Abstract
Hypocotyl elongation is considered an important typical seedling trait contributing directly to an increase in and stabilization of the yield in Brassica napus, but its molecular genetic mechanism is poorly understood. In the present study, hypocotyl lengths of 210 lines were measured in an illuminated culture room. A genome-wide association study (GWAS) was performed with 23,435 single nucleotide polymorphisms (SNPs) for hypocotyl length. Three lines with long hypocotyl length and three lines with short hypocotyl length from one doubled haploid line (DH) population were used for transcriptome sequencing. A GWAS followed by transcriptome analysis identified 29 differentially expressed genes associated with significant SNPs in B. napus. These genes regulate hypocotyl elongation by mediating flowering morphogenesis, circadian clock, hormone biosynthesis, or important metabolic signaling pathways. Among these genes, BnaC07g46770D negatively regulates hypocotyl elongation directly, as well as flowering time. Our results indicate that a joint GWAS and transcriptome analysis has significant potential for identifying the genes responsible for hypocotyl elongation; The extension of hypocotyl is a complex biological process regulated by a polygenic network.
Rapeseed (Brassica napus L., 2n = 4x = 38; genome AACC) is one of the most important oilseed crops in the world and the genetic control of yield and yield-related traits has been studied extensively. However, less attention has been focused on elucidating the genetic mechanism of seedling traits. Well-developed seedling traits can contribute directly to an increase in and stabilization of yield and its heterosis, mineral nutrient absorption, drought tolerance, and salinity tolerance in crops1,2,3,4,5,6,7,8,9,10. Thus, understanding the seedling traits will be conducive to breeding programs in B. napus.
In rapeseed, early seedling development traits have been shown to significantly correlate with agronomic traits2. Many heterosis-related quantitative trait loci (QTLs) for seedling traits in B. napus are common with yield-related QTLs1. Several promising positional and functional candidate genes have been associated with germination speed, absolute germination rate, and radicle growth in B. napus6. The number of loci detected for 14 seedling development traits, and candidate genes GER1, AILP1, PECT, and FBP were reported to strongly relate to the seedling development traits in B. napus7. Hypocotyl elongation is thought to be an importantly typical seedling trait in plants. However, almost all studies on hypocotyl elongation have focused on Arabidopsis. Global transcriptome analysis has revealed circadian regulation of key pathways in rhythmic hypocotyl elongation11. The transcriptional regulators CIRCADIAN CLOCK ASSOCIATED1 (CCA1), EARLY FLOWERING3 (ELF3), ELF4, and LUX ARRHYTHMO (LUX) appear to link the circadian clock to diurnal control of hypocotyl growth12,13. Therefore, hypocotyl elongation has a fiercely complex genetic mechanism, but little knowledge is available about hypocotyl elongation in B. napus.
Expressed sequence tag (EST) sequence data, array analysis, amplicon resequencing, sequences, and next-generation sequencing technologies efficiently allow genome-wide association studies (GWASs) and transcriptome analysis to be novel strategies for dissecting complex traits in plants14,15,16,17,18,19,20,21. In the present study, a GWAS for hypocotyl elongation was carried out with a panel of 210 B. napus accessions genotyped for 23,435 SNPs. We also performed transcriptome sequencing of long and short hypocotyl phenotypes. The main objectives of this study were to obtain a better understanding of hypocotyl elongation and its relationship with yield-related traits or heterosis, dissect the genetic basis of hypocotyl elongation by combining GWAS and transcriptome analysis, and perform Gene Ontology (GO) and KEGG pathway analysis for associated genes in B. napus.
Results
Phenotypic variations and correlation analysis
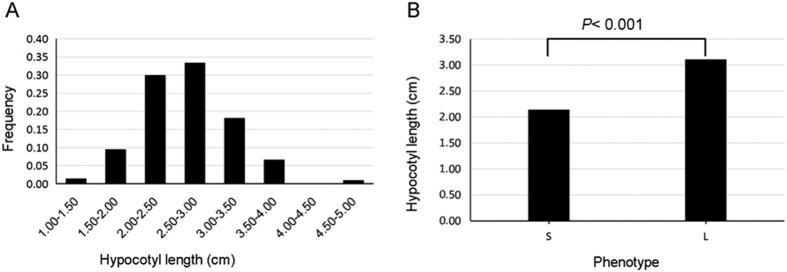
Extensive phenotypic variations in hypocotyl elongation were observed in the 210 rapeseed lines (Fig. 1A and Supplementary Tables S1 and S2). The hypocotyl elongation of the lines was normally distributed (average = 2.66, range 1.38 to 4.81), and 63.33% of hypocotyl elongation values were between 2.00 and 3.00.
Figure 1. Phenotypic variation in hypocotyl length.
(A) Frequency of phenotypic variation in 210 accessions. (B) Comparison of two phenotypes by t-test.
The correlation coefficients between hypocotyl elongation and yield-related traits showed that hypocotyl elongation positive correlated with seed yield per plant (0.29) and biomass yield per plant (0.21) at P = 0.01 and plant height (0.19) at P = 0.05 (Supplementary Table S3). Linear regression analysis of the correlated traits indicated that hypocotyls elongation can explain 3.28% of the total seed yield per plant (P < 0.05), 4.49% of the total biomass yield per plant (P < 0.01), 3.59% of the total plant height (P < 0.05), respectively.
Genetic diversity, population structure, and relative kinship analysis
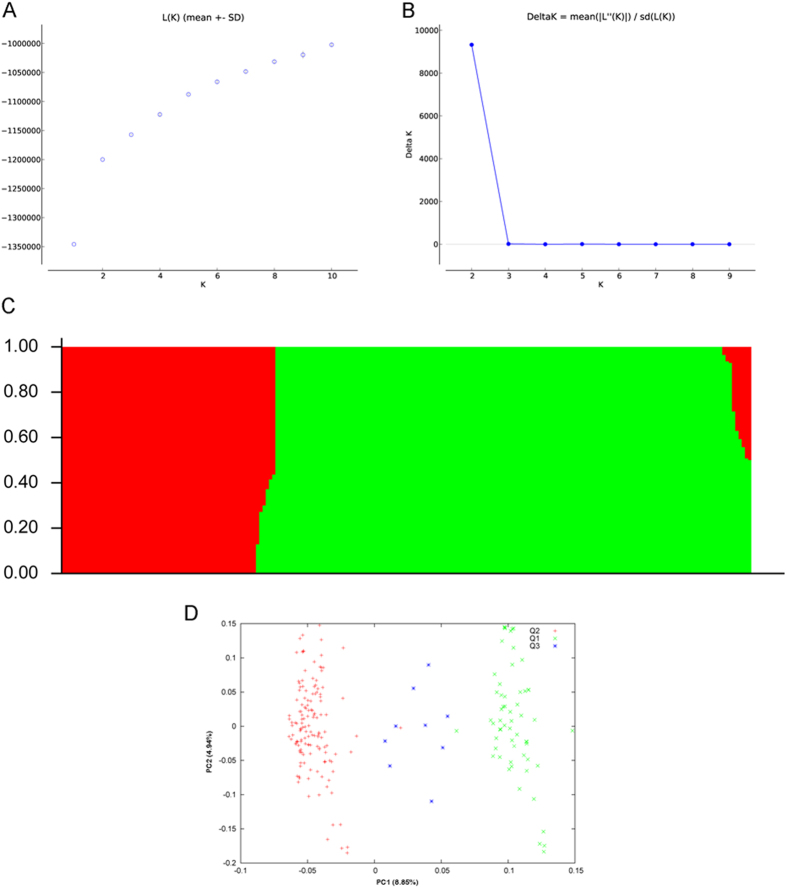
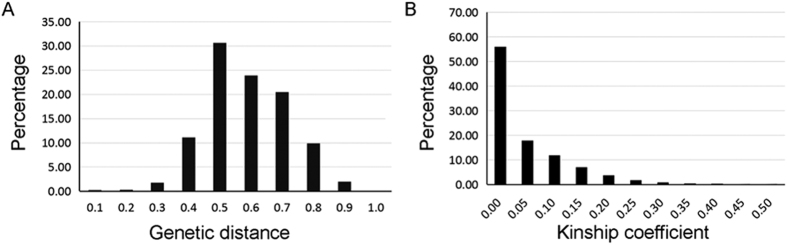
The genetic diversity and population structure of the 210 accessions were analyzed using 5,334 SNPs (Supplementary Table S4). Clustering inference showed that the most significant change in likelihood occurred when K increased from 2 to 3, and the highest Δk value was observed at K = 2 (Fig. 2A–C). Considering the probability of membership threshold of 0.70, 61 and 140 accessions were assigned to subgroups Q1 and Q2, respectively (Supplementary Table S1). The remaining nine accessions were assigned to a mixed group (Q3). The PCA also provided a pattern for the genetic structure of the GWAS population (Fig. 2D). The top two principal components clearly separated these subpopulations and explained 8.85% and 4.94% of the total SNP variations in the rapeseed panel, respectively. All of the parameters suggest that the three-group model (subgroups Q1, Q2, and Q3) sufficiently explained the genetic structure among the 210 accessions. The mean genetic distance (GD) between lines was 0.54, and 74.85% of pairs had a GD ranging from 0.5 to 0.7 (Fig. 3A). The average kinship coefficient identity by descent (IBD) within the total diversity set was 0.06 (Fig. 3B). A total of 55.93% of the pairwise kinship estimates were equal to 0, and 17.85% of pairwise kinship coefficients varied from 0 (excluding 0) to 0.05.
Figure 2. Analysis of the population structure of 210 rapeseed accessions using STRUCTURE.
(A) Estimated L(K) of possible clusters (k) from 1 to 10. (B) Delta K based on the rate of change of L(K) between successive K values. (C) Population structure based on k = 2. Red represents subgroup Q1; green represents subgroup Q2. (D) Principal component (PC) analysis.
Figure 3. Genetic distance and kinship coefficient analysis between pairs of accessions.
LD analysis
All 23,435 SNPs in the total panel were used for LD analysis. The distributions of r2 with respect to the physical distance from each chromosome are shown in the Supplementary data (Fig. S1 and Supplementary Table S5). As expected, the mean r2 between 0 and 500 kb decreased rapidly and continuously, followed by much slower decay at increased physical distance for both the A genome and C genome. The overall LD decay distance was 893.84 Kb when the r2 cutoff was set to 0.1. The rate of LD decay varied over different chromosomes in both the A genome and C genome, with the shortest LD decays of 459.03 kb on chromosome A07 and 602.91 kb on chromosome C08 and the longest LD decays of 968.17 kb on chromosome A09 and 3,190.79 kb on chromosome C09. Obviously, the LD of the A genome decayed significantly faster than the LD of the C genome.
Association mapping and candidate gene prediction
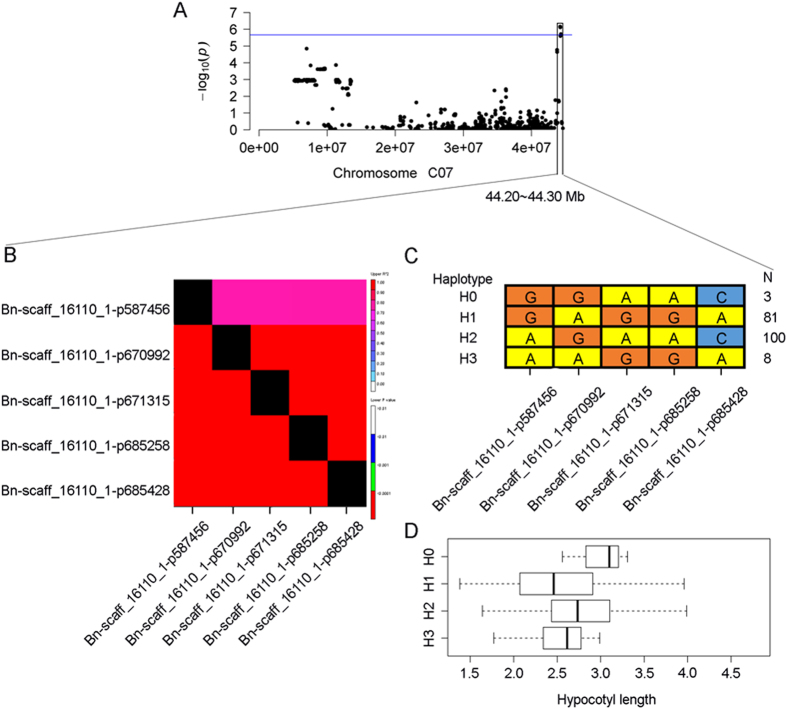
Total 23,435 polymorphisms with minor allele frequency (MAF) ≥ 0.05 were selected for association mapping of hypocotyl elongation using the BLUP value across multiple replications (Supplementary Table S1). Model comparison analyses indicated that P-values from the PCA + K model were nearer the expected P-values than those of the GLM, Q, PCA, and Q + K models (Fig. 4A). Thus, the PCA + K model was selected for association mapping of hypocotyl elongation. Five SNPs on C07 were highly significantly associated with hypocotyl elongation at P < 2.13 E−06, with a FDR of 1.0% (Fig. 4B and Table 1). All detected SNPs were located between 42.15 and 42.25 Mb on C07 and could explain 4.82% of the total phenotypic variance. Thus, the development of hypocotyl is controlled by a minor-effect polygene. In LD analyses, the r2 values were > 0.79 for all pairs of associated SNPs, suggesting that the associated SNPs were in high LD with each other (Fig. 5A and B).
Figure 4. Association analyses of hypocotyl elongation.
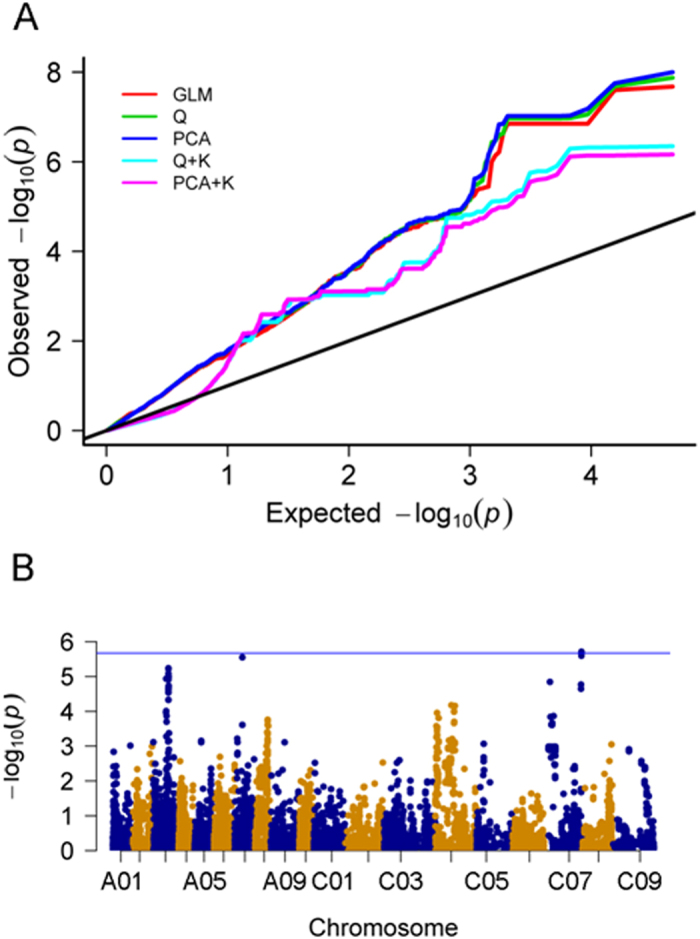
(A) Quantile–quantile plots of estimated −log10(P) from the association analysis of hypocotyl elongation. The black line represents expected P-values with no associations. The red line represents observed P-values using the GLM model. The green line represents observed P-values using the Q model. The blue line represents observed P-values using the PCA model. The cyan line represents observed P-values using the Q + K model. The pink line represents observed P-values using the PCA + K model (color figure online). (B) Manhattan and quantile–quantile plots generated from the genome-wide association analysis of hypocotyl elongation. The blue horizontal line depicts the Bonferroni significance threshold (2.13 E-6).
Table 1. Summary of SNPs significantly associated with hypocotyl elongation.
| SNP | Chromosome | Allele | Position | MAF | P-value | R2(%) |
|---|---|---|---|---|---|---|
| Bn-scaff_16110_1-p587456 | C07 | A/G | 44303399 | 0.49 | 1.94E-06 | 4.82 |
| Bn-scaff_16110_1-p670992 | C07 | G/A | 44218661 | 0.50 | 7.25E-07 | |
| Bn-scaff_16110_1-p671315 | C07 | A/G | 44218337 | 0.49 | 7.66E-07 | |
| Bn-scaff_16110_1-p685258 | C07 | A/G | 44204592 | 0.50 | 6.87E-07 | |
| Bn-scaff_16110_1-p685428 | C07 | C/A | 44204201 | 0.50 | 7.24E-07 |
MAF, minor allele frequency; R2(%), amount of phenotypic variation for each trait explained by multiple SNPs.
Figure 5. LD and haplotype analysis for five associated SNPs.
(A) Association peak on chromosome C07. (B) LD analysis among the associated SNPs. (C) Haplotype analysis with associated SNPs in the population. (D) Phenotypic variation of hypocotyl length in each haplotype.
According to the associated SNP variations, four haplotypes (H0, H1, H2, and H3) were identified from these B. napus accessions (Fig. 5C). H0, H1, H2, and H3 were observed in 3, 81, 100, and 8 lines, respectively. Approximately 94.27% of accessions comprised H1 and H2. Therefore, H0 and H3 are rare variations, whereas H1 and H2 are conserved by artificial selection during the genetic improvement of modern B. napus breeding accessions. Further analysis showed that H0, H1, H2, and H3 have mean hypocotyl elongations of 2.99, 2.52, 2.79, and 2.52, respectively (Fig. 5D). H2 had a significantly (P = 0.001) greater hypocotyl elongation than H1. H0 and H3 were not analyzed because they are extremely rare. Thus, H2 may be a favorable haplotype and facilitate the selection of better genotypes for hypocotyl elongation in breeding B. napus.
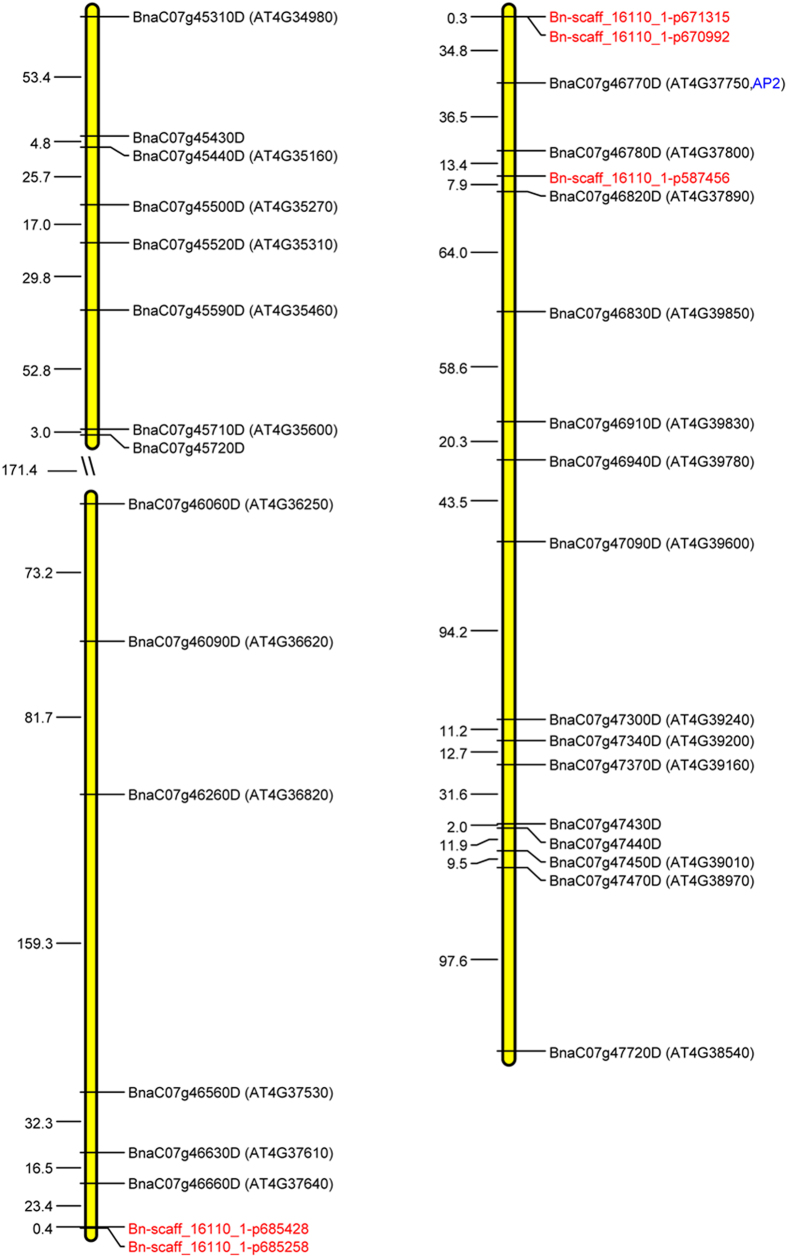
Candidate genes were predicted along the ~100 Kb region between two associated SNPs (Bn-scaff_16110_1-p685428 and Bn-scaff_16110_1-p587456) according to the newly released B. napus genome sequence22. Only five genes (BnaC07g46740D, BnaC07g46760D, BnaC07g46770D, BnaC07g46780D, and BnaC07g46800D) were detected in the candidate region (Supplementary Table S6). Of these genes, BnaC07g46770D was previously identified to regulate the flowering time in rapeseed23. The closest distance between BnaC07g46770D and a significant SNP (Bn-scaff_16110_1-p670992) was 34 Kb. Considering the LD decay of 754.95 Kb in C07, candidate genes were also predicted in the region between 754.95 Kb upstream and downstream of the associated peak; 196 genes were obtained in the enlarged candidate region (Supplementary Table S6). All of the genes were blasted against A. thaliana genome data, but none of the predicted genes were homologous to the genes directly controlling hypocotyl elongation in Arabidopsis.
Transcriptome sequencing analysis
The DH-6004 population had considerably variable flowering time when grown at Hezheng, Gansu province, in the 2015 growing season and Wuhan, Hubei province, in the 2015–2016 growing season. Three lines with extremely early-flowering and three lines with extremely late-flowering exhibited long hypocotyls and short hypocotyls, respectively. The mean hypocotyl elongation in the S and L groups was 2.14 and 3.11, respectively (P < 0.001, t test; Fig. 1B and Supplementary Table S1). Haplotype analyses indicated that DH2 and DH3 in S group showed H1, and DH4, DH5 and DH6 in L group showed H2. The DH1 could not been distributed to any of the four haplotypes as it possessed heterozygosity loci. RNA from the three S lines and three L lines was pooled with two biological replications to generate S1, S2, L1, and L2. A total of 26.82, 54.07, 55.43, and 28.87 million raw sequence reads were generated from the four libraries (Supplementary Table S7). After removing low-quality reads and adaptor sequences, 23.61, 49.17, 49.97, and 25.88 million clean reads were obtained for S1, S2, L1, and L2, respectively. More than 70% of the reads were successfully mapped to the reference genome; the unique and multiple reads that aligned with the genome accounted for 73.09% in L2 to 86.49% in S2.
Of the 196 genes located within the candidate region determined by the GWAS analysis, 29 significant DEGs were identified in the two groups based on the criteria |log2(L/S)| ≥ 1 and P < 0.05 (Table 2). Compared to the S group, 16 (53.33%) DEGs were up-regulated and 13 (46.67%) DEGs down-regulated in the L group.
Table 2. Differentially expressed genes detected by transcriptome sequencing and genome-wide association.
|
B. napus |
A. thaliana |
|||||
|---|---|---|---|---|---|---|
| Gene ID | Position | log2(L/S) | P-value | L change | Homologous | Description |
| BnaC07g45310D | 43459847 | 1.957 | 0.001 | Up | AT4G34980 | Serine protease similar to subtilisin |
| BnaC07g45430D | 43513273 | 6.841 | 0 | Up | — | |
| BnaC07g45440D | 43518096 | 3.658 | 0 | Up | AT4G35160 | Encodes a cytosolic N-acetylserotonin O-methyltransferase |
| BnaC07g45500D | 43543746 | −1.789 | 0.038 | Down | AT4G35270 | Plant regulator RWP-RK family protein |
| BnaC07g45520D | 43560727 | 5.151 | 0 | Up | AT4G35310 | calmodulin-domain protein kinase |
| BnaC07g45590D | 43590520 | 2.228 | 0 | Up | AT4G35460 | NADPH-dependent thioredoxin reductase 1 |
| BnaC07g45710D | 43643363 | −1.57 | 0.033 | Down | AT4G35600 | Encodes a receptor-like cytoplasmic kinase |
| BnaC07g45720D | 43646363 | −4.048 | 0 | Down | — | |
| BnaC07g46060D | 43817797 | 1.646 | 0.029 | Up | AT4G36250 | Encodes a putative aldehyde dehydrogenase |
| BnaC07g46090D | 43890978 | 2.268 | 0.005 | Up | AT4G36620 | Encodes a member of the GATA factor family of zinc finger transcription factors |
| BnaC07g46260D | 43972670 | 3.074 | 0.017 | Up | AT4G36820 | Protein of unknown function |
| BnaC07g46560D | 44131930 | −1.645 | 0.013 | Down | AT4G37530 | Peroxidase superfamily protein |
| BnaC07g46630D | 44164232 | −2.414 | 0 | Down | AT4G37610 | BTB and TAZ domain protein |
| BnaC07g46660D | 44180768 | −1.175 | 0.044 | Down | AT4G37640 | Encodes a calmodulin-regulated Ca(2+)-pump |
| BnaC07g46770D | 44253480 | −1.725 | 0.037 | Down | AT4G37750 | Encodes a putative transcriptional regulator similar to AP2 |
| AT4G36920 (AP2) | Encodes a floral homeotic gene | |||||
| BnaC07g46780D | 44290014 | 1.138 | 0.02 | Up | AT4G37800 | Xyloglucan endotransglucosylase/hydrolase 7 |
| BnaC07g46820D | 44311339 | 1.123 | 0.038 | Up | AT4G37890 | Embryo sac development arrest 40 |
| BnaC07g46830D | 44375293 | −1.362 | 0.028 | Down | AT4G39850 | Encodes a peroxisomal protein of the ATP binding cassette |
| BnaC07g46910D | 44433889 | −4.869 | 0 | Down | AT4G39830 | Cupredoxin superfamily protein |
| BnaC07g46940D | 44454217 | 1.424 | 0.016 | Up | AT4G39780 | Protein contains one AP2 domain |
| BnaC07g47090D | 44497714 | −4.227 | 0 | Down | AT4G39600 | Galactose oxidase/kelch repeat superfamily protein |
| BnaC07g47300D | 44591928 | −1.42 | 0.046 | Down | AT4G39240 | Galactose oxidase/kelch repeat superfamily protein |
| BnaC07g47340D | 44603110 | 1.861 | 0.004 | Up | AT4G39200 | Ribosomal protein S25 family protein |
| BnaC07g47370D | 44615843 | −2.426 | 0.042 | Down | AT4G39160 | Homeodomain-like superfamily protein |
| BnaC07g47430D | 44647393 | 2.13 | 0.002 | Up | — | |
| BnaC07g47440D | 44649385 | 1.179 | 0.044 | Up | — | |
| BnaC07g47450D | 44661277 | 3.187 | 0.001 | Up | AT4G39010 | Glycosyl hydrolase 9B18 |
| BnaC07g47470D | 44670819 | 1.54 | 0.004 | Up | AT4G38970 | Response to ABA |
| BnaC07g47720D | 44768381 | −5.589 | 0 | Down | AT4G38540 | FAD/NAD(P)-binding oxidoreductase family protein |
A dash indicates no homolog has been identified in the respective genome.
Functional classification of DEGs
To monitor the gene expression pattern, GO enrichment analysis of DEGs was performed for two genotypes (Fig. S2). The 29 DEGs were finally classified into 10, 6, and 19 main GO categories according to the cellular component (CC), molecular function (MF), and biological process (BP), respectively. The CC categories, such as cell, cell part, and organelle, were overrepresented. Most of the DEGs function in catalytic activity and binding. The BP category occurring in metabolic processes was significantly overrepresented and included approximately 83% of the DEGs. Further cluster analysis according to BP indicated that eight DEGs (BnaC07g45520D, BnaC07g45710D, BnaC07g45720D, BnaC07g46090D, BnaC07g46630D, BnaC07g46660D, BnaC07g46770D, and BnaC07g47470D) were associated with the response to hormone and flower morphogenesis (Supplementary Table S8).
To explore the function of DEGs in the biosynthesis and metabolite pathways, KEGG pathway analysis was performed in two phenotypic groups (Supplementary Table S9). Six DEGs (BnaC07g45590D, BnaC07g45710D, BnaC07g46060D, BnaC07g46560D, BnaC07g46660D, and BnaC07g47470D) acted in the 30 pathways by encoding corresponding enzymes. For example, BnaC07g46060D and BnaC07g46560D regulate the lignins and phenylpropanoid biosynthesis in phenylpropanoid metabolic pathways by encoding dehydrogenase and lactoperoxidase, respectively. Furthermore, BnaC07g46060D and BnaC07g47470D participate in glycolysis/gluconeogenesis and nitrogen metabolism and carbon fixation in photosynthetic organisms by encoding dehydrogenase and aldolase, respectively. A global examination of gene expression demonstrated that genes encoding dehydrogenase regulate the phenylpropanoid and lignin biosynthetic pathways and are clock-controlled in the same manner as the pathways involved in the assimilation of mineral nutrients and carbon fixation in the process of photosynthesis11. However, no direct evidence is available regarding the detected genes regulating metabolic pathways to affect hypocotyl elongation in relation to circadian rhythm.
Comparative analysis
Of the 29 DEGs detected by combining transcriptome sequencing analysis and a GWAS, 26 homologous genes were identified using the Brassica Genome Browser database and A. thaliana Genome Browser database (Fig. 6 and Table 2). BnaC07g46770D and BnaC07g46780D were located within 84.7 Kb of two significant SNPs: Bn-scaff_16110_1-p670992 and Bn-scaff_16110_1-p587456. BnaC07g46770D was previously reported to regulate flowering time and is orthologous to A. thaliana AP2 and AT4G37750. AP2 belongs to the AP2/ERF gene family and is involved in plant development, in turning leaves into floral organs24. AT4G37750 belongs to the AP2/EREBP gene family and directly regulates a key clock gene (CCA1) that provides molecular links between different signaling modules and the circadian clock25. BnaC07g46780D is orthologous to AT4G37800, one member of the complex endotransglucosylase/hydrolase (XTH) gene family acting within floral stages to strengthen or loosen cell walls26,27.
Figure 6. The distribution pattern of candidate genes and SNPs associated with hypocotyl elongation.
The abbreviations for orthologous genes in A. thaliana are shown in parentheses. SNPs are marked in red. Numbers represent the relative distance in the reference genome in kilobases.
BnaC07g46660D, BnaC07g46630D, and BnaC07g46060D were located 23.4 Kb, 39.9 Kb, and 386.4 Kb upstream of associated SNP Bn-scaff_16110_1-p685428, and are orthologous to AT4G37640, AT4G37610, and AT4G36250, respectively. AT4G37640 functions in a complex process of pollen germination and tube growth28. AT4G37610, which encodes TAZ domain protein, could act as the master clock control gene CCA1 to regulate the organic nitrogen-responsive genes29. AT4G36250 contains five TGTG sites and one HUD site and could been regulated by TOC1(TIMING OF CAB EXPRESSION1), which acts as a general transcriptional repressor to negatively regulate CCA1/LHY30.
BnaC07g46830D, BnaC07g46910D, BnaC07g46940D, BnaC07g47470D, and BnaC07g47720D, which are orthologous to AT4G39850, AT4G39830, AT4G39780, AT4G38970, and AT4G38540, were detected 71.9 Kb, 130.5 Kb, 150.8 Kb, 367.3 Kb, and 465.1 Kb downstream from significant SNP Bn-scaff_16110_1-p587456. AT4G39850 and AT4G38540 include TGTG sites and ME sites, which could also be regulated by TOC130. AT4G39830 showed significant changes in expression during pollen germination and tube growth and, thus, regulate the process of reproduction in Arabidopsis28. AT4G39780 belongs to the Arabidopsis ERF gene family, a part of the AP2/ERF superfamily, which have important roles in the transcriptional regulation of a variety of biological processes related to growth and development, as well as various responses to environmental stimuli31. AT4G38970 is expressed in the regulation of biochemical pathways during photomorphogenesis32. However, to date, little knowledge is available about the function of the other 15 homologous genes in A. thaliana.
Discussion
Optimal seedling development of plants leads to a promising yield, and hypocotyl elongation is considered a typical seedling trait. Seedling traits measured at an early stage of development significantly correlate with agronomic traits in B.napus2. Here, we evaluated the phenotypic variation of hypocotyl elongation, which exhibited continuous variation and approximated a normal distribution. Correlation analysis indicated that hypocotyl length positively correlates with seed yield per plant, biomass yield per plant, and plant height. Five SNPs explaining 4.82% of the total phenotypic variance were highly significantly associated with hypocotyl elongation, and 196 genes were obtained in the enlarged candidate region. The results imply that hypocotyl elongation is a complex quantitative trait controlled by a minor-effect polygene.
Genome-wide association study, also known as LD mapping, has emerged as very promising strategies for understanding naturally occurring phenotypic variation33,34,35,36. Recently, more and more studies tended to identify the candidate genes by combining GWAS and linkage mapping in rice37, maize38, sunflower39 and wheat40. However, it is extremely laborious and time-consuming to develop large-scale linkage mapping populations or linkage–LD mapping populations, such as nested association mapping41 and multi-parent advanced generation inter-cross42. In rapeseed, combined SNP-trait association and transcriptome sequencing analyses successfully identified twenty-four genes associated with the resistance to Sclerotinia stem rot43. In the present study, a GWAS followed by transcriptome analysis confirmed 29 genes mainly related to circadian clock, flowering morphogenesis, hormone biosynthesis, or important metabolic signaling pathways regulating hypocotyl elongation in B. napus. Therefore, joint genome-wide association and transcriptome sequencing is an alternate method of dissecting the genetic and biochemical basis of hypocotyl elongation in B. napus.
Of the 29 genes, transcriptome sequencing assays revealed that six genes responsible for hormone (Table S8). This may correspond to the variation of hypocotyl elongation, because hormone regulates many aspects of growth and development containing hypocotyl elongation in plants. The light-mediated photomorphogenesis triggered by hormone biosynthetic factors directly affects hypocotyl elongation in Arabidopsis44. Likewise, overexpressing auxin biosynthetic genes could increase hypocotyl elongation in Arabidopsis45. In addition, six genes were detected to act in the 30 pathways by encoding corresponding enzymes, implying that these genes probably regulate the hypocotyl elongation by affecting important metabolites biosynthesis in B.napus. Furthermore, 25 homologs of the Arabidopsis genes were identified in the B. napus genome through homologous alignment. Among of them, BnaC07g46770D was previously found to directly relate to flower time23 and is orthologous to A. thaliana AP2 and AT4G37750. AP2 is involved in the development of floral organs24 and AT4G37750 directly regulates a key clock gene (CCA1) controlled the hypocotyl elongation in Arabidopsis. We supposed that BnaC07g46770D may regulate circadian gene or floral development to affect the flowering time and hypocotyl elongation in B.napus, which at least partially explains the correlation between flowering time and hypocotyl elongation. Similarly, BnaC07g46630D is orthologous to A. thaliana AT4G37610, which acts as the master clock control gene CCA129. BnaC07g46060D, BnaC07g46830D and BnaC07g47720D are orthologous to A. thaliana AT4G36250, AT4G39850 and AT4G38540, respectively, regulated by TOC130. TOC1 is an important component of the circadian clock in Arabidopsis with a crucial function in the integration of light signals to control hypocotyl elongation46. The results indicated that these genes may affect hypocotyl elongation by interacting with circadian clock genes in B. napus. BnaC07g47470D is orthologous to Arabidopsis AT4G38970 which expressed in the regulation of biochemical pathways during photomorphogenesis32. Photomorphogenesis is linked to photoperiod, an important challenging factors affected hypocotyl elongation by regulating cell elongation47. In addition, BnaC07g46660D and BnaC07g46910D are orthologous to Arabidopsis AT4G37640 and AT4G39830 acting within floral morphogenesis, but it needs to further study of their roles in the development of hypocotyl elongation in B.napus.
In summary, this study is the first to study the hypocotyl elongation by integrating GWAS and transcriptome sequencing in B.napus. We demonstrated that the genes mediated by circadian clock, hormone biosynthesis, floral morphogenesis, or other metabolic signaling pathways may regulate the hypocotyl elongation in B. napus. These findings reveal that the phenotypic variation of the hypocotyl is a complex biological process regulated by a polygenic network in B.napus. Over the past decade, circadian clock and hormone effects had been linked to agronomic traits in plant48,49. Hypocotyl elongation represents the best-studied model of plant circadian clock and hormone response system. Therefore, modification of these areas may have the potential for systemic effects that produce beneficial yield trait in B.napus.
Materials and Methods
Plant materials and trait collection
A set of 210 elite inbred rapeseed lines with abundant phenotypic variation were collected to construct an association panel (Supplementary Table S1); 55 lines (X001-X055) were used to isolate and characterize the sucrose transporter (SUT) gene50, and 155 lines (X056-X210) were derived from an association mapping population genotyped using the 60 K Illumina® Infinium SNP array51. The yield-related traits of these lines were measured in a previous study50,51. The 210 lines were grown with 20 replications in 10 × 10 culture plates. When cotyledons were fully developed, all of the lines were sprayed with nutrient solution as described previously52. To control environmental conditions, the seedlings were grown in an illuminated culture room under 16 L:8D conditions at 20 °C and measurements performed on day 20. Photographs of seedlings were analyzed using AutoCAD software (http://www.autodesk.com.cn/education/free-software/featured). Three long hypocotyl (L) and three short hypocotyl (S) lines were used for transcriptome sequencing. These lines were selected from a doubled haploid (DH) population (DH-6004) developed from 2011–5515–137 × Gui01A10 F1 (field code 9–6004), in which ‘2011–5515–137’ exhibits early flower and ‘Gui01A10’ moderate flower.
SNP genotyping
Fifty-five lines (X001-X055) and six DHs (DH1, DH2, DH3, DH4, DH5 and DH6) were genotyped using the Brassica 60 K Illumina ® Infinium SNP array. Combined with genotype information obtained previously for the other 155 lines, 26,016 SNPs were mapped in silico using the probe sequences of 52,157 SNPs to perform a Blast N search against B. napus genome sequences53. Only the top hits, using an E-value cut-off of 1E-15 against the B. napus genome sequences, were considered. Hits with AA or BB frequency equal to zero (i.e., monomorphic), call frequency <0.8, or minor frequency <0.05 were excluded. Thus, a total of 23,435 SNPs were filtered for association analysis (Supplementary Table S4). Genetic diversity and Nei’s genetic distance54 were estimated using PowerMarker version 3.2555.
Population structure, relative kinship, and linkage disequilibrium
The population structure was inferred using the software package STRUCTURE v2.3.456 based on 5,334 SNPs with AA or BB frequency >0.05, call frequency ≥0.9 and minor frequency >0.2. Five independent runs were performed with a K-value (the putative number of genetic groups) from 1 to 10, with both the length of the burning period and the number of Markov Chain Monte Carlo (MCMC) replications after burning set to 100,000 iterations under the ‘admixture model’. The most likely k-value was determined by the log probability of data [LnP(D)] and ad hoc statistic Δk based on the rate of change of LnP(D) between successive k values as described previously57. Accessions with a probability of membership >0.7 were assigned to corresponding clusters, and those with a probability of membership <0.7 were assigned to a mixed group. The relative kinship matrix comparing all pairs of accessions was calculated using the software package SPAGeDi58. Negative values between two individuals were set to 059. Principal component analysis (PCA) based on SNPs was carried out using the EIGENSTRAT tool60. The linkage disequilibrium (LD) parameter r2 was calculated using the software TASSEL 3.0 with 1,000 permutations61.
GWAS and statistical analysis
The effects of population structure (Q, PC) and kinship (K) on the traits were evaluated by a GWAS using five models (GLM, Q, PCA, PCA + K, and Q + K). Significant loci were identified by comparing P-values with the Bonferroni threshold (0.05/23,435 = 2.13E-06). Quantile-quantile plots of the estimated –log10 (P) values in the association mapping model were created using the observed P-values from marker-trait associations versus the expected P-values. In addition, false discovery rates (FDRs) were calculated as [(m × P)/n] × 100%, where m is the total number of SNPs (23,435 in this study), P is the P-value threshold for detecting a significant association, and n is the total number of significant associations per trait62.
Phenotypic variation, correlation and linear regression analyses were performed using SPSS version 19.0 (IBM Corp., Armonk, NY, USA).
Nuclear RNA extraction and RNA sequencing
When the second cotyledons were fully expanded in the illuminated culture room, the seedlings of three S lines (DH1, DH2 and DH3) and three L lines (DH4, DH5 and DH6) were pooled to long hypocotyl bulk and short hypocotyl bulk, respectively, then immediately frozen in liquid nitrogen and stored at −80 °C. Total nuclear RNA was extracted from ~100 mg of frozen plants using the RNAprep Pure Plant Kit (TIANGEB BIOTECH, Beijing, China) according to the manufacturer’s instructions in two biological replicates. NanoDrop ND 1000 (NanoDrop technologies) was used to evaluate the quality of the extracted RNA. RNA with an RNA Integrity Number (RIN) > 8 as assessed by Agilent Technologies 2100 Bioanalyzer (Agilent) was used to prepare the c-DNA library. The sequencing library was generated using the Illumina RNA Library Prep Kit (NASDAQ: ILMN, America) following the manufacturer’s recommendations. The library preparations were sequenced on an Illumina Hiseq 200 platform, and 100-bp paired-end reads were generated.
DEG identification and gene annotations
The sequenced data were trimmed by removing adapters, low-quality sequences or bases, and contaminations or overrepresented sequences using Trimmomatic software version 0.33. The clean reads were aligned to the B. napus reference genome22 using Hisat software version 0.1.6 and then assembled using TopHat 2.0.0 and Cufflinks63. Fragments per kilobase million (FPKM) was determined to estimate gene expression levels. Differentially expressed genes (DEGs) between two genotypes were identified by Cuffdiff based on the criteria P < 0.05 and |log2 (L/S)| > 1. To identify possible homologous genes, DEGs were blasted against the A. thaliana genome database (http://www.arabidopsis.org/). The GO enrichment analysis for DEGs was implemented by Blast2GO and significantly enriched GO terms (P < 0.05) displayed using the online tool WEGO (http://wego.genomics.org.cn). The enrichment of DEGs was determined by KEGG pathway analysis using the KOBAS2.0 website (http://kobas.cbi.pku.edu.cn/home.do). To analyze the metabolic pathway and functional classification of DEGs, expression data were mapped to metabolic pathways using MapMan software64.
Additional Information
How to cite this article: Luo, X. et al. Joint genome-wide association and transcriptome sequencing reveals a complex polygenic network underlying hypocotyl elongation in rapeseed (Brassica napus L.). Sci. Rep. 7, 41561; doi: 10.1038/srep41561 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Key Research and Development Program of China (No.2016YFD0100803) and the National Natural Science Foundation of China (31571706).
Footnotes
The authors declare no competing financial interests.
Author Contributions C.M. and X.L. conceived and designed the experiments. X.L., Z.X., Z.Z. and S.D performed the experiments. X.L., Z.X. and K.H. analyzed the data. C.M., J.T., J.S., B.Y. and T.F contributed materials. X.L. and C.M. wrote the paper. All authors reviewed the manuscript.
References
- Basunanda P. et al. Comparative mapping of quantitative trait loci involved in heterosis for seedling and yield traits in oilseed rape (Brassica napus L.). TAG. Theoretical and applied genetics. Theoretische und angewandte Genetik 120, 271–281, doi: 10.1007/s00122-009-1133-z (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber N. et al. Seedling development in a Brassica napus diversity set and its relationship to agronomic performance. TAG. Theoretical and applied genetics. Theoretische und angewandte Genetik 125, 1275–1287, doi: 10.1007/s00122-012-1912-9 (2012). [DOI] [PubMed] [Google Scholar]
- Liu S. et al. Genome-wide analysis of ZmDREB genes and their association with natural variation in drought tolerance at seedling stage of Zea mays L. PLoS Genet 9, e1003790, doi: 10.1371/journal.pgen.1003790 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adolf V. I., Jacobsen S.-E. & Shabala S. Salt tolerance mechanisms in quinoa (Chenopodium quinoa Willd.). Environmental and Experimental Botany 92, 43–54 (2013). [Google Scholar]
- Ahmad M., Shahzad A., Iqbal M., Asif M. & Hirani A. H. Morphological and molecular genetic variation in wheat for salinity tolerance at germination and early seedling stage. Australian Journal of Crop Science 7, 66 (2013). [Google Scholar]
- Hatzig S. V. et al. Genome-wide association mapping unravels the genetic control of seed germination and vigor in Brassica napus. Frontiers in plant science 6, doi: 10.3389/fpls.2015.00221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Körber N. et al. Seedling development traits in Brassica napus examined by gene expression analysis and association mapping. BMC plant biology 15, 1 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao H. et al. A transposable element in a NAC gene is associated with drought tolerance in maize seedlings. Nature Communications 6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace J., Gardner C., Romay C., Ganapathysubramanian B. & Lubberstedt T. Genome-wide association analysis of seedling root development in maize (Zea mays L.). BMC genomics 16, 47, doi: 10.1186/s12864-015-1226-9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P. C. et al. ArabidopsisYL1/BPG2 Is Involved in Seedling Shoot Response to Salt Stress through ABI4. Scientific reports 6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer S. L. et al. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290, 2110–2113 (2000). [DOI] [PubMed] [Google Scholar]
- Lu S. X. et al. CCA1 and ELF3 Interact in the Control of Hypocotyl Length and Flowering Time in Arabidopsis. Plant Physiology 158, 1079–1088, doi: 10.1104/pp.111.189670 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusinow D. A. et al. The ELF4–ELF3–LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475, 398–402, doi: 10.1038/nature10182 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X. et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nature genetics 42, 961–967, doi: 10.1038/ng.695 (2010). [DOI] [PubMed] [Google Scholar]
- Huang X. et al. Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nature genetics 44, 32–39, doi: 10.1038/ng.1018 (2012). [DOI] [PubMed] [Google Scholar]
- Li H. et al. Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat Genet 45, 43–50, doi: 10.1038/ng.2484 (2013). [DOI] [PubMed] [Google Scholar]
- Ganal M. W., Altmann T. & Roder M. S. SNP identification in crop plants. Curr Opin Plant Biol 12, 211–217, doi: 10.1016/j.pbi.2008.12.009 (2009). [DOI] [PubMed] [Google Scholar]
- Wei G. et al. A transcriptomic analysis of superhybrid rice LYP9 and its parents. Proceedings of the National Academy of Sciences 106, 7695–7701, doi: 10.1073/pnas.0902340106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G. et al. Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. The Plant cell 22, 17–33, doi: 10.1105/tpc.109.072041 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. et al. Genomic, Transcriptomic, and Phenomic Variation Reveals the Complex Adaptation of Modern Maize Breeding. Molecular Plant 8, 871–884, doi: 10.1016/j.molp.2015.01.016 (2015). [DOI] [PubMed] [Google Scholar]
- Raman H. et al. SNP markers-based map construction and genome-wide linkage analysis inBrassica napus. Plant biotechnology journal 12, 851–860, doi: 10.1111/pbi.12186 (2014). [DOI] [PubMed] [Google Scholar]
- Chalhoub B. et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950–953, doi: 10.1126/science.1253435 (2014). [DOI] [PubMed] [Google Scholar]
- Wang N. et al. Association Mapping of Flowering Time QTLs and Insight into Their Contributions to Rapeseed Growth Habits. Frontiers in plant science 7, 338, doi: 10.3389/fpls.2016.00338 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen G. & Saedler H. Plant biology: floral quartets. Nature 409, 469–471 (2001). [DOI] [PubMed] [Google Scholar]
- Pruneda-Paz J. L. et al. A genome-scale resource for the functional characterization of Arabidopsis transcription factors. Cell reports 8, 622–632 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S. & Lashbrook C. C. Stamen abscission zone transcriptome profiling reveals new candidates for abscission control: enhanced retention of floral organs in transgenic plants overexpressing Arabidopsis ZINC FINGER PROTEIN2. Plant Physiology 146, 1305–1321 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose J. K., Braam J., Fry S. C. & Nishitani K. The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant and Cell Physiology 43, 1421–1435 (2002). [DOI] [PubMed] [Google Scholar]
- Wang Y. et al. Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis. Plant Physiology 148, 1201–1211 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez R. A. et al. Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proceedings of the National Academy of Sciences 105, 4939–4944 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron J. M. et al. Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proceedings of the National Academy of Sciences 109, 3167–3172 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T., Suzuki K., Fujimura T. & Shinshi H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiology 140, 411–432 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghassemian M. et al. Integrative analysis of transcript and metabolite profiling data sets to evaluate the regulation of biochemical pathways during photomorphogenesis. Archives of Biochemistry and Biophysics 448, 45–59 (2006). [DOI] [PubMed] [Google Scholar]
- Aranzana M. J. et al. Genome-wide association mapping in Arabidopsis thaliana identifies previously known genes responsible for variation in flowering time and pathogen resistance. PLoS Genetics preprint, e60, doi: 10.1371/journal.pgen.0010060.eor (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. et al. Genome-wide association study dissects the genetic architecture of seed weight and seed quality in rapeseed (Brassica napus L.). DNA research: an international journal for rapid publication of reports on genes and genomes 21, 355–367, doi: 10.1093/dnares/dsu002 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V. et al. Genome-wide association mapping of salinity tolerance in rice (Oryza sativa). DNA Research 22, 133–145, doi: 10.1093/dnares/dsu046 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L. et al. Genome-wide association study reveals the genetic architecture of flowering time in rapeseed (Brassica napus L.). DNA Research, dsv035, doi: 10.1093/dnares/dsv035 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Famoso A. N. et al. Genetic architecture of aluminum tolerance in rice (Oryza sativa) determined through genome-wide association analysis and QTL mapping. PLoS Genet 7, e1002221, doi: 10.1371/journal.pgen.1002221 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y. et al. Joint linkage-linkage disequilibrium mapping is a powerful approach to detecting quantitative trait loci underlying drought tolerance in maize. Proceedings of the National Academy of Sciences of the United States of America 107, 19585–19590, doi: 10.1073/pnas.1006105107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadic E. et al. Combined linkage and association mapping of flowering time in Sunflower (Helianthus annuus L.). TAG. Theoretical and applied genetics. Theoretische und angewandte Genetik 126, 1337–1356, doi: 10.1007/s00122-013-2056-2 (2013). [DOI] [PubMed] [Google Scholar]
- Mir R. R. et al. Genetic dissection of grain weight in bread wheat through quantitative trait locus interval and association mapping. Molecular Breeding 29, 963–972, doi: 10.1007/s11032-011-9693-4 (2012). [DOI] [Google Scholar]
- Tian F. et al. Genome-wide association study of leaf architecture in the maize nested association mapping population. Nature genetics 43, 159–162, doi: 10.1038/ng.746 (2011). [DOI] [PubMed] [Google Scholar]
- Kover P. X. et al. A Multiparent Advanced Generation Inter-Cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS genetics 5, e1000551, doi: 10.1371/journal.pgen.1000551 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L. et al. Genome-wide association analysis and differential expression analysis of resistance to Sclerotinia stem rot in Brassica napus. Plant Biotechnol J 14, 1368–1380, doi: 10.1111/pbi.12501 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon Y. et al. A novel Arabidopsis MYB-like transcription factor, MYBH, regulates hypocotyl elongation by enhancing auxin accumulation. Journal of experimental botany 64, 3911–3922, doi: 10.1093/jxb/ert223 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. et al. Trp-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes & development 16, 3100–3112 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas P. Dual Role of TOC1 in the Control of Circadian and Photomorphogenic Responses in Arabidopsis. The Plant Cell Online 15, 223–236, doi: 10.1105/tpc.006734 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche F., Verbelen J.-P. & Van Der Straeten D. Of light and length: Regulation of hypocotyl growth inArabidopsis. BioEssays 27, 275–284, doi: 10.1002/bies.20199 (2005). [DOI] [PubMed] [Google Scholar]
- Murai N. Review: Plant Growth Hormone Cytokinins Control the Crop Seed Yield. American Journal of Plant Sciences 05, 2178–2187, doi: 10.4236/ajps.2014.514231 (2014). [DOI] [Google Scholar]
- Bendix C., Marshall C. M. & Harmon F. G. Circadian Clock Genes Universally Control Key Agricultural Traits. Molecular plant 8, 1135–1152, doi: 10.1016/j.molp.2015.03.003 (2015). [DOI] [PubMed] [Google Scholar]
- Li F. et al. Characterization of sucrose transporter alleles and their association with seed yield-related traits in Brassica napus L. BMC plant biology 11, 168 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X. et al. Unravelling the complex trait of harvest index in rapeseed (Brassica napus L.) with association mapping. BMC Genomics 16, 379, doi: 10.1186/s12864-015-1607-0 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi T. et al. Brassica napus root mutants insensitive to exogenous cytokinin show phosphorus efficiency. Plant and Soil 358, 61–74, doi: 10.1007/s11104-012-1219-2 (2012). [DOI] [Google Scholar]
- Chalhoub B. et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950–953 (2014). [DOI] [PubMed] [Google Scholar]
- Nei M. & Takezaki N. Estimation of genetic distances and phylogenetic trees from DNA analysis. Proc 5th World Cong Genet Appl Livestock Prod 21, 405–412 (1983). [Google Scholar]
- Liu K. & Muse S. V. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21, 2128–2129 (2005). [DOI] [PubMed] [Google Scholar]
- Pritchard J. K., Stephens M. & Donnelly P. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno G., Regnaut S. & Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular ecology 14, 2611–2620 (2005). [DOI] [PubMed] [Google Scholar]
- Hardy O. J. & Vekemans X. SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Molecular ecology notes 2, 618–620 (2002). [Google Scholar]
- Yu J. et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nature genetics 38, 203–208 (2006). [DOI] [PubMed] [Google Scholar]
- Price A. L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nature genetics 38, 904–909, doi: 10.1038/ng1847 (2006). [DOI] [PubMed] [Google Scholar]
- Bradbury P. J. et al. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635 (2007). [DOI] [PubMed] [Google Scholar]
- Benjamini Y. & Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological), 289–300 (1995). [Google Scholar]
- Trapnell C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature Protocols 7, 562–578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O. et al. mapman: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant Journal for Cell & Molecular Biology 37, 914–939 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.