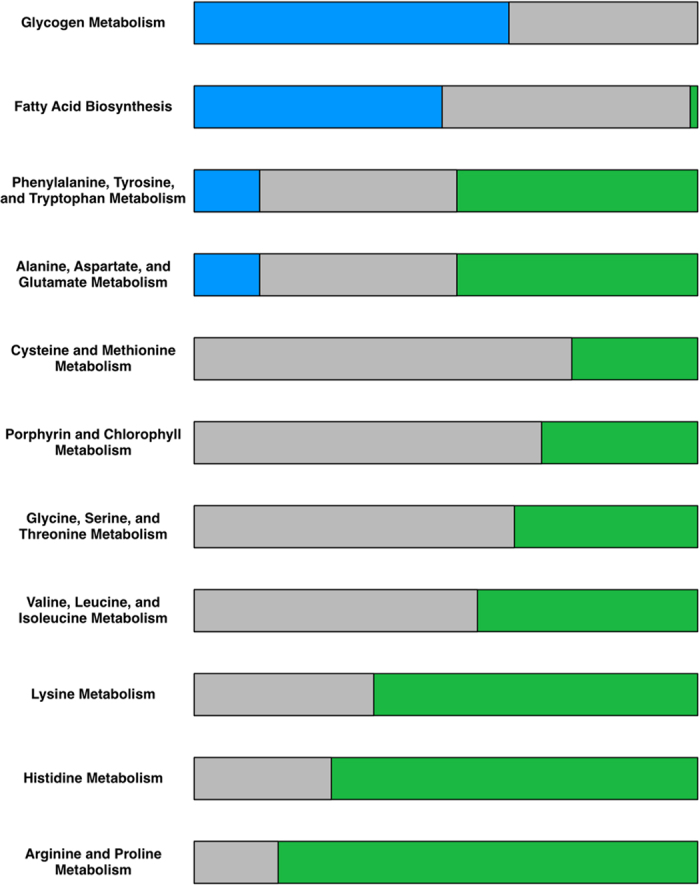
Pathway annotations from iSyn731 and SEED were used. All flux ranges were standardized by the amount of carbon taken up. Grey represented the percentage of reactions with that pathway annotation whose flux ranges overlapped. Blue and green represent the percentage of reactions with non-overlapping ranges where Synechococcus 7942 and Synechococcus 2973 had higher flux respectively.
With the supplied CO2 flux, both models can achieve growth rates higher than experimentally reported, with Synechococcus 2973 achieving a growth rate of 0.4569 hr−1 (doubling time: 1.5 hours) as compared to its experimentally reported optimal rate of 0.3014 hr−1 (doubling time: 2.3 hours), and Synechococcus 7942 with a growth rate of 0.2256 hr−1 (doubling time: 3.1 hours) as compared to the experimental value of 0.1415 hr−1 (doubling time: 4.9 hours). When the flux through the biomass reaction was fixed to the experimentally reported values, the model identified the main destinations of excess carbon as TCA cycle intermediates or metabolites that are several reactions removed from the TCA cycle (i.e. pyruvate, acetate, and glutamate). In total 33.4% of the carbon taken up is exported for 2973 and 37.2% is exported for 7942. This increased fraction of carbon allocated to biomass in 2973 could in part contribute to the divergent growth rates.
Comparing Synechococcus with Model Organisms
Given Synechococcus 2973’s potential as a production strain, it is important to compare its capabilities with that of model cyanobacteria, with pairwise genome-wide BLAST comparisons performed between Synechococcus 7942 and Synechocystis PCC 6803, Synechococcus PCC 7002, Cyanothece ATCC 51142, Anabaena PCC 7120, and Anabaena variabilis. A set of 696 genes in 7942 does not have satisfactory hits in the other five genomes (See Materials and Methods for BLAST hit criteria). 47 of these genes are in the 7942 GPR relationships and are essential for 21 reactions that can carry flux at max biomass production. These reactions are spread across a variety of pathways including amino acid and deoxyribonucleotide biosynthesis. As these genes comprise only 6.8% of the genes within the Synechococcus 7942 GPR relationships, a majority of the metabolic genes have homologs in other cyanobacterial strains. Therefore insights gained into the metabolism of these strains can more readily inform work in the metabolism of the fast-growing strain. Expanding past metabolism, gene ontology (GO) terms provide a means to contextualize the remaining genes. Of these 696 genes, 304 have annotations including GO terms in the Uniprot database. This fraction of genes with GO terms (43.7%) is much lower than that of the entire 7942 genome (70.7%), following a trend seen in the percent of reviewed annotations (3.3% in the subset compared to 18.1% overall) and percent of genes annotated as uncharacterized (70.4% in the subset and 39.2% overall). Two GO terms have at least ten genes in the subset and greater than 50% of the genes in the organism with that GO term within the subset. There are 14 genes within this subset annotated with the term GO:0000155 – phosphorelay sensor kinase activity, and 10 genes annotated with the term GO:0043565 – sequence specific DNA binding. This suggests that most of the differences between these strains are related to the response to external stimuli and transcription regulation. These differences are consistent with the comparison of a number of thioredoxins involved in light dependent protein regulation37. The BLAST search identified three homologs in Synechococcus 7942 in contrast to four and eight thioredoxin genes in Synechocystis 6803 and Anabaena 712038.
Another key difference between cyanobacterial strains is the classification of some organisms as obligate photoautotrophs. Schmetterer et al. showed that the deletion of a three gene cytochrome c oxidase (coxBAC) operon from Anabaena variabilis ATCC 29413 resulted in a strain capable of unhindered photoautotrophic growth but which was no longer capable of chemoheterotrophic growth39. BLAST searches were performed between A. variabilis 29413’s five operons and seven other cyanobacteria with information on their ability to grow heterotrophically. An independent annotation search of each organism was also performed, which identified the same operon sets for each organism as were identified through the BLAST search. Table 2 shows the results of these comparisons, with obligate photoautotrophs all having a single operon, as compared to two or more for each organism capable of heterotrophic growth. However, organisms capable of nitrogen fixation have at least three operons while the non-diazotrophic heterotrophs are restricted to two operons. Several specialized roles have been identified for specific coxBAC operons including an operon required for growth on sugar39 and separate operons that are heterocyst specific and required for nitrogen fixation40. These roles suggest a possible cause for the varying number of operons across cyanobacterial strains, and provide a promising avenue for further comparison among cyanobacteria.
Table 2. Number of cytochrome c oxidase operons in model cyanobacteria.
| Organism | Obligate Photoautotroph? | Capable of Fixing Nitrogen? | Cyt C Oxidase operons present |
|---|---|---|---|
| Anabaena 29413 | No | Yes | 5 |
| Nostoc punctiforme | No | Yes | 4 |
| Anabaena 7120 | No | Yes | 3 |
| Cyanothece 51142 | No | Yes | 3 |
| Synechocystis 6803 | No | No | 2 |
| Synechococcus 7002 | No | No | 2 |
| Synechococcus 7942 | Yes | No | 1 |
| Synechococcus 6301 | Yes | No | 1 |
| Synechococcus 6312 | Yes | No | 1 |
Conclusions
In this paper we presented a GSM model for both Synechococcus 7942 and Synechococcus 2973 with GPR relationships that were tested and refined using gene essentiality data for Synechococcus 7942. This model was used to compare the two strains and identify possible reasons for the diverging fast-growth phenotype. The GSM model allowed for the identification of the pathways where flux was allocated to differing levels between the two strains along with the identification of four SNPs as possible contributors to modified kinetic parameters of metabolic enzymes and candidates for further study. Several differences between the two strains were identified including the fraction of carbon allocated to biomass and the weight fractions of different biomass precursors, with the most prominent being the difference in carbon uptake rates, which contributed to the different in silico growth rates. Finally, a genomic comparison between Synechoccocus 7942 and model organisms highlighted a number of differences including regulatory genes without identified homologs and a variation in the number of copies of the cytochrome c oxidase operon as related to heterotrophic and diazotrophic capabilities.
Methods
GSM Model Generation
The composite model was generated using the workflow previously described in Mueller et al.23. This workflow integrates and prioritizes information from both a curated GSM model of a related organism and reviewed gene annotations to expedite the generation of a GSM model for the organism of interest. The iSyn731 model of Synechocystis PCC 680324 was used as the reference GSM model for both organisms. This organism was selected using sequence alignments of the 16 s sequences of the cyanobacterial strains targeted in the genome comparisons using Clustal2.141. This comparison revealed a similar percent identity between previously modeled cyanobacteria Synechococcus 7002 and Synechocystis 680324,26 and Synechococcus 7942, resulting in the selection of our previously developed iSyn731 model of Synechocystis 680324. The workflow uses homolog pairs between Synechocystis and the Synechococcus strains to identify which reactions should be transferred to the new GSM model. To identify homolog pairs a bidirectional BLAST search between each organism pair was performed using an e value cutoff of 10−2. A result was determined to be a hit if the alignment length was at least 75% of the length of both the query and target genes, and if the raw score was at least 25% of the self-hit score of the query gene. A homolog pair was identified if both genes had only each other as a hit42. This process was repeated to identify homologs between Synechococcus 7942 and 2973. Given that the organisms are closely related, the score cutoff was increased to 75% of the self-hit score. This reduced the number of occurrences of multiple hits for a gene and identified only the best hit, increasing the number of homolog pairs from 2274 to 2538. These homolog pairs were used to map reviewed annotations from Synechococcus 7942 to Synechococcus 2973. The composite model is available in both table (Supplementary File S1) and Systems Biology Markup Language (SBML) formats (Supplementary Files S2 and S3)43. For the comparison with current industrial strains the requirements for a BLAST hit were reduced in order to account for the greater phylogenetic distance between the species (i.e. 10% of self-hit score, 50% of length).
The biomass equation previously developed for the iSyn731 model was updated for the Synechococcus strains using experimental measurements for both strains. All biomass measurements for both strains were taken from Abernathy et al. (Abernathy et al., in review). These measurements were used to determine biomass equation stoichiometric coefficients for lipids, glycogen, amino acids, and chlorophyll. The coefficients for the unmeasured precursors were updated by scaling the coefficients from the iSyn731 biomass equation so that all biomass precursors added up to 1 g per gDW of biomass. This standardization facilitates comparison between the two strains.
Experimental Measurements of Carbon Uptake
Experiemental measurements of the rate of carbon dioxide uptake were taken for both strains. Both Synechococcus strains were grown in a MC-1000 multicultivator (Photon Systems Instruments) in standard BG11 medium at 38 C, and supplemented with 3% CO2 to an OD720 of 1.0. Synechococcus 2973 was grown at 500 μE·m−2·s−1 light and 7942 was grown at 300 μE·m−2·s−1 light, the light intensities at which each strain achieved its maximal growth rate19, allowing for comparison of factors contributing to those optimal growth rates. Two 1 mL samples were sealed in 13 mL hungate tubes with rubber septa and sparged with 3% CO2 for 5 minutes. One tube for each strain was placed on its side on a shaker under their respective white LED illumination at 38 C for 1 hour after which time total CO2 was measured. The second tube was measured immediately. Total CO2 was calculated as the sum of the dissolved + gaseous CO2 within a sealed system. Total CO2 of the system was determined after injecting 100uL 10 N HCl into the tube to gas out dissolved CO2, followed by quantification of the headspace CO2 content on a HP 5980 gas chromatograph under the following conditions: porapak N column; temperature, 100 °C; carrier gas, helium at a flow rate of 40 ml min−1; and TCD detector. These uptake rates were given as umol CO2 mg−1 chl hr−1. Using the molecular weight and the biomass stoichiometry (mmol chl gDW−1) of chlorophyll A these rates were converted to the units of CO2 uptake flux within the model (mmol CO2 gDW−1 hr−1). The original measurements and their conversion to fluxes are in Supplemental File S5.
GSM Model Implementation
FBA was performed as described by Orth et al.27 to determine maximum flux through the biomass equation. The carbon dioxide uptake fluxes calculated from the experimental data were used as constraints on the exchange flux for carbon dioxide. Flux variability analysis (FVA)44 was used to identify the flux ranges of each reaction by iteratively minimizing and maximizing the flux of a single reaction subject to the FBA constraints, along with constraints on biomass and photon uptake. In the context of the model, photons are treated as two separate metabolites consumed by photosystems 1 and 2. The minimum required photons to achieve a biomass level were determined by fixing biomass, retaining the FBA constraints, and minimizing the sum of the exchange fluxes for photons for photosystems 1 and 2.
In order to compare in silico and in vivo essentiality, the GPR for each reaction was reviewed upon the removal of each individual gene. If the GPR logic statement was no longer true (i.e. if the gene was an essential subunit of a protein complex or the gene did not have any homologs) then the reaction was not allowed to carry flux for the in silico mutant. If the model could not produce at least 10% of the wild-type biomass the mutation was classified as lethal. Gene knockouts were performed in silico for every gene in the model. The in vivo results were taken from those genes that were identified as essential or non-essential from the transposon library of Synechococcus 7942 published by Rubin et al.31. These results were categorized using the GrowMatch terminology, where either the in silico and in vivo results for growth or no growth match (i.e. GG and NGNG) or conflict (i.e. GNG and NGG)30.
Additional Information
How to cite this article: Mueller, T. J. et al. Identifying the Metabolic Differences of a Fast-Growth Phenotype in Synechococcus UTEX 2973. Sci. Rep. 7, 41569; doi: 10.1038/srep41569 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
Supported by funding from the Department of Energy DE-SC0012722.
Footnotes
The authors declare no competing financial interests.
Author Contributions T.J.M. created the GSM model and performed all computational studies. J.U. measured carbon uptake. T.J.M. and C.D.M. wrote the paper. All authors read and approved of the final manuscript.
References
- Thajuddin N. & Subramanian G. Cyanobacterial biodiversity and potential applications in biotechnology. Curr Sci India 89, 47–57 (2005). [Google Scholar]
- Flombaum P. et al. Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. Proc Natl Acad Sci USA 110, 9824–9829, doi: 10.1073/pnas.1307701110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Raouf N., Al-Homaidan A. A. & Ibraheem I. B. Microalgae and wastewater treatment. Saudi J Biol Sci 19, 257–275, doi: 10.1016/j.sjbs.2012.04.005 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dismukes G. C., Carrieri D., Bennette N., Ananyev G. M. & Posewitz M. C. Aquatic phototrophs: efficient alternatives to land-based crops for biofuels. Curr Opin Biotechnol 19, 235–240, doi: 10.1016/j.copbio.2008.05.007 (2008). [DOI] [PubMed] [Google Scholar]
- Ono E. & Cuello J. L. Carbon dioxide mitigation using thermophilic cyanobacteria. Biosyst Eng 96, 129–134, doi: 10.1016/j.biosystemseng.2006.09.010 (2007). [DOI] [Google Scholar]
- Sayre R. Microalgae: The Potential for Carbon Capture. Bioscience 60, 722–727, doi: 10.1525/bio.2010.60.9.9 (2010). [DOI] [Google Scholar]
- Verseux C. et al. Sustainable life support on Mars - the potential roles of cyanobacteria. Int J Astrobiol 15, 65–92, doi: 10.1017/S147355041500021x (2016). [DOI] [Google Scholar]
- Menezes A. A., Cumbers J., Hogan J. A. & Arkin A. P. Towards synthetic biological approaches to resource utilization on space missions. J R Soc Interface 12, doi: UNSP 2014071510.1098/rsif.2014.0715 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angermayr S. A., Rovira A. G. & Hellingwerf K. J. Metabolic engineering of cyanobacteria for the synthesis of commodity products. Trends Biotechnol 33, 352–361, doi: 10.1016/j.tibtech.2015.03.009 (2015). [DOI] [PubMed] [Google Scholar]
- Ducat D. C., Way J. C. & Silver P. A. Engineering cyanobacteria to generate high-value products. Trends Biotechnol 29, 95–103, doi: 10.1016/j.tibtech.2010.12.003 (2011). [DOI] [PubMed] [Google Scholar]
- Burja A. M., Banaigs B., Abou-Mansour E., Burgess J. G. & Wright P. C. Marine cyanobacteria - a prolific source of natural products. Tetrahedron 57, 9347–9377, doi: 10.1016/S0040-4020(01)00931-0 (2001). [DOI] [Google Scholar]
- Singh R. K., Tiwari S. P., Rai A. K. & Mohapatra T. M. Cyanobacteria: an emerging source for drug discovery. J Antibiot 64, 401–412, doi: 10.1038/ja.2011.21 (2011). [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay A., Stockel J., Min H., Sherman L. A. & Pakrasi H. B. High rates of photobiological H2 production by a cyanobacterium under aerobic conditions. Nat Commun 1, 139, doi: 10.1038/ncomms1139 (2010). [DOI] [PubMed] [Google Scholar]
- Lan E. I., Ro S. Y. & Liao J. C. Oxygen-tolerant coenzyme A-acylating aldehyde dehydrogenase facilitates efficient photosynthetic n-butanol biosynthesis in cyanobacteria. Energ Environ Sci 6, 2672–2681, doi: 10.1039/c3ee41405a (2013). [DOI] [Google Scholar]
- Oliver J. W. K., Machado I. M. P., Yoneda H. & Atsumi S. Cyanobacterial conversion of carbon dioxide to 2,3-butanediol. P Natl Acad Sci USA 110, 1249–1254, doi: 10.1073/pnas.1213024110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savakis P. E., Angermayr S. A. & Hellingwerf K. J. Synthesis of 2,3-butanediol by Synechocystis sp PCC6803 via heterologous expression of a catabolic pathway from lactic acid- and enterobacteria. Metab Eng 20, 121–130, doi: 10.1016/j.ymben.2013.09.008 (2013). [DOI] [PubMed] [Google Scholar]
- Lindberg P., Park S. & Melis A. Engineering a platform for photosynthetic isoprene production in cyanobacteria, using Synechocystis as the model organism. Metab Eng 12, 70–79, doi: 10.1016/j.ymben.2009.10.001 (2010). [DOI] [PubMed] [Google Scholar]
- Yu Y. et al. Development of Synechocystis sp PCC 6803 as a Phototrophic Cell Factory. Mar Drugs 11, 2894–2916, doi: 10.3390/md11082894 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J. J. et al. Synechococcus elongatus UTEX 2973, a fast growing cyanobacterial chassis for biosynthesis using light and CO2. Sci Rep-Uk 5, doi: 10.1038/srep08132 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt K. E., Ungerer J., Cobb R. E., Zhao H. & Pakrasi H. B. CRISPR/Cas9 mediated targeted mutagenesis of the fast growing cyanobacterium Synechococcus elongatus UTEX 2973. Microb Cell Fact 15, 115, doi: 10.1186/s12934-016-0514-7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele I. & Palsson B. O. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat Protoc 5, 93–121, doi: 10.1038/nprot.2009.203 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry C. S. et al. High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat Biotechnol 28, 977–U922, doi: 10.1038/nbt.1672 (2010). [DOI] [PubMed] [Google Scholar]
- Mueller T. J., Berla B. M., Pakrasi H. B. & Maranas C. D. Rapid construction of metabolic models for a family of Cyanobacteria using a multiple source annotation workflow. Bmc Syst Biol 7, doi: 10.1186/1752-0509-7-142 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha R. et al. Reconstruction and Comparison of the Metabolic Potential of Cyanobacteria Cyanothece sp ATCC 51142 and Synechocystis sp PCC 6803. Plos One 7, doi: 10.1371/journal.pone.0048285 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoop H. et al. Flux balance analysis of cyanobacterial metabolism: the metabolic network of Synechocystis sp. PCC 6803. PLoS Comput Biol 9, e1003081, doi: 10.1371/journal.pcbi.1003081 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu T. T. et al. Computational evaluation of Synechococcus sp. PCC 7002 metabolism for chemical production. Biotechnol J 8, 619–630, doi: 10.1002/biot.201200315 (2013). [DOI] [PubMed] [Google Scholar]
- Orth J. D., Thiele I. & Palsson B. O. What is flux balance analysis? Nat Biotechnol 28, 245–248, doi: 10.1038/nbt.1614 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash S., Mueller T. J., Venkataramanan K. P., Papoutsakis E. T. & Maranas C. D. Capturing the response of Clostridium acetobutylicum to chemical stressors using a regulated genome-scale metabolic model. Biotechnol Biofuels 7, doi: 10.1186/s13068-014-0144-4 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganathan S. et al. Using OptForce to customize metabolic interventions for the overproduction of fatty acids C6 through C16 in Escherichia coli. Abstr Pap Am Chem S 243 (2012). [Google Scholar]
- Kumar V. S. & Maranas C. D. GrowMatch: An Automated Method for Reconciling In Silico/In Vivo Growth Predictions. Plos Computational Biology 5, doi: 10.1371/journal.pcbi.1000308 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin B. E. et al. The essential gene set of a photosynthetic organism. Proc Natl Acad Sci USA, doi: 10.1073/pnas.1519220112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology C. Gene Ontology Consortium: going forward. Nucleic Acids Res 43, D1049–1056, doi: 10.1093/nar/gku1179 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau E., Takahashi Y., Lemieux C., Turmel M. & Rochaix J. D. The chloroplast ycf3 and ycf4 open reading frames of Chlamydomonas reinhardtii are required for the accumulation of the photosystem I complex. EMBO J 16, 6095–6104, doi: 10.1093/emboj/16.20.6095 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde A. et al. Inactivation of a Synechocystis sp strain PCC 6803 gene with homology to conserved chloroplast open reading frame 184 increases the photosystem II-to-photosystem I ratio. Plant Cell 7, 649–658, doi: 10.1105/tpc.7.5.649 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhut M. et al. The photorespiratory glycolate metabolism is essential for cyanobacteria and might have been conveyed endosymbiontically to plants. Proc Natl Acad Sci USA 105, 17199–17204, doi: 10.1073/pnas.0807043105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoop H., Zilliges Y., Lockau W. & Steuer R. The metabolic network of Synechocystis sp. PCC 6803: systemic properties of autotrophic growth. Plant Physiol 154, 410–422, doi: 10.1104/pp.110.157198 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Perez M. E., Martin-Figueroa E. & Florencio F. J. Photosynthetic regulation of the cyanobacterium Synechocystis sp. PCC 6803 thioredoxin system and functional analysis of TrxB (Trx x) and TrxQ (Trx y) thioredoxins. Mol Plant 2, 270–283, doi: 10.1093/mp/ssn070 (2009). [DOI] [PubMed] [Google Scholar]
- Florencio F. J., Perez-Perez M. E., Lopez-Maury L., Mata-Cabana A. & Lindahl M. The diversity and complexity of the cyanobacterial thioredoxin systems. Photosynth Res 89, 157–171, doi: 10.1007/s11120-006-9093-5 (2006). [DOI] [PubMed] [Google Scholar]
- Schmetterer ,. G. et al. The coxBAC operon encodes a cytochrome c oxidase required for heterotrophic growth in the cyanobacterium Anabaena variabilis strain ATCC 29413. J Bacteriol 183, 6429–6434, doi: 10.1128/JB.183.21.6429-6434.2001 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valladares A., Herrero A., Pils D., Schmetterer G. & Flores E. Cytochrome c oxidase genes required for nitrogenase activity and diazotrophic growth in Anabaena sp. PCC 7120. Mol Microbiol 47, 1239–1249 (2003). [DOI] [PubMed] [Google Scholar]
- Larkin M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948, doi: 10.1093/bioinformatics/btm404 (2007). [DOI] [PubMed] [Google Scholar]
- Mueller T. J., Welsh E. A., Pakrasi H. B. & Maranas C. D. Identifying regulatory changes to facilitate nitrogen fixation in the non-diazotroph Synechocystis sp. PCC 6803. ACS Synthetic Biology (2015). [DOI] [PubMed] [Google Scholar]
- Schellenberger J. et al. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox v2.0. Nat Protoc 6, 1290–1307, doi: 10.1038/nprot.2011.308 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan R. & Schilling C. H. The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab Eng 5, 264–276 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


