Abstract
Monitoring of neuronal activity within circuits facilitates integrated responses and rapid changes in behavior. We have identified a system in Caenorhabditis elegans where neuropeptide expression is dependent on the ability of the BAG neurons to sense carbon dioxide. In C. elegans, CO2 sensing is predominantly coordinated by the BAG-expressed receptor-type guanylate cyclase GCY-9. GCY-9 binding to CO2 causes accumulation of cyclic GMP and opening of the cGMP-gated TAX-2/TAX-4 cation channels; provoking an integrated downstream cascade that enables C. elegans to avoid high CO2. Here we show that cGMP regulation by GCY-9 and the PDE-1 phosphodiesterase controls BAG expression of a FMRFamide-related neuropeptide FLP-19 reporter (flp-19::GFP). This regulation is specific for CO2-sensing function of the BAG neurons, as loss of oxygen sensing function does not affect flp-19::GFP expression. We also found that expression of flp-19::GFP is controlled in parallel to GCY-9 by the activity-dependent transcription factor CREB (CRH-1) and the cAMP-dependent protein kinase (KIN-2) signaling pathway. We therefore show that two parallel pathways regulate neuropeptide gene expression in the BAG sensory neurons: the ability to sense changes in carbon dioxide and CREB transcription factor. Such regulation may be required in particular environmental conditions to enable sophisticated behavioral decisions to be performed.
C. elegans performs multiple sensory modalities when navigating its environment. Such behaviors include sensing and responding to changes in temperature, atmospheric gases, food and mating1,2,3,4,5,6,7,8. Coordination of these complex behavioral responses requires constant monitoring of the status of the nervous system. Neuropeptide and neuropeptide receptor expression levels have previously been linked to the activity of neurons4,9,10,11,12,13. For example, insulin and neuropeptide signaling have been shown to report feeding status14. More recently, the regulation of expression of the serpentine receptor srh-234 was linked with starvation, and dependent on the neuropeptide Y receptor NPR-1 and insulin-like growth factor receptor DAF-212.
Despite having a small nervous system, the C. elegans genome encodes many neuropeptides15. These neuropeptides are classified into three main families: insulin-like family, neuropeptide-like protein family (NLPs) and FMRFamide-related peptides (FLPs). For FLP neuropeptides, the function for only a few are known suggesting that they work redundantly or are involved in fine-tuning of the nervous system in combination with neurotransmitter systems. However, certain functions have been elucidated for FLPs: FLP-11 has been identified as a regulator of a sleep-like state16, FLP-13 acts in the ALA neurons to regulate quiescence after heat-stress17, FLP-10 and FLP-17 inhibit egg laying18 and FLP-21 regulates social feeding behavior through the NPR-1 receptor19.
The C. elegans nervous system consists of 302 neurons, some of which function in non-overlapping circuits to regulate distinct behaviors. The BAG neurons for example are major regulators of acute CO2 avoidance behavior5,6, the regulation of pharyngeal pumping rate when animals are exposed to high levels of CO220, and the control of egg laying18. The BAG neurons also act in conjunction with the URX neurons to coordinate behavioral responses to changes in O2 gradients. The BAG and URX neurons are activated when O2 levels drop and increase respectively3,4,21,22,23. However, the relationship between the BAG and URX neurons is not exclusive to O2 sensing: BAG-ablated animals live longer, while ablation of the URX neurons results in shorter lifespan, and these physiological changes are regulated by O2-sensing guanylate cyclases expressed in these neurons24. With such divergent and sophisticated roles for the BAG and URX neurons, prioritization and integration of information to guide behavioral responses may require modular regulation of neuropeptide expression.
Here we describe a system where the activity of the BAG neurons is crucial for the expression of a transcriptional reporter for the FLP-19 neuropeptide (flp-19::GFP). Furthermore, we show that the expression of flp-19::GFP is regulated via two parallel modules: 1) cGMP signaling through GCY-9 and PDE-1 and 2) the CREB transcription factor.
Results
flp-19::GFP Expression in the BAG Neurons Requires Cilia Function
It has previously been shown that neuropeptide expression is highly plastic, and can be controlled by neuronal activity4,10,13. The flp-19::GFP transcriptional reporter drives expression in the BAG, URX, AIN, AWA and HSN neurons in the hermaphrodite25. Our previous studies showed that flp-19::GFP expression in the BAG neurons is exquisitely sensitive to perturbations in parallel transcriptional pathways that control BAG cell fate and function26,27,28. We hypothesized that neuronal activity may underpin the transcriptional regulation of flp-19::GFP in the BAG neurons. We therefore tested if the expression of flp-19::GFP is compromised when the activity of the BAG neurons is reduced or abolished.
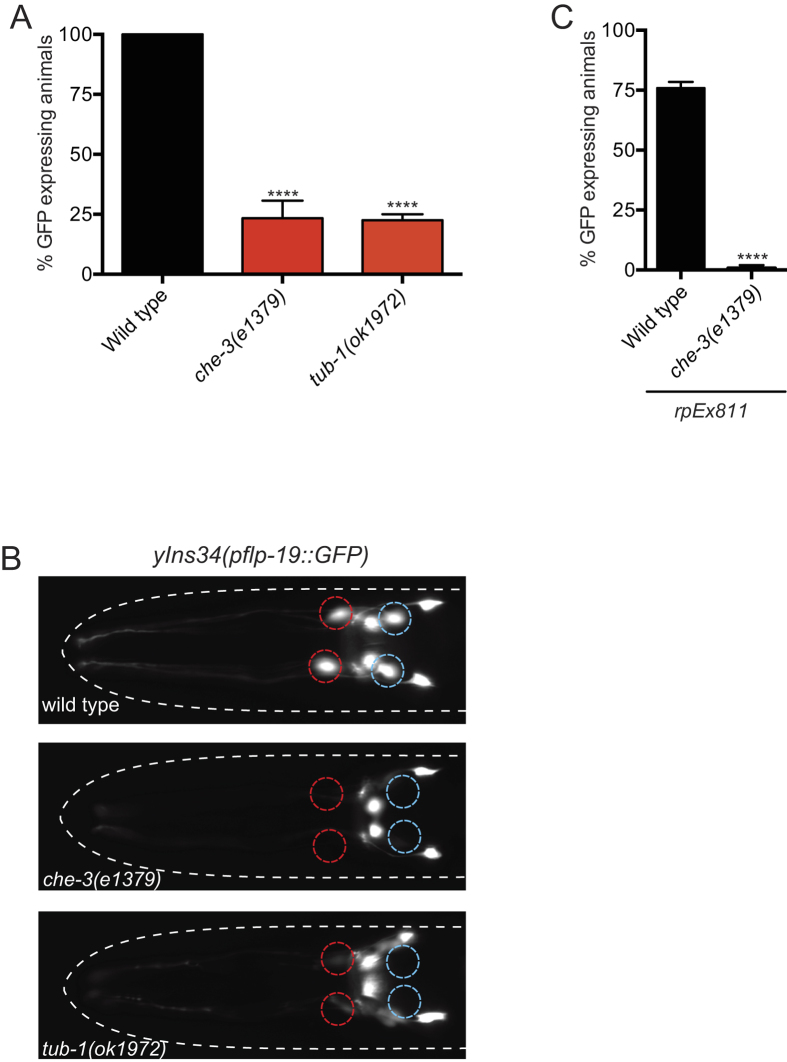
As the BAG neurons are ciliated, we first examined if disruption of cilia structure, which is required for several behaviors in C. elegans7,29,30, would affect flp-19::GFP expression. We crossed the flp-19::GFP reporter (ynIs34) with a mutant of che-3, which encodes a dynein that is required for structural integrity of sensory cilia. We observed that che-3(e1379) mutant animals exhibit strong defects in the expression of flp-19::GFP in the ciliated BAG and AWA neurons but not in the non-ciliated neurons (Fig. 1A,B). To verify this regulation, we crossed che-3(e1379) mutant animals with an independent flp-19::GFP reporter (rpEx811) and observed a similar phenotype (Fig. 1C). We next asked if disturbance of cilia transport would produce a similar effect on flp-19::GFP expression. TUB-1, homolog of TUBBY in mammals, is required for correct localization of G protein coupled receptors to cilia31,32,33. We found that removal of TUB-1 function phenocopied the che-3 mutant loss of flp-19::GFP expression in the BAG neurons (Fig. 1A,B), indicating that correct cilia function is required for expression of flp-19::GFP.
Figure 1. Cilia Function is Required for Expression of flp-19::GFP in the BAG Gas-sensing Neurons.
(A) Quantification of the expression of the flp-19::GFP fluorescent reporter (ynIs34) in wild type, che-3(e1379) and tub-1(ok1972) mutant animals. n > 50. ****P < 0.0001. See Materials and Methods for neuronal scoring criteria used. (B) Micrographs show representative pictures of flp-19::GFP(ynIs34) expression in wild type, che-3(e1379) and tub-1(ok1972) strains. BAG positions are marked with red dashed circles. Note - expression of flp-19::GFP is also lost in the ciliated AWA neurons in che-3(e1379) and tub-1(ok1972) mutant animals (blue dotted circles). Anterior to the left. Scale bar = 20 μm. (C) Quantification of the expression of the flp-19::GFP reporter (rpEx811) in wild type and che-3(e1379) backgrounds. These data confirm the results shown in Fig. 1A. n > 50. ****P < 0.0001.
GCY-9 Regulates flp-19::GFP Expression Cell-autonomously in the BAG Neurons
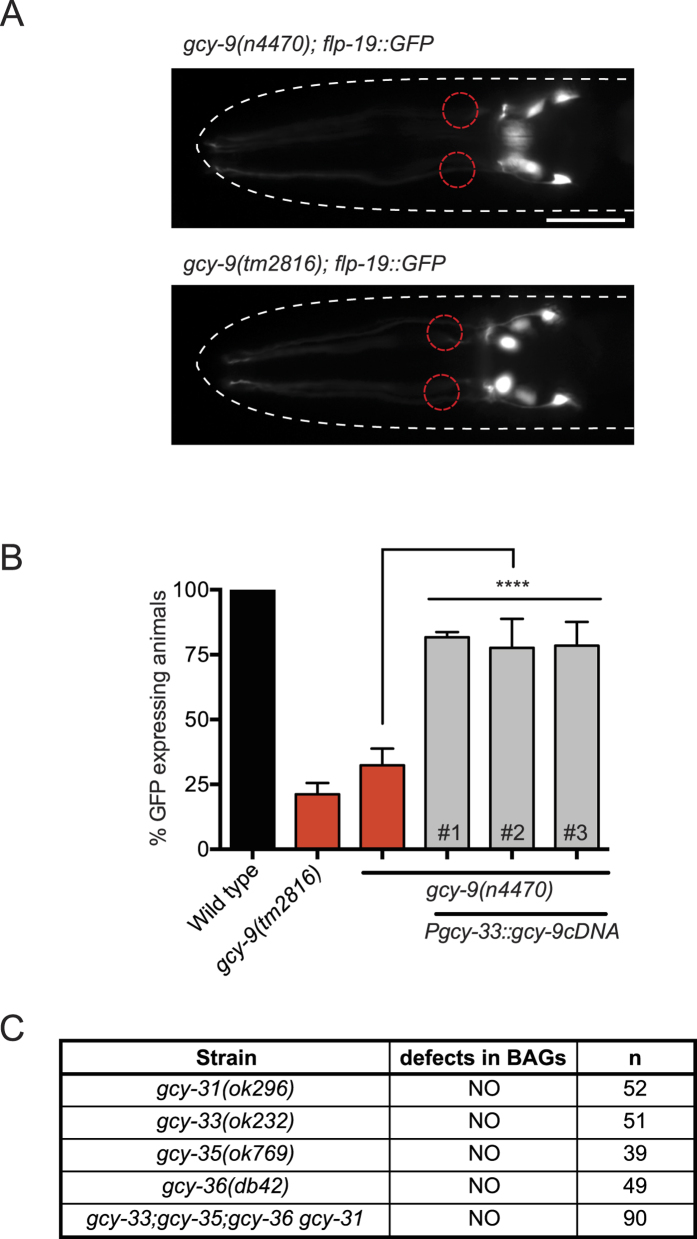
Acute responses to CO2 are regulated by a neuronal circuit that includes the BAG neurons. The BAG neurons sense carbon dioxide through the GCY-9 receptor-type guanylate cyclase, homolog of the human GC-D26,34,35. As such, gcy-9 mutants are unable to sense and respond to changes in CO2 concentration34. We asked whether the CO2-sensing function of the BAG neurons is required for flp-19::GFP expression. To this end, we crossed two independent deletion alleles of gcy-9 (n4470, tm2816) with the flp-19::GFP reporter and observed a reduced number of animals expressing GFP in the BAG neurons (Fig. 2A,B). When we resupplied gcy-9 cDNA driven by a BAG specific gcy-33 promoter into gcy-9(n4470) mutant animals, flp-19::GFP expression was restored (Fig. 2B). The BAG neurons are also involved in sensing downshifts in O2 concentration, through expression of the soluble guanylate cyclases GCY-31 and GCY-3322. To ask whether O2-sensing function of the BAG neurons is also required to regulate flp-19::GFP expression, we crossed gcy-31(ok296) and gcy-33(ok232) mutants with the flp-19::GFP reporter. We found no detectable change in expression of flp-19::GFP when BAG O2-sensing function was ablated (Fig. 2C). As it has been previously shown that the URX and BAG communicate with each other23,24, we asked whether removal of O2-sensing from the URX neurons would affect the expression of flp-19::GFP. We therefore crossed the flp-19::GFP reporter into gcy-35(ok769) and gcy-36(db42) mutant strains, in which soluble guanylate cyclases required for URX O2-sensing function are mutated3,4,22,36. However, we observed no defect in the expression of flp-19::GFP in the BAG or URX neurons (Fig. 2C and data not shown). Furthermore, animals which lack O2 sensing function in both the BAG and URX neurons (gcy-31 gcy-36; gcy-33; gcy-35 quadruple mutant) exhibit wild type expression of flp-19::GFP (Fig. 2C). Taken together, these data show that CO2 sensing function, and not O2 sensing function, regulates the expression of the flp-19::GFP reporter in the BAG neurons.
Figure 2. Autonomous CO2 Sensing is Required for flp-19::GFP Expression in the BAG Neurons.
(A) gcy-9(n4470) and gcy-9(tm2816) mutants exhibit reduced expression of the flp-19::GFP reporter in the BAG neurons. Micrographs show representative images of gcy-9 mutant hermaphrodites expressing the flp-19::GFP reporter (compare to Fig. 1B, top panel). BAG positions are marked with red dashed circles. Anterior to the left. Scale bar = 20 μm. (B) Quantification of flp-19::GFP expression in gcy-9(n4470) and gcy-9(tm2816) mutant animals. Transgenic expression of gcy-9 cDNA under the control of a BAG-specific gcy-33 promoter rescues gcy-9(n4470) mutant phenotype. #1-3 indicate independent transgenic rescue lines. See Materials and Methods for neuronal scoring criteria used. n > 50. ****P < 0.0001. (C) Table showing the effect of loss of O2-sensing guanylate cyclases on flp-19::GFP expression in the BAG neurons. BAG guanylate cyclase functional nulls gcy-31(ok296) and gcy-33(ok232) do not exhibit defects in expression of flp-19::GFP in the BAG neurons. Mutants for URX guanylate cyclases gcy-35(ok769) and gcy-36(db42) do not exhibit defects in expression of flp-19::GFP in the BAG neurons. The quadruple gcy-31 gcy-36; gcy-33; gcy-35 also does not exhibit any defects in BAG expression suggesting that flp-19::GFP expression in the BAG neurons is dependent on CO2 sensing and not O2 sensing.
To ask whether CO2 sensing has a general effect on neuropeptide expression in the BAG neurons, we analyzed the expression of other neuropeptides in gcy-9(n4470) mutant animals. We crossed the gcy-9(n4470) mutant with reporters for flp-17(ynIs64) and flp-13(ynIs37) and observed no change of expression compared to wild type (Figure S1A). Furthermore we tested other reporters expressed in the BAG neurons (soluble guanylate cyclases gcy-31(rpIs29) and gcy-33(rpIs7) and the transcription factor egl-13(rpIs32)) in the gcy-9 mutant background and we observed no change of expression (Figure S1A). Therefore, the regulation of flp-19::GFP expression by GCY-9 is somewhat specific.
cGMP Levels Regulate flp-19::GFP Expression in the BAG Neurons
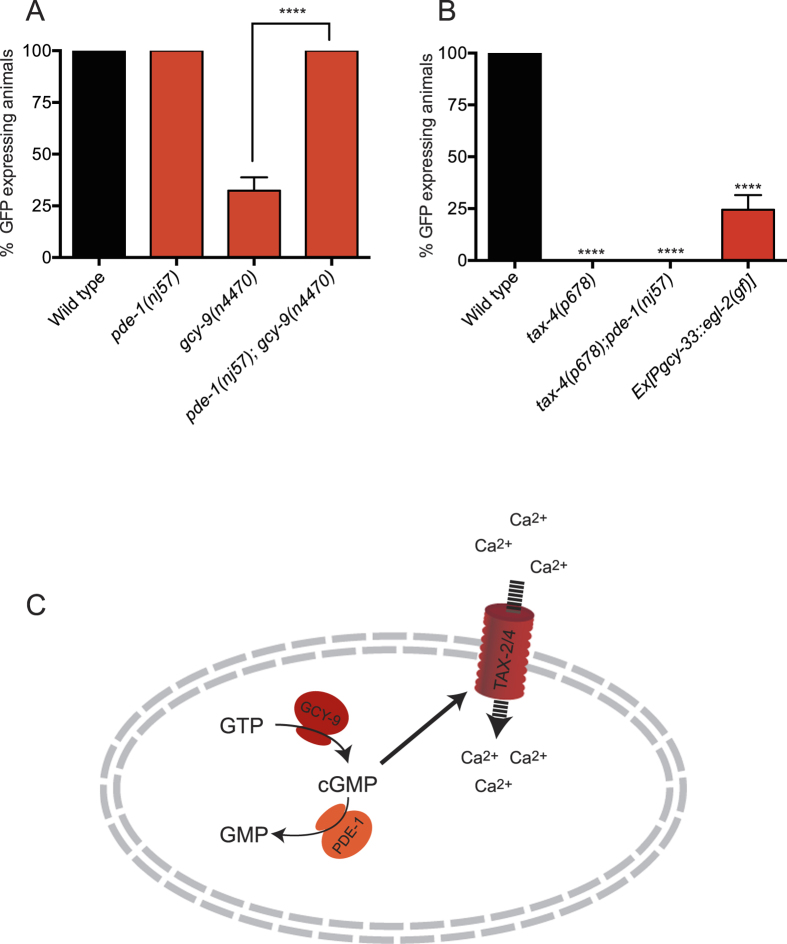
gcy-9 encodes a receptor-type guanylate cyclase. The role of these enzymes is to generate cGMP for gating downstream cyclic nucleotide-gated TAX-2/TAX-4 cation channels. As such GCY-9 regulates the activity of the BAG neurons through the control of cGMP levels. We speculated therefore that cGMP is a key regulator of flp-19::GFP expression in the BAG neurons. In order to test this hypothesis, we genetically manipulated cGMP levels. Phosphodiesterases are enzymes that catalyse the breakdown of cGMP to GMP and it has previously been shown that the phosphodiesterase PDE-1 is expressed in the BAG neurons34. We therefore asked whether the predicted increase in cGMP levels in pde-1 mutant animals would affect flp-19::GFP expression. We crossed the pde-1(nj57) mutant with the flp-19::GFP reporter and observed that BAG expression of flp-19::GFP was unaffected (Fig. 3A). Next we tested if loss of pde-1, and therefore increase of cGMP in the BAG neurons, was sufficient to derepress flp-19::GFP expression in the gcy-9 mutant. We therefore examined flp-19::GFP expression in the pde-1(nj57); gcy-9(n4470) double mutant and found that expression in the BAG neurons was fully restored (Fig. 3A). This suggests that reduced levels of cGMP in the BAG neurons causes transcriptional downregulation of flp-19::GFP.
Figure 3. cGMP Levels Regulate flp-19::GFP Expression in the BAG Neurons.
(A) Quantification of the flp-19::GFP reporter suggests that the guanylate cyclase GCY-9 and the phosphodiesterase PDE-1 exhibit opposing regulation of cGMP levels in the BAG neurons. The pde-1(nj57) mutant exhibits wild type expression of flp-19::GFP in the BAG neurons while gcy-9(n4470) mutants exhibit a strong defect in expression. In gcy-9(n4470); pde-1(nj57) double mutant animals, flp-19::GFP expression in the BAG neurons is restored, suggesting that cGMP levels in the BAG neurons regulate flp-19::GFP expression. See Materials and Methods for neuronal scoring criteria used. n > 50. ****P < 0.0001. (B) flp-19::GFP in the BAG neurons is completely abrogated in tax-4(p678) mutant animals. Furthermore, the tax-4(p678) mutant defect in flp-19::GFP expression cannot be rescued by mutation in pde-1 indicating that tax-4 acts downstream of cGMP signaling. Expression of an constitutively active form of EGL-2 in the BAGs also reduces flp-19::GFP expression indicating that flp-19::GFP expression is activity dependent. See Materials and Methods for neuronal scoring criteria used. n > 50. ****P < 0.0001. (C) Schematic model of cGMP regulation in the BAG neurons. GCY-9 converts GTP into cGMP, while PDE-1 catalyzes the conversion of cGMP into GMP. cGMP opens the TAX-2/TAX-4 channels leading to an influx of calcium and activation of the neuron.
Activation of GCY-9 by CO2 normally triggers the conversion of cGMP from GTP35. Subsequently, cGMP opens the cyclic nucleotide-gated channels TAX-2/TAX-4, through which the neuron is activated by calcium influx5,6. We therefore hypothesized that GCY-9-mediated regulation of the flp-19::GFP reporter was through this canonical pathway. Indeed, we found that in tax-4(p678) mutant animals flp-19::GFP expression is undetectable in the BAG neurons (Fig. 3B). In addition, we observed loss of flp-19::GFP expression in the URX O2-sensing neurons of tax-4(p678) mutant animals, suggesting that similar mechanisms of regulation may exist in these functionally-related neurons. However, we did not observe loss of flp-19::GFP expression in the gcy-35 or gcy-36 mutants or the quadruple gcy-31 gcy-36; gcy-33; gcy-35 mutant strain (data not shown), suggesting that another pathway of regulation, independent of O2 sensing, regulates the expression of flp-19::GFP through the TAX-2/TAX-4 channels in the URX neurons. To confirm that the BAG neurons are present in tax-4 mutant animals, we crossed tax-4(p678) mutant animals with an independent reporter for BAG neurons (egl-13::GFP), and observed no loss of expression (Figure S1B). Furthermore, we found that loss of pde-1 was not able to restore the tax-4(p678) mutant flp-19::GFP expression, indicating that tax-4 acts downstream of cGMP regulation, and that the channels are necessary for the expression of flp-19::GFP regardless of the levels of cGMP (Fig. 3B).
The data we have presented suggest that activity of the BAG neurons is important for the expression of flp-19::GFP. To reinforce this assertion, we inactivated the BAG neurons by expressing a constitutively-active EGL-2(GF) potassium channel using the gcy-33 promoter4,37,38. We found that animals carrying the Pgcy-33::egl-2(gf) transgene exhibit a decrease in the expression of flp-19::GFP in the BAGs (Fig. 3B).
Taken together, our data show that GCY-9 and the downstream cGMP-regulated TAX-2/TAX-4 channels are required for the expression of flp-19::GFP in the BAG neurons. These data suggest that the conversion of GTP to cGMP by GCY-9 triggers opening of the TAX-2/TAX-4 channels, resulting in calcium influx and that this change in activity regulates the transcription of the neuropeptide reporter flp-19::GFP (Fig. 3C).
flp-19::GFP Expression Does not Require Neuropeptide or Neurotransmitter Signaling
We have shown that the expression of flp-19::GFP is sensitive to GCY-9 and TAX-4-controlled activity. We next asked whether the expression of flp-19::GFP was regulated by neuropeptide and neurotransmitter signaling, through either an autocrine or paracrine fashion. We first tested whether flp-19::GFP expression was affected by abolishing neuropeptide signaling. To this end, we crossed the flp-19::GFP reporter with two mutants that do not have proper neuropeptide signaling: egl-3(nr2090), responsible for maturation of neuropeptides, and unc-31(e169), involved in dense core vesicle function39,40,41. The expression of flp-19::GFP in the BAG neurons in egl-3 and unc-31 mutants was unchanged when compared to wild type (Table 1). The BAG neurons are glutamatergic as they express the vesicular glutamate transporter EAT-442,43. We found that the glutamatergic function of the BAG neurons is not required for the regulation of flp-19::GFP as expression is unchanged in an eat-4(ky5) mutant (Table 1). Furthermore, we asked if neurotransmitter release through synaptic vesicle exocytosis was required for flp-19::GFP expression. We crossed flp-19::GFP with the unc-13(e1091) mutant, defective in neurotransmission due to compromised synaptic vesicle fusion and the snb-1(md247) mutant, defective in synaptic transmission44,45,46. We found that unc-13 and snb-1 mutant animals exhibit wild type expression of flp-19::GFP in the BAG neurons (Table 1). These data suggest that the regulation of flp-19::GFP in the BAG neurons occurs through a BAG-intrinsic mechanism.
Table 1. Genes Tested for Expression of flp-19::GFP in the BAG Neurons.
| Strain | Defects in BAGs | n |
|---|---|---|
| Neuropeptide | ||
| egl-3(nr2090) | NO | 62 |
| unc-31(e169) | NO | 56 |
| Neurotransmitter | ||
| unc-13(e1091) | NO | 51 |
| eat-4(ky5) | NO | 58 |
| snb-1(md247) | NO | 67 |
| Activity-related genes | ||
| crh-1(tz2) | YES | 111 |
| crh-1(n3315) | YES | 75 |
| kin-2(ce179) | YES | 193 |
| jun-1(gk557) | NO | 90 |
| egl-4(n479) | NO | 41 |
| cmk-1(ok287) | NO | 58 |
| ckk-1(ok1033) | NO | 51 |
| rgs-3(ok2288) | NO | 51 |
We crossed the flp-19::GFP reporter with mutants for neuropeptide function, neurotransmission and synaptic regulators and other activity-related genes. From the screen, we found that crh-1 and kin-2 are necessary for the proper expression of the flp-19::GFP reporter.
CRH-1/CREB, an Activity-dependent Transcription Factor Acts in Parallel to GCY-9 to Regulate flp-19::GFP Expression in the BAG Neurons
In order to better understand the mechanism through which flp-19::GFP is regulated, we studied mutants of various genes involved in activity-dependent expression in the nervous system (Table 1). Our screen found that both crh-1 and kin-2 mutants display defects in the expression of the flp-19::GFP reporter in the BAG neurons. CRH-1 is the C. elegans homolog of CREB and functions in neurons to regulate various behaviors. CRH-1 regulates lifespan, tap habituation and it has been linked with the control of thermotaxis behavior from the AFD neurons47,48,49. KIN-2 is the homolog of the regulatory subunit of cAMP-dependent protein kinase (PKA) and it can regulate CREB activity through a cascade of phosphorylation events50,51,52,53,54. We found that two independent null alleles of crh-1 (tz2 and n3315) exhibit reduced expression of flp-19::GFP in the BAG neurons (Fig. 4A). In addition, kin-2(ce179) mutants show a similar phenotype to crh-1 mutants, and the crh-1(tz2); kin-2(ce179) double mutant is not significantly different from either single mutant, suggesting that they function in the same genetic pathway (Fig. 4A). We speculated that CRH-1 might be the downstream effector in the GCY-9 cascade. To ask whether gcy-9 and crh-1 act in the same genetic pathway to control flp-19::GFP expression we constructed a gcy-9(n4470); crh-1(tz2) double mutant. Surprisingly, we found that these double mutant animals exhibited complete abrogation of flp-19::GFP expression (Fig. 4B). We confirmed that the BAG neurons are present in the gcy-9(n4470); crh-1(tz2) double mutant by examining a flp-13::GFP transgene (Figure S1C). Together, these data suggest that gcy-9 and crh-1 act in two separate genetic pathways to regulate flp-19::GFP expression (Fig. 5).
Figure 4. CREB/CRH-1 Acts in Parallel to GCY-9 to Regulate flp-19::GFP Expression in the BAG Neurons.
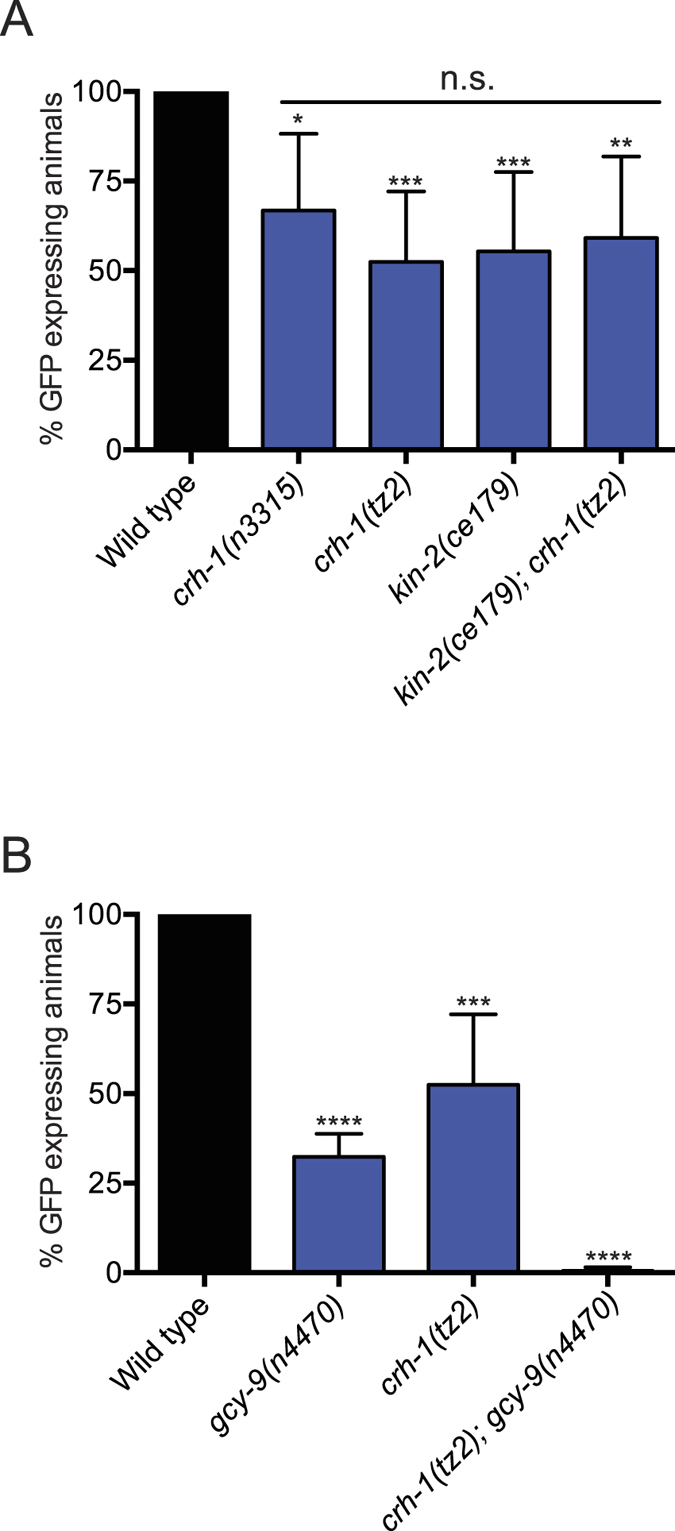
(A) Quantification of the defects in flp-19::GFP expression in the BAG neurons in crh-1 mutant alleles tz2 and n3315 and the kin-2(ce179) mutant. Both crh-1 and kin-2 mutants show a similar penetrance of defects in the expression of flp-19::GFP in the BAG neurons. Furthermore, the kin-2(ce179); crh-1(tz2) double mutant defect is not significantly different to either single mutant, suggesting that they act in the same genetic pathway. See Materials and Methods for neuronal scoring criteria used. n > 50. *P < 0.05, **P < 0.01, ****P < 0.0001, n.s. = not significantly different from wild type. (B) Quantification of the defects in flp-19::GFP expression in the BAG neurons in gcy-9(n4470), crh-1(tz2) and the gcy-9(n4470); crh-1(tz2) double mutant. While crh-1 mutants show a partial defect in flp-19::GFP expression, in the crh-1(tz2); gcy-9(n4470) double mutant flp-19::GFP expression is undetectable. This indicates that crh-1 acts independently to the gcy-9 regulatory pathway. See Materials and Methods for neuronal scoring criteria used. n > 50. ***P < 0.001, ****P < 0.0001.
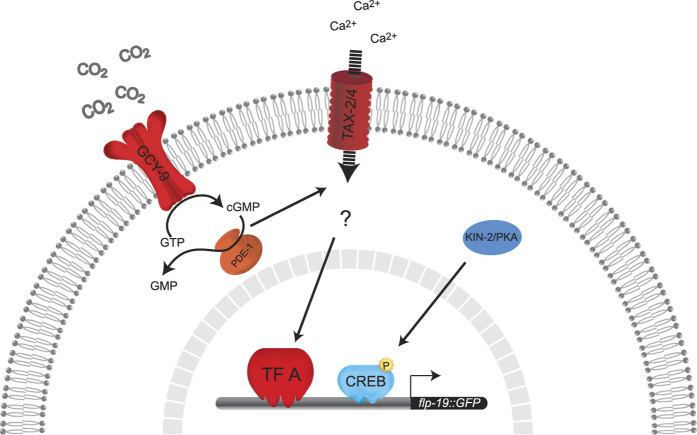
Figure 5. Expression of flp-19::GFP in the BAG Neurons is Controlled by Parallel Pathways.
Schematic representation of the proposed model of regulation of flp-19::GFP expression in BAG neurons. The GCY-9 receptor binds CO2 converting GTP into cGMP. cGMP triggers opening of the cyclic nucleotide-gated TAX-2/TAX-4 cation channels, allowing Ca2+ to enter the neuron, leading to regulation of an unknown transcription factor (TF A) that positively regulates the expression of flp-19::GFP. PDE-1 reduces the levels of cGMP, opposing the action of GCY-9. In parallel, CRH-1 regulates the expression of flp-19::GFP by a signaling pathway involving the cAMP-dependent protein kinase KIN-2.
In summary, we propose a model where flp-19::GFP expression is regulated by two parallel pathways: 1) GCY-9 regulation of the TAX-2/TAX-4 cation channels through the control of cGMP levels, signaling to an unknown transcription factor and 2) CRH-1/CREB regulation by the kinase KIN-2. likely through a CO2-independent signaling pathway (Fig. 5).
Discussion
Activity-dependent regulation is common occurrence in sensory neurons and can contribute in changes in behavior. Chemoreceptor genes can be regulated by different mechanisms such as developmental changes, neuronal activation, or in a paracrine fashion through pheromones. Transcriptional changes of chemoreceptors might be a strategy to modulate external responses. For example, the TAX-2/TAX-4 calcium channel regulates the expression of chemoreceptors such as STR-2 and SRD-1 in the AWC and ASI neurons respectively11,55. In addition, TRPV channels control the biosynthesis of serotonin through regulation of tryptophan hydroxylase expression in the ADF neurons56.
We have presented data to show that flp-19::GFP expression in the BAG neurons is regulated by two parallel pathways. The GCY-9 pathway controls flp-19::GFP expression by modulating the levels of cGMP, counterbalanced by the phosphodiesterase PDE-1. PDEs and GCYs are known to function together in other neurons to regulate activity. For example, gcy-12 and pde-2 control cGMP levels to determine body size in C. elegans57. Similarly, in the AFD neurons, the opposing roles of GCY-8 and PDE-2, control C. elegans thermotaxis behavior58. It might be possible that PDE-1 is involved in sensing the levels of CO2 in the BAG neurons; potentially to set a quantitative threshold or temporal window of TAX-2/TAX-4 channel opening. We have not identified the effector downstream the TAX-2/TAX-4 signaling, but we speculate that it may be a transcription factor regulated by calcium or calmodulin dependent activation. In parallel to GCY-9, CRH-1/CREB also controls the level of flp-19::GFP through the activity of the PKA kinase. As PKA is a cAMP regulated kinase, this may suggest that a cAMP-regulated pathway controls flp-19::GFP expression through CRH-1.
It is necessary to further study the implications of the two parallel pathways we have identified to understand the advantages they may provide in the natural habitat of C. elegans. Interestingly worms are attracted to CO2 as dauers while L4 larvae avoid CO25,6,34. Furthermore it has been shown how in juvenile infective stages of parasitic worms (related to C. elegans) BAG neurons are involved in the attraction to CO259. It might be possible that this change in the attraction/repulsion to CO2 is regulated by the expression of neuropeptides such as FLP-19. When worms are grown in low O2 levels, they become attracted to lower levels of O2, instead of being repelled. This change of behavior is due to changes in the expression level of guanylate cyclases that detect O2. It might be possible that similar adaptation occurs when worms are grown in hypercapnic conditions, and the transcriptional regulation of flp-19 could be involved in such adaptation. However, the role of flp-19 may not be directly related to CO2 sensing, but to other broader functions. Indeed, the BAG neurons are involved in lifespan regulation, as is the CRH-1 transcription factor, therefore it would be interesting to examine whether flp-19 mutant animals display defects in longevity.
flp-19 is expressed in a subset of neurons that are distinct in class and function: from oxygen (URX/BAG) and carbon dioxide sensing (BAG), chemotaxis (AWA), egg laying (HSN) or pheromone sensing (CEM). Similar means of flp-19 activity-dependent regulation maybe present in these other neurons through different molecular pathways, providing this neuropeptide with multiple layers of control that may be required in particular ephemeral habitats.
Methods
Strains used in this study
Strains were grown using standard growth conditions on NGM agar at 20 °C on Escherichia coli OP5060. Transgenic animals were created as previously described61. Strain information is detailed in Table S1.
Constructs and generation of transgenic worms
The Pgcy-33::gcy-9 cDNA rescue construct was generated by cloning the 1 kb gcy-33 promoter using HindIII and BamHI and gcy-9cDNA, using KpnI and NheI into pPD49.26 vector into multiple cloning sites 1 and 2 respectively (Fire Vector Kit). Transgenic animals were obtained through microinjection61. The construct was injected into young adult hermaphrodites as a simple array (20 ng/ul Pgcy-33::gcy-9 cDNA, 10 ng/ul myo-3::RFP as co-injection marker).
The BAG::egl-2(gf) construct was generated by cloning the 1 kb gcy-33 promoter using HindIII and BamHI and egl-2(gf)cDNA (kind gift of Mario De Bono) using KpnI into the pPD49.26 vector into multiple cloning sites 1 and 2 respectively (Fire Vector Kit). The construct was injected as simple array (50 ng/ul egl-2(gf) and 5 ng/ul myo-3::RFP as co-injection marker).
Microscopy
Worms were anesthetized in 20 mM NaN3 on 5% agarose on glass slides and images were taken using an upright fluorescence microscope (Zeiss, AXIO Imager M2) and ZEN software (version 2.0). Neuronal scoring: Neurons were given a numerical value according to their expression levels. Wild-type expression scored 1, decreased expression scored 0.5 and abolished expression scored 0. Percentage of GFP expressing animals was then correlated to the theoretical maximum score using the equation below.
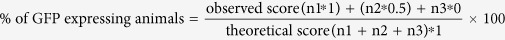 |
Statistical analysis
Statistical analysis was performed in GraphPad Prism 6 using one-way ANOVA with Newman-Keuls Multiple Comparison Test. Values are expressed as mean +/− s.d. Differences with a P value < 0.05 were considered significant.
Additional Information
How to cite this article: Romanos, T. R. et al. Control of Neuropeptide Expression by Parallel Activity-dependent Pathways in Caenorhabditis elegans. Sci. Rep. 7, 38734; doi: 10.1038/srep38734 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank members of Pocock laboratory for comments on the manuscript. Some strains used in this study were provided by the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440) and by Shohei Mitani at the National Bioresource Project (Japan). We also thank Mario De Bono for the gift of egl-2(gf) cDNA. This work was supported by a grant from the Lundbeck Foundation (R140–2013–13330) and veski innovation fellowship (VIF#23) to RP.
Footnotes
The authors declare no competing financial interests.
Author Contributions T.R.R. and J.G.P. performed the experiments, T.R.R. and R.P. wrote the manuscript.
References
- Hedgecock E. M. & Russell R. L. Normal and mutant thermotaxis in the nematode Caenorhabditis elegans. Proc Natl Acad Sci USA 72, 4061–4065 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori I. & Ohshima Y. Neural regulation of thermotaxis in Caenorhabditis elegans. Nature 376, 344–348 (1995). [DOI] [PubMed] [Google Scholar]
- Gray J. M. et al. Oxygen sensation and social feeding mediated by a C. elegans guanylate cyclase homologue. Nature 430, 317–322 (2004). [DOI] [PubMed] [Google Scholar]
- Cheung B. H., Cohen M., Rogers C., Albayram O. & de Bono M. Experience-dependent modulation of C. elegans behavior by ambient oxygen. Curr Biol 15, 905–917 (2005). [DOI] [PubMed] [Google Scholar]
- Bretscher A. J., Busch K. E. & de Bono M. A carbon dioxide avoidance behavior is integrated with responses to ambient oxygen and food in Caenorhabditis elegans. Proc Natl Acad Sci USA 105, 8044–8049, doi: 10.1073/pnas.0707607105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallem E. A. & Sternberg P. W. Acute carbon dioxide avoidance in Caenorhabditis elegans. Proc Natl Acad Sci USA 105, 8038–8043, doi: 10.1073/pnas.0707469105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S. Chemotaxis by the nematode Caenorhabditis elegans: identification of attractants and analysis of the response by use of mutants. Proc Natl Acad Sci USA 70, 817–821 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croll N. A. Components and patterns in the behavior of the nematode Caenorhabditis elegans. J. Zool. 176, 159–176 (1975). [Google Scholar]
- Lakhina V. et al. Genome-wide functional analysis of CREB/long-term memory-dependent transcription reveals distinct basal and memory gene expression programs. Neuron 85, 330–345, doi: 10.1016/j.neuron.2014.12.029 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhl G. R. & Nishimori T. Neuropeptide gene expression and neural activity: assessing a working hypothesis in nucleus caudalis and dorsal horn neurons expressing preproenkephalin and preprodynorphin. Cell Mol Neurobiol 10, 73–98 (1990). [DOI] [PubMed] [Google Scholar]
- Peckol E. L., Troemel E. R. & Bargmann C. I. Sensory experience and sensory activity regulate chemosensory receptor gene expression in Caenorhabditis elegans. Proc Natl Acad Sci USA 98, 11032–11038. (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruner M. et al. Feeding state, insulin and NPR-1 modulate chemoreceptor gene expression via integration of sensory and circuit inputs. PLoS Genet 10, e1004707, doi: 10.1371/journal.pgen.1004707 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent P. et al. Decoding a neural circuit controlling global animal state in C. elegans. Elife 4, doi: 10.7554/eLife.04241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon J., Holden-Dye L., O’Connor V. & Hopper N. A. Context-dependent regulation of feeding behaviour by the insulin receptor, DAF-2, in Caenorhabditis elegans. Invert Neurosci 16, 4, doi: 10.1007/s10158-016-0187-2 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Nelson L. S., Kim K., Nathoo A. & Hart A. C. Neuropeptide gene families in the nematode Caenorhabditis elegans. Ann N Y Acad Sci 897, 239–252 (1999). [DOI] [PubMed] [Google Scholar]
- Turek M., Besseling J., Spies J. P., Konig S. & Bringmann H. Sleep-active neuron specification and sleep induction require FLP-11 neuropeptides to systemically induce sleep. Elife 5, doi: 10.7554/eLife.12499 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M. D. et al. FMRFamide-like FLP-13 neuropeptides promote quiescence following heat stress in Caenorhabditis elegans. Curr Biol 24, 2406–2410, doi: 10.1016/j.cub.2014.08.037 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringstad N. & Horvitz H. R. FMRFamide neuropeptides and acetylcholine synergistically inhibit egg-laying by C. elegans. Nat Neurosci 11, 1168–1176, doi: 10.1038/nn.2186 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers C. et al. Inhibition of Caenorhabditis elegans social feeding by FMRFamide-related peptide activation of NPR-1. Nat Neurosci 6, 1178–1185, doi: 10.1038/nn1140nn1140 (2003). [DOI] [PubMed] [Google Scholar]
- Sharabi K. et al. The response to high CO2 levels requires the neuropeptide secretion component HID-1 to promote pumping inhibition. PLoS Genet 10, e1004529, doi: 10.1371/journal.pgen.1004529 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers C., Persson A., Cheung B. & de Bono M. Behavioral motifs and neural pathways coordinating O2 responses and aggregation in C. elegans. Curr Biol 16, 649–659, doi: S0960-9822(06)01316-910.1016/j.cub.2006.03.023 (2006). [DOI] [PubMed] [Google Scholar]
- Zimmer M. et al. Neurons detect increases and decreases in oxygen levels using distinct guanylate cyclases. Neuron 61, 865–879, doi: S0896-6273(09)00158-510.1016/j.neuron.2009.02.013 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo M. A., Guillermin M. L., Rengarajan S., Okubo R. P. & Hallem E. A. O2-sensing neurons control CO2 response in C. elegans. J Neurosci 33, 9675–9683, doi: 10.1523/JNEUROSCI.4541-12.2013 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T. & Cai D. Counterbalance between BAG and URX neurons via guanylate cyclases controls lifespan homeostasis in C. elegans. EMBO J 32, 1529–1542, doi: 10.1038/emboj.2013.75 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. & Li C. Expression and regulation of an FMRFamide-related neuropeptide gene family in Caenorhabditis elegans. J Comp Neurol 475, 540–550 (2004). [DOI] [PubMed] [Google Scholar]
- Brandt J. P. et al. A single gene target of an ETS-family transcription factor determines neuronal CO2-chemosensitivity. PLoS One 7, e34014, doi: 10.1371/journal.pone.0034014 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramstrup Petersen J. et al. EGL-13/SoxD specifies distinct O2 and CO2 sensory neuron fates in Caenorhabditis elegans. PLoS Genet 9, e1003511, doi: 10.1371/journal.pgen.1003511 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo Romanos T., Petersen J. G., Riveiro A. R. & Pocock R. A novel role for the zinc-finger transcription factor EGL-46 in the differentiation of gas-sensing neurons in Caenorhabditis elegans. Genetics 199, 157–163, doi: 10.1534/genetics.114.172049 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins L. A., Hedgecock E. M., Thomson J. N. & Culotti J. G. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev Biol 117, 456–487 (1986). [DOI] [PubMed] [Google Scholar]
- Wicks S. R., de Vries C. J., van Luenen H. G. & Plasterk R. H. CHE-3, a cytosolic dynein heavy chain, is required for sensory cilia structure and function in Caenorhabditis elegans. Dev Biol 221, 295–307 (2000). [DOI] [PubMed] [Google Scholar]
- Brear A. G., Yoon J., Wojtyniak M. & Sengupta P. Diverse cell type-specific mechanisms localize G protein-coupled receptors to Caenorhabditis elegans sensory cilia. Genetics 197, 667–684, doi: 10.1534/genetics.114.161349 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay A., Deplancke B., Walhout A. J. & Tissenbaum H. A. C. elegans tubby regulates life span and fat storage by two independent mechanisms. Cell Metab 2, 35–42, doi: 10.1016/j.cmet.2005.06.004 (2005). [DOI] [PubMed] [Google Scholar]
- Ikeda A. et al. Microtubule-associated protein 1A is a modifier of tubby hearing (moth1). Nat Genet 30, 401–405, doi: 10.1038/ng838 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallem E. A. et al. Receptor-type guanylate cyclase is required for carbon dioxide sensation by Caenorhabditis elegans. Proc Natl Acad Sci USA 108, 254–259, doi: 10.1073/pnas.1017354108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E. S., Martinez-Velazquez L. & Ringstad N. A chemoreceptor that detects molecular carbon dioxide. J Biol Chem 288, 37071–37081, doi: 10.1074/jbc.M113.517367 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A. J., Chronis N., Karow D. S., Marletta M. A. & Bargmann C. I. A distributed chemosensory circuit for oxygen preference in C. elegans. PLoS Biol 4, e274, doi: 06-PLBI-RA-0332R310.1371/journal.pbio.0040274 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coates J. C. & de Bono M. Antagonistic pathways in neurons exposed to body fluid regulate social feeding in Caenorhabditis elegans. Nature 419, 925–929, doi: 10.1038/nature01170nature01170 (2002). [DOI] [PubMed] [Google Scholar]
- Weinshenker D., Wei A., Salkoff L. & Thomas J. H. Block of an ether-a-go-go-like K(+) channel by imipramine rescues egl-2 excitation defects in Caenorhabditis elegans. J Neurosci 19, 9831–9840 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery L., Bargmann C. I. & Horvitz H. R. The Caenorhabditis elegans unc-31 gene affects multiple nervous system-controlled functions. Genetics 134, 455–464 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speese S. et al. UNC-31 (CAPS) is required for dense-core vesicle but not synaptic vesicle exocytosis in Caenorhabditis elegans. J Neurosci 27, 6150–6162, doi: 27/23/615010.1523/JNEUROSCI.1466-07.2007 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass J., Jacob T. C., Kim P. & Kaplan J. M. The EGL-3 proprotein convertase regulates mechanosensory responses of Caenorhabditis elegans. J Neurosci 21, 9265–9272 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Saiz E. et al. Modular control of glutamatergic neuronal identity in C. elegans by distinct homeodomain proteins. Cell 155, 659–673, doi: 10.1016/j.cell.2013.09.052 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R. Y., Sawin E. R., Chalfie M., Horvitz H. R. & Avery L. EAT-4, a homolog of a mammalian sodium-dependent inorganic phosphate cotransporter, is necessary for glutamatergic neurotransmission in caenorhabditis elegans. J Neurosci 19, 159–167 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet M. L., Saifee O., Zhao H., Rand J. B. & Wei L. Synaptic transmission deficits in Caenorhabditis elegans synaptobrevin mutants. J Neurosci 18, 70–80 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond J. E., Davis W. S. & Jorgensen E. M. UNC-13 is required for synaptic vesicle fusion in C. elegans. Nat Neurosci 2, 959–964 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K. G. et al. A genetic selection for Caenorhabditis elegans synaptic transmission mutants. Proc Natl Acad Sci USA 93, 12593–12598 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair W. et al. Lifespan extension induced by AMPK and calcineurin is mediated by CRTC-1 and CREB. Nature 470, 404–408, doi: 10.1038/nature09706 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timbers T. A. & Rankin C. H. Tap withdrawal circuit interneurons require CREB for long-term habituation in Caenorhabditis elegans. Behav Neurosci 125, 560–566, doi: 10.1037/a0024370 (2011). [DOI] [PubMed] [Google Scholar]
- Nishida Y., Sugi T., Nonomura M. & Mori I. Identification of the AFD neuron as the site of action of the CREB protein in Caenorhabditis elegans thermotaxis. EMBO Rep 12, 855–862, doi: 10.1038/embor.2011.120 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez G. A. & Montminy M. R. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell 59, 675–680 (1989). [DOI] [PubMed] [Google Scholar]
- Brindle P., Linke S. & Montminy M. Protein-kinase-A-dependent activator in transcription factor CREB reveals new role for CREM repressors. Nature 364, 821–824, doi: 10.1038/364821a0 (1993). [DOI] [PubMed] [Google Scholar]
- Quinn P. G. Distinct activation domains within cAMP response element-binding protein (CREB) mediate basal and cAMP-stimulated transcription. J Biol Chem 268, 16999–17009 (1993). [PubMed] [Google Scholar]
- Mayr B. & Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol 2, 599–609, doi: 10.1038/35085068 (2001). [DOI] [PubMed] [Google Scholar]
- Pastok M. W. et al. Structural diversity of the cAMP-dependent protein kinase regulatory subunit in Caenorhabditis elegans. Cell Signal 25, 168–177, doi: 10.1016/j.cellsig.2012.09.006 (2013). [DOI] [PubMed] [Google Scholar]
- Troemel E. R., Sagasti A. & Bargmann C. I. Lateral signaling mediated by axon contact and calcium entry regulates asymmetric odorant receptor expression in C. elegans. Cell 99, 387–398 (1999). [DOI] [PubMed] [Google Scholar]
- Zhang S., Sokolchik I., Blanco G. & Sze J. Y. Caenorhabditis elegans TRPV ion channel regulates 5HT biosynthesis in chemosensory neurons. Development 131, 1629–1638 (2004). [DOI] [PubMed] [Google Scholar]
- Fujiwara M. et al. The Importance of cGMP Signaling in Sensory Cilia for Body Size Regulation in Caenorhabditis elegans. Genetics 201, 1497–1510, doi: 10.1534/genetics.115.177543 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., O’Halloran D. & Goodman M. B. GCY-8, PDE-2, and NCS-1 are critical elements of the cGMP-dependent thermotransduction cascade in the AFD neurons responsible for C. elegans thermotaxis. J Gen Physiol 142, 437–449, doi: 10.1085/jgp.201310959 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallem E. A. et al. A sensory code for host seeking in parasitic nematodes. Curr Biol 21, 377–383, doi: 10.1016/j.cub.2011.01.048 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D. & Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. Embo J 10, 3959–3970 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.