Abstract
Isoleucyl-tRNA synthetase (IleRS) is an aminoacyl-tRNA synthetase whose essential function is to aminoacylate tRNAIle with isoleucine. Like some other aminoacyl-tRNA synthetases, IleRS can mischarge tRNAIle and correct this misacylation through a separate post-transfer editing function. To explore the biological significance of this editing function, we created a ileS(T233P) mutant of Bacillus subtilis that allows tRNAIle mischarging while retaining wild-type Ile-tRNAIle synthesis activity. As seen in other species defective for aminoacylation quality control, the growth rate of the ileS(T233P) strain was not significantly different from wild-type. When the ileS(T233P) strain was assessed for its ability to promote distinct phenotypes in response to starvation, the ileS(T233P) strain was observed to exhibit a significant defect in formation of environmentally resistant spores. The sporulation defect ranged from 3-fold to 30-fold and was due to a delay in activation of early sporulation genes. The loss of aminoacylation quality control in the ileS(T233P) strain resulted in the inability to compete with a wild-type strain under selective conditions that required sporulation. These data show that the quality control function of IleRS is required in B. subtilis for efficient sporulation and suggests that editing by aminoacyl-tRNA synthetases may be important for survival under starvation/nutrient limitation conditions.
An essential step for the accuracy of mRNA translation is the charging of tRNAs with their cognate amino acid. Aminoacyl-tRNA synthetases (aaRS) are the enzymes that catalyze this reaction, and are a family of ancient proteins that have been conserved throughout evolution, due to their essential cellular function1. AaRSs aminoacylate tRNAs through a two-step mechanism: 1) formation of an aminoacyl-adenylate (e.g. Ile-AMP by IleRS), and 2) aminoacylation of tRNA (e.g. Ile-tRNAIle). Due to the need for accuracy in protein synthesis, some aaRSs contain a quality control (QC) function along with their aminoacylation function2. QC functions can hydrolyze either the aminoacyl-adenylate (pre-transfer editing) or the aminoacyl bond of charged tRNA (post-transfer editing). While aaRS editing functions are highly conserved in bacteria, archaea, and eukaryotes, QC is not essential for cellular growth under most laboratory conditions, and few studies have identified cellular functions that rely on aaRS editing3,4,5,6,7.
Isoleucine-tRNA synthetase (IleRS) possesses QC functions that discriminate between isoleucine, the non-cognate amino acid valine, and the non-proteinogenic amino acids, norvaline, a byproduct of branched-chain amino acid synthesis, and homocysteine (Hcy), a by-product from degradation of S-adenosylhomocysteine by LuxS in bacteria8. The aminoacylation site of IleRS is only able to discriminate Ile from Val with an accuracy of ~1/2009. IleRS improves this accuracy with two QC activities, pre- and post-transfer editing. Pre-transfer editing hydrolyzes Val-AMP in the synthetic site where aminoacylation also occurs10,11, and pre-transfer editing similarly hydrolyzes Hcy-AMP12. Post-transfer editing in contrast deacylates Val-tRNAIle in a domain, the connective peptide 1 (CP1) domain, which is distinct from the site at which aminoacylation occurs13,14,15,16. Several studies have identified substitutions in CP1 that disrupt post-transfer editing, but retain close to wild-type levels of aminoacylation activity13,14,17.
In vivo studies of IleRS mutants have begun to reveal potential cellular functions of QC by aaRSs. One primary function of quality control is to prevent misincorporation in proteins of the non-cognate and non-proteinogenic amino acids, such as valine and norvaline, respectively. Loss of IleRS QC in E. coli results in decreased growth when the cells are grown in high concentrations of valine or norvaline18. The E. coli IleRS QC mutant strain is also more sensitive to stressful conditions, including high temperatures and antibiotics, although the mechanism by which these mutant cells increase their sensitivity to stress is unknown19. Loss of quality control by IleRS may also be beneficial under certain conditions. In E. coli and Acinetobacter baylyi, loss of IleRS QC functions resulted in an improved growth rate when concentrations of isoleucine were limiting, but moderate concentrations of valine or norvaline were present18,20. Streptococcus pneumoniae IleRS naturally lacks a post-transfer editing function, and mischarged tRNAIle serves as a substrate for peptidoglycan biosynthesis21. Potentially beneficial is the higher level of mutations observed in colonies of E. coli IleRS QC-defective cells that are allowed to age; the full mechanism by which DNA mutations arise in response to loss of quality control is also unknown22. Overall, while a number of phenotypes have been described for QC mutants, the mechanism underlying the observed changes and their broader importance remain unknown.
To address the question of what conditions require quality control for fitness, we choose to address how loss of QC by IleRS affects the function of the model Gram-positive organism, Bacillus subtilis. Here, we show that QC by IleRS is required for B. subtilis, to form environmentally-resistant endospores. These endospores are formed in response to starvation through a developmental process. This process is initiated by a master regulator, Spo0A, and gene expression activated by this transcription factor is severely delayed in cells that lacking QC by IleRS. Furthermore, the QC-defective IleRS strain was outcompeted by wild-type strains under conditions involving periodic sporulation and outgrowth, supporting a general role for quality control by aaRS under starvation or nutrient-limited conditions.
Results
Cell expressing IleRS(T233P) are defective for quality control of mischarged tRNAIle
IleRS was chosen for this study as it has a well characterized post-transfer editing activity, and substitutions that disrupt this editing without affecting aminoacylation have been made10,11,23. While several studies have looked at the role of QC by IleRS in E. coli, relatively few studies have examined the importance of QC in other organisms. As we had previously proposed that QC maybe more critical under conditions of stress that limit cellular growth3, we chose to analyze the role of QC by IleRS in B. subtilis, which undergoes several distinct phenotypic or developmental shifts in response to starvation or slow growth, including the formation of environmentally-resistant endospores24.
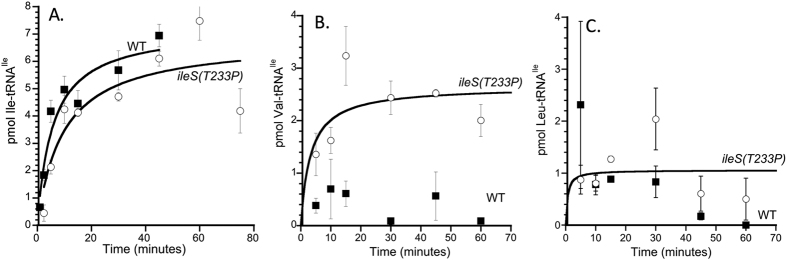
To determine the function of QC by IleRS in B. subtilis, a single point mutation was introduced into the ileS gene on the chromosome of B. subtilis strain JH64225. The point mutation introduced causes a substitution of Thr at codon 233 to Pro. This T233P substitution was chosen because a similar substitution in IleRS of Escherichia coli was shown to disrupt QC by this enzyme, and while other substitutions in E. coli IleRS disrupt QC, the T233P substitution has virtually no effect on aminoacylation activity14,23. To test whether the cells with the ileS(T233P) allele lacked tRNAIle QC activity, cell lysates of the wild-type ileS and mutant ileS(T233P) were tested for their ability to charge tRNAIle with either the cognate Ile or the non-cognate amino acids, Val and Leu. Lysates from the ileS(T233P) strain showed Ile-tRNAIle production at a similar rate as lysates from wild-type cells (Fig. 1A), indicating that the ileS(T233P) mutation did not affect the overall level of Ile-tRNAIle formation in cells. Similarly, the ileS(T233P) lysate showed only low levels of Leu-tRNAIle production, similar to that seen with lysates from wild-type cells, and consistent with the Leu not being an aminoacylation substrate for either wild-type IleRS or IleRS(T233P) (Fig. 1C). Production of Val-tRNAIle however was significantly different between wild-type and ileS(T233P) cell lysates, with the ileS(T233P) lysate producing significantly more Val-tRNAIle than the wild-type lysate (Fig. 1B). Increased Val-tRNAIle synthesis for IleRS(T233P) may either indicate improved Val recognition and/or reduced hydrolysis of either misactivated Val-AMP or mischarged Val-tRNAIle compared with wild-type. Importantly, whatever the precise mechanism by which the T233P substitution increased Val-tRNAIle synthesis, this did not adversely affect the ability of cells to aminoacylate tRNAIle with its cognate amino acid and did disrupt QC by IleRS.
Figure 1. The ileS(T233P) strain mischarges tRNAIle with valine.
Cell extracts of BAL4574 (WT; filled squares) and BAL4571(ileS(T233P); open circles) were assayed for their ability to aminoacylate tRNAIle with 14C versions of isoleucine (A), valine (B), or leucine (C). Plotted is the average pmoles of the radiolabeled tRNA from three reactions versus the time after the addition of tRNAIle and the radiolabeled amino acid. Error bars are standard error of the mean. Lines are best fits of the data to a Michaelis-Menton equation, with the exception of WT in panels B and C, as the basal level of aminoacylation observed could not be fit.
Cells expressing IleRS(T233P) are defective in sporulation
Having created a mutant strain of B. subtilis that expresses an QC-defective version of IleRS, we sought to determine whether ileS(T233P) cells had any phenotypic differences from wild-type. Any phenotypes altered in these mutant cells would suggest pathways that are sensitive to QC by IleRS. The exponential growth rate (∆Ln OD600/∆time(min) ± standard deviation) in rich medium, Difco Sporulation Medium (DSM), of wild-type cells and cells expressing IleRS(T233P) were not significantly different (0.020 ± 0.002 and 0.021 ± 0.002, respectively). Similarly, in minimal medium, the exponential growth rate of these two strains was also not significantly different (0.011 ± 0.002 for wild-type cells and 0.010 ± 0.003 for the ileS(T233P) cells). These data are consistent with other observations of QC-defective aaRS bacterial strains are not affected for growth under standard laboratory conditions3.
As the editing function of IleRS is conserved through evolution3, and cells in natural environments are characterized by slow growth, the wild-type and ileS(T233P) mutant B. subtilis cells were assessed for their ability to form biofilms and spores, two major phenotypic states entered by this bacterium under slow-growth conditions26. While wild strains of B. subtilis form more robust biofilms, laboratory strain JH642 used in this study is able to form biofilms that adhere to the wells of a microtiter plate and to form flat, unstructured pellicle biofilms at air-liquid interfaces27,28,29,30. Both the ileS(T233P) strain and its isogenic wild-type strain formed morphologically indistinguishable pellicles that were observed as films of cells at the top of media in test tubes. In microtiter plates, there was at best a small difference in how the two strains adhered to the plates, and the A570 ± standard deviation was 1.4 ± 0.3 for the wild-type strain and 0.87 ± 0.02 for the ileS(T233P) strain. These data support a conclusion that the QC by IleRS is not essential for biofilm formation, although we cannot rule out a more prominent role for editing in wild strains of B. subtilis.
To assess the ability of the ileS(T233P) mutant cells to sporulate, these cells and the isogenic wild-type control cells were grown in DSM and incubated until sixteen hours after entry into stationary phase. At this time, the frequency of sporulation was determined as the number of heat-resistant colony-forming units (CFU)/ml relative to the total viable CFU/ml (Table 1). While the fold difference in the sporulation frequency of wild-type and the ileS(T233P) cells was variable, ranging from 1.5- to 15-fold, the ileS(T233P) cells showed a statistically significant (p = 0.015) defect in sporulation. The variability in the sporulation phenotype is not unexpected given the stochasticity in frequency of mischarging of a tRNA.
Table 1. Sporulation Frequency of Wild-Type and IleRS(T233P) Cells.
| Trial | Wild-Type |
IleRS (T233P) |
||||
|---|---|---|---|---|---|---|
| Viablea | Sporesb | Frequencyc | Viablea | Sporesb | Frequencyc | |
| Difco Sporulation Media | ||||||
| 1 | 5.9 × 108 | 3.5 × 108 | 60% | 3.6 × 108 | 1.3 × 108 | 35% |
| 2 | 6.8 × 108 | 3.8 × 108 | 56% | 3.5 × 108 | 1.2 × 108 | 34% |
| 3 | 4.9 × 108 | 3.8 × 108 | 78% | 3.2 × 108 | 1.6 × 107 | 5% |
| 4 | 4.6 × 108 | 3.6 × 108 | 77% | 6.0 × 108 | 7.1 × 107 | 12% |
| 5 | 3.4 × 108 | 3.5 × 108 | 100% | 3.5 × 108 | 2.5 × 107 | 71% |
| Glucose Minimal Media | ||||||
| 1 | 2.7 × 108 | 1.8 × 108 | 67% | 1.6 × 108 | 3.4 × 106 | 2.1% |
| 2 | 2.7 × 108 | 2.3 × 108 | 85% | 1.8 × 108 | 1.3 × 107 | 7.2% |
| 3 | 9.1 × 107 | 9.2 × 107 | 100% | 1.1 × 108 | 3.1 × 107 | 27% |
aReported are the CFU/ml of the cultures after 16 hours of incubation in stationary phase.
bAfter testing for viable cells, the cultures were heat treated at 80 °C for 20 minutes and then plated for CFU/ml counts.
cThe frequency of sporulation was calculated as [(spores CFU/ml)/(viable CFU/ml)*100].
To determine whether this sporulation defect was specific to growth in DSM, cells were grown in minimal media containing 0.1% glucose. Cells in this media exhaust glucose and transition to stationary phase at an OD600 of ~1.0. After 16 hours in stationary phase, the frequency of sporulation was assayed (Table 1). Similar to DSM conditions, the ileS(T233P) mutant exhibited a statistically significant (p > 0.001) defect in sporulation, except that the ileS(T233P) cells showed a larger defect in sporulation, ranging from a 4- to 30-fold defect.
The ileS(T233P) mutant is outcompeted by wild-type cells during cycles of sporulation
To assess the importance of the QC function of IleRS for B. subtilis, we asked how well the ileS(T233P) mutant was able to compete with the wild-type strain. We introduced into the wild-type and ileS(T233P) strains either a thrC::erm allele that encodes erythromycin resistance (ErmR) or a thrC::cat allele that encodes chloramphenicol resistance (CmR). Cultures were inoculated with an equal mixture of both wild-type and ileS(T233P) cells of different antibiotic-resistance markers. In the repeats of the cultures, the antibiotic-resistance markers were swapped between the two strains to ensure that any differences observed were due to the presence of the ileS(T233P) mutation. The cultures were grown in DSM and passaged through a cycle of exponential growth, sporulation, and heat-kill of non-sporulated cells. A 1/10th fraction of the spores that remained after the heat-kill step were used to inoculate the next culture that was similarly passaged through a cycle of exponential growth, sporulation, and heat-kill of non-sporulated cells. This was repeated for a total of four cycles. At the end of each cycle, the percent of the spores that were from wild-type and ileS(T233P) cells was measured through CmR and ErmR CFU/ml, as appropriate. For each independent culture started, there was a similar number of cells of each strain at the end of the first passage, consistent with no significant differences in the growth rate of these two strains. Subsequently, the number of ileS(T233P) cells dropped relative to the number of wild-type cells by the second or third passage, with the level of ileS(T233P) mutant after the fourth passage being 105-fold lower than wild-type (Fig. 2). Interestingly, under these competition experiments, the ileS(T233P) mutant consistently showed a greater that 10-fold defect in sporulation (Fig. 2), unlike what was observed for this strain when grown individually (Table 1). These data predict that the ileS(T233P) mutant would go to extinction after five cycles and strongly suggests that sporulation is one of the evolutionary pressures maintaining the QC function of IleRS in B. subtilis.
Figure 2. The ileS(T233P) strain is outcompeted by wild-type cells.
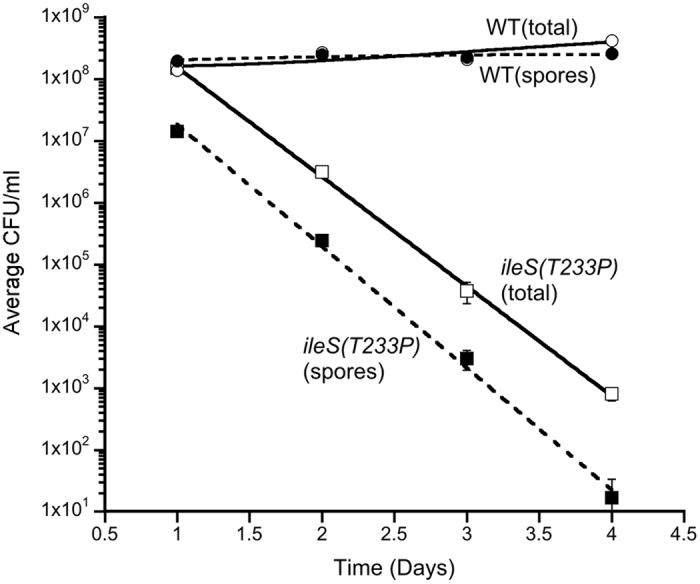
Derivatives of BAL4574 (WT; circles) and BAL4571 (ileS(T233P); squares), which carried either erythromycin-resistant or chloramphenicol-resistant genes, were mixed in equal numbers in DSM and incubated until 16 hours post the onset of stationary phase. At that time, the total CFU/ml (open symbols) was assessed for both cell types. The cultures were then heated to kill vegetative cells, and the CFU/ml of spores (filled symbols) was assessed for both cell types. A 1/10th volume of the spores was then transferred to a new DSM culture, and the process was repeated for four days. Plotted is the average CFU/ml of four separate cultures versus the day of the competition. Error bars are standard error of the mean.
The QC-defective IleRS mutant is defective in expressing genes activated by the master regulator of sporulation, Spo0A
A major question from this work is why sporulation is sensitive to mistakes made by IleRS. The process of sporulation can be divided into distinct genetic and morphological stages31. The sporulation defect of the ileS(T233P) could stem from mistranslation of Ile codons, which could result in a non-functional protein that is required for sporulation. If a particular protein is negatively affected in the ileS(T233P) strain, we would predict the ileS(T233P) cells to be specifically delayed at one stage of sporulation, resulting in a strong reduction in expression of those stage-specific genes. However, mistranslation is predicted to happen randomly at Ile codons, with each cell having a different mixture of mistranslated proteins, and as a result each cell in a population could be delayed at a different stage in sporulation, and we would not observe a strong effect on any particular stage-specific gene expression at the population level.
To determine whether there is a strong decrease in stage-specific sporulation gene expression in the ileS(T233P) mutant, we monitored gene expression of Spo0A-controlled genes. Spo0A is the first transcription factor to become active, through phosphorylation by a phosphorelay, in the sporulation regulatory cascade32, and if Spo0A-dependent genes have strongly reduced expression, then all subsequent stage-specific sporulation genes will also have reduced expression. One of the operons activated by Spo0A is spoIIE, which encodes the phosphatase required for activation of the forespore specific sigma factor, Sigma-F33. Thus, we introduced into our isogenic wild-type and ileS(T233P) mutant a spoIIE-lacZ fusion. Cells were grown in DSM, and samples were collected for ß-galactosidase assay at time points just prior to and for several hours after the entry into stationary phase. As seen in Fig. 3A, while wild-type cells induced spoIIE expression one hour after entry into stationary phase, spoIIE expression was delayed in the ileS(T233P) cells for five hours, after which point spoIIE is induced. This late induction of spoIIE expression in the ileS(T233P) mutant may explain why sporulation does occur in the mutant, albeit at a lower efficiency than wild-type cells.
Figure 3. Expression of a Spo0A-activated gene is delayed in the ileS(T233P) mutant.
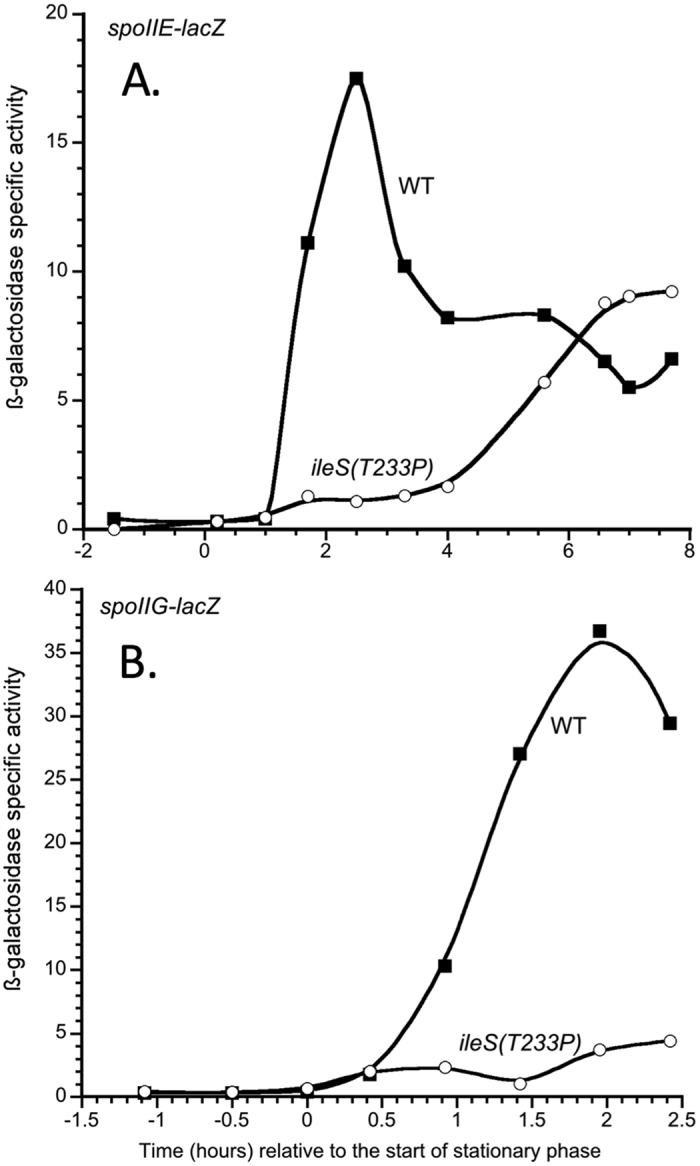
Strains carrying a spoIIE-lacZ (A) or spoIIG-lacZ (B) fusion were grown in DSM. Samples were removed periodically, and the ß-galactosidase specific activity ((ΔA420 per minute per ml of culture per OD600) × 1000) was measured and plotted versus the time the sample was harvested relative to the time at which stationary phase for the culture commenced. The strains grown in panel A were BAL4575 (WT; closed squares) and BAL4576 (ileS(T233P); open circles), and strains grown in panel B were BAL4418 (WT; closed squares) and BAL4419 (ileS(T233P); open circles). The data shown are representative of at least three independent experiments.
Altered spoIIE expression in the ileS(T233P) mutant strain strongly suggests that Spo0A activity is reduced in this mutant strain. To confirm this, we assessed expression of a second Spo0A-activated promoter, spoIIG, as a lacZ fusion34. The spoIIG operon encodes the mother cell-specific sigma factor, sigma-E35. Similar to what was seen with spoIIE expression, expression of spoIIG was defective for several hours after entry into sporulation in the ileS(T233P) strain (Fig. 3B), confirming the general defect in Spo0A-controlled gene expression.
Discussion
Here, we present the first strain of B. subtilis to lack QC for a mischarged tRNA, specifically tRNAIle. This strain has a point mutation in the endogenous copy of ileS, and whole cell extracts of this mutant strain mischarged tRNAIle with valine unlike the wild-type strain. While this QC-defective strain showed no defect in exponential growth, it has a significantly reduced ability to sporulate. The sporulation defect of the QC-defective IleRS strain appears to be due to weak and/or delayed activation of the first transcription factor in the sporulation cascade, Spo0A. Competition through periods of exponential growth, sporulation, and lethal challenge reveal that the ileS(T233P) strain was rapidly outcompeted by the wild-type strain, suggesting that starvation conditions that induce sporulation have selected for the maintenance of the IleRS QC function in B. subtilis.
The loss of QC by IleRS in B. subtilis leads to a delay in expression of sporulation genes controlled by Spo0A, the first transcription factor that is activated in the sporulation cascade. Spo0A also regulates genes involved in other processes including biofilm formation26, but the ileS(T233P) strain exhibited a less than two-fold defect in the ability to form biofilms, much smaller than the defect observed for sporulation. Phosphorylation of Spo0A is the rate-limited step for the initiation of sporulation36,37, and expression of sporulation genes requires a higher level of phosphorylated Spo0A than biofilm genes38,39, suggesting that the ileS(T233P) strain may be defective in forming high levels of phosphorylated Spo0A. Spo0A is activated by phosphorylation through a phosphorelay40, and there are several known negative regulators of this phosphorelay. Future research will address what step along this Spo0A activation pathway is affected in the ileS(T233P) mutant or whether there is a Spo0A-independent mechanism that is affecting the expression of these spoII genes. Understanding how B. subtilis controls sporulation has allowed researchers to identify how cells sense cell cycle problems, which are amplified by checkpoints that delay sporulation, including DNA replication, chromosome partitioning, and tricarboycylic cycle metabolism41,42, and research into how loss of QC by IleRS affects sporulation could reveal a novel checkpoint. It is not known at this time if the requirement of QC for sporulation is specific to IleRS or whether loss of QC by other aaRSs will similarly disrupt sporulation in B. subtilis. Intriguingly, the ileS gene of B. subtilis and other endospore-forming species is adjacent to a gene, ylyA, required for spore germination43.
What causes sporulation to be so sensitive to the loss of QC by IleRS? IleRS could encounter conditions that promote a higher rate of mischarging of tRNAIle in stationary phase, through a higher ratio of non-cognate Val to cognate Ile. Alternatively, the rate of mischarging, and subsequent mistranslation, may be identical between exponential growth and stationary phase, and a protein(s) involved in sporulation may be particularly sensitive to mistranslation. A further possibility is that there is a higher ratio of charged to uncharged tRNAIle as a result of loss of QC by IleRS, and this altered ratio may delay activation of the sporulation pathway. E. coli cells expressing an editing-defective form of the phenylalanine tRNA synthetase (PheRS), have a higher ratio of charged to uncharged tRNAPhe under amino acid starvation and are unable to mount a stringent response44. In B. subtilis, the stringent response is induced by uncharged tRNAs and has been shown to activate sporulation45. The higher ratio of charged to uncharged tRNAIle in B. subtilis may also affect the putative ILE-T-Box (i.e. riboswitch) of ileS, which could alter the levels of IleRS and possibly exacerbate the problems caused by loss of QC46,47. Biosynthesis of Ile and other branched-chain amino acids (BCAA) is not predicted to be directly affected in the IleRS QC-defective mutant, as aaRS are highly specific for their cognate tRNA and the BCAA biosynthesis operon ilvBHC-leuABCD is controlled by tRNALeu and a LEU-T-Box48. Future experiments to address the potential models outlined here for why quality control by IleRS is critical for sporulation, but not exponential growth, will require measuring cellular amino acid pools, the level of the stringent response alarmone, ppGpp, mistranslation, and charged tRNAIle in actively growing and sporulating cells.
Sporulation by B. subtilis is a developmental process induced by starvation, specifically limitation for carbon, nitrogen, or phosphorous49,50,51, and it is the major state in which B. subtilis is isolated from the environment52. Cells lacking QC by IleRS are rapidly outcompeted by wild-type cells under conditions requiring sporulation, which strongly argues that sporulation is the physiological condition that selects for the maintenance of QC by IleRS in B. subtilis. These data further suggest that the primary role of QC by IleRS in other species, and possibly QC by other aaRSs, may be in allowing survival of cells that have a severely reduced growth rate. Consistent with this, QC by alanine tRNA synthetase (AlaRS) was shown to be required in terminally differentiated mouse neuronal cells and cardiomyocytes, non-growing or slowly-growing cell populations5,53. Intriguingly, QC by some aaRSs has been lost from mitochondria and Mycoplasma species, many of which are intracellular parasites54,55,56,57. This latter finding suggests that, in the absence of changing environmental conditions, QC by aaRSs may not be required and is consistent with the suggestion of the work presented here that QC is required by cells that must transition from rapid growth to slow-growth/stationary phase induced by nutrient limitation.
In summary, we have shown for the first time that the quality control function of a tRNA synthetase is critical for sporulation in B. subtilis. The IleRS QC was required for activating the genes controlled by the master transcription factor, Spo0A, and cells lacking IleRS QC were rapidly lost from a mixed population when a selection for sporulation was applied. These data suggest that the QC function of tRNA synthetases may be conserved to deal with particular stress conditions that are encountered by cells as their growth slows or they enter stationary phase in response to nutrient limitation.
Experimental Procedures
tRNA Preparation
Bacillus subtilis tRNAIle was transcribed in vitro as previously described58.
Cell Free Extract Preparation
B. subtilis tRNA-free cell free extracts were prepared as described59, using strains containing either a wild-type ileS gene or an editing deficient allele of ileS (ileS(T233P)), except EDTA-free protease inhibitor (Roche) was used during lysis to replace PMSF. Briefly, the supernatant following ultracentrifugation was loaded onto DEAE Sephacel (GE) resin. Cell free extracts were quantified using a BCA Protein Assay kit (Thermo Scientific Pierce).
Aminoacylation Assays
Aminoacylation assays using cell free extracts of wild-type ileS and ileS(T233P) strains were performed as previously described21. Each reaction contained 450 μg extract protein, 1070 ng/μL of B. subtilis tRNAIle, 0.1 mM cognate [14C]Ile at 200 cpm/pmol (Moravek Biochemicals), 0.15 mM non-cognate [14C]Val at 120 cpm/pmol (PerkinElmer), and 0.3 mM non-cognate [14C]Leu at 120 cpm/pmol (Moravek Biochemicals).
Growth conditions
E. coli and B. subtilis strains were routinely grown in Luria-Bertani (LB) medium. For sporulation assays, B. subtilis strains were grown in either Difco sporulation medium (DSM)60 or S750 minimal medium with 0.1% glucose61. Both E. coli and B. subtilis strains were grown at 37 °C. Antibiotics were used as required at the following concentrations: ampicillin 100 μg/ml, chloramphenicol 5 μg/ml, erythromycin 0.5 μg/ml, and tetracycline 12.5 μg/ml. The growth rate was measured as the slope of the line from the natural log of the OD600 readings versus the time of incubation in minutes. Only points in exponential growth were used for a line fit, and only line fits that gave an R-value greater than 0.99 were considered valid.
Strain construction
E. coli strain Top10 (Invitrogen) was used for the routine construction and maintenance of plasmids. The B. subtilis strains used in this study are all derivatives of the JH642 strain, which contains the trpC2 and pheA1 mutations25 and were constructed by transformation of genetically competent cells with chromosomal DNA or plasmids using standard protocols60. Gene replacement was verified using PCR.
The B. subtilis ileS(T233P) strain was constructed by transformation with a plasmid, pBL4566, containing the ileS(T233P) gene. The pBL4566 plasmid is a derivative of pJet1.2 (Thermo Fisher) that contains a restriction site for the I-SceI meganuclease62 and the cat gene for chloramphenicol resistance. The ileS gene was PCR amplified with primers ileS-F (5′-GCATGGATCCAAGGAATAAATTCTCTGATTA) and ileS-R (5′- GCATAGATCTGCATATCGGTCAACTGAACGC) and ligated to pJet1.2 according to the instructions of the CloneJET PCR Cloning Kit (ThermoFisher Scientific). An I-SceI-cat cassette was constructed by PCR amplification with pJH10163 as a template and primers I-SceI-cat-A (5′-ATGCCTGCAGTAGGGATAACAGGGTAATTATTGGGCGCTCTTCCGCTAAGCATGCGTTACCCTTATTATCAAGA) and I-SecI-cat-B (5′-ATGCCTGCAGGCGAGTCAGTGAGCGAGGAAGCAAGCATGCGGAGCTGTAATATAAAAAC), in which the underlined sequence corresponds to the cat gene, the I-SecI site is bold, and PstI restriction sites are italicized. Both the I-SceI-cat PCR and the pJet1.2-ileS plasmid were digested with PstI and ligated, to yield a pJet-ileS-I-SceI-cat plasmid. This plasmid was subjected to site-directed mutagenesis using primers T233P-F (5′- GCATCATCATTTGGACACCAACGCCGTGGACAATT) and its reverse complement (the bold, underlined nucleotide is the introduced mutation), according to directions of Quikchange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies). The presence of the mutation was confirmed by DNA sequencing, and the resulting plasmid was named pBL4566.
To transfer the ileS(T233P) allele on pBL4566 to the chromosome of B. subtilis, a wild-type strain of B. subtilis, JH64225, was transformed with pBL4566, and transformants were selected on plates containing chloramphenicol. The resulting strain, BL4568, which has pBL4566 integrated into its chromosome at the ileS locus, has two alleles of ileS, one wild-type and one ileS(T233P), flanking the pJet1.2 plasmid backbone and the I-Sce1-cat cassette. To return the chromosome to just one copy of ileS and remove the pBL4566 plasmid, the pBKJ223 plasmid that expresses the I-Sce1 meganuclease64 was transformed into BAL4568, and transformants were selected on plates containing tetracycline. I-Sce1 cleaves the chromosome at the I-SceI restriction site on the pBL4566 plasmid, and repair of this DNA break by homologous recombination results in a strain that has removed the plasmid sequences and restored the native ileS chromosomal structure, with the ileS allele present being either wild-type or ileS(T233P). To screen for cells that have lost the plasmid sequence, transformants were screened for chloramphenicol sensitivity. The ileS allele present in the chloramphenicol-sensitive clones were screened by PCR amplification and DNA sequencing, including the genes to either side of ileS, ylyA and divIVA. One strain with the ileS(T233P) allele and one with the wild-type ileS allele were selected. To remove pBKJ223 from the desired clones, cells were grown in LB liquid cultures lacking antibiotic selection. Colonies from these cultures were streak-purified on LB agar plates and then screened for tetracycline sensitivity. Two tetracycline-sensitive clones, one wild-type ileS and one ileS(T233P), were named BAL4574 and BAL4571, respectively. To assay expression of sporulation genes, thrC::(spoIIE-lacZ, erm)65,66 and Spß::(spoIIG-lacZ, cat)67 were introduced into BAL4574 (wild-type) and BAL4571 (ileS(T233P)).
Sporulation Assays
To assess the ileS(T233P) mutant for sporulation, strains were assessed for the presence of heat-resistant spores, as previously described60. Briefly, cells were grown in liquid Difco Sporulation Medium (DSM) until 16 hours after the onset of stationary phase. At this time, the total viable CFU/ml was measured and then the cultures were heated at 80 °C for 20 minutes, and the heat-resistant CFU/ml was measured as the number of spores present.
β-Galactosidase Assays
Cells were grown as described above for sporulation assays, except that samples were harvested throughout the exponential growth phase and several hours past the entry of the cultures into stationary phase. The cells were then harvested by centrifugation, and resuspended in Z-Buffer, and ß-galactosidase activity was measured as previously described68. β-galactosidase-specific activity is presented as (ΔA420 per minute per ml of culture per OD600) × 1000).
Biofilm formation Assays
Cells were grown for 24 hours in polyvinyl chloride microtiter plates in Biofilm Growth Medium, a modified LB based medium supplemented with 124 mM potassium phosphate pH 7.0, 15 mM ammonium sulphate, 3.4 mM sodium citrate, 0.01 mM MgSO4, and 0.1% glucose as previously described28,29,30. After incubation, the wells were washed twice and subsequently stained with crystal violet. The wells were rinsed to remove any non-adherent crystal violet and treated with a solubilizing solution of 80% ethanol, 20% acetic acid; the A570 of the solution was then measured. The A570 of 16–24 wells were averaged. When the SEM (standard error of the mean) of the A570 measurements obtained for the 16–24 wells was <10% of the mean, the assay was considered valid. Each assay was repeated on at least three independent occasions, and the averages from all valid assays were averaged to determine the level of biofilm formation for a strain.
Growth of pellicle biofilms
Bacterial cells were inoculated at OD600 0.025 into Biofilm Growth Medium and grown with shaking until OD600 ~1.0. The cells were diluted into fresh Biofilm Growth Medium to an OD600 of 0.01 and 5 ml were added to culture test tubes. The tubes were incubated at 37 °C for 48 hours with no shaking, at which times the pellicles were imaged.
Additional Information
How to cite this article: Kermgard, E. et al. Quality Control by Isoleucyl-tRNA Synthetase of Bacillus subtilis Is Required for Efficient Sporulation. Sci. Rep. 7, 41763; doi: 10.1038/srep41763 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Acknowledgments
We thank members of the Lazazzera and Ibba labs for comments on the manuscript. We particularly thank Sharon Hoover, Ph.D. for her suggestions for and assistance with experiments. Plasmid pBKJ223 was a kind gift from Scott Stibitz, Ph.D. (NIH). This research was supported by National Science Foundation (MCB 1412773 to B.L.; MCB 1412611 to M.I.). J.W. was supported through a Biomedical Research Summer Scholarship from UCLA.
Footnotes
The authors declare no competing financial interests.
Author Contributions E.K., M.I. and B.A.L. contributed to the conception or design of the study, E.K., Z.Y., A.-M.M., R.S. and J.W. contributed to the acquisition, analysis, and interpretation of the data, and E.K. and B.A.L. were involved in writing of the manuscript.
References
- Perona J. J. & Gruic-Sovulj I. Synthetic and editing mechanisms of aminoacyl-tRNA synthetases. Top Curr Chem 344, 1–41, doi: 10.1007/128_2013_456 (2014). [DOI] [PubMed] [Google Scholar]
- Yadavalli S. S. & Ibba M. Quality control in aminoacyl-tRNA synthesis its role in translational fidelity. Adv Protein Chem Struct Biol 86, 1–43, doi: 10.1016/B978-0-12-386497-0.00001-3 (2012). [DOI] [PubMed] [Google Scholar]
- Reynolds N. M., Lazazzera B. A. & Ibba M. Cellular mechanisms that control mistranslation. Nat Rev Microbiol 8, 849–856, doi: 10.1038/nrmicro2472 (2010). [DOI] [PubMed] [Google Scholar]
- Schimmel P. Development of tRNA synthetases and connection to genetic code and disease. Protein Sci 17, 1643–1652, doi: 10.1110/ps.037242.108 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. et al. Deficiencies in tRNA synthetase editing activity cause cardioproteinopathy. Proc Natl Acad Sci USA 111, 17570–17575, doi: 10.1073/pnas.1420196111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvetesic N., Palencia A., Halasz I., Cusack S. & Gruic-Sovulj I. The physiological target for LeuRS translational quality control is norvaline. EMBO J 33, 1639–1653, doi: 10.15252/embj.201488199 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullwinkle T. J. et al. Oxidation of cellular amino acid pools leads to cytotoxic mistranslation of the genetic code. Elife 3, doi: 10.7554/eLife.02501 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei D. & Zhu J. Mechanism of action of S-ribosylhomocysteinase (LuxS). Curr Opin Chem Biol 8, 492–497, doi: 10.1016/j.cbpa.2004.08.003 (2004). [DOI] [PubMed] [Google Scholar]
- Schmidt E. & Schimmel P. Mutational isolation of a sieve for editing in a transfer RNA synthetase. Science 264, 265–267 (1994). [DOI] [PubMed] [Google Scholar]
- Dulic M., Perona J. J. & Gruic-Sovulj I. Determinants for tRNA-dependent pretransfer editing in the synthetic site of isoleucyl-tRNA synthetase. Biochemistry 53, 6189–6198, doi: 10.1021/bi5007699 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulic M., Cvetesic N., Perona J. J. & Gruic-Sovulj I. Partitioning of tRNA-dependent editing between pre- and post-transfer pathways in class I aminoacyl-tRNA synthetases. J Biol Chem 285, 23799–23809, doi: 10.1074/jbc.M110.133553 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikora M. & Jakubowski H. Homocysteine editing and growth inhibition in Escherichia coli. Microbiology 155, 1858–1865, doi: 10.1099/mic.0.026609-0 (2009). [DOI] [PubMed] [Google Scholar]
- Hendrickson T. L. et al. Mutational separation of two pathways for editing by a class I tRNA synthetase. Mol Cell 9, 353–362 (2002). [DOI] [PubMed] [Google Scholar]
- Hendrickson T. L., Nomanbhoy T. K. & Schimmel P. Errors from selective disruption of the editing center in a tRNA synthetase. Biochemistry 39, 8180–8186 (2000). [DOI] [PubMed] [Google Scholar]
- Bishop A. C., Beebe K. & Schimmel P. R. Interstice mutations that block site-to-site translocation of a misactivated amino acid bound to a class I tRNA synthetase. Proc Natl Acad Sci USA 100, 490–494, doi: 10.1073/pnas.0237335100 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvetesic N. et al. Naturally Occurring Isoleucyl-tRNA Synthetase without tRNA-dependent Pre-transfer Editing. J Biol Chem 291, 8618–8631, doi: 10.1074/jbc.M115.698225 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop A. C., Nomanbhoy T. K. & Schimmel P. Blocking site-to-site translocation of a misactivated amino acid by mutation of a class I tRNA synthetase. Proc Natl Acad Sci USA 99, 585–590, doi: 10.1073/pnas.012611299 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezo V. et al. Artificially ambiguous genetic code confers growth yield advantage. Proc Natl Acad Sci USA 101, 8593–8597, doi: 10.1073/pnas.0402893101 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacher J. M., de Crecy-Lagard V. & Schimmel P. R. Inhibited cell growth and protein functional changes from an editing-defective tRNA synthetase. Proc Natl Acad Sci USA 102, 1697–1701, doi: 10.1073/pnas.0409064102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacher J. M., Waas W. F., Metzgar D., de Crecy-Lagard V. & Schimmel P. Genetic code ambiguity confers a selective advantage on Acinetobacter baylyi. Journal of bacteriology 189, 6494–6496, doi: 10.1128/JB.00622-07 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd J. & Ibba M. Relaxed substrate specificity leads to extensive tRNA mischarging by Streptococcus pneumoniae class I and class II aminoacyl-tRNA synthetases. MBio 5, e01656–01614, doi: 10.1128/mBio.01656-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacher J. M. & Schimmel P. An editing-defective aminoacyl-tRNA synthetase is mutagenic in aging bacteria via the SOS response. Proc Natl Acad Sci USA 104, 1907–1912, doi: 10.1073/pnas.0610835104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga R., Fukai S., Ishitani R., Nureki O. & Yokoyama S. Crystal structures of the CP1 domain from Thermus thermophilus isoleucyl-tRNA synthetase and its complex with L-valine. J Biol Chem 279, 8396–8402, doi: 10.1074/jbc.M312830200 (2004). [DOI] [PubMed] [Google Scholar]
- Higgins D. & Dworkin J. Recent progress in Bacillus subtilis sporulation. FEMS Microbiol Rev 36, 131–148, doi: 10.1111/j.1574-6976.2011.00310.x (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perego M., Spiegelman G. B. & Hoch J. A. Structure of the gene for the transition state regulator, abrB: regulator synthesis is controlled by the spo0A sporulation gene in Bacillus subtilis. Molecular microbiology 2, 689–699 (1988). [DOI] [PubMed] [Google Scholar]
- Lopez D., Vlamakis H. & Kolter R. Generation of multiple cell types in Bacillus subtilis. FEMS Microbiology Reviews 33, 152–163, doi: 10.1111/j.1574-6976.2008.00148.x (2009). [DOI] [PubMed] [Google Scholar]
- Hamon M. A. & Lazazzera B. A. The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Molecular microbiology 42, 1199–1209 (2001). [DOI] [PubMed] [Google Scholar]
- Hamon M. A., Stanley N. R., Britton R. A., Grossman A. D. & Lazazzera B. A. Identification of AbrB-regulated genes involved in biofilm formation by Bacillus subtilis. Molecular microbiology 52, 847–860, doi: 10.1111/j.1365-2958.2004.04023.x (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley N. R. & Lazazzera B. A. Defining the genetic differences between wild and domestic strains of Bacillus subtilis that affect poly-gamma-dl-glutamic acid production and biofilm formation. Molecular microbiology 57, 1143–1158, doi: 10.1111/j.1365-2958.2005.04746.x (2005). [DOI] [PubMed] [Google Scholar]
- Terra R., Stanley-Wall N. R., Cao G. & Lazazzera B. A. Identification of Bacillus subtilis SipW as a bifunctional signal peptidase that controls surface-adhered biofilm formation. Journal of bacteriology 194, 2781–2790, doi: 10.1128/jb.06780-11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stragier P. & Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet 30, 297–241, doi: 10.1146/annurev.genet.30.1.297 (1996). [DOI] [PubMed] [Google Scholar]
- Stephenson K. & Lewis R. J. Molecular insights into the initiation of sporulation in Gram-positive bacteria: new technologies for an old phenomenon. FEMS Microbiology Reviews 29, 281–301, doi: 10.1016/j.fmrre.2004.10.003 (2005). [DOI] [PubMed] [Google Scholar]
- Yudkin M. D. & Clarkson J. Differential gene expression in genetically identical sister cells: the initiation of sporulation in Bacillus subtilis†. Molecular microbiology 56, 578–589, doi: 10.1111/j.1365-2958.2005.04594.x (2005). [DOI] [PubMed] [Google Scholar]
- Satola S., Kirchman P. A. & Moran C. P. Jr. Spo0A binds to a promoter used by sigma A RNA polymerase during sporulation in Bacillus subtilis. Proc Natl Acad Sci USA 88, 4533–4537 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney T. J. & Moran C. P. Jr. Organization and regulation of an operon that encodes a sporulation-essential sigma factor in Bacillus subtilis. Journal of bacteriology 169, 3329–3339 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastanet A. et al. Broadly heterogeneous activation of the master regulator for sporulation in Bacillus subtilis. Proc Natl Acad Sci USA 107, 8486–8491, doi: 10.1073/pnas.1002499107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswaramoorthy P. et al. The threshold level of the sensor histidine kinase KinA governs entry into sporulation in Bacillus subtilis. Journal of bacteriology 192, 3870–3882, doi: 10.1128/JB.00466-10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M. & Losick R. Evidence that entry into sporulation in Bacillus subtilis is governed by a gradual increase in the level and activity of the master regulator Spo0A. Genes Dev 19, 2236–2244, doi: 10.1101/gad.1335705 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M., Gonzalez-Pastor J. E. & Losick R. High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. Journal of bacteriology 187, 1357–1368, doi: 10.1128/jb.187.4.1357-1368.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggot P. J. & Hilbert D. W. Sporulation of Bacillus subtilis. Current opinion in microbiology 7, 579–586, doi: 10.1016/j.mib.2004.10.001 (2004). [DOI] [PubMed] [Google Scholar]
- Grossman A. D. Genetic networks controlling the initiation of sporulation and the development of genetic competence in Bacillus subtilis. Annu Rev Genet 29, 477–508, doi: 10.1146/annurev.ge.29.120195.002401 (1995). [DOI] [PubMed] [Google Scholar]
- Errington J. Regulation of endospore formation in Bacillus subtilis. Nat Rev Microbiol 1, 117–126, doi: 10.1038/nrmicro750 (2003). [DOI] [PubMed] [Google Scholar]
- Traag B. A., Pugliese A., Eisen J. A. & Losick R. Gene conservation among endospore-forming bacteria reveals additional sporulation genes in Bacillus subtilis. Journal of bacteriology 195, 253–260, doi: 10.1128/JB.01778-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullwinkle T. J. & Ibba M. Translation quality control is critical for bacterial responses to amino acid stress. Proc Natl Acad Sci USA 113, 2252–2257, doi: 10.1073/pnas.1525206113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tojo S., Hirooka K. & Fujita Y. Expression of kinA and kinB of Bacillus subtilis, necessary for sporulation initiation, is under positive stringent transcription control. Journal of bacteriology 195, 1656–1665, doi: 10.1128/jb.02131-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy F. J. et al. The Staphylococcus aureus ileS gene, encoding isoleucyl-tRNA synthetase, is a member of the T-box family. Journal of bacteriology 179, 3767–3772 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood A. V., Grundy F. J. & Henkin T. M. T box riboswitches in Actinobacteria: translational regulation via novel tRNA interactions. Proc Natl Acad Sci USA 112, 1113–1118, doi: 10.1073/pnas.1424175112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandoni J. A., Fulmer S. B., Brizzio V., Zahler S. A. & Calvo J. M. Regions of the Bacillus subtilis ilv-leu operon involved in regulation by leucine. Journal of bacteriology 175, 7581–7593 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freese E. In Sporulation & Germination - Proceedings of the VIII International Spores Conference (eds H. S. Levinson, A. L. Sonenshein, & D. J. Tipper) 1-12 (American Society for Microbiology, 1981).
- Losick R. & Youngman P. In Microbial Development (eds R. Losick & L. Shapiro) 63–68 (Cold Spring Harbor Laboratory Press, 1984). [Google Scholar]
- Sonenshein A. L. In Molecular biology of microbial differentiation (eds J. A. Hoch & P. Setlow) 185–193 (American Society of Microbiology, 1985). [Google Scholar]
- Nicholson W. L. Roles of Bacillus endospores in the environment. Cellular and molecular life sciences: CMLS 59, 410–416 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. W. et al. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature 443, 50–55, doi: 10.1038/nature05096 (2006). [DOI] [PubMed] [Google Scholar]
- Ye Q. et al. Degenerate CP1 Domain from Human Mitochondrial Leucyl-tRNA Synthetase. Journal of Biological Chemistry, doi: 10.1074/jbc.M115.672824 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy H., Ling J., Alfonzo J. & Ibba M. Loss of editing activity during the evolution of mitochondrial phenylalanyl-tRNA synthetase. J Biol Chem 280, 38186–38192, doi: 10.1074/jbc.M508281200 (2005). [DOI] [PubMed] [Google Scholar]
- Li L. et al. Naturally occurring aminoacyl-tRNA synthetases editing-domain mutations that cause mistranslation in Mycoplasma parasites. Proc Natl Acad Sci USA 108, 9378–9383, doi: 10.1073/pnas.1016460108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadavalli S. S. & Ibba M. Selection of tRNA charging quality control mechanisms that increase mistranslation of the genetic code. Nucleic Acids Res 41, 1104–1112, doi: 10.1093/nar/gks1240 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy H., Ling J., Irnov M. & Ibba M. Post-transfer editing in vitro and in vivo by the beta subunit of phenylalanyl-tRNA synthetase. EMBO J 23, 4639–4648, doi: 10.1038/sj.emboj.7600474 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curnow A. W., Tumbula D. L., Pelaschier J. T., Min B. & Soll D. Glutamyl-tRNA(Gln) amidotransferase in Deinococcus radiodurans may be confined to asparagine biosynthesis. Proc Natl Acad Sci USA 95, 12838–12843 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood C. R. & Cutting S. M. Molecular biological methods for Bacillus. (Wiley, 1990). [Google Scholar]
- Solomon J. M., Lazazzera B. A. & Grossman A. D. Purification and characterization of an extracellular peptide factor that affects two different developmental pathways in Bacillus subtilis. Genes Dev 10, 2014–2024 (1996). [DOI] [PubMed] [Google Scholar]
- Viret J. F. Meganuclease I-Scei as a Tool for the Easy Subcloning of Large DNA Fragments Devoid of Selection Marker. Biotechniques 14, 325–326 (1993). [PubMed] [Google Scholar]
- Ferrari F. A., Nguyen A., Lang D. & Hoch J. A. Construction and properties of an integrable plasmid for Bacillus subtilis. Journal of bacteriology 154, 1513–1515 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janes B. K. & Stibitz S. Routine markerless gene replacement in Bacillus anthracis. Infect Immun 74, 1949–1953, doi: 10.1128/iai.74.3.1949-1953.2006 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arigoni F., Guerout-Fleury A. M., Barak I. & Stragier P. The SpoIIE phosphatase, the sporulation septum and the establishment of forespore-specific transcription in Bacillus subtilis: a reassessment. Molecular microbiology 31, 1407–1415 (1999). [DOI] [PubMed] [Google Scholar]
- Prepiak P. et al. MecA dampens transitions to spore, biofilm exopolysaccharide and competence expression by two different mechanisms. Molecular microbiology 80, 1014–1030, doi: 10.1111/j.1365-2958.2011.07627.x (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney T. J., York K., Youngman P. & Moran C. P. Jr. Genetic evidence that RNA polymerase associated with sigma A factor uses a sporulation-specific promoter in Bacillus subtilis. Proc Natl Acad Sci USA 86, 9109–9113 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. H. Experiments in molecular genetics. (Cold Spring Harbor Laboratory, 1972). [Google Scholar]