Abstract
Resistance against new hepatitis C virus (HCV) antivirals is an area of increasing interest. Resistance-associated substitutions (RASs) have been identified in treatment-naïve individuals, but pressures driving treatment-independent RAS emergence are poorly understood. We analysed the longitudinal evolution of RASs in twelve participants with early acute HCV infections. Full-genome deep sequences were analysed for changes in RAS frequency within NS3, NS5A and NS5B-coding regions over the course of the infection. Emergence of RASs relevant only to the polymerase non-nucleoside inhibitors (NNI) was detected, and these lay within CD8+ T-cell epitopes. Conversely, the loss of NNI RASs over time appeared likely to be driven by viral fitness constraints. These results highlight the importance of monitoring CD8+ T cell epitope-associated RASs in populations with dominant HLA types.
Antiviral therapy for hepatitis C virus (HCV) has undergone a recent revolution with the approval of several direct-acting antivirals (DAA) targeting the HCV protease (NS3), phosphoprotein (NS5A) and polymerase (NS5B). Interferon-free regimens, which contain multiple DAAs, have been approved in several countries to treat infections with different HCV genotypes1. Each agent has been shown to promote emergence of resistance-associated substitutions (RASs) when studied in vitro, and depending on the DAA, the presence of these substitutions could reduce the efficacy of antiviral treatment in vivo2. Monitoring the emergence of HCV RASs in vivo is therefore of particular importance to both clinical practice and public health strategies.
As a consequence of an error-prone viral polymerase, and a high replication rate, a swarm of mutations constantly get generated during HCV infection. Some of these natural mutations encode for RASs and have been reported in many studies in treatment-naïve patients3,4,5,6. Such naturally occurring RASs could negatively impact treatment success rates, particularly in patients with cirrhosis, those infected with genotype 3, and those who have failed interferon-based therapies. In the context of antiviral treatment, the emergence of RASs is initially limited by the genetic barrier to resistance, defined by the number and type of nucleotide changes required for amino acid substitutions7. The potential for particular RASs to persist in the host ultimately depends on the trade-off between the loss of replicative fitness and the survival advantage under strong antiviral drug selection pressure. In the absence of treatment, RASs may be retained if they increase viral fitness, are unable to revert back to wild-type2, or potentially if the relevant sites are under other selective pressures including host immune responses. For instance, some RAS sites within NS3 and NS5B have been shown to fall within experimentally-confirmed or predicted CD8+ T cell epitopes6,8,9. In addition, the prevalence of an NS5A RAS was recently shown to be associated with the host interferon λ4 (IFNL4) genotype10. These findings therefore suggest that both innate and adaptive immune responses play a role in the emergence of HCV DAA resistance in the absence of antiviral treatment.
As primary HCV infection is typically asymptomatic, it has been challenging to characterise the evolution of HCV mutations, and hence RASs, in the early phase of infection. Furthermore, the lack of well-characterised samples collected over the course of infection has limited longitudinal analyses of the interplay between the host immune response and viral fitness in relation to RAS development. Characterisation of the emergence of RASs in the absence of antiviral pressures is critical to our understanding of their stability within the host, and their potential influence on treatment options. The aim of this study was to examine the longitudinal emergence of RASs in the early phase of primary HCV infection. Specifically, the aim was to understand the interplay between viral replicative fitness and the host T cell responses in driving the emergence of RASs in the absence of antiviral treatment.
Methods
Subjects and samples
Viremic blood samples were obtained from prospective cohorts of high-risk, HCV-uninfected subjects recruited between 2005 and 2014 in NSW Australia. Sample were obtained from two cohorts; the Hepatitis C Incidence and Transmission Study in prisons (HITS-p) and in the community (HITS-c)11,12. Participants were tested every three to six months for HCV seroconversion, and then followed regularly post-infection until spontaneous clearance or persistence was determined when antiviral treatment was offered if they remained infected. For this study, twelve participants with early acute HCV infection were included. An early infection case was defined by the availability of at least one viremic sample prior to seroconversion. The estimated date of infection was calculated for each subject by subtracting the recognized mean pre-seroconversion window period of 51 days from the midpoint between the last HCV RNA-positive/HCV Ab-negative time point and the first seropositive time point11. Ethical approvals were obtained from the Human Research Ethics Committees of Justice Health (reference number G304/11), the New South Wales Department of Corrective Services (reference number 05/0884), and the University of New South Wales (reference numbers 05094, 08081, 13237, 09075, 14170). Written informed consent was obtained from the participants. All methods were also performed in accordance with the relevant guidelines and regulations.
Viral genome sequencing and analysis
Viral RNA was extracted from plasma samples and amplicons covering the full HCV genome were generated either as single near-full length products, or as three overlapping fragments, as described previously13,14. Next-generation sequencing of the amplicons was conducted using either the Roche 454 FLX, or the Illumina MiSeq sequencing platform, as previously reported11,13. Sequence alignments were generated using Bowtie 215 against the corresponding genotype reference sequence: GT1a (GenBank accession numbers AF009606), GT1b (AJ238799), GT2b (AB030907) and GT3a (D17763). Single-nucleotide polymorphism (SNP) analysis was performed using the Geneious software package version 816 with minimum variant frequency threshold of 0.001, maximum variant P-value of 10−6, and a minimum coverage of 1,000 (Illumina) or 400 (Roche 454). Sixty eight positions across the genome where RASs have previously been reported2 were analysed. These covered RASs impacting NS3, NS5A and NS5B inhibitors. Consensus sequences for each HCV genotype were obtained from the Los Alamos Hepatitis C Sequence Database (http://hcv.lanl.gov).
Analysis of autologous T cell responses across known RAS sites
HCV residue positions where RASs were gained over the course of infection were examined to determine whether their site of occurrence fell within potential CD8+ T cell epitopes. Potential HLA-I restricted epitopes were identified from previous experimental validation, and via bioinformatic predictions using NetMHC (www.iedb.org net). Predictions were obtained from HLA-I typing of the subject as well as from the autologous viral consensus sequence, generated via next generation sequencing. As predicted epitopes could represent false positives, strict selection criteria were applied as previously described13. Selected autologous HLA-1 restricted peptides were synthesised and epitope-specific interferon gamma (IFN-γ) production assessed via enzyme-linked immunospot (ELISpot) assays as previously described11. A positive response towards an autologous peptide was defined as >20 spot-forming units (SFU)/million peripheral blood mononuclear cells (PBMC).
Results and Discussion
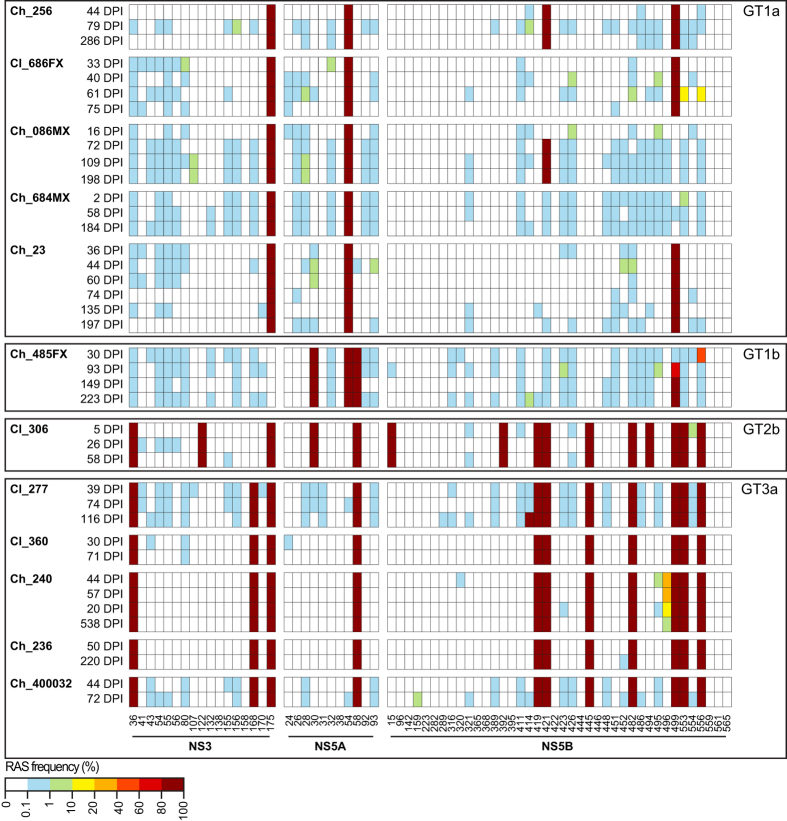
The longitudinal prevalence of RASs was analysed in 12 participants, each with 2–6 time-points (Fig. 1). The median number of days post-infection (DPI) at the earliest time-points was estimated at 34 (range 2–50). Four of these participants demonstrated spontaneous clearance (Cl), and eight developed chronic (Ch) infection (Fig. 1). HCV genotype distribution was GT1a (n = 5), GT1b (n = 1), GT2b (n = 1) and GT3a (n = 5).
Figure 1. Longitudinal analysis of HCV resistance-associated substitutions (RAS) in early HCV infections.
RASs are shown in longitudinally sampled infections across HCV NS3, NS5A and NS5B. Rows represent individual samples and columns indicate amino acid positions, grouped by different coding regions and numbered at the bottom. Sample identification numbers are shown together with the estimated days post infection (DPI) to the left of each row. Samples are grouped by HCV genotype (GT). The frequency of RASs is indicated by the color according to the scale bar. RASs that are fixed within an HCV genotype34 (i.e. consensus RASs) are presented with 100% frequency across time-points.
Longitudinal analysis of HCV RASs
Sixty-eight previously known RAS sites within NS3, NS5A and NS5B were screened in a total of 40 samples sequenced by next generation sequencing. Across the entire dataset, RASs were detected in 30/68 genome positions at frequencies >1% spanning NS3, NS5A and NS5B-coding regions (Fig. 1). While low-frequency variants (<1%) were detected within 51/68 (75%) of the examined sites at the earliest time-points, longitudinal analysis revealed that RASs within the majority of these sites (49/51, 96%) did not reach frequencies >10% over the course of the infection (Fig. 1).
Within NS3 and NS5A, all RASs detected at the consensus-level in the earliest time-points were conserved over time, and no new RASs emerged at high frequency or were lost over the course of the infection (Fig. 1). By contrast, within NS5B non-synonymous mutations resulting in emergence of consensus-level RASs M414T, A421V and V499A were observed in three participants infected with HCV GT3a, GT1a and GT1b, respectively (Fig. 1 and Table 1). Additionally, loss of consensus NS5B RASs S556G and P496S was observed in two participants infected with HCV GT1b and GT3a, respectively (Fig. 1 and Table 1). Interestingly, two NS5B RASs, A553V and S556G, were transiently detected at 61 DPI with frequencies 17–18% in a participant infected with GT1a (Cl_686FX), but these RASs were both lost within two weeks (Fig. 1). In summary, RASs affecting HCV DAAs targeting NS3 and NS5A are stable over the course of primary infection in this study, but emergence and loss of RASs were detected within NS5B of multiple genotypes.
Table 1. Summary of the RAS analysis in this study with corresponding ELISpot responses.
| Participant | HCV Genotype | Substitution detected | RAS outcomea | Positionb | Reversion | CD8 T cell epitope analysis | |||
|---|---|---|---|---|---|---|---|---|---|
| Epitope sequence | HLA | Result (SFU/106 PBMC)c | Time-point (DPI)d | ||||||
| Ch_086MX | 1a | A421V | Gain | 2841 | No | 2841ARMVMMTHF2849 | HLA-B27:05 | 25 | 72 |
| Ch_485FX | 1b | V499A | Gain | 2918 | No | 2913GVPPLRVWR2921 | HLA-A01:01 | Negative | 79 |
| S556G | Loss | 2975 | Yes | — | — | — | — | ||
| Cl_277 | 3a | M414T | Gain | 2844 | No | 2838WLGNIIMYA2846 | HLA-A02:01 | 45 | 116 |
| Ch_240 | 3a | P496S | Loss | 2926 | Yes | — | — | — | — |
| Cl_686FX | 1a | A553V | Gain/Loss | 2973 | Yes | 2967LSGWFTAGY2975 | HLA-A01:01 | Negative | 117 |
| S556G | Gain/Loss | 2976 | Yes | — | — | — | — | ||
aRAS; resistance-associated substitution. Gain or loss or RAS is determined longitudinally with reference to the earliest sample.
bAmino acid position with reference to strain H77 (AF009606) for GT1a, Con1 (AJ238799) for GT1b, and NZL1 (D17763) for GT3a.
cSpot-forming units per million peripheral blood mononuclear cells.
dEstimated days post infection (DPI) at which analysis was performed.
Factors associated with the emergence of HCV RASs
In order to examine selective pressures that could be associated with the gain of NS5B RASs, we assessed whether these substitutions fell within known, or predicted, CD8+ T cell epitopes. For the three subjects with evidence of RAS emergence and reaching fixation over time, autologous CD8+ T cell responses were tested via IFN-γ ELISpot assays. In parallel, selection by replication fitness was defined as the emergence of a mutation that was identical to the consensus genome for that sample’s corresponding genotype, which was termed to be a reversion event.
In all three cases where fixation events resulted in the gain of RASs, M414T (Cl_277), A421V (Ch_86MX) and V499A (Ch_485FX), the mutations were not reversion events, suggesting that viral fitness was not driving selection of these residues (Table 1). In relation to T cell driven pressures, M414T was identified to lie within the published HLA-A2 restricted epitope 2838WLGNIIMYA2846 (RAS site underlined)17. When this peptide was examined by ELISpot, a moderate IFN-γ response was detected for participant Cl_277 at 74 DPI, with 20 SFU/million PBMC. At 116 DPI, the time-point at which the RAS was detected, a slightly stronger T cell response was detected, with 45 SFU/million PBMC. These findings support the hypothesis that CD8+ T cell responses exerted selective pressure on this RAS site (Table 1).
The second RAS, A421V, reached fixation in participant Ch_086MX at 72 DPI, and is recognised to lie within an HLA-B27-restricted CD8+ T cell epitope 2841ARMILMTHF284918,19. In the subject analysed here (Ch_086MX), the autologous epitope present in the transmitted/founder virus that established the infection differed from this known epitope at two positions, 2844 and 2845 (2841ARMVMMTHF2849). Experimental validation of this epitope was positive at 72 DPI, with a low CD8+ T cell frequency response of 25 SFU/million PBMCs (Table 1). When the putative escape variant epitope, 2841VRMVMMTHF2849 was tested using ELISpot, no response could be detected.
Despite the lack of longitudinal evidence of an ongoing T cell response against this epitope, this immunodominant epitope is reported to be targeted by almost all subjects with the HLA-B27 genotype that are infected with HCV, and the A421V substitution has been shown to be HLA-driven18,19. These data support the hypothesis that the A421V substitution in subject Ch_086MX could be driven by T cell responses.
To further validate the hypothesis that the evolution of RASs within a single infection could be driven by the T cell response, two participants, infected with the same transmitted founder virus and with longitudinal information on viral mutations were examined. Participants Ch_086MX and Ch_684MX were siblings who concurrently became infected. The consensus viral sequences for these subjects shared 99.9% nucleotide similarity across the transmitted founder virus genome (Fig. 1). At the earliest time-points analysed (16 DPI and 2 DPI for Ch_086MX and Ch_684MX, respectively), both subjects carried the variant sequence 2841ARMVMMTHF2849 (Fig. 1), which is the known HLA-B27-restricted CD8+ T cell epitope. Notably, only the virus in the the subject carrying the HLA-B27:05 allele developed the A421V substitution, while the sibling carried the HLA-B27:10, for which the predicted binding score for the same epitope is significantly lower (157 μM and 2,619 μM, respectively). Despite this, the 2841ARMVMMTHF2849 peptide was tested via ELISpot in Ch_684MX and found to be weakly positive (30 SFU/million PBMC). A number of factors could explain the lack of emergence of A421V in subject Ch_684MX, including differences in peptide affinity or T cell exhaustion20. Investigation of these possibilities was beyond the scope of this study.
The RAS V499A was detected in participant Ch_485FX, a site which falls within the known HLA-B57-restricted epitope, 2912LGVPPLRAWR292121. However, the autologous circulating virus carried a substitution at position 2919, and the predicted epitope was the nonamer 2913GVPPLRVWR2921. This subject was not HLA-B57 positive, and the only HLA allele of Ch_485FX predicted to bind to this peptide was HLA-A01:01, which had an IC50 score of 22 mM. The ELISpot response against the predicted transmitted founder derived, HLA-A01:01-restricted autologous epitope, 2913GVPPLRVWR2921 was negative (Table 1). Therefore, the role of T cell responses in driving the emergence of this RAS could not be ascertained.
Overall, these results suggest that RASs M414T and A421V emerged in the early phase of HCV infection as a result of T cell responses in participants carrying relevant HLA-I alleles.
Factors associated with the loss of HCV RASs
Unlike emerging RASs, all mutations which resulted in the loss of HCV RASs in this study were reversion events (Table 1). The loss of RAS S556G was observed as a result of a fixation event around 93 DPI in the same GT1b infected participant in whom another RAS, V499A, emerged (Ch_485FX, Fig. 1). Peptides encompassing this RAS were screened for CD8 T cell responses in previous studies, and found to be negative22,23. The loss of the second RAS, P496S, was observed in a GT3a infected participant (Ch_240) as a gradual decrease in frequency from 33% to 1.5% between 44 DPI and 538 DPI (Fig. 1). The site of this RAS, P496, was found to be conserved across all HCV GTs. This suggests that the loss of the RAS could be driven by fitness costs of the substitution. In the participant where two RASs emerged at low frequency, Cl_686FX, both RASs also reverted to the consensus sequence (Table 1). One of these RASs, S556G, overlapped with that detected in another participant, Ch_485FX (Fig. 1). The second RAS, A553V, fell within a predicted HLA-A01:01 restricted epitope, however, no response could be detected against this epitope in the ELISpot assays (Table 1). Overall, these results indicate that T cell responses do not appear to be driving the loss of HCV RASs detected in early acute HCV infection. Instead, reversion of residues carried by the virus towards consensus sequences indicates that the observed loss of RASs could be driven by reduced viral fitness.
The occurrence of RASs in the transmitted founder sequences within these subjects could be attributed to immune responses within the original host (i.e the transmission source) that were not investigated in this study, or to bottleneck selection in the new host (i.e the recipient) during viral transmission. They could also be due to transmission of drug-resistance variants from a treatment-experienced source. However, the participants in this study were recruited and sampled prior to the availability of HCV NNIs, and while the participants were not part of DAA clinical trials, these were being conducted concurrently in the Australian community, and there remains the possibility that the source was treatment-experienced.
Relevance of detected evolving RASs
All six RASs with longitudinal evolution in this study are relevant to one class of DAAs; non-nucleoside inhibitors (NNI) of the HCV NS5B. The RAS V499A, causes a minor reduction to some thumb-binding HCV NNIs24 but both none of those in development. A421V is associated with resistance to thumb-binding NS5B inhibitors, such as beclabuvir25; A421V, was identified upon treatment of GT1 patients with beclabuvir25, and with the combination daclatasvir/asunaprevir/beclabuvir, but had no significant association with virologic outcome26. M414T confers resistance to palm-binders, including the recently approved NNI dasabuvir27. Viral breakthrough and relapse after treatment in an IFN-free combination including dasabuvir have been associated with substitutions with M414I/T28,29,30. It should be noted, however, that the M414T substitution is observed in a GT3a infected participant and dasabuvir is known to have significantly reduced activity against GT3a in general27.
The fitness of these RASs in cell culture models is variable, and depends on the examined genotypes. A421V does not seem to impact the replication of GT1a31 or GT1b32 replicons, whilst M414T affects the fitness of GT1b but not GT1a in replicon studies27. V499A was shown to impact the replication of GT1b in vitro24. This RAS, however, is not uncommon in GT1b infections, and therefore its impact on viral fitness could not be ascertained.
Of the RASs that were observed to be lost over the analysed course of infection, A553T (in GT1a), A553V (in GT1b) and S556G (in both GT1a and GT1b) are key substitutions associated with reduced response to dasabuvir in vitro and in vivo27. These substitutions have been associated with reduced fitness of GT1 HCV in vitro27, potentially explaining their loss over time.
Summary
Selection of HCV RASs via immune factors has been proposed in a number of studies. Emergence and loss of an NS3 RAS, R155K, has been previously reported in a single patient co-infected with both HIV-1 and GT1a HCV when sampled over time33. This site was later confirmed to fall within a CD8+ T cell epitope and the RAS was proposed to be associated with loss of recognition by these cells9. Resistance to other HCV DAAs has been proposed to arise through immune-driven selection in some putative HLA-restricted epitopes8, but these responses were not verified experimentally. Our study describes the evolution of six RASs during early acute HCV infections with three different genotypes, and proposes that immune selection pressures contribute to the emergence of HCV RASs in early HCV infections, particularly within NS5B, even in the absence of treatment. This study also supports previous reports highlighting the incurred fitness cost of RASs, and suggests that this cost contributes to their loss. These results suggest that populations with particular HLA dominant types should be closely monitored for changes in prevalence of immune-related RASs.
Additional Information
How to cite this article: Eltahla, A. A. et al. Dynamic evolution of hepatitis C virus resistance-associated substitutions in the absence of antiviral treatment. Sci. Rep. 7, 41719; doi: 10.1038/srep41719 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Acknowledgments
Research support for the HITS-p cohort includes National Health and Medical Research Council of Australia (NHMRC) - Project No. 222887, Partnership No. 1016351, Program Nos. 510488 and 1053206; The HITS-c cohort is supported by the UNSW Hepatitis C Vaccine Initiative and NHMRC Project Grant No. 630483. The Kirby Institute is funded by the Australian Government Department of Health and Ageing. AAE, JG, ARL and RAB are supported by NHMRC research fellowships. The views expressed in this publication do not necessarily represent the position of the Australian Government.
Footnotes
Jason Grebely is a consultant/advisor and has received research grants from AbbVie, Bristol–Myers Squibb, Gilead Sciences and Merck/MSD. Andrew Lloyd has received grants for investigator-initiated research from Gilead and Merck/MSD. The remaining authors have no conflicts of interest to disclose.
Author Contributions Authors A.A.E., M.R.P., C.R. and R.A.B. completed the laboratory work. A.E., P.L. and F.L. performed the computational data analysis. J.G. and T.A. contributed to the study design. Authors A.R.L. and L.M. contributed samples and reagents. The first manuscript was drafted by A.A.E., F.L., A.R.L. and R.A.B. All authors contributed to and have approved the final manuscript.
References
- Dore G. J. & Feld J. J. Hepatitis C virus therapeutic development: in pursuit of “perfectovir”. Clin. Infect. Dis. 60, 1829–1836 (2015). [DOI] [PubMed] [Google Scholar]
- Pawlotsky J.-M. & Hepatitis C. Virus Resistance to Direct-Acting Antiviral Drugs in Interferon-Free Regimens. Gastroenterology (2016). [DOI] [PubMed] [Google Scholar]
- Eltahla A. et al. Analysis of resistance‐associated substitutions in acute hepatitis C virus infection by deep sequencing across six genotypes and three continents. J. Viral Hepat. (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applegate T. L. et al. Naturally occurring dominant drug resistance mutations occur infrequently in the setting of recently acquired hepatitis C. Antivir. Ther. 20, 199–208, doi: 10.3851/IMP2821 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels D. J. et al. Hepatitis C virus variants with decreased sensitivity to direct-acting antivirals (DAAs) were rarely observed in DAA-naive patients prior to treatment. J. Virol. 87, 1544–1553, doi: 10.1128/JVI.02294-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuntzen T. et al. Naturally occurring dominant resistance mutations to hepatitis C virus protease and polymerase inhibitors in treatment-naive patients. Hepatology 48, 1769–1778, doi: 10.1002/hep.22549 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powdrill M. H. et al. Contribution of a mutational bias in hepatitis C virus replication to the genetic barrier in the development of drug resistance. Proc. Natl. Acad. Sci. USA. 108, 20509–20513, doi: 10.1073/pnas.1105797108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudieri S. et al. Hepatitis C virus drug resistance and immune-driven adaptations: relevance to new antiviral therapy. Hepatology 49, 1069–1082, doi: 10.1002/hep.22773 (2009). [DOI] [PubMed] [Google Scholar]
- Salloum S., Kluge S. F., Kim A. Y., Roggendorf M. & Timm J. The resistance mutation R155K in the NS3/4A protease of hepatitis C virus also leads the virus to escape from HLA-A* 68-restricted CD8 T cells. Antiviral Res. 87, 272–275 (2010). [DOI] [PubMed] [Google Scholar]
- Peiffer K. H. et al. Interferon lambda 4 genotypes and resistance‐associated variants in patients infected with hepatitis C virus genotypes 1 and 3. Hepatology 63, 63–73 (2016). [DOI] [PubMed] [Google Scholar]
- Bull R. A. et al. Transmitted/Founder Viruses Rapidly Escape from CD8+ T Cell Responses in Acute Hepatitis C Virus Infection. J. Virol. 89, 5478–5490, doi: 10.1128/JVI.03717-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teutsch S. et al. Incidence of primary hepatitis C infection and risk factors for transmission in an Australian prisoner cohort. BMC Public Health 10, 633, doi: 10.1186/1471-2458-10-633 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull R. A. et al. Sequential bottlenecks drive viral evolution in early acute hepatitis C virus infection. PLoS Pathog. 7, e1002243, doi: 10.1371/journal.ppat.1002243 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull R. A. et al. A method for near full-length amplification and sequencing for six hepatitis C virus genotypes. BMC Genomics 17, 247, doi: 10.1186/s12864-016-2575-8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B. & Salzberg S. L. Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359, doi: 10.1038/nmeth.1923 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M. et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649, doi: 10.1093/bioinformatics/bts199 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra S. et al. Peptide-pulsed dendritic cells induce the hepatitis C viral epitope-specific responses of naive human T cells. Vaccine 32, 3285–3292, doi: 10.1016/j.vaccine.2014.03.083 (2014). [DOI] [PubMed] [Google Scholar]
- Nitschke K. et al. HLA-B*27 subtype specificity determines targeting and viral evolution of a hepatitis C virus-specific CD8+ T cell epitope. J. Hepatol. 60, 22–29, doi: 10.1016/j.jhep.2013.08.009 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timm J. et al. Human leukocyte antigen–associated sequence polymorphisms in hepatitis C virus reveal reproducible immune responses and constraints on viral evolution. Hepatology 46, 339–349 (2007). [DOI] [PubMed] [Google Scholar]
- Bengsch B. et al. Coexpression of PD-1, 2B4, CD160 and KLRG1 on exhausted HCV-specific CD8+ T cells is linked to antigen recognition and T cell differentiation. PLoS Pathog. 6, e1000947 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer G. M. et al. High resolution analysis of cellular immune responses in resolved and persistent hepatitis C virus infection. Gastroenterology 127, 924–936 (2004). [DOI] [PubMed] [Google Scholar]
- Smyk-Pearson S. et al. Differential antigenic hierarchy associated with spontaneous recovery from hepatitis C virus infection: implications for vaccine design. J. Infect. Dis. 194, 454–463 (2006). [DOI] [PubMed] [Google Scholar]
- Tester I. et al. Immune evasion versus recovery after acute hepatitis C virus infection from a shared source. The Journal of experimental medicine 201, 1725–1731 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukolj G. et al. Binding site characterization and resistance to a class of non-nucleoside inhibitors of the hepatitis C virus NS5B polymerase. J. Biol. Chem. 280, 39260–39267, doi: 10.1074/jbc.M506407200 (2005). [DOI] [PubMed] [Google Scholar]
- McPhee F. et al. Characterization of Viral Escape in Hcv Genotype 1-Infected Patients Treated with Bms-791325 and Pegylated Interferon-Alfa and Ribavirin. J. Hepatol. 56, S473–S473 (2012). [Google Scholar]
- Muir A. J. et al. Daclatasvir in combination with asunaprevir and beclabuvir for hepatitis C virus genotype 1 infection with compensated cirrhosis. JAMA 313, 1736–1744, doi: 10.1001/jama.2015.3868 (2015). [DOI] [PubMed] [Google Scholar]
- Kati W. et al. In vitro activity and resistance profile of dasabuvir, a nonnucleoside hepatitis C virus polymerase inhibitor. Antimicrob. Agents Chemother. 59, 1505–1511, doi: 10.1128/AAC.04619-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poordad F. et al. ABT-450/r–ombitasvir and dasabuvir with ribavirin for hepatitis C with cirrhosis. N. Engl. J. Med. 370, 1973–1982 (2014). [DOI] [PubMed] [Google Scholar]
- Zeuzem S. et al. Retreatment of HCV with ABT-450/r–ombitasvir and dasabuvir with ribavirin. N. Engl. J. Med. 370, 1604–1614 (2014). [DOI] [PubMed] [Google Scholar]
- Poordad F. et al. 12-week interferon-free regimen of ABT-450/R+ABT-333+ ribavirin achieved SVR 12 in more than 90% of treatment-naive HCV genotype-1-infected subjects and 47% of previous non-responders. J. Hepatol. 56, S549–S550 (2012). [Google Scholar]
- Lemm J. A. et al. Preclinical characterization of BMS-791325, an allosteric inhibitor of hepatitis C Virus NS5B polymerase. Antimicrob. Agents Chemother. 58, 3485–3495, doi: 10.1128/AAC.02495-13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe A. Y. et al. Molecular mechanism of a thumb domain hepatitis C virus nonnucleoside RNA-dependent RNA polymerase inhibitor. Antimicrob. Agents Chemother. 50, 4103–4113, doi: 10.1128/AAC.00365-06 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A. Y. et al. Temporal dynamics of a predominant protease inhibitor-resistance mutation in a treatment-naive, hepatitis C virus-infected individual. J. Infect. Dis. 199, 737–741, doi: 10.1086/596657 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCloskey R. M. et al. Global origin and transmission of hepatitis C virus NS3 Q80 K polymorphism. J. Infect. Dis. jiu613 (2014). [DOI] [PubMed] [Google Scholar]